Abstract
With the advent of gene therapies for amyotrophic lateral sclerosis (ALS), there is a surge in gene testing for this disease. Although there is ample experience with gene testing for C9orf72, SOD1, FUS and TARDBP in familial ALS, large studies exploring genetic variation in all ALS-associated genes in sporadic ALS (sALS) are still scarce. Gene testing in a diagnostic setting is challenging, given the complex genetic architecture of sALS, for which there are genetic variants with large and small effect sizes. Guidelines for the interpretation of genetic variants in gene panels and for counselling of patients are lacking.
We aimed to provide a thorough characterization of genetic variability in ALS genes by applying the American College of Medical Genetics and Genomics (ACMG) criteria on whole genome sequencing data from a large cohort of 6013 sporadic ALS patients and 2411 matched controls from Project MinE.
We studied genetic variation in 90 ALS-associated genes and applied customized ACMG-criteria to identify pathogenic and likely pathogenic variants. Variants of unknown significance were collected as well. In addition, we determined the length of repeat expansions in C9orf72, ATXN1, ATXN2 and NIPA1 using the ExpansionHunter tool.
We found C9orf72 repeat expansions in 5.21% of sALS patients. In 50 ALS-associated genes, we did not identify any pathogenic or likely pathogenic variants. In 5.89%, a pathogenic or likely pathogenic variant was found, most commonly in SOD1, TARDBP, FUS, NEK1, OPTN or TBK1. Significantly more cases carried at least one pathogenic or likely pathogenic variant compared to controls (odds ratio 1.75; P-value 1.64 × 10−5). Isolated risk factors in ATXN1, ATXN2, NIPA1 and/or UNC13A were detected in 17.33% of cases. In 71.83%, we did not find any genetic clues. A combination of variants was found in 2.88%.
This study provides an inventory of pathogenic and likely pathogenic genetic variation in a large cohort of sALS patients. Overall, we identified pathogenic and likely pathogenic variants in 11.13% of ALS patients in 38 known ALS genes. In line with the oligogenic hypothesis, we found significantly more combinations of variants in cases compared to controls. Many variants of unknown significance may contribute to ALS risk, but diagnostic algorithms to reliably identify and weigh them are lacking. This work can serve as a resource for counselling and for the assembly of gene panels for ALS. Further characterization of the genetic architecture of sALS is necessary given the growing interest in gene testing in ALS.
Keywords: complex genetic disease, oligogenic inheritance, motor neuron disease
The development of gene therapies for amyotrophic lateral sclerosis (ALS) has led to increased interest in genetic testing, including for patients with the sporadic form of the disease. Van Daele et al. characterize variability in ALS-associated genes, and search for likely pathogenic variants, in more than 6000 patients with sporadic ALS.
Introduction
Amyotrophic lateral sclerosis (ALS) is a motor neuron disorder characterized by upper and lower motor neuron degeneration, which leads to progressive muscle weakness and wasting.1 Up to 50% of patients develop extramotor symptoms, such as cognitive or behavioural dysfunction, as seen in frontotemporal dementia (FTD).1 The disease is relentlessly progressive and most people die between 2 and 5 years after disease onset, as effective treatments are lacking.1,2 ALS has a strong genetic component. In 5–10%, there is a familial history of ALS (fALS). Highly penetrant causal variants are found in ∼70% of fALS patients, most commonly in C9orf72, SOD1, TARDPB and FUS, which are responsible for about 40%, 20%, 4% and 3% of familial cases in Western populations, respectively.3 The remaining 90–95% of patients present with apparently sporadic ALS (sALS),4 but mutations in the same genes are found at lower frequencies.5 Twin studies suggest a heritability in sALS patients of around 60%.6 Both rare variants with a variable effect size, common variants with small effect size and combinations of such variants are thought to confer genetic risk in sALS patients, but convincing data showing this are still lacking.7,8 Nevertheless, much of the genetic architecture of ALS remains unknown. Over the past few years, many efforts have been made to unravel the missing heritability. Genetic research has linked a considerable number of genes and variants to ALS through various techniques.7,9 However, strong evidence for association is variable, and some findings have failed to be replicated in subsequent studies.10 Furthermore, the clinical significance of individual variants is often unclear (e.g. monogenetic with high penetrance, modifier, risk factor or in linkage with causal variant), especially in sALS patients. As the risk of ALS is age-dependent, the life-time risk is in the order of 1/400 for males and 1/550 for females and since many of the reported disease-associated variants have been associated with incomplete penetrance (in fALS pedigrees), such variants will invariably also be found in control populations.11 One of the difficulties in the interpretation of variants in complex genetic diseases like ALS is how to weigh the pathogenicity of variants, since consensus criteria are not available and different methods for scoring genetic variants are used.12,13
With the advent of precision medicine for genetic subtypes of ALS, the interest in gene testing is increasing. It is common practice to offer gene testing for fALS, but much less so for sALS. Large studies systematically describing the genetic variation in sALS are rare.14,15 However, a better insight in the level of pathogenicity of single variants is essential for the interpretation of variants in a diagnostic setting, for counselling of patients and their families and for use in clinical drug trials.
We analysed whole genome data of a large cohort of 6013 sporadic ALS patients and focused on a set of 90 genes reported to be associated with causing ALS.16 We implemented an automatic pipeline for classification of variants using modified criteria of the American College of Medical Genetics and Genomics (ACMG) to adjudicate the variants.17 Here, we present a comprehensive overview of the pathogenic and likely pathogenic variants encountered in these ALS-associated genes, simultaneously validating the association of some of these genes with ALS in our cohort. In light of the oligogenic inheritance model of ALS, we also searched for the occurrence of combinations of such variants.
Materials and methods
Patient selection
This study used whole genome sequencing (WGS) data of a total of 9600 samples of ALS cases and age- and sex- matched controls from Project MinE. Samples were mostly of people of European descent and collected from 17 centres across 13 countries: Belgium, The Netherlands, Switzerland, Spain, France, UK, Ireland, Israel, Italy, Portugal, Sweden, Turkey, and USA. The diagnosis of ALS was made based on the revised El Escorial criteria in the including centre, which also provided clinical information. Case/control ratio was two to one. For more detailed information on patient and control selection we refer to the pilot study of Project MinE.16 A set of 8940 genomes passed quality control, of which 6529 unrelated ALS index patients and 2411 unrelated healthy controls. fALS patients, defined as having a family member with ALS in the first or second degree, were excluded for this analysis (n = 301). Patients with primary lateral sclerosis (n = 74), progressive muscular atrophy (n = 90) and progressive bulbar palsy (n = 51) were also excluded. Our final cohort consisted therefore of 6013 sALS patients and 2411 controls.
