Abstract
This scientific commentary refers to ‘Transactive response DNA-binding protein 43 is enriched at the centrosome in human cells’ by Bodin et al. (https://doi.org/10.1093/brain/awad228).
This scientific commentary refers to ‘Transactive response DNA-binding protein 43 is enriched at the centrosome in human cells’ by Bodin et al. (https://doi.org/10.1093/brain/awad228).
TDP-43 is a highly conserved, ubiquitously expressed RNA-binding protein (RBP) that serves critical functions in RNA splicing, stability and transport. While normally concentrated in the nucleus, TDP-43 is mislocalized to cytosolic inclusions in most amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) patients. Thus, TDP-43 localization and the mechanisms regulating its distribution may be highly relevant for native TDP-43 function as well as disease pathogenesis. In this issue of Brain, Bodin and colleagues1 identify a new and unanticipated localization of TDP-43 within the centrosome, hinting at a novel function for TDP-43 in microtubule organization.
TDP-43 contains two RNA-recognition motifs (RRMs), enabling recognition of UG-rich sequences in approximately one-third of all transcribed RNAs.2 A low-complexity domain (LCD) located at the C-terminus mediates protein-protein interactions and facilitates TDP-43 liquid-liquid phase separation (LLPS), a reversible de-mixing of biomolecules into droplet-like, membraneless organelles (MLOs).3 MLOs are key players in RNA and protein dynamics, effectively concentrating molecules within specialized compartments and promoting functional interactions independently of lipid membrane-bound organelles.
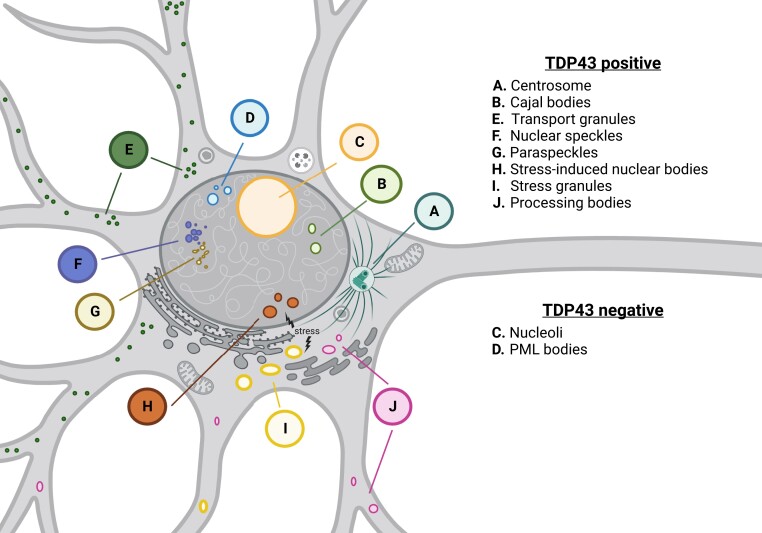
As illustrated in a growing body of work, TDP-43 is an active component of several MLOs (Fig. 1). Consistent with its contribution to transcription and alternative splicing, TDP-43 localizes to nuclear speckles4 and Cajal bodies,5 structures that are essential for RNA splicing, ribosome biogenesis and telomere maintenance. Nuclear paraspeckles are found adjacent to nuclear speckles, yet are morphologically and functionally distinct. They are maintained by the long non-coding RNA, NEAT1, which acts as a scaffold for the nucleation of RNA edited transcripts and RBPs such as TDP-43. Super-resolution microscopy confirmed the presence of TDP-43 within the shells of paraspeckles,5 while unbiased studies revealed a dramatic increase in the affinity of TDP-43 for NEAT1 in FTD patient tissue compared to controls,2 suggesting a disease-specific shift in paraspeckle content. In response to stress, TDP-43 relocalizes from these MLOs to droplet-like nuclear bodies that partially overlap with speckles and paraspeckles.6 While the native function of these stress-induced nuclear bodies remains elusive, they may modulate RNA processing under stressful conditions by sequestering splicing factors such as TDP-43, eliciting a rapid and marked change in alternative splicing that is reversible upon removal of the inciting stress.
Figure 1.
TDP-43 is a component of membraneless organelles (MLOs). Consistent with its role in RNA processing, TDP-43 is present in nuclear speckles, paraspeckles, and Cajal bodies. In response to stress, TDP-43 relocalizes to both nuclear bodies and cytosolic stress granules. TDP-43 has also been detected in cytosolic processing bodies involved in mRNA degradation, and transport bodies that relocate mRNAs to distal compartments for local translation. Bodin et al.1 add to this work by providing novel evidence for TDP-43 within the centrosome, an MLO critical for microtubule organization.
