Abstract
Banana Xanthomonas wilt (BXW) caused by Xanthomonas campestris pv. musacearum (Xcm) is a severe bacterial disease affecting banana production in East and Central Africa, where banana is cultivated as a staple crop. Classical breeding of banana is challenging because the crop is clonally propagated and has limited genetic diversity. Thus, genetic engineering serves as a viable alternative for banana improvement. Studies have shown that transfer of the elongation factor Tu receptor gene (AtEFR) from Arabidopsis thaliana to other plant species can enhance resistance against bacterial diseases. However, AtEFR activity in banana and its efficacy against Xcm has not been demonstrated. In this study, transgenic events of banana (Musa acuminata) cultivar dwarf Cavendish expressing the AtEFR gene were generated and evaluated for resistance against Xcm under greenhouse conditions. The transgenic banana events were responsive to the EF-Tu-derived elf18 peptide and exhibited enhanced resistance to BXW disease compared to non-transgenic control plants. This study suggests that the functionality of AtEFR is retained in banana with the potential of enhancing resistance to BXW under field conditions.
Introduction
Banana (Musa spp.) is a staple food for about 400 million people, a crucial food security crop, and a source of income, especially in low-income countries [1]. The Great Lakes region, including Burundi, Rwanda, Uganda, Kenya, Tanzania, and the Democratic Republic of Congo, is the largest banana producer in Africa. Bananas in this region are produced by small holder farmer mainly for local consumption. The daily consumption is estimated to be 147 kcal daily per person, which is 15 times higher than the global average and six times higher than Africa’s average [2]. Unfortunately, banana production in this region is hindered by banana Xanthomonas wilt (BXW), a systemic bacterial disease caused by Xanthomonas campestris pv. musacearum (Xcm). The disease is the biggest threat to banana production in the region, with economic losses estimated to be US$ 2–8 billion over a decade [3]. Symptoms of BXW disease manifest as premature ripening and rotting of fruits, shriveling of inflorescence, wilting, and yellowing of leaves leading to the death of the infected plant. The bacterial pathogen spread by insect vectors, use of contaminated tools and infected planting materials. The disease is managed through cultural practices, including removing male buds to eliminate insect vectors, removing diseased plants, use of clean pathogen-free planting material, and disinfecting farm tools [4]. Conventional breeding of clonally propagated crops like banana is limited by the lack of genetic diversity and availability of important traits in the gene pool [5]. All cultivated banana varieties are susceptible to BXW disease; however, resistance has been observed in one of the wild banana progenitor Musa balbisiana belonging to the BB genome [3, 6]. Unfortunately, the B genome is laced with banana streak virus (BSV) sequences; during hybridization, recombination of integrated virus sequences sometimes results in BSV infection [7]. This has limited the use of M. balbisiana in conventional breeding. Notably, tolerance to Xcm has also been reported in Musa acuminata subsp. Zebrina belonging to AA genome, suggesting that tolerant traits could be present in existing banana germplasm [8]. Further, the molecular basis of disease resistance in banana progenitor Musa balbisiana against Xcm was investigated [6]. Comparative transcriptome analysis of BXW-resistant genotype Musa balbisiana and BXW-susceptible banana cultivar ‘Pisang Awak’ challenged with Xcm showed differentially expressed genes associated with response to biotic stress, which were mapped to the biotic stress pathways to identify genes associated with defense mechanisms. This study identified several genes involved in the activation of pathogen-associated molecular patterns (PAMP)-triggered basal defense and disease resistance (R) protein-mediated defense in Musa balbisiana as an early response to Xcm infection. These Musa defense genes can be overexpressed in the BXW-susceptible cultivars using biotechnological tools such as transgenic or genome editing for develing resistance against Xcm.
One of the approaches to enhancing plant disease resistance is by boosting their immune system [9]. Plants have evolved two main mechanisms for evading pathogens. One involves recognizing potential phytopathogens before they establish and is achieved through pattern recognition receptors (PRRs), which recognize conserved pathogen-associated molecular patterns (PAMPs), resulting in PAMP-triggered immunity (PTI). In addition, plants utilize resistance (R) proteins, that are mainly nucleotide-binding site leucine-rich repeat receptors (NLRs) that sense specific pathogen effectors leading to effector-triggered immunity (ETI) [10]. Resistance through ETI is typically race-specific and limited to plant varieties with a particular R-gene and specific pathogens with corresponding virulence effector. Meanwhile, PAMPs are conserved among a wide range of microbes; thus, plants tend to exhibit broad-spectrum resistance to pathogens. As PAMPs are essential for microbial survival, their evolution is slower than virulence effectors [11]. Therefore, PTI has the potential of conferring durable and broad-spectrum disease resistance compared to ETI.
Several plant PRRs have been identified and characterized. The best studied PRR is FLAGELLIN SENSING 2 (FLS2), a leucine-rich repeat receptor kinase (LRR-RK), which recognizes a N-terminal 22-amino acid motif, flg22, of bacterial flagellin [12]. Another PRR, related to FLS2, that has been extensively studied is the Brassicaceae-specific ELONGATION FACTOR TU RECEPTOR (EFR), which recognizes the N-terminal acetylated motif elf18 of bacterial elongation factor thermal unstable (EF-Tu), thereby activating plant defense response against many bacteria [13]. EF-Tu is the most copious protein in bacteria and plays an indispensable role in protein synthesis by catalyzing the binding of aminoacyl transfer RNA to the ribosome [14].
EFR plays a significant role in plant response to bacterial infection, thus, it has been widely applied to engineer crops against diverse bacterial diseases. Heterologous expression of Arabidopsis thaliana EFR (AtEFR) in Nicotiana benthamiana and Solanum lycopersicum (tomato) conferred responsiveness to elf18 peptide from diverse bacterial genera, including Agrobacterium, Xanthomonas, Pseudomonas, and Ralstonia. Furthermore, the N. benthamiana and tomato transgenic events exhibited enhanced resistance to different genera of bacterial pathogens [15]. So far, transgenic expression of AtEFR has effectively enhanced the resistance of major crops against various bacterial pathogens, including wheat (Triticum aestivum) against Pseudomonas syringae pv. oryzae [16], rice (Oryza sativa) against Xanthomonas oryzae pv. oryzae [17], potato (S. tuberosum) against Ralstonia solanacearum [18], apple (Malus malus) against Erwinia amylovora [19], and sweet orange (Citrus sinensis) against Xanthomonas citri and Xylella fastidiosa [20].