Whole genome sequencing and bioinformatics analysis
DNA was extracted and sequenced on the Illumina Hiseq 2000 and Hiseq X platforms. Sequencing data was then aligned to GRCh37 using the iSAAC Aligner, and variants called using the iSAAC variant caller; both the aligner and caller are standard to Illumina’s aligning and calling pipeline. Liftover to GRCh38 was performed using picard LiftoverVcf (v2.20.2-SNAPSHOT). Quality control was performed at sample and variant level as described previously.18
Gene selection
A list of ‘ALS-associated genes’ was selected from four databases (search conducted on 28 April 2021): OMIM (search term ‘amyotrophic lateral sclerosis’), the neuromuscular homepage (NMHP), the online version of the gene table and ALSoD (genes with definitive and strong evidence).19–22 All genes were included to provide an unbiased selection, regardless of the evidence that was available for an association with ALS. This resulted in a set of 91 genes (Supplementary Table 1). For one gene, NOTCH2NLC, the liftover to GRCh38 failed because of mappability issues, excluding this gene from further analysis. This brought the total number of eligible genes to 90. Additionally, we randomly selected 90 genes with expression in brain as obtained from the GTEx Portal for comparison (Supplementary Table 2).23
The exact genomic regions corresponding to these genes were extracted from Ensembl (version 96), to which we added a buffer region of 2500 base pairs both upstream and downstream.24 For each gene, we selected the canonical transcript (Supplementary Table 1).
Variant extraction and annotation
We extracted all sequencing variants located in the genomic region of the selected genes from WGS data of patients and controls. Variants were annotated using nsemble-vep (v96.0-0). Vcfanno (v0.3.1) was used to add precalculated scores of the dbnsfp (v4.1a) and dbscsnv (v1.1) databases and add the allele frequency from Gnomad (v2.1.1 liftover_grch38).
Repeat expansions in C9orf72 (the GGGGCC-repeat sequence in intron 1a) and NIPA1, as well as intermediate repeat expansions in ATXN1 and ATXN2, were identified in our WGS data using the ExpansionHunter tool.25 Cut-offs used were repeat lengths: 30 for C9orf72, 39 for ATXN1, 33 for ATXN2 and 10 for NIPA1. Expanded means that the lower bound of the estimated repeat length range is higher or equal to the cut-off. Normal means that the upper bound of the range is lower than the cut-off. Intermediate means that the result is in between ‘expanded’ and ‘normal’. Where available, results from ExpansionHunter were compared with wet lab results (repeat primed PCR). Inconsistent means that a discrepancy between the result from ExpansionHunter and the wet lab was found. Unknown means the length of the repeat could not be determined. The single nucleotide polymorphism (SNP) rs12608932 in UNC13A is a known risk factor for ALS.26 Frequency of the CC genotype was distilled from our data.
Attribution of variant pathogenicity
To predict variant pathogenicity we used the software tool CharGer (Characterization of Germline variants), an open-source framework conducting a fully automated variant interpretation based on the ACMG-criteria, enriched with custom modules and a user-adjustable scoring system.27 We partially customized the criteria and included some changes to make them more suitable for complex diseases. A summary of how we customized the ACMG criteria can be found in Supplementary Figs 1 and 2 and Supplementary Tables 1 and 3. After applying these modified criteria on the extracted sequencing variants, we selected all variants with a classification of pathogenic (class V) or likely pathogenic (class IV).
Expansions in ATXN1, ATXN2 and NIPA1 and the SNP rs12608932 in UNC13A were not considered pathogenic but were logged as risk factors.
Known mutations associated with ALS
We extracted variants associated with ALS in the 90 genes of interest from two databases: ClinVar and the NMHP.20,28 We reclassified these variants as described in Supplementary Fig. 1 and selected the variants with a final classification as pathogenic and likely pathogenic as ‘known ALS-associated mutations’. An overview of the number of variants reclassified per gene is provided in Supplementary Table 4.
Statistical analysis
R (v3.6.3) was used to perform Fisher exact tests to compare variant frequency between cases and controls in our dataset assuming a dominant model. Odds ratios (ORs) with zero cell counts in the control population were estimated by assuming the control population had a count of one, these ORs are denoted with a tilde. Multiple testing correction was performed using the Holm method as implemented in p.adjust. For comparison between patients with and patients without pathogenic and likely pathogenic variants χ2 test was performed for categorical variables and a t-test for continuous variables.
GnomAD
To calculate ORs compared to GnomAD v3, the variants that passed quality control were downloaded from https://gnomad.broadinstitute.org and processed through our modified ACMG criteria, retaining only the pathogenic and likely pathogenic variants.29 These were compared to the pathogenic and likely pathogenic variants present in our ALS cohort after removing the variants that were also removed in GnomAD. Fisher’s exact test was then used as described above.
Graphical representation
Graphical representations called ‘lolliplots’ were created in R (version 3.6.0) using the Bioconductor package trackViewer (version 1.20.5).27,30
Ethical approval
The study was approved by the national ethical committees of the participating centres. All participants signed an informed consent, which also included permission to publish scientific results at the group level. The Commission of medical ethics of UZ KU Leuven gave permission for sample collections (S50354) and for exome/genome sequencing (S52853).
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.
Results
Our final cohort consisted of 6013 sALS patients and 2411 controls. The MinE project was set up to discover new ALS genes, so patients with known causal mutations might be underrepresented. The main characteristics of the cohort are summarized in Table 1.
Table 1.
Summary of the main characteristics of the MinE cohort
| Cases (n = 6013) | Controls (n = 2411) | |
|---|---|---|
| Sex | ||
| Female, n | 2399 (39.90%) | 1140 (47.28%) |
| Male, n | 3614 (60.10%) | 1271 (52.72%) |
| Age at onset | ||
| Number of patients for whom data was available | 5878 | – |
| Mean age at onset, years | 60.35 | – |
| Age at time of blood sample | ||
| Number of patients for whom data was available | 4883 | 2197 |
| Mean age at time of blood sample, years | 62.20 | 61.30 |
| Site of onset | ||
| Bulbar | 1617 (26.89%) | – |
| Spinal | 3916 (65.13%) | – |
| Generalized | 201 (3.34%) | – |
| Thoracic/respiratory | 107(1.78%) | – |
| FTD | 7 (0.12%) | – |
| Unknown | 165 (2.74%) | – |
| El Escorial (most recent value) | ||
| Definite | 1385 (23.03%) | – |
| Probable | 2309 (38.40%) | – |
| Possible | 515 (8.56%) | – |
| Suspected | 57(0.95%) | – |
| Unknown | 1747 (29.05%) | – |
| C9orf72 RE status | ||
| Expanded | 313 (5.21%) | 7 (0.29%) |
| Intermediate | 18 (0.30%) | 4 (0.17%) |
| Normal | 5659 (94.11%) | 2397 (99.42%) |
| Inconsistent | 22 (0.37%) | 0 (0%) |
| Unknown | 1 (0.02%) | 3 (0.12%) |
Numbers in parentheses refer to percentage of total. RE = repeat expansion.