Although TDP-43 is concentrated primarily in the nucleus of healthy cells, it is also found in several cytosolic MLOs at baseline, under stress and in disease. TDP-43 is a peripheral component of stress granules, dynamic cytosolic organelles that assemble in response to various stressors.5 Much like nuclear bodies, stress granules sequester RBPs, RNA and ribosome subunits, temporarily isolating these factors in the face of stress. In keeping with its relevance to RNA stability, TDP-43 is also concentrated within processing (P)-bodies, cytosolic MLOs that function in mRNA degradation.7 Additionally, TDP-43 may shuttle RNA within transport granules to distal sites within compartmentalized cells such as neurons for local protein synthesis.8
Bodin and colleagues1 add to this considerable collection of TDP-43-positive MLOs by demonstrating the consistent localization of TDP-43 to the centrosome, a complex structure composed of two centrioles wrapped in phase-separated pericentriolar material (PCM). Conventionally, the centrosome serves to organize microtubules for processes such as cell division, signalling, establishment of polarity, and motility. However, recent investigations have uncovered novel centrosome constituents seemingly unrelated to traditional concepts of centrosome function, including ribosomes, RBPs and a subset of protein-encoding mRNAs.9 Together, these results imply that centrosomes may be sites for local translation, allowing for rapid replacement of principal centrosome components. These findings also drove Bodin et al.1 to explore a potential relationship between TDP-43 and the centrosome. Through a robust combination of high-resolution imaging and biochemical techniques in several distinct cell lines, the authors confirmed that TDP-43 does in fact localize to the centrosome.1 These results have intriguing and potentially important implications for the functions of TDP-43 and the centrosome, and how these might be disrupted in TDP-43 proteinopathies such as ALS and FTD.
Many questions remain unanswered, however. Perhaps the most pressing of these relates to the role of TDP-43 within the centrosome. Is TDP-43 required for the formation of the centrosome? Analogous to its proposed function in transport granules, it is possible that TDP-43 coats essential mRNAs encoding centrosomal components, concentrating them within the PCM and enabling local translation of constituent proteins on demand. Alternatively, by virtue of its RRMs and LCD, TDP-43 could act as a scaffold for the PCM, similar to the proposed role of the PCM nucleating factor SPD-5.10 Either or both of these functions are consistent with the fact that TDP-43 remains enriched at the centrosome in all phases of the cell cycle, as shown by Bodin and colleagues.1 Under prolonged stress or in disease conditions, cytosolic MLOs such as stress granules and their component proteins and RNA become less dynamic, perhaps forming a nidus around which pathogenic inclusions form. Could centrosomes also act as a seed for the evolution of cytosolic protein aggregates in ALS, FTD and other TDP-43 proteinopathies such as Alzheimer’s disease and limbic predominant age-related TDP-43 encephalopathy (LATE)? Additional studies are required to address these fundamental questions.
The mechanism responsible for localizing TDP-43 to the centrosome also remains mysterious. Bodin and co-authors1 collated data on centrosomal proteins and RNAs, several of which were also identified as TDP-43 interactors or substrates, respectively. One or more of these factors may be required for TDP-43 localization within centrosomes, similar to NEAT1 lncRNA in paraspeckles. Separately, the localization of TDP-43 to the centrosome may be driven by microtubule-dependent motor proteins, akin to its movement through neuronal processes as a component of neuronal transport granules.
As with most compelling science, the study by Bodin and colleagues1 raises more questions than anticipated. Rather than adding to the complexity of TDP-43 function, however, this investigation may exemplify a larger role for TDP-43 in shielding mRNA from unwarranted processing, be it splicing (within the nucleus), degradation or untimely translation (in the cytoplasm). Such a notion predicts that TDP-43 would follow the RNA, so to speak: any cytoplasmic MLO that contains sufficient quantities of RNA—including the centrosome—may be expected to harbour TDP-43. This possibility may explain the localization of TDP-43 to nearly every mRNA-rich cytosolic MLO (Fig. 1). It is also consistent with the broad array of TDP-43 substrates identified in prior work, as well as the extensive abnormalities in RNA processing that ensue upon loss of TDP-43 and/or its cytosolic accumulation in disease. Future investigations building on the work by Bodin and colleagues1 are needed to explore and extend these hypotheses to healthy cells as well as those affected by ALS, FTD and other TDP-43-related conditions.
Contributor Information
Megan Dykstra, Neuroscience Graduate Program, University of Michigan, Ann Arbor, MI 48109, USA; Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Sami J Barmada, Neuroscience Graduate Program, University of Michigan, Ann Arbor, MI 48109, USA; Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Funding
This work was supported by National Institutes of Health (R01 NS097542 and R01 NS113943 to S.J.B.; P30 AG072931 to the Alzheimer’s Disease Research Center; and 1F31NS134123-01 to M.D.).
Competing interests
The authors report no competing interests.
References
- 1. Bodin A, Greibill L, Gouju J, et al. . Transactive response DNA-binding protein 43 is enriched at the centrosome in human cells. Brain. 2023;146:3624–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tollervey JR, Curk T, Rogelj B, et al. . Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 4. Casafont I, Bengoechea R, Tapia O, Berciano MT, Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J Struct Biol. 2009;167:235–241. [DOI] [PubMed] [Google Scholar]
- 5. Pérez-Berlanga M, Wiersma VI, Zbinden A, et al. . Loss of TDP-43 oligomerization or RNA binding elicits distinct aggregation patterns. EMBO J. Published online 11 July 2023. 10.15252/embj.2022111719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang C, Duan Y, Duan G, et al. . Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle LncRNA NEAT1-mediated liquid-liquid phase separation. Mol Cell. 2020;79:443–458.e7. [DOI] [PubMed] [Google Scholar]
- 7. Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. [DOI] [PubMed] [Google Scholar]
- 8. Alami NH, Smith RB, Carrasco MA, et al. . Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Safieddine A, Coleno E, Salloum S, et al. . A choreography of centrosomal mRNAs reveals a conserved localization mechanism involving active polysome transport. Nat Commun. 2021;12:1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017;169:1066–1077.e10. [DOI] [PubMed] [Google Scholar]