Previously, the transgenic banana expressing sweet pepper hypersensitive response-assisting protein (Hrap) or plant ferredoxin-like protein (Pflp) genes were developed and tested [21, 22]. These transgenic bananas showed enhanced resistance to BXW through successive crop cycles and agronomic performance comparable to non-transgenic bananas under field trials in Uganda [23]. Transgenic bananas with resistance against BXW were also developed by transforming embryogenic cell suspensions of banana cultivar ‘Sukali Ndiizi’ with Xa21 gene [24]. The Xa21 transgenic banana events exhibited enhanced resistance against BXW under greenhouse conditions. Transgenic banana expressing rice NH1 gene also showed enhanced resistance against BXW disease [25]. The bacterial pathogen can evolve resistance mechanisms to overcome the defense conferred by most single genes or single mode of action. To avoid or minimize the chances of breaking down of disease resistance, there is a need to stack several transgenes with different mode of actions in the banana event for enhanced and durable resistance to BXW disease.
In this study, transgenic banana cultivar dwarf Cavendish (Musa spp., AAA genomic group) constitutively expressing AtEFR under the control of the constitutive CaMV35S promoter were developed and evaluated against Xcm under controlled greenhouse conditions. The transgenic banana events showed enhanced resistance to BXW disease, as exhibited by significantly reduced wilting incidences and a lower bacterial population compared to the non-transgenic control plants. Assessment of defense-related gene expression and oxidative burst assay further revealed that the transgenic events gained responsiveness to elf18, indicating that AtEFR activity is retained after its transfer and integration into the banana genome.
Materials and methods
Plant material
Embryogenic cell suspensions (ECS) of banana cultivar dwarf Cavendish were used for transformation. The ECS were initiated from meristematic shoot tissues using the protocol described by Tripathi et al. [26] and maintained at 28±2°C on a rotary shaker at 95 rpm in the dark.
Generation of transgenic plants
The plasmid construct pBIN19g-35S::AtEFR [15], harboring ELONGATION FACTOR TU RECEPTOR gene from Arabidopsis thaliana (AtEFR) and neomycin phosphotransferase II (nptII) gene for kanamycin selection was transformed into Agrobacterium tumefaciens strain EHA105 by electroporation. The plasmid construct was validated through restriction enzyme digestion and PCR analysis using AtEFR-specific primers before using it for transformation. Banana ECSs were transformed as per the method described previously [26]. Agrobacterium infected ECSs were regenerated on a selective medium supplemented with 100 mg/l kanamycin following the protocol described by Tripathi et al. [26]. All regenerated putative transgenic events were maintained and multiplied on proliferation medium at 28±2°C for a 16-/ 8-h light/dark photoperiod under fluorescent tube lights. The putative transgenic events were characterized for the presence of the transgene. The transgenic shoots were transferred to the rooting medium. The well-rooted events were transferred to soil in the greenhouse for disease evaluation.
Molecular analysis of transgenic events
Polymerase chain reaction analysis
Genomic DNA was extracted from the putatively transformed events using a cetyltrimethylammonium bromide (CTAB) method [27]. PCR was performed in a 25-μL reaction volume containing 2.5 μL 10X PCR buffer, 0.3 μL dNTPs, 1μL of 10 μM reverse and forward AtEFR primers, 2 μL genomic DNA (100ng/μL), 0.2 μL Taq DNA polymerase (Qiagen, Germany), and 18 μL nuclease-free water. Thermocycler conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s, then final extension at 72°C for 7 min. Genomic DNA from non-transgenic control plant and pBIN19g-35S::AtEFR plasmid were used as negative and positive controls, respectively. The primers [forward 5’CGGGAATCTTGTAAGCCTGC 3’and reverse 5’GCACCCTTCCCTCAAACTTG 3’] amplifying 635 base pairs (bp) region within the AtEFR gene were used for PCR analysis. The amplified products were run on a 1% agarose gel (Duchefa, Netherlands) stained with GelRed® (Biotium, San Francisco, USA) and visualized under ultraviolet light.
Southern blot analysis
Southern blot analysis was performed as per the method described by Tripathi et al. [23]. Briefly, 10 μg of genomic DNA from each sample was restricted with BamH1 for 12 h. The DNA samples, including plasmid pBIN19g-35S::AtEFR and genomic DNA sample from a control non-transgenic plant, were run for a 0.8% agarose gel at 50 V. The gel stained with GelRed® was viewed under ultraviolet light to confirm the digestion. The restricted DNA was denatured, then blotted onto a positively charged membrane (Roche Diagnostics, West Sussex, UK) and fixed using ultraviolet cross-linking. The blots were then hybridized with a digoxigenin (DIG) PCR-labeled 635-bp AtEFR-specific probe. Hybridization and probe detection was performed using a DIG Luminescent Detection Kit for Nucleic Acids (Roche Diagnostics, UK) as per the manufacturer’s protocol.
Plant growth analysis
Eight PCR positive transgenic events and control plants were randomly selected for the plant growth analysis. Three replicates of well-rooted plants for each event were transferred to soil in small plastic cups and acclimatized for 30 days in a humidity chamber, then transferred to bigger pots and grown in the greenhouse for 90 days at 25–30°C. The growth parameter data, including plant height, pseudostem girth, number of functional leaves, and length and width of the middle leaf, were recorded from 90-day-old plants. The total leaf area was calculated using the formula below [28].
Total leaf area = 0.8 ⊆ L⊆ W ⊆N
In which, L = Length of the middle leaf, W = width of the middle leaf, and N = total number of leaves in the plant.