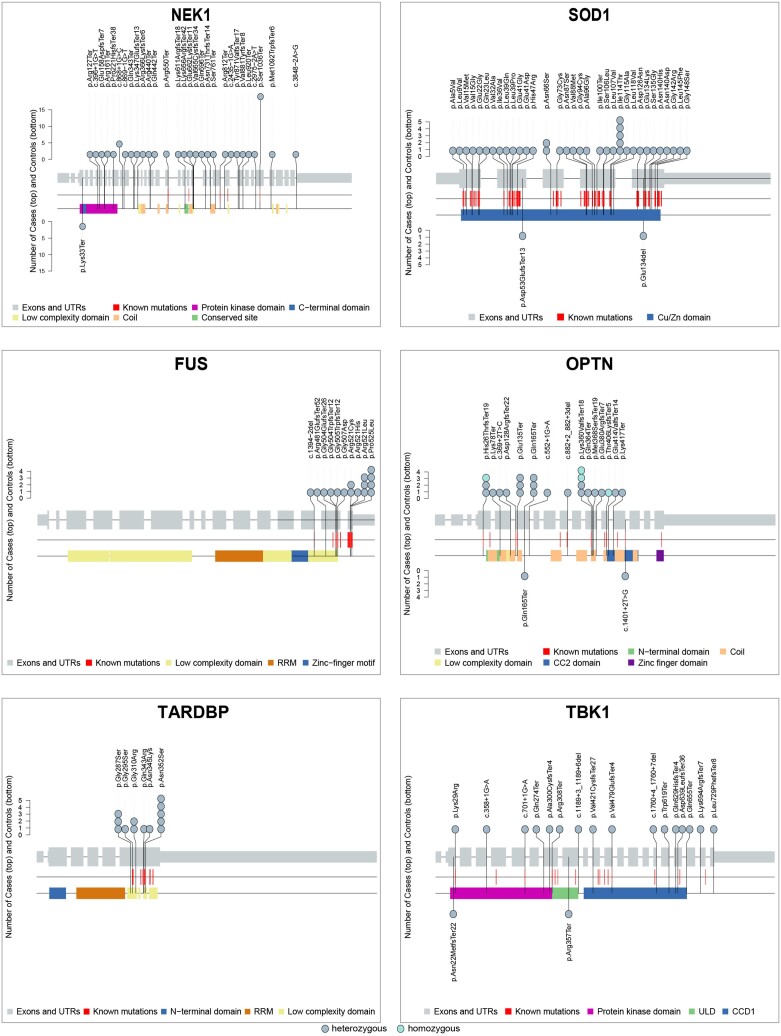
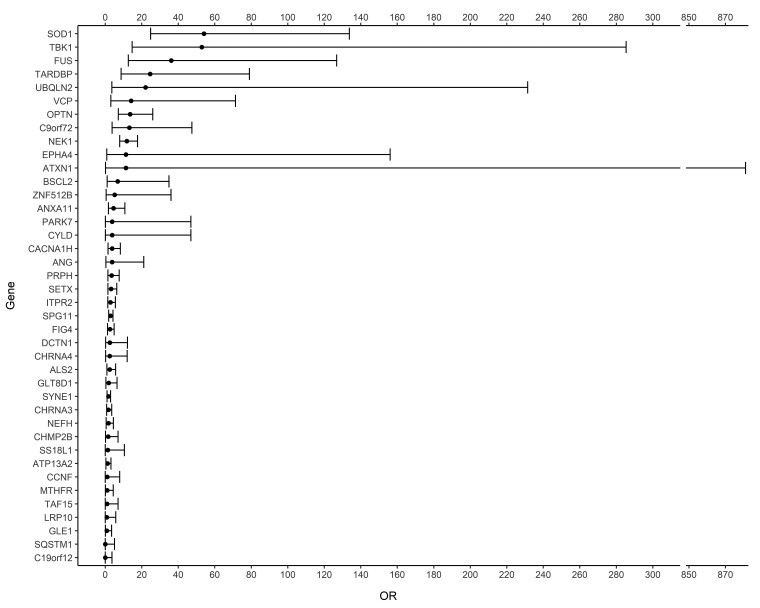
Using the ExpansionHunter tool, we identified 313 cases (5.21%) with a C9orf72 hexanucleotide repeat expansion. Looking at the 90 genes, we identified 365 pathogenic and likely pathogenic sequencing variants in 354 (5.89%) cases and 87 pathogenic and likely pathogenic sequencing variants in 83 controls (3.44%) (Table 2, Supplementary Table 5 and Supplementary Table 6). Significantly more cases carried at least one pathogenic or likely pathogenic sequencing variant compared to controls (OR 1.75; P-value 1.64 × 10−5). Pathogenic and likely pathogenic sequencing variants were observed in 40 of the 90 genes, that is in 38 genes in cases and in 24 genes in controls (Table 2, Supplementary Table 5, Fig. 1 and Supplementary Fig. 3). In 25 genes, a higher proportion of patients carried at least one pathogenic or likely pathogenic variant compared to controls. For 10 genes, the OR for carrying a pathogenic or likely pathogenic variant was higher than two, with a P-value below the significance threshold of 0.05 for the genes NEK1 (OR 21.84; P-value 2.57 × 10−7), SOD1 (OR 7.66; P-value 3.15 × 10−4), FUS (OR ∼6.43; P-value 9.40 × 10−3), OPTN (OR 4.83; P-value 1.58 × 10−2) and TARDBP (OR ∼5.22; P-value 2.59 × 10−2). P-values remained below the adjusted significance threshold only for NEK1 and SOD1 after correcting for multiple testing. However, by increasing the size of the control population by taking data from gnomAD, we could confirm these top five genes, obtaining even lower P-values. Some other genes also reached P-values below the significance threshold when compared to gnomAD after correction for multiple testing. However, only TBK1 could be added to the list of significant genes using a more stringent genome wide correction for the ±20 000 genes in the genome. The ORs and their 95% confidence interval after comparison to gnomAD are illustrated in Fig. 2. The known Asp91Ala (D91A) variant in SOD1 was discarded by our pipeline, because rule BS1 was triggered (allele frequency higher than expected for the disorder). We extracted the frequencies in a separate analysis and found 19 sALS patients with a heterozygous SOD1 D91A variant, 21 with a homozygous variant and five controls with a heterozygous variant. In 15 genes, a higher proportion of controls carried at least one pathogenic or likely pathogenic variant. The P-value was below the significance threshold of 0.05 only in NEFH (OR 0.30; P-value 3.26 × 10−2) but did not survive correction for multiple testing and was also not significant when controls from gnomAD were added. In 50 genes, we did not find any pathogenic or likely pathogenic variant. Variants of unknown significance (VUS) are listed in Supplementary Table 7. Patients with pathogenic and likely pathogenic variants were more often diagnosed with FTD (32.41% versus 21.56% of patients with data available, P-value 1.70 × 10−2) and had younger age of onset (58.62 years versus 60.56 years, P-value 1.17 × 10−4) (Supplementary Table 8). In the set of 90 randomly selected genes, we did not find significantly more pathogenic variants in cases or controls, neither in total nor at the individual gene level (OR 1.46; P-value 3.39 × 10−1) (Supplementary Table 9). Of the variants associated with ALS in ClinVar and the NMHP, 367 were reclassified as likely pathogenic or pathogenic by applying our modified ACMG criteria. These variants were considered ‘known ALS-associated mutations’. In our cohort, 60 (23.0%) of the unique identified variants in cases were known ALS-associated mutations, the majority were novel variants.
Table 2.
Genes with pathogenic or likely pathogenic variants in cases and/or controls
| GnomAD | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Case (%) | Control (%) | P-value | Adj. P-value | Est. OR | 95% CI | GnomAD (%) | P-value | Adj. P-value | Est. OR | 95% CI |
| NEK1 | 54 (0.90) | 1 (0.04) | 2.57 × 10−7 | 1.03 × 10−5 | 21.84 | 3.75–875.45 | 52 (0.04) | 9.19 × 10−31 | 3.58 × 10−29 | 11.84 | 7.93–17.68 |
| SOD1 | 38 (0.63) | 2 (0.08) | 3.15 × 10−4 | 1.23 × 10−2 | 7.66 | 1.98–65.66 | 8 (0.01) | 4.47 × 10−34 | 1.79 × 10−32 | 54.09 | 24.83–133.75 |
| FUS | 16 (0.27) | 0 (0.00) | 9.40 × 10−3 | 0.36 | ∼6.43 | 1.55–Inf | 5 (0.00) | 5.01 × 10−14 | 1.85 × 10−12 | 36.19 | 12.67–126.77 |
| OPTN | 24 (0.40) | 2 (0.08) | 1.58 × 10−2 | 0.58 | 4.83 | 1.2–42.2 | 20 (0.01) | 2.29 × 10−15 | 8.72 × 10−14 | 13.62 | 7.2–26.01 |
| TARDBP | 13 (0.22) | 0 (0.00) | 2.59 × 10−2 | 0.93 | ∼5.22 | 1.22–Inf | 6 (0.00) | 1.12 × 10−10 | 3.92 × 10−9 | 24.57 | 8.71–79.01 |
| NEFH | 6 (0.10) | 8 (0.33) | 3.26 × 10−2 | 1.00 | 0.30 | 0.09–0.99 | 32 (0.02) | 0.23 | 1.00 | 1.73 | 0.53–4.48 |
| PRPH | 9 (0.15) | 0 (0.00) | 6.79 × 10−2 | 1.00 | ∼3.61 | 0.79–Inf | 29 (0.02) | 2.85 × 10−3 | 7.40 × 10−2 | 3.51 | 1.46–7.63 |
| TBK1 | 16 (0.27) | 2 (0.08) | 0.12 | 1.00 | 3.21 | 0.75–28.84 | 3 (0.00) | 2.88 × 10−13 | 1.04 × 10−11 | 52.92 | 14.76–285.4 |
| ITPR2 | 12 (0.20) | 1 (0.04) | 0.13 | 1.00 | 4.82 | 0.71–205.93 | 47 (0.03) | 2.48 × 10−3 | 6.70 × 10−2 | 2.89 | 1.4–5.54 |
| SPG11 | 32 (0.53) | 7 (0.29) | 0.16 | 1.00 | 1.84 | 0.8–4.94 | 126 (0.09) | 1.28 × 10−6 | 4.35 × 10−5 | 2.88 | 1.89–4.28 |
| C19orf12 | 0 (0.00) | 1 (0.04) | 0.29 | 1.00 | 0.00 | 0–15.64 | 13 (0.01) | 0.62 | 1.00 | 0.00 | 0–3.68 |
| SQSTM1 | 0 (0.00) | 1 (0.04) | 0.29 | 1.00 | 0.00 | 0–15.64 | 10 (0.01) | 1.00 | 1.00 | 0.00 | 0–5.05 |
| CHRNA4 | 2 (0.03) | 2 (0.08) | 0.32 | 1.00 | 0.40 | 0.03–5.53 | 9 (0.01) | 0.23 | 1.00 | 2.48 | 0.26–11.99 |
| VCP | 5 (0.08) | 0 (0.00) | 0.33 | 1.00 | ∼2.01 | 0.37–Inf | 4 (0.00) | 3.36 × 10−4 | 1.04 × 10−2 | 14.15 | 3.05–71.34 |
| ALS2 | 7 (0.12) | 5 (0.21) | 0.34 | 1.00 | 0.56 | 0.15–2.24 | 32 (0.02) | 3.57 × 10−2 | 0.81 | 2.48 | 0.92–5.71 |
| SYNE1 | 18 (0.30) | 10 (0.41) | 0.41 | 1.00 | 0.72 | 0.32–1.75 | 116 (0.09) | 3.72 × 10−2 | 0.81 | 1.76 | 1.01–2.9 |
| CACNA1H | 29 (0.48) | 15 (0.62) | 0.41 | 1.00 | 0.77 | 0.4–1.56 | 27 (0.02) | 1.91 × 10−3 | 5.55 × 10−2 | 3.77 | 1.56–8.27 |
| LRP10 | 1 (0.02) | 1 (0.04) | 0.49 | 1.00 | 0.40 | 0.01–31.48 | 13 (0.01) | 1.00 | 1.00 | 0.87 | 0.02–5.8 |
| TAF15 | 1 (0.02) | 1 (0.04) | 0.49 | 1.00 | 0.40 | 0.01–31.48 | 11 (0.01) | 1.00 | 1.00 | 1.02 | 0.02–7 |
| CHRNA3 | 9 (0.15) | 5 (0.21) | 0.56 | 1.00 | 0.72 | 0.22–2.74 | 58 (0.04) | 0.12 | 1.00 | 1.76 | 0.76–3.57 |
List of the first 20 of 40 genes in which pathogenic or likely pathogenic variants were found in cases and/or controls (sorted on increasing P-value). On the left, the absolute number of ALS patients and controls of our cohort with minimal one pathogenic or likely pathogenic variant in that gene is indicated along with the proportion of total cases (n = 6013) and controls (n = 2411) as a percentage in brackets. P-value, adjusted (Adj.) P-value after correction for multiple testing, estimated odds ratio and 95% confidence interval are shown in separate columns. To the right, a comparison with controls of the gnomAD database. Repeat expansions and SNP rs12608932 in UNC13A are not included in this table. The full table with all 40 genes can be found in the Supplementary material (Supplementary Table 5). Adj = adjusted; CI = confidence interval; Est = estimated; Inf = infinity; NMHP = neuromuscular homepage; OR = odds ratio.