Greenhouse evaluation of transgenic events for resistance to BXW disease
Three replicates of each transgenic event and non-transgenic control were evaluated for resistance against Xcm under greenhouse conditions. A total of 31 transgenic events were used for the disease assay. The transgenic events and non-transgenic control plants were arranged in a completely randomized design. The culture of Xcm (Ugandan isolate, sublineage 2) that met the four Koch postulates were cultured in YPGA medium (0.5% yeast extract, 0.5% peptone, 1% glucose, and 0.8% micro agar) for 48 h at 28°C. A single colony was aseptically isolated and further cultured for 48 h in 50 mL of YPG medium at 28°C in an incubator shaker (200 rpm). Subsequently, the liquid culture was centrifuged at 4000 rpm for 15 min and the pellet resuspended in sterile distilled water. The suspension concentration was adjusted to OD600nm of 1 using sterile distilled water. The second open functional leaf of 90-day-old potted plant was inoculated with 100 μL of the bacterial suspension using an insulin syringe. The plants were maintained in the greenhouse under observation, and symptoms were recorded as they occurred for 60 days post-inoculation (dpi). The data collected included number of days for appearance of the first symptom, number of days for complete wilting, disease severity, and the number of leaves showing symptoms at 60 dpi. The data was used to calculate percent resistance compared to control non-transgenic plants using the formula below [27]:
Resistance (%) = (Reduction in wilting of transgenic event/number of leaves wilted in the control plant) ⊆100
In which, reduction in wilting was the total number of leaves minus the number of leaves wilted.
Disease severity was rated on a scale of 0–5 (0- no signs of the disease, 1- a single leaf showing symptom, 2-two to three leaves with symptoms, 3- four to five leaves with symptoms, 4- all the leaves have symptoms but plant not dead, 5- complete death).
Plants were categorized as resistant if they did not show any symptom, partially resistant if the symptoms did not spread to all the leaves, and susceptible if the symptoms spread to all the leaves, causing complete wilting or death of the plant.
In planta bacterial population analysis
For the bacterial population study, two transgenic events (T5 and T7) that exhibited enhanced resistance against Xcm under greenhouse conditions, along with control non-transgenic plants, were used for this experiment. Three replicates of 90-day-old potted plants were inoculated with Xcm culture as described previously [21]. Leaf-mid rib sections (1 cm) of the inoculated leaves were collected at 0, 3, 6, 9, 12, and 15 pdi. The samples were ground in 15-mL falcon tubes after adding 2 mL of YPG medium and incubated in an incubator shaker (200 rpm) for 1 h at 28°C. Five serial dilutions of the sample suspensions were spread on YPGA medium supplemented with 50 mg/L cephalexin to select for Xcm and cultured at 28°C for 48 h. Samples from each line were cultured in triplicate for every time point. The bacterial population was determined by counting colonies for each dilution and described using growth curve analysis.
Transgene expression analysis
To determine the relative gene expression of various transgenic events compared to control non-transgenic plant, total RNA was extracted from the leaves of two-week-old shoots using RNeasy plant mini kit (Qiagen, Germany) according to the manufacturer’s instructions. The RNA quality and concentration were determined using NanoDrop 2000 (Thermo Fisher Scientific). And 1 μg of the total RNA was reverse transcribed into cDNA using Luna script RT Supermix (New England Biolabs) as per the manufacturer’s protocol. The cDNA templates were then used for PCR amplification of the AtEFR gene transcript. The PCR was performed in a 25-μL reaction volume containing 2.5 μL10X PCR buffer, 0.3 μL dNTPs, 1μL of 10 μM reverse and forward AtEFR primers (S1 Table), 2 μL cDNA, 0.2 μL Taq DNA polymerase, and 18 μL nuclease-free water under the following conditions: initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and extension at 72°C for 1 min, then final extension at 72°C for 7 min. Musa 25s ribosomal transcript was used as an internal control to check the quality of the cDNA. qRT-PCR was performed using Luna® Universal qPCR Master Mix (New England Biolabs) on a Quanta Studio Real-Time PCR System (Applied Biosystems, Foster City, CA) under the following conditions: initial denaturation at 95°C for 5 min followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. The reaction mixture was in a 20 μl reaction volume consisting of 10 μl Luna® Universal qPCR Master Mix, 0.2 μl forward and reverse primers (S1 Table), 5 μl cDNA, and 4.6 μl nuclease-free water. Each sample consisted of three biological replicates. Musa 25s ribosomal transcript served as an internal control for the normalization of gene expression. The relative levels of the AtEFR gene were analysed using Livak & Schmittgen method [29].
Expression analysis of defense-related genes
The leaves of two-week-old shoots were infiltrated with 250 nM elf18 peptide solution for 60, 120, and 180 min. Total RNA was extracted and qRT-PCR was performed as described in the above section to check the relative expression of defense-related genes (MaWRKY-22 like, MaPR1-like, MaPR2-like, MaPR3-like, MaPR4-like, and MaPR5-like) in the transgenic events compared to control non-transgenic events. qRT-PCR was performed with initial denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. Each sample consisted of three biological replicates. Musa 25s ribosomal transcript was used as an internal control for the normalization of gene expression. The primers used are presented in S1 Table.
ROS production assay
Hydrogen peroxide production in the transgenic events after elf18 infiltration was determined through histochemical staining assay using DAB (3,3’-diaminobenzidine) solution [30]. Briefly, freshly opened leaves of 14-day-old in vitro plantlets were infiltrated with 100 μL of 250 nM elf18 peptide dissolved in sterile distilled water. The leaves were left to incubate for 2 h, after which they were harvested and submerged in DAB solution in 15-mL falcon tubes wrapped with aluminum foil. The samples were incubated in DAB solution for 5 h on a shaker (70 rpm) at room temperature. Following incubation, DAB solution was removed, and samples were bleached with a solution (ethanol: acetic acid: glycerol = 3:1:1) for 15 min in the water bath at 95°C to remove chlorophyll. The bleaching solution was refreshed thrice, and the samples were further incubated for 30 min at room temperature. The samples were then removed from the bleaching solution, dried on paper towels, and photographed using SMZ1500 stereomicroscope (Carlsbad, CA, USA) attached with high zoom Nikon camera. The photomicrographs were analyzed using image J software (National Institutes of Health, USA) to compare the browning intensities.
Statistical analysis
Data analysis was performed using Minitab Statistical Software, version 17 (Pennsylvania, USA). Differences in disease resistance and plant growth characteristics between various transgenic events and non-transgenic control plants were analyzed using one-way analysis of variance (ANOVA) and means separated by Fisher’s HSD test. Statistical significance was determined at p≤0.05.
Results
Generation of transgenic events
Embryogenic cell suspensions (ECSs) of banana cultivar dwarf Cavendish were co-cultivated with Agrobacterium tumefaciens strain EHA105 harboring the binary vector pBIN19g:35S::AtEFR [15] for three days. Agrobacterium-infected ECSs turned from cream to brown color during co-cultivation. Browning intensified when the cells were transferred to embryo development medium (EDM) supplemented with 100 mg/L kanamycin for selecting transformed embryogenic cells. After about 30 days of culture on the same medium, white embryos began to appear on the surface of the dark heap of cells (Fig 1A). The white embryos increased in number and size when the cells were transferred to fresh EDM. After 14 days of culture on selective embryo maturation medium (EMM), a few embryos germinated into plantlets, but most continued to expand on the same medium without developing any organized structures (Fig 1B). Most embryos germinated into complete plantlets on a selective embryo germination medium (EGM) after 20–30 days of culture (Fig 1C). After germination, the shoots elongated when transferred to proliferation medium (PM) supplemented with 100 mg/L kanamycin (Fig 1D). In total, 32 putative transgenic events were regenerated from three separate experiments.
Fig 1. Generation of transgenic banana events expressing AtEFR gene.
(A) Embryogenic cells in selective embryo development medium (EDM), (B) Embryos maturing and germinating on selective embryo maturation medium (EMM), (C) Germination of embryos in selective embryo germination medium (EGM), (D) Complete plantlets on selective proliferation medium (PM).
Molecular characterization of transgenic events
The regenerated putative transgenic events were validated for the presence and integration of the transgene by PCR and Southern blot analysis, respectively. The PCR analysis using AtEFR-specific primers revealed an amplicon of the expected size (635 bp) (Fig 2A), confirming the presence of the AtEFR gene in the transgenic events. No amplification was observed in control non-transgenic plants. Further, 14 PCR-positive events were assessed using Southern blot analysis to confirm transgene integration and copy number. Southern hybridization of BamH1-digested genomic DNA using an AtEFR-specific probe confirmed the integration of the transgene in the plant genome with different hybridization profiles, indicating random insertion of the transgene in the genome of the tested events. The copy number of the transgene incorporated in the different events ranged from at least one to multiple (Fig 2B). No transgene integration was detected in the non-transgenic control plants.
Fig 2. Molecular analysis of putatively transgenic banana events expressing AtEFR gene.
A) Polymerase chain reaction assay to confirm the presence of AtEFR gene, B) Southern blot analysis to confirm the integration of AtEFR gene. M; molecular marker, C; Control, P; plasmid.
Plant growth analysis
To assess growth parameters of the generated transgenic events, plant height, pseudostem girth, total leaf area and number of functional leaves were evaluated in eight randomly selected transgenic events and non-transgenic control plants. All plants showed normal growth with no morphological abnormalities. No significant differences (p<0.05) in plant height, pseudostem girth, and total leaf area were observed between the transgenic events and the non-transgenic control plants (Fig 3A–3D), indicating that overexpression of the AtEFR gene in transgenic banana did not alter plant growth.
Fig 3. Growth analysis of transgenic banana events expressing AtEFR gene compared to non-transgenic control plants.
(A) Plant height, B) Pseudostem girth, (C) Number of functional leaves, (D) Total leaf area. Data are presented as means ± standard deviation. Bars with different letters are significantly different at p<0.05 according to Fisher’s HSD test (n = 3).
Evaluation of transgenic banana events for resistance to banana Xanthomonas wilt
Thirty-one transgenic events were evaluated through a greenhouse bioassay for disease resistance compared to non-transgenic control plants. Significant differences were observed among the transgenic and non-transgenic control events regarding the number of days to the appearance of the first symptom and the number of days to complete wilting of plants. BXW disease symptoms, such as necrosis and wilting of leaves, appeared in non-transgenic control plants at 16 dpi, whereas transgene-positive plants showed symptoms at only 18 to 42 dpi (Table 1). None of the transgene-positive plants showed complete resistance to BXW disease. Even though several transgene-positive plants showed disease symptoms, disease progression was significantly slower than in control plants. Complete wilting was observed in control plants after an average of 22 dpi, compared to transgene-positive plants, which started to collapse 25 dpi, with the majority dying 30 dpi. Out of the 31 events evaluated, 18 events were found to exhibit partial resistance (50–75%) compared to control non-transgenic plants (Table 1, Fig 4A & 4B).
Table 1. Greenhouse evaluation of transgenic events of banana cultivar ‘dwarf Cavendish’ expressing AtEFR gene for resistance against Xanthomonas campestris pv. musacearum.