Figure 1.
Lolliplots showing pathogenic and likely pathogenic variants encountered in NEK1, SOD1, FUS, OPTN, TARDBP and TBK1 in cases and controls. Lolliplots showing the mutational spectra of the displayed genes in cases (n = 6013) and controls (n = 2411). For each gene, exons and untranslated regions are represented as grey bars, connected by introns shown as grey lines (reduced with a factor 1/100). Protein domains and motifs are shown as coloured bars. Each lollipop represents a pathogenic or likely pathogenic variant encountered in cases (top) or controls (bottom). The absolute number of cases and controls with the variant is reflected by the height of the lollipop. Known ALS-associated mutations (from ClinVar and the NMHP) are shown as red lines. AXH = ataxin-1/HBP1 domain; CCD1 = coiled-coil domain 1; NMHP = neuromuscular homepage; RRM = RNA recognition motif; ULD = ubiquitin-like domain; UTR = untranslated region.
Figure 2.
Odds ratios and their 95% confidence intervals. Illustration of the odds ratios and their 95% confidence intervals for the 40 genes with pathogenic or likely pathogenic variants in. Odds ratios are calculated compared to gnomAD v3.
Taken together, 313 cases (5.21%) were explained by a C9orf72 hexanucleotide repeat expansion. A pathogenic or likely pathogenic sequencing variant was observed in SOD1, FUS, TARDBP, NEK1, OPTN and TBK1 in 38 (0.63%), 16 (0.27%), 13 (0.22%), 54 (0.90%) and 24 (0.40%) and 16 (0.27%), respectively. Of the remaining cases, 179 (2.98%) had a mutation in one of the outstanding genes. In the remaining part of the cohort, we detected one or more susceptibility factors in 1042 (17.33%) cases. In the other 4319 (71.83%) cases, we did not find any genetic clue. In the control cohort, a C9orf72 hexanucleotide repeat expansion was observed in seven (0.29%) controls. Pathogenic or likely pathogenic sequencing variants were observed in SOD1, NEK1, OPTN and TBK1 in two (0.08%), one (0.04%), one (0.04%) and one (0.04%) control, respectively. Pathogenic or likely pathogenic sequencing variants in the other genes were found in 76 (3.15%) controls [not statistically different from the proportion of 2.98% in cases (P-value 0.72)]. We found only susceptibility factors in 387 (16.05%) controls. No variants of interest were found in 1934 (80.22%) controls.
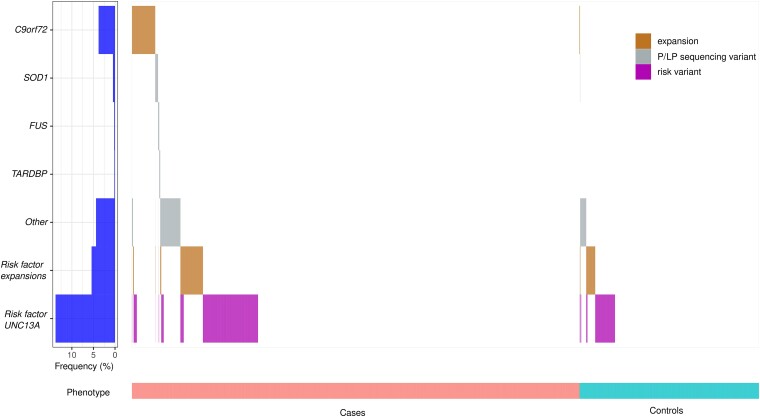
To evaluate the incidence of combinations of variants, we took following variants into account: pathogenic and likely pathogenic sequencing variants, C9orf72 repeat expansions and ALS susceptibility factors including ATXN1 and ATXN2 intermediate repeat expansions, NIPA1 repeat expansions and homozygosity for the C-allele of rs12608932 in UNC13A. A combination of two of these variants was found in 164 (2.73%) cases and 34 (1.41%) controls (OR 1.96; P-value 2.31 × 10−4) and a combination of three variants in nine (0.15%) cases and in two (0.08%) controls (OR 1.81; P-value 0.74) (Fig. 3 and Supplementary Table 10).
Figure 3.
Waterfall plot: combinations of variants. A waterfall plot shows for all cases (left) and controls (right) which variants they carry: repeat expansions in C9orf72; pathogenic or likely pathogenic sequencing variants in NEK1, SOD1, FUS, OPTN, TARDBP, TBK1 or in one of the other ALS-associated genes; risk factors such as (intermediate) repeat expansions in ATXN1, ATXN2 and NIPA1 or the CC-genotype of the SNP rs12608932 in UNC13A. Brown = repeat expansions; grey = pathogenic (P) and likely pathogenic (LP) sequencing variants; purple = non-repeat expansion risk variants.
Discussion
We here present a comprehensive genetic profile of a large sALS cohort. In total, 10.85% of the studied cases with sALS diagnosis had a pathogenic variant—5.21% had a pathogenic repeat expansion in C9orf72 and 2.66% a pathogenic or likely pathogenic mutation in either SOD1, TARDBP, FUS, NEK1, OPTN or TBK1. We observed potentially causative variants in other ALS-associated genes in an additional 3.26% of sALS patients (in 2.98% when patients carrying a combination with a C9orf72 hexanucleotide repeat expansion or with variants in SOD1, FUS, TARDBP, NEK1, OPTN and TBK1 were excluded). In 17.33%, we found isolated risk factors, but of importance, in 71.83%, no genetic significant variants were detected.