| Line number | Mean number of days for appearance of first symptoms | Mean number of days for complete wilting | Percent resistance | Disease rating |
|---|---|---|---|---|
| T1 | 22.5±7.8bc | NCW | 75.4±5.4bc | PR |
| T2 | 27.0±2.83b | NCW | 66.7±9.5b | PR |
| T3 | 29.5±0.7b | 42.5±6.4a | 33.3±10.5a | S |
| T4 | 36.0±2.83a | NCW | 66.7±10.5b | PR |
| T5 | 20.5±2.12bc | NCW | 75.0±6.9bc | PR |
| T6 | 42.5±2.12a | NCW | 58.3±7.8ab | PR |
| T7 | 20±1.5bc | NCW | 66.7±9.5b | PR |
| T8 | 18±2.1c | NCW | 50 ±10.5ab | PR |
| T9 | 21.0±2.83bc | 33.5±2.12ab | 33.3±10.5a | S |
| T10 | 22.0±0.7bc | NCW | 71.4±9.0bc | PR |
| T11 | 27.0±0.0b | NCW | 66.7±10.5b | PR |
| T12 | 28.5±14.8b | 31.0±2.6abc | 38.1±9.5a | S |
| T13 | 18.3±1.5c | 26.3±2.1bc | 0a | S |
| T14 | 21.0±2.8bc | NCW | 66.7±10.5b | PR |
| T15 | 24.0±1.4bc | 38.0±2.6ab | 47.6±9.9ab | S |
| T16 | 21.5±2.1bc | 29.0±2.8bc | 33.3±10.5a | S |
| T17 | 20±3.1bc | NCW | 50±9.9ab | PR |
| T18 | 21.0±2.8bc | NCW | 66.7±10.5b | PR |
| T19 | 22.0±2.1bc | NCW | 66.7±10.5b | PR |
| T20 | 20.5±3.5bc | 33.0±2.8ab | 33.3±9.5a | S |
| T21 | 28.5±10.6b | 43.0±14.1a | 33.3±9.9a | S |
| T22 | 19.0±0.0bc | 25.0±2.8bc | 33.3±7.5a | S |
| T23 | 29.0±9.9b | NCW | 66.7±9.5b | PR |
| T24 | 24.0±4.2bc | NCW | 66.7±6.5b | PR |
| T25 | 23.0±0.0bc | 34±2.6ab | 41.7±9.0ab | S |
| T26 | 28.3±2.1b | NCW | 66.7±10.5b | PR |
| T27 | 25.5±3.1b | NCW | 66.7±10.5b | PR |
| T28 | 21.0±1.8bc | NCW | 66.7±10.5b | PR |
| T29 | 25.5±2.8b | 35±2.0ab | 33.3±9.5a | S |
| T30 | 27.0±0.0b | 32±2.8ab | 33.3±9.5a | S |
| T31 | 22.5±0.7bc | NCW | 66.7±10.5b | PR |
| Control | 16.3±1.2c | 22.0±2.0c | 0a | S |
Means followed by the same superscript letter in the same column are not significantly different according to Fisher HSD test at p<0.05
Data are presented as means±standard diviation (SD). NS; no symptoms
NCW; no complete wilting
S; suseptible
PR; partial resistance.
Fig 4. Evaluation of transgenic banana events expressing AtEFR gene and non-transgenic control for resistance to BXW disease.
(A) Transgenic banana event T7 showing partial resistance to BXW disease, (B) Non-transgenic control plants showing complete wilting due to BXW disease. Photographs were taken at 60 days post inoculations (dpi).
Testing transgenic banana events for oxidative burst
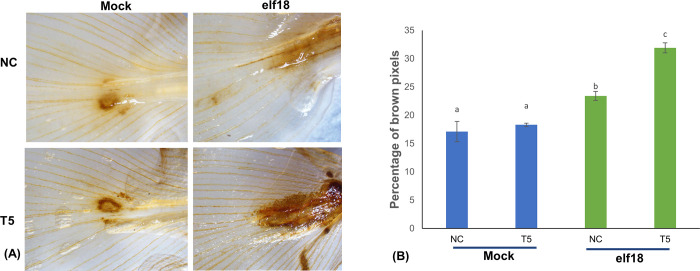
To assess whether the transgenic events exhibited enhanced production of reactive oxygen species upon pathogen infection, the leaves of transgenic event T5 and non-transgenic control were infiltrated with either sterile water or 250 nM elf18 for 2 hpi, followed by diaminobenzidine (DAB) staining to detect the accumulation of hydrogen peroxide. Positive-transgenic plants infiltrated with elf18 exhibited more intense browning than the mock-treated control and transgenic plants (Fig 5A & 5B) suggesting that the expression of AtEFR confers elf18 perception and subsequent activation of early signaling immune outputs in the transgenic plants.
Fig 5. ROS production assay in transgenic event T5 and non-transgenic control (NC) after elf18 infiltration followed by diaminobenzidine (DAB) staining.
A) Photomicrographs showing the difference in browning intensity between the leaves of transgenic events compared to non-transgenic control plants, (B) Graphical representation of browning intensity of non-transgenic control and transgenic leaves after elf18 infiltration followed by DAB staining. Error bars represent the standard error of the mean of three independent biological replicates.
Relative expression of AtEFR gene among transgenic events
The relative expression of the AtEFR transgene was assessed through reverse-transcriptase (RT) quantitative PCR (RT-qPCR) assays in seven transgenic events compared to non-transgenic control plants. The seven transgenic events were selected based on their level of disease resistance in the glasshouse, ranging from 33% resistance in T3 event to 75% resistance in T1 and T5 event (Table 1). Variations in transcript levels were observed between the various transgenic events tested. For example, T5 event showed the highest level of AtEFR gene expression, whereas the lowest gene expression was observed in T3 event (Fig 6A–6C). No EFR expression was detected in non-transgenic control plants.
Fig 6. Relative expression of AtEFR gene in transgenic banana events.
(A) Reverse-transcriptase (RT)-PCR products corresponding to 100 bp AtEFR gene fragment, (B) RT-PCR products corresponding to Musa 25s gene (internal control), (C) Transcription level of AtEFR in the transgenic bananas as determined using qRT-PCR. Data shown are fold induction relative to the non-transgenic control and were normalized against the internal control gene Musa 25s. Data presented are mean± standard deviation (SD) of three replicate experiments. Bars with different letters are significantly different at p<0.05 according to Fisher’s HSD test. M; 100-base pair marker; T; Transgenic event expressing AtEFR, C; non-transgenic control.
Relative expression of defense-related genes in transgenic banana events
The transgenic T5 event with 75% resistance was infiltrated with 250 nM elf18, and the relative expression of six selected defense marker genes (MaWRKY-22 like, MaPR1-like, MaPR2-like, MaPR3-like, MaPR4-like, and MaPR5-like) was evaluated at 1, 2, and 3 h time points, relative to the water-treated replicates. Among the tested defense-related genes, MaWRKY-22-like, MaPR1-like and MaPR3-like were significantly upregulated in AtEFR transgene-positive plants relative to non-transgenic control plants. MaWRKY-22-like was highly expressed mainly at 1h post-infiltration (hpi), after which its expression reduced significantly. At 3 hpi, its expression was not substantially different from that of the non-transgenic control. Similarly, MaPR3-like was upregulated at all the time points, but significantly at 1 hpi. Meanwhile, the expression of MaPR1-like and MaPR2-like was significantly higher at 3 hpi. Notably, no significant increase in the expression of MaPR4-like and MaPR5-like was observed at all time points, compared with the non-transgenic control (Fig 7A–7C).
Fig 7. Relative expression of defense related genes in transgenic bananas expressing AtEFR upon activation with elf18 peptide.
Expression profile of defense-related genes induced at (A) 1 h post inoculation (hpi), (B) 2 hpi, and (C) 3 hpi following infiltration with 250 nM elf18 peptide in transgenic line T5 and non-transgenic control as revealed by qRT-PCR. Results are presented as fold induction relative to water treatment and normalized to internal control Musa 25s gene. Data presented are mean values ± standard deviation (SD) from three replicate experiments. Bars with different letters are significantly different at p<0.05 according to Fisher’s HSD test.
Bacterial population analysis of transgenic banana events
In planta bacterial population analysis was performed with two transgene-positive plants (T5 and T7) and non-transgenic control plants to determine differences in the Xcm growth rate post inoculation. Samples were collected from each plant at specific time points following inoculation with Xcm and cultured on a YPGA medium to examine the bacterial population. No significant difference in bacterial titers were observed between the control and transgenic plants up to 3 dpi (Fig 8A). However, a reduction in bacterial titers was observed in transgene-positive plants compared to non-transgenic control at 6, 9, 12, and 15 dpi (Fig 8A). Also, significant differences in the bacterial population were observed between the two transgenic events assayed. Transgenic event T7 exhibited a significantly lower level of bacterial population compared to event T5 at 15 dpi (Fig 8B).
Fig 8. Bacterial population analysis in two transgenic events (T5 and T7) and non-transgenic control plants post inoculation with Xanthomonas campestris pv. musacearum (Xcm).
(A) Bacterial population analysis at 0 to 15 dpi with Xcm, (B) Bacterial population at 15 dpi. Data are presented as means ± standard deviations. Bars with different letters are significantly different at p<0.05 according to Fisher’s HSD test (n = 3).
Discussion
BXW disease continues to be the primary threat to banana production in the Great Lakes region of East Africa since its first report in Ethiopia. Considering that most bananas grown in these regions are mainly for subsistence, the impact can be severe. Developing banana varieties with disease resistance through conventional breeding is possible; however, the process has many limitations [31]. For example, most breeding programs use fertile diploids to hybridize triploid varieties or tetraploid intermediates. However, given that most cultivated varieties are triploids with low female and male fertility levels, it is not viable to repeatedly backcross the primary hybrid to the original variety. As such, the introgression of new traits into elite varieties is not practical. Also, being a clonally propagated crop, there are limited desirable traits in banana gene pool. In this case, there are no fertile diploids with resistance to BXW, except Musa balbisiana, which is not preferred due to its association with banana streak virus susceptibility [5].
The exploitation of natural plant defenses for crop improvement through genetic engineering is increasingly becoming an attractive option for improving clonally propagated crops like banana. Transferring PRRs across plant species that would not naturally interbreed is relatively a recent crop improvement approach for developing disease resistance [32]. Herein, the EFR gene from A. thaliana (AtEFR) was overexpressed in the susceptible banana cultivar ‘dwarf Cavendish,’ and the transgenic events were evaluated for their response against Xcm in greenhouse conditions. These assays revealed that transgenic bananas expressing AtEFR exhibited enhanced resistance against Xcm compared to non-transgenic controls.
Heterologous expression of some defense-related genes has been sometimes associated with constitutive expression of defense-related genes, which can have adverse effects on plant growth and development, and usually manifest as shoot and root growth inhibition, necrosis, or both [33, 34]. Such effects directly translate to yield losses. In this study, transgenic bananas expressing AtEFR were phenotypically similar to non-transgenic control plants, indicating no limitation to their normal growth characteristics. This further implies that constitutive expression of AtEFR in banana is not likely to affect yield, which is consistent with results previously reported in tomato [15, 35], rice [17, 36], potato [18], Medicago [37], apple [19] and sweet orange [20].
Pattern recognition receptors, such as EFR, confer basal resistance against adapted and non-adapted pathogens, and thus contribute to both basal and non-host disease resistances. The resistance usually is quantitative, but in some instances, it can lead to qualitative or complete resistance [38]. In this study, all the transgenic events showed quantitative resistance manifested by delay in symptom appearance and disease progression, as well as lower disease severity compared with the wildtype controls.
Substantial evidence shows that PRRs confer more broad-spectrum resistance to pathogens compared to R-genes, mainly because they recognize PAMPs, which are often conserved across a wide range of genera and play essential roles in the survival of the pathogens. PRRs are therefore also predicted to confer more durable resistance [32, 39]. However, in rare cases, variations in PAMP genes have been observed in adapted plant pathogens or commensals to evade recognition [40–42]. Also, some pathogens produce effectors that can suppress PTI in some plants [40, 43], but this has not been very effective as some level of PTI is usually retained, as exemplified by enhanced susceptibility to virulent pathogens in mutants of PRRs [44, 45].
EFR sense bacterial EF-Tu through recognition of the N-acetylated elf18 motif, thereby activating PTI [13]. In this study, transgenic bananas expressing AtEFR gained responsiveness to elf18 as exhibited by ROS accumulation and upregulation of defense marker genes following elf18 peptide infiltration. These findings indicate that AtEFR activity was retained after its transfer to banana and that necessary downstream signaling components are conserved between Arabidopsis and the monocotyledonous plant banana. Accordingly, we observed a significant reduction in the bacterial population at various time points following Xcm inoculation compared to control plants, indicating that the activation of early immune outputs (e.g. ROS, defense gene expression) mediated by the recognition of Xcm EF-Tu by EFR leads to enhanced resistance to this otherwise adapted pathogen.