Until now, similar projects have mainly focused on fALS patients. Here we describe the genetic landscape of a large cohort of sporadic ALS patients.13 An overview of the genetic findings in ALS patients is a first step to get better understanding of the genetic architecture of ALS. Especially in the era of next generation sequencing and with the advent of gene-specific therapies, genetic screening is offered to an increasing number of patients. Therefore, a comprehensive genetic characterization of sALS patients is indispensable for informed counselling of these patients. Evidence on the strength of association of genes with ALS is also valuable for the development and analysis of gene panels.
Overall, when we analysed genetic variation in 90 genes for which association with ALS has been reported, we observed an enrichment of pathogenic and likely pathogenic sequencing variants in cases compared to controls. In NEK1, SOD1, FUS, OPTN and TARDBP enrichment was significant. That these genes end up as top genes is not surprising since it is known that 2–3% of sALS patients carry pathogenic mutations in these genes.5 That only NEK1 and SOD1 survived multiple testing is probably due to the low numbers of variants per individual gene. This strongly reduces statistical power, even though this is the largest sALS cohort described to date. Using data from the gnomAD database to supplement our control data, all five originally significant genes were confirmed with higher significance levels. The low statistical power might also provide an explanation for the non-significant results in some of the 33 other genes with pathogenic and likely pathogenic variants, as illustrated by TBK1. For most of these genes without significant enrichment however, especially for the ones with ORs close to 1, we suspect that the probability of an association with ALS is rather low. The same is true for the 52 genes without pathogenic or likely pathogenic variants detected in cases. Nevertheless, we cannot exclude that some of these genes harbour truly pathogenic mutations in only a small minority of sALS patients which are therefore not detected in our cohort. Genes conferring moderate risk for ALS are also not reliably identified by our pipeline, as is illustrated by the known polymorphism in UNC13A.
The yield of WGS (combined gene panel analysis and C9orf72 repeat expansion detection in WGS data) in sALS patients was 11.09% overall and 7.87% when only taking C9orf72 repeat expansions, SOD1, FUS, TARDBP, NEK1, OPTN and TBK1 into consideration. This is lower than what is reported by others, but in those studies less stringent variant selection was performed (e.g. all novel and rare variants and/or ATXN2 intermediate expansions were reported in the yield) and a different number of genes was sequenced.15,31 In 71.83% of sALS patients, we did not find any pathogenic or likely pathogenic variants, so genetic determinants in this patient group remain unknown. Possible explanations are the presence of variants in as yet unidentified genes, variants in noncoding regions of the genome, multiple polygenic additive contributions or epigenetic variation. Another possible reason is an oligogenic architecture where multiple less penetrant variants together constitute the risk of ALS in these patients. Such variants are not easily identified using the ACMG criteria. However, the fact that we found a significantly higher incidence of combinations in cases compared to controls could be a hint to a complex inheritance of (risk) variants in multiple genes.8 Furthermore, the presence of pathogenic and likely pathogenic variants in our control cohort points towards a reduced penetrance, which also fits with the oligogenic model. Even in SOD1, we found two controls carrying a pathogenic variant (0.08%), which is higher than the expected frequency (0.0025%) in controls, assuming a lifetime risk for ALS of 1/400 and a frequency of 1% SOD1 mutations in sporadic patients.32 This illustrates that the penetrance at the population level is lower than what is expected based on smaller clinical cohorts, as has been shown for other neurodegenerative disorders before.33,34 Probably other genetic and environmental factors play a role and explain why some mutation carriers seem more resistant to developing a clinical phenotype. That we found a higher frequency of variants in controls (3.15%) compared to cases (2.98%) in the remaining genes after excluding repeat expansions in C9orf72 and variants in SOD1, FUS, TARDBP, NEK1, OPTN and TBK1 can have multiple explanations: not all genes are robustly associated with ALS, some variants have a low penetrance or there is an overrepresentation of some variants because of populations stratification in a small control cohort. The occurrence of pathogenic and likely pathogenic variants in 3.44% of controls might thus be indicative for reduced penetrance and oligogenic inheritance but should be interpreted with caution because of the reasons mentioned above. It is important to acknowledge that the finding of a likely pathogenic or pathogenic variant in an ALS gene does not automatically imply Mendelian inheritance of ALS. It would be interesting to follow up controls with these variants longitudinally, as well as controls with a C9orf72 repeat expansion, however this is not intended in the design of Project MinE. Further exploration of the penetrance of variants and the meaning of the presence of variants in the control population is of great importance since this dramatically influences genetic counselling in patients.
One of the problems in the analysis and interpretation of variants in complex genetic disorders such as ALS is the lack of adequate scoring systems to assess the pathogenicity of variants. Frequently used criteria such as the ACMG guidelines are designed for Mendelian disorders and less applicable for diseases where variants might have low penetrance, be risk factors or might only be pathogenic in certain combinations.17,35 Furthermore, these criteria are often vaguely defined, and standard algorithms for implementing them are lacking, which leads to leeway and subjectivity in their practical implementation.17,36 This makes an automated implementation on large datasets challenging. Since better classification systems are lacking, there is no consensus on how to assess the pathogenicity for variants in complex disorders, and different methods are used through literature.12,13 International attempts to correct this, such as ClinGen Gene Curation Expert Panels and Variant Curation Expert Panels, may represent a way forward. For this project, we used the software tool CharGer applying modified ACMG-criteria.27 Despite our modifications, our criteria still favour the identification of more deleterious variants (e.g. nonsense and truncating variants) that are more often encountered in Mendelian disorders. On the other hand, some of the identified variants are equally present in cases and controls. Although the classification of pathogenic and likely pathogenic variants already entails many challenges, the interpretation of VUS variants is even more complicated. This underlines the need for better classification systems for variants in complex diseases.
There are some limitations to this study. Since Project MinE is a multicentre study, our cohort is composed of samples from different countries. Patients are mostly of European descent, which makes it a good reference work for this population, but caution is needed for the extrapolation to other populations. Although the control population was matched for the different populations included, the size of the control population was too small to calculate robust ORs for every single variant. Furthermore, the samples were not included according to strict criteria other than the revised El Escorial criteria, which might have led to small differences in inclusion criteria per centre. Lastly, the definition of ‘familial ALS’ has been slippery over the years.37 Extensive data about the familial history was not (always) available (e.g. occurrence of FTD in the family, ALS in third degree family members), so only the more ‘obvious’ fALS cases were excluded in this study, possibly leading to an overinflation of sALS cases.