Induction of pathogenesis-related (PR) genes is associated with systemic acquired resistance (SAR), a form of plant defense mechanism that begins from a localized region of a plant and spreads throughout the entire plant [46]. Induction of PR gene following elf18 treatment has been reported in transgenic rice expressing AtEFR [36]. However, it is not clear whether the upregulation of PR genes observed in this study is a secondary response to constitutive expression of AtEFR in banana. Therefore, further studies should be conducted to verify these findings.
In conclusion, this study indicates that AtEFR retains its activity in banana when ectopically expressed and confers resistance to Xcm. This further confirms that immune signaling networks downstream of PRRs are conserved across various plant families and thus interfamily transfer of PRRs can be used to engineer disease resistance. Given that the resistance observed herein was however quantitative, future studies on banana improvement against bacterial diseases should focus on combining AtEFR with other defense genes like Pflp and Hrap, which could lead to stronger and more durable resistance. The transgenic banana expressing AtEFR gene should be further tested under fields conditions for several generations to confirm the durable resistance.
Supporting information
(DOCX)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the 2 Blades Foundation, the Gatsby Charitable Foundation, and the United States Agency for International Development (USAID). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Voora V, Larrea C, Bermudez S. Global Market Report: Bananas.:12. [Google Scholar]
- 2.Ainembabazi JH, Tripathi L, Rusike J, Abdoulaye T, Manyong V. Ex-Ante Economic Impact Assessment of Genetically Modified Banana Resistant to Xanthomonas Wilt in the Great Lakes Region of Africa. PloS One. 2015;10(9):e0138998. doi: 10.1371/journal.pone.0138998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripathi L, Mwangi M, Abele S, Aritua V, Tushemereirwe WK, Bandyopadhyay R. Xanthomonas Wilt: A Threat to Banana Production in East and Central Africa. Plant Dis. 2009. Apr 6;93(5):440–51. doi: 10.1094/PDIS-93-5-0440 [DOI] [PubMed] [Google Scholar]
- 4.Kikulwe EM, Kyanjo JL, Kato E, Ssali RT, Erima R, Mpiira S, et al. Management of Banana Xanthomonas Wilt: Evidence from Impact of Adoption of Cultural Control Practices in Uganda. Sustainability. 2019. Jan;11(9):2610. [Google Scholar]
- 5.Dale J, Paul JY, Dugdale B, Harding R. Modifying Bananas: From Transgenics to Organics? Sustainability. 2017. Mar;9(3):333. [Google Scholar]
- 6.Tripathi L, Tripathi JN, Shah T, Muiruri KS, Katari M. Molecular Basis of Disease Resistance in Banana Progenitor Musa balbisiana against Xanthomonas campestris pv. musacearum. Sci Rep. 2019. May 7;9(1):7007. doi: 10.1038/s41598-019-43421-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iskra-Caruana M line Duroy PO, Chabannes M, Muller E. The common evolutionary history of badnaviruses and banana. Infect Genet Evol. 2014. Jan 1;21:83–9. doi: 10.1016/j.meegid.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 8.Nakato GV, Christelová P, Were E, Nyine M, Coutinho TA, Doležel J, et al. Sources of resistance in Musa to Xanthomonas campestris pv. musacearum, the causal agent of banana xanthomonas wilt. Plant Pathol. 2019;68(1):49–59. [Google Scholar]
- 9.Nicaise V. Boosting innate immunity to sustainably control diseases in crops. Curr Opin Virol. 2017. Oct 1;26:112–9. doi: 10.1016/j.coviro.2017.07.030 [DOI] [PubMed] [Google Scholar]
- 10.Ngou BPM, Ding P, Jones JDG. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell. 2022. Apr 26;34(5):1447–78. doi: 10.1093/plcell/koac041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014. Jul;35(7):345–51. doi: 10.1016/j.it.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 12.Gómez-Gómez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000. Jun;5(6):1003–11. doi: 10.1016/s1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- 13.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006. May 19;125(4):749–60. doi: 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- 14.Sprinzl M. Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem Sci. 1994. Jun 1;19(6):245–50. doi: 10.1016/0968-0004(94)90149-x [DOI] [PubMed] [Google Scholar]
- 15.Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol. 2010. Apr;28(4):365–9. doi: 10.1038/nbt.1613 [DOI] [PubMed] [Google Scholar]
- 16.Schoonbeek H jan Wang HH, Stefanato FL, Craze M, Bowden S, Wallington E, et al. Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 2015;206(2):606–13. doi: 10.1111/nph.13356 [DOI] [PubMed] [Google Scholar]
- 17.Lu F, Wang H, Wang S, Jiang W, Shan C, Li B, et al. Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR. J Integr Plant Biol. 2015. Jul;57(7):641–52. doi: 10.1111/jipb.12306 [DOI] [PubMed] [Google Scholar]
- 18.Boschi F, Schvartzman C, Murchio S, Ferreira V, Siri MI, Galván GA, et al. Enhanced Bacterial Wilt Resistance in Potato Through Expression of Arabidopsis EFR and Introgression of Quantitative Resistance from Solanum commersonii. Front Plant Sci [Internet]. 2017. [cited 2023 Jun 9];8. Available from: https://www.frontiersin.org/articles/10.3389/fpls.2017.01642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piazza S, Campa M, Pompili V, Costa LD, Salvagnin U, Nekrasov V, et al. The Arabidopsis pattern recognition receptor EFR enhances fire blight resistance in apple. bioRxiv. 2021. Jan 22;2021.January.22.427734. doi: 10.1038/s41438-021-00639-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitre LK, Teixeira-Silva NS, Rybak K, Magalhães DM, Souza-Neto RR de, Robatzek S, et al. The Arabidopsis immune receptor EFR increases resistance to the bacterial pathogens Xanthomonas and Xylella in transgenic sweet orange. bioRxiv. 2021. Jan 22;2021.January.22.427732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi L, Mwaka H, Tripathi JN, Tushemereirwe WK. Expression of sweet pepper Hrap gene in banana enhances resistance to Xanthomonas campestris pv. musacearum. Mol Plant Pathol. 2010. Nov;11(6):721–31. doi: 10.1111/j.1364-3703.2010.00639.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namukwaya B, Tripathi L, Tripathi JN, Arinaitwe G, Mukasa SB, Tushemereirwe WK. Transgenic banana expressing Pflp gene confers enhanced resistance to Xanthomonas wilt disease. Transgenic Res. 2012. Aug;21(4):855–65. doi: 10.1007/s11248-011-9574-y [DOI] [PubMed] [Google Scholar]
- 23.Tripathi L, Tripathi JN, Kiggundu A, Kori S, Shotkoski F, Tushemereirwe WK. Field trial of Xanthomonas wilt disease-resistant bananas in East Africa. Nature Biotechnology 2014; 32:868–870. doi: 10.1038/nbt.3007 [DOI] [PubMed] [Google Scholar]
- 24.Tripathi JN, Lorenzen J, Bahar O, Ronald P, Tripathi L. Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotechnol J. 2014. Aug;12(6):663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi JN. Genetic Engineering of Banana Using Nonexpressor of Pathogenesis Related Gene for Resistance to Xanthomonas Wilt and Fusarium Wilt Diseases [Internet] [Thesis]. Kenyatta University; 2018. [cited 2023 Jul 17]. Available from: https://ir-library.ku.ac.ke/handle/123456789/19013 [Google Scholar]
- 26.Tripathi JN, Oduor RO, Tripathi L. A High-Throughput Regeneration and Transformation Platform for Production of Genetically Modified Banana. Front Plant Sci. 2015;6:1025. doi: 10.3389/fpls.2015.01025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart VIA LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Rapid CTAB DNA Isol Tech Useful RAPD Fingerprinting PCR Appl. 1993;14(5):748–51. [PubMed] [Google Scholar]
- 28.Kumar N, Krishnamoorthy V., Nalina L., Soorianathasundharam K. A new factor for estimating total leaf area in banana [Internet]. 2002. [cited 2020 Dec 17]. Available from: https://www.musalit.org/seeMore.php?id=14204 [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 30.Daudi A, O’Brien JA. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio-Protoc [Internet]. 2012. [cited 2020 Dec 19];2(18). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4932902/ [PMC free article] [PubMed] [Google Scholar]
- 31.Batte M, Swennen R, Uwimana B, Akech V, Brown A, Tumuhimbise R, et al. Crossbreeding East African Highland Bananas: Lessons Learnt Relevant to the Botany of the Crop After 21 Years of Genetic Enhancement. Front Plant Sci [Internet]. 2019. [cited 2020 Dec 22];10. Available from: https://www.frontiersin.org/articles/10.3389/fpls.2019.00081/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boutrot F, Zipfel C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu Rev Phytopathol. 2017. Aug 4;55:257–86. doi: 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- 33.Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: what are we going to express? Trends Biotechnol. 2005. Jun;23(6):275–82. doi: 10.1016/j.tibtech.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 34.Hammond-Kosack KE, Parker JE. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol. 2003. Apr;14(2):177–93. doi: 10.1016/s0958-1669(03)00035-1 [DOI] [PubMed] [Google Scholar]
- 35.Kunwar S, Iriarte F, Fan Q, Evaristo da Silva E, Ritchie L, Nguyen NS, et al. Transgenic Expression of EFR and Bs2 Genes for Field Management of Bacterial Wilt and Bacterial Spot of Tomato. Phytopathology®. 2018. Jun 20;108(12):1402–11. doi: 10.1094/PHYTO-12-17-0424-R [DOI] [PubMed] [Google Scholar]
- 36.Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, et al. Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses. PLOS Pathog. 2015. Mar 30;11(3):e1004809. doi: 10.1371/journal.ppat.1004809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeilmeier S, George J, Morel A, Roy S, Smoker M, Stransfeld L, et al. Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol J. 2019. Mar;17(3):569–79. doi: 10.1111/pbi.12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranf S. Pattern Recognition Receptors—Versatile Genetic Tools for Engineering Broad-Spectrum Disease Resistance in Crops. Agronomy. 2018. Aug;8(8):134. [Google Scholar]
- 39.van Esse HP, Reuber TL, van der Does D. Genetic modification to improve disease resistance in crops. New Phytol. 2020;225(1):70–86. doi: 10.1111/nph.15967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buscaill P, van der Hoorn RAL. Defeated by the nines: nine extracellular strategies to avoid microbe-associated molecular patterns recognition in plants. Plant Cell. 2021. Jul 1;33(7):2116–30. doi: 10.1093/plcell/koab109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colaianni NR, Parys K, Lee HS, Conway JM, Kim NH, Edelbacher N, et al. A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe. 2021. Apr 14;29(4):635–649.e9. doi: 10.1016/j.chom.2021.02.006 [DOI] [PubMed] [Google Scholar]
- 42.Parys K, Colaianni NR, Lee HS, Hohmann U, Edelbacher N, Trgovcevic A, et al. Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host Microbe. 2021. Apr 14;29(4):620–634.e9. doi: 10.1016/j.chom.2021.02.008 [DOI] [PubMed] [Google Scholar]
- 43.Cui H, Xiang T, Zhou JM. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol. 2009. Oct;11(10):1453–61. doi: 10.1111/j.1462-5822.2009.01359.x [DOI] [PubMed] [Google Scholar]
- 44.Nguyen QM, Iswanto ABB, Son GH, Kim SH. Recent Advances in Effector-Triggered Immunity in Plants: New Pieces in the Puzzle Create a Different Paradigm. Int J Mol Sci. 2021. Jan;22(9):4709. doi: 10.3390/ijms22094709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thordal-Christensen H. A holistic view on plant effector-triggered immunity presented as an iceberg model. Cell Mol Life Sci. 2020;77(20):3963–76. doi: 10.1007/s00018-020-03515-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421 [DOI] [PubMed] [Google Scholar]