In conclusion, this paper constitutes an inventory of pathogenic variation in a large cohort of European-ancestry sALS patients, which can serve as a valuable resource for genetic counselling and for the design of ALS gene panels. We could not provide evidence for association with ALS for all genes. However, further research is needed to exclude very rare pathogenic variants or variants with moderate risk factors in these genes. To better explore the oligogenic hypothesis, even larger cohorts are needed to reach acceptable statistical power. A challenging journey lies ahead to obtain reliable comprehensive gene testing and counselling of sALS patients. However, it is a priority for clinical trial design and therapy development in ALS.
Supplementary Material
Acknowledgements
P.V.D. holds a senior clinical investigatorship of FWO-Vlaanderen and is supported by the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders, the ALS Liga België and the KU Leuven funds ‘Een Hart voor ALS’, ‘Laeversfonds voor ALS Onderzoek’ and the ‘Valéry Perrier Race against ALS Fund’. Several authors of this publication are members of the European Reference Network for Rare Neuromuscular Diseases (ERN-NMD). A.A.-C. is an NIHR Senior Investigator (NIHR202421). This is an EU Joint Programme—Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organisations under the aegis of JPND—www.jpnd.eu (United Kingdom, Medical Research Council (MR/L501529/1; MR/R024804/1) and Economic and Social Research Council (ES/L008238/1)) and through the Motor Neurone Disease Association, My Name’5 Doddie Foundation, and Alan Davidson Foundation. This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. Samples used in this research were in part obtained from the UK National DNA Bank for MND Research, funded by the MND Association and the Wellcome Trust. We would like to thank people with MND and their families for their participation in this project. We acknowledge sample management undertaken by Biobanking Solutions funded by the Medical Research Council at the Centre for Integrated Genomic Medical Research, University of Manchester.
Contributor Information
Sien Hilde Van Daele, Department of Neurosciences, Experimental Neurology, KU Leuven—University of Leuven, and Leuven Institute for Neuroscience and Disease (LIND), 3000 Leuven, Belgium; VIB, Center for Brain & Disease Research, Laboratory of Neurobiology, 3000 Leuven, Belgium; Department of Neurology, University Hospitals Leuven, 3000 Leuven, Belgium; Department of Human genetics, University Hospitals Leuven, 3000 Leuven, Belgium.
Matthieu Moisse, Department of Neurosciences, Experimental Neurology, KU Leuven—University of Leuven, and Leuven Institute for Neuroscience and Disease (LIND), 3000 Leuven, Belgium; VIB, Center for Brain & Disease Research, Laboratory of Neurobiology, 3000 Leuven, Belgium.
Joke J F A van Vugt, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Ramona A J Zwamborn, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Rick van der Spek, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Wouter van Rheenen, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Kristel Van Eijk, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Kevin Kenna, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Philippe Corcia, Centre SLA, CHRU de Tours, 37044 Tours, France; UMR 1253, iBrain, Université de Tours, Inserm, 37032 Tours, France.
Patrick Vourc'h, UMR 1253, iBrain, Université de Tours, Inserm, 37032 Tours, France.
Philippe Couratier, Centre SLA, CHU Limoges, 87042 Limoges, France.
Orla Hardiman, Academic Unit of Neurology, Trinity College Dublin, Trinity Biomedical Sciences Institute, Dublin D02 PN40, Republic of Ireland.
Russell McLaughin, Complex Trait Genomics Laboratory, Smurfit Institute of Genetics, Trinity College Dublin, Dublin D02 PN40, Republic of Ireland.
Marc Gotkine, The Agnes Ginges Center for Human Neurogenetics, Hadassah Medical Organization and Faculty of Medicine, Hebrew University of Jerusalem, 91120 Jerusalem, Israel.
Vivian Drory, Department of Neurology, Tel-Aviv Sourasky Medical Centre, 64239 Tel Aviv, Israel.
Nicola Ticozzi, Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milano, Italy; Department of Pathophysiology and Transplantation, ‘Dino Ferrari’ Center, Università degli Studi di Milano, 20122 Milan, Italy.
Vincenzo Silani, Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milano, Italy; Department of Pathophysiology and Transplantation, ‘Dino Ferrari’ Center, Università degli Studi di Milano, 20122 Milan, Italy.
Antonia Ratti, Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milano, Italy; Department of Medical Biotechnology and Translational Medicine, Università degli Studi di Milano, 20133 Milano, Italy.
Mamede de Carvalho, Instituto de Fisiologia, Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, 1649-028 Lisbon, Portugal.
Jesús S Mora Pardina, ALS Unit, Hospital University San Rafael, 28016 Madrid, Spain.
Monica Povedano, Servei de Neurologia, HUB-IDIBELL, 08908 Barcelona, Spain.
Peter M Andersen, Department of Clinical Science, Neurosciences, Umeå University, 901 87 Umeå, Sweden.
Markus Weber, Neuromuscular Diseases Unit/ALS Clinic, Kantonsspital St. Gallen, 9007 St. Gallen, Switzerland.
Nazli A Başak, Koç University, School of Medicine, KUTTAM-NDAL, 34010 Istanbul, Turkey.
Chris Shaw, Maurice Wohl Clinical Neuroscience Institute, King's College London, Department of Basic and Clinical Neuroscience, London SE5 9RT, UK.
Pamela J Shaw, Sheffield Institute for Translational Neuroscience (SITraN), University of Sheffield, Sheffield S10 2HQ, UK.
Karen E Morrison, School of Medicine, Dentistry and Biomedical Sciences, Queen’s University Belfast, Belfast BT9 7BL, UK.
John E Landers, Department of Neurology, University of Massachusetts Medical School, Worcester, MA 01655, USA.
Jonathan D Glass, Department Neurology, Emory University School of Medicine, Atlanta, GA 30322, USA.
Michael A van Es, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Leonard H van den Berg, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Ammar Al-Chalabi, Maurice Wohl Clinical Neuroscience Institute, King's College London, Department of Basic and Clinical Neuroscience, London SE5 9RT, UK.
Jan Veldink, Department of Neurology, UMC Utrecht Brain Center, Utrecht University, 3584 CX Utrecht, The Netherlands.
Philip Van Damme, Department of Neurosciences, Experimental Neurology, KU Leuven—University of Leuven, and Leuven Institute for Neuroscience and Disease (LIND), 3000 Leuven, Belgium; VIB, Center for Brain & Disease Research, Laboratory of Neurobiology, 3000 Leuven, Belgium; Department of Neurology, University Hospitals Leuven, 3000 Leuven, Belgium.
Funding
This work was supported by grants from the KU Leuven (C1—C14-17-107), FWO-Vlaanderen (G0B2819N), the IWT (Project MinE), the Belgian National Lottery and the ALS Liga België. S.H.V.D. is funded by a PhD fellowship of the Research Foundation—Flanders (FWO) (1164018N, file number 40900). Research is furthermore funded by the Fund for Clinical Academic studies of the university hospitals of Leuven, Belgium (S61184). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no 772376—EScORIAL). The collaboration project is co-funded by the PPP Allowance made available by Health∼Holland, Top Sector Life Sciences & Health, to stimulate public-private partnerships. This study was supported by the ALS Foundation Netherlands.
Competing interests
J.V. reports to have sponsored research agreements with Biogen and Astra Zeneca. A.A.-C reports consultancies or advisory boards for Amylyx, Apellis, Biogen, Brainstorm, Cytokinetics, GenieUs, GSK, Lilly, Mitsubishi Tanabe Pharma, Novartis, OrionPharma, Quralis, Sano, Sanofi and Wave Pharmaceuticals. P.V.D. has served in advisory boards for Biogen, CSL Behring, Alexion Pharmaceuticals, Ferrer, QurAlis, Cytokinetics, Argenx, UCB, Muna Therapeutics, Alector, Augustine Therapeutics, VectorY and Zambon.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Hardiman O, Al-Chalabi A, Chio A, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;3:17085. [DOI] [PubMed] [Google Scholar]
- 2. Volk AE, Weishaupt JH, Andersen PM, Ludolph AC, Kubisch C. Current knowledge and recent insights into the genetic basis of amyotrophic lateral sclerosis. Med Genet. 2018;30:252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Byrne S, Heverin M, Elamin M, et al. Aggregation of neurologic and neuropsychiatric disease in amyotrophic lateral sclerosis kindreds: a population-based case-control cohort study of familial and sporadic amyotrophic lateral sclerosis. Ann Neurol. 2013;74:699–708. [DOI] [PubMed] [Google Scholar]
- 5. Van Damme P, Robberecht W, Van Den Bosch L. Modelling amyotrophic lateral sclerosis: progress and possibilities. Dis Model Mech. 2017;10:537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leblond CS, Kaneb HM, Dion PA, Rouleau GA. Dissection of genetic factors associated with amyotrophic lateral sclerosis. Exp Neurol. 2014;262(Pt B):91–101. [DOI] [PubMed] [Google Scholar]
- 8. van Blitterswijk M, van Es MA, Hennekam EAM, et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:3776–3784. [DOI] [PubMed] [Google Scholar]
- 9. Al-Chalabi A, van den Berg LH, Veldink J. Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat Rev Neurol. 2017;13:96–104. [DOI] [PubMed] [Google Scholar]
- 10. He J, Mangelsdorf M, Fan D, Bartlett P, Brown MA. Amyotrophic lateral sclerosis genetic studies: from genome-wide association mapping to genome sequencing. Neuroscientist. 2015;21:599–615. [DOI] [PubMed] [Google Scholar]
- 11. Johnston CA, Stanton BR, Turner MR, et al. Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. J Neurol. 2006;253:1642–1643. [DOI] [PubMed] [Google Scholar]
- 12. Morgan S, Shatunov A, Sproviero W, et al. A comprehensive analysis of rare genetic variation in amyotrophic lateral sclerosis in the UK. Brain. 2017;140:1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Müller K, Brenner D, Weydt P, et al. Comprehensive analysis of the mutation spectrum in 301 German ALS families. Journal of neurology. Neurosurgery & Psychiatry. 2018;89:817–827. [DOI] [PubMed] [Google Scholar]
- 14. Debray S, Race V, Crabbé V, et al. Frequency of C9orf72 repeat expansions in amyotrophic lateral sclerosis: a Belgian cohort study. Neurobiol Aging. 2013;34:2890.e7–e2890.e12. [DOI] [PubMed] [Google Scholar]
- 15. Couthouis J, Raphael AR, Daneshjou R, Gitler AD. Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS Genet. 2014;10:e1004704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Project Mine ALS Sequencing Consortium . Project MinE: study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet. 2018;26:1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015; 17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moisse M, Zwamborn RAJ, van Vugt J, et al. The effect of SMN gene dosage on ALS risk and disease severity. Ann Neurol. 2021;89:686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Online Mendelian Inheritance in Man, OMIM®. Accessed 28 April 2021. https://omim.org/
- 20. Neuromuscular Disease Centre . Accessed 28 April 2021. https://neuromuscular.wustl.edu/
- 21. Benarroch L, Bonne G, Rivier F, Hamroun D. The 2021 version of the gene table of neuromuscular disorders (nuclear genome). Neuromuscul Disord. 2020; 30:1008–1048. [DOI] [PubMed] [Google Scholar]
- 22. Abel O, Powell JF, Andersen PM, Al-Chalabi A. ALSod: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum Mutat. 2012;33:1345–1351. [DOI] [PubMed] [Google Scholar]
- 23. GTEx Portal. Accessed 25 November 2021. https://gtexportal.org/home/
- 24. Yates AD, Achuthan P, Akanni W, et al. Ensembl 2020. Nucleic Acids Res. 2020;48(D1):D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dolzhenko E, Deshpande V, Schlesinger F, et al. Expansionhunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics. 2019;35:4754–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Es MA, Veldink JH, Saris CGJ, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–1087. [DOI] [PubMed] [Google Scholar]
- 27. Scott AD, Huang KL, Weerasinghe A, et al. Charger: clinical characterization of germline variants. Bioinformatics. 2019;35:865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landrum MJ, Lee JM, Benson M, et al. Clinvar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ou J, Zhu LJ. Trackviewer: A bioconductor package for interactive and integrative visualization of multi-omics data. Nat Methods. 2019;16:453–454. [DOI] [PubMed] [Google Scholar]
- 31. Cady J, Allred P, Bali T, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. [DOI] [PubMed] [Google Scholar]
- 33. Kay C, Collins JA, Miedzybrodzka Z, et al. Huntington disease reduced penetrance alleles occur at high frequency in the general population. Neurology. 2016;87:282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gardiner SL, Boogaard MW, Trompet S, et al. Prevalence of carriers of intermediate and pathological polyglutamine disease-associated alleles among large population-based cohorts. JAMA Neurol. 2019;76:650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lattante S, Marangi G, Doronzio PN, et al. High-Throughput genetic testing in ALS: the challenging path of variant classification considering the ACMG guidelines. Genes (Basel). 2020;11:1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Q, Wang K. Intervar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Byrne S, Bede P, Elamin M, et al. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:157–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy restrictions.