Abstract
Nerve injury to peripheral somatosensory system causes refractory neuropathic pain. Maladaptive changes of gene expression in primary sensory neurons are considered molecular basis of this disorder. Long non-coding RNAs (lncRNAs) are key regulators of gene transcription; however, their significance in neuropathic pain remains largely elusive.Here, we reported a novel lncRNA, named sensory neuron-specific lncRNA (SS-lncRNA), for its expression exclusively in dorsal root ganglion (DRG) and trigeminal ganglion. SS-lncRNA was predominantly expressed in small DRG neurons and significantly downregulated due to a reduction of early B cell transcription factor 1 in injured DRG after nerve injury. Rescuing this downregulation reversed a decrease of the calcium-activated potassium channel subfamily N member 1 (KCNN1) in injured DRG and alleviated nerve injury-induced nociceptive hypersensitivity. Conversely, DRG downregulation of SS-lncRNA reduced the expression of KCNN1, decreased total potassium currents and afterhyperpolarization currents and increased excitability in DRG neurons and produced neuropathic pain symptoms.Mechanistically, downregulated SS-lncRNA resulted in the reductions of its binding to Kcnn1 promoter and heterogeneous nuclear ribonucleoprotein M (hnRNPM), consequent recruitment of less hnRNPM to the Kcnn1 promoter and silence of Kcnn1 gene transcription in injured DRG.These findings indicate that SS-lncRNA may relieve neuropathic pain through hnRNPM-mediated KCNN1 rescue in injured DRG and offer a novel therapeutic strategy specific for this disorder.
Keywords: sensory neuron-specific long noncoding RNA, EBF1, hnRNPM, KCNN1, dorsal root ganglion, neuropathic pain
Wang et al. identify a sensory neuron-specific long non-coding RNA expressed predominantly in small neurons of the dorsal root ganglion, and show that this RNA contributes to the development and maintenance of neuropathic pain by regulating KCNN1 expression.
Introduction
Neuropathic pain is a chronic, refractory clinical condition affecting approximately 6.9–10% of the world population.1,2 Current treatments for this disorder are highly limited.3 Although opioids are the last option for pharmacological management of neuropathic pain, they have unwanted side effects.4 Specifically, prescription opioid abuse recently in the USA has been accompanied by huge increases in the incidences of addition and opioid-related mortality.5 Thus, identifying new targets and mechanisms for neuropathic pain may open the door for non-opioid treatments for this disorder.
Persistent noxious stimuli arising from somatic damage or injury to the peripheral nervous system are detected by primary sensory neurons in the dorsal root ganglion (DRG).6 These sensory neuronal bodies, like neuromas at the injured sites, exhibit abnormal ectopic firing and hyperexcitability that are generally considered as one of the common primary causes of peripheral neuropathic pain.6–8 Nerve injury-induced reduction of potassium channels in the DRG may contribute to this abnormal spontaneous activity.9 Kv1.2, an α-pore-forming subunit of a voltage-gated potassium channel, is enriched highly in most medium and large DRG neurons.10 Peripheral nerve injury downregulates its expression in injured DRG.10,11 Rescuing this downregulation attenuates nerve injury-induced nociceptive hypersensitivity.10–14 Mimicking this downregulation reduces total Kv currents, depolarizes the resting membrane potential and decreases the current threshold for action potential activation in injured DRG neurons and produces augmented responses to noxious stimuli.11–14 DRG Kv1.2 is likely a key player in neuropathic pain. KCNN1 (also called SK1, KCa2.1, SKCA1 or hSK1), is a member of small conductance calcium-activated potassium channels (KCNN1–3) that mediate action potential afterhyperpolarization (AHP) and gate neuronal excitability.15 Unlike Kv1.2, KCNN1 is expressed predominantly in small DRG neurons.16 Expression of KCNN1 was reduced in the DRG avulsed from neuropathic pain patients.17 However, whether KCNN1 plays a pivotal role in neuropathic pain is still elusive.9
Long non-coding RNAs (lncRNAs) are classified as non-protein coding RNAs longer than 200 nucleotides and regulate gene expression at both transcriptional and translational levels in physiological and pathological processes.18–20 The majority of lncRNAs have tissue specificity in the brain,21,22 implying that they function in a tissue-specific manner. Nevertheless, whether and how these tissue-specific lncRNAs participate in neuropathic pain remains unclear.23–25 Here we identified a unique and native lncRNA and named it sensory neuron-specific lncRNA (SS-lncRNA), because it is expressed exclusively in sensory neurons of DRG and trigeminal ganglion, particularly in small nociceptive neurons of DRG. We found that SS-lncRNA mitigates the development and maintenance of neuropathic pain through heterogeneous nuclear ribonucleoprotein M (hnRNPM, an RNA-binding protein) mediated rescue of KCNN1 expression in injured DRG. SS-lncRNA is likely a new potential target for treatment of this disorder.
Materials and methods
Animals
Wild-type male and female CD-1 mice were purchased from Charles River Laboratories. SS-lncRNAfl/fl mice and Rosa26SS-lncRNA knock-in (KI) mice were generated by Biocytogen. AdvillinCreERT2/+ mice were purchased from the Jackson Laboratory (Stock No: 032027). SS-lncRNAfl/fl mice or Rosa26SS-lncRNA KI mice were backcrossed onto a C57BL/6 background for at least three generations to generate heterozygotes in our facility. These heterozygotes female mice were then mated with male AdvillinCreERT2/+ mice to generate sensory neuron-specific conditional knockdown (cKD) or knock-in (cKI) mice. All mice were kept under standard housing conditions (12/12-h dark-light cycle and food and water ad libitum) at Rutgers New Jersey Medical School. Animal procedures were approved by the Institutional Animal Care and Use Committee of Rutgers New Jersey Medical School and consistent with the ethical guidelines of the US National Institutes of Health and the International Association for the Study of Pain.
Chronic pain models
Spinal nerve ligation (SNL) and chronic constriction injury (CCI)-induced chronic neuropathic pain models in mice were carried out according to previously published methods.14,24,26,27 Briefly, for the SNL model, unilateral L4 spinal nerve was exposed after removal of the L4 transverse process and then ligated with 7–0 silk thread with transection distal to this site. For the CCI model, the unilateral sciatic nerve was exposed and loosely ligated with 7–0 silk thread at three sites, separated by ∼1 mm. Sham mice were subjected to the same surgery except for ligation and/or transection of the respective nerve. For chronic inflammatory pain models in mice, 20 μl of undiluted complete Freund’s adjuvant (CFA, Sigma-Aldrich) was injected subcutaneously into the plantar surface of one hind paw or 5 µl of mono-iodoacetate (MIA, Millipore Sigma) at a concentration of 50 µg/µl (diluted in 0.9% saline) was injected into the unilateral knee joint, as previously described.28–31
Behavioural testing
The evoked behavioural testing including mechanical, heat and cold tests was carried out in sequential order at 1-h intervals. Conditional place preference (CPP) testing was performed 2 or 4 weeks after surgery. Locomotor function testing was carried out prior to tissue collection. All mice were habituated for 1 to 2 h every day for 2–3 days before basal behavioural testing.
Mechanical nociceptive hypersensitivity was quantified by measuring paw withdrawal frequency to two calibrated von Frey filaments (0.07 and 0.4 g, Stoelting Co), as previously described.14,24,26,27 Briefly, mice were placed in a Plexiglas chamber on an elevated wire mesh screen to habituate for 30 min. Animals received 10 repeated applications (10–20 s apart) of each von Frey filament to the middle of the plantar surface of the hind paws. A quick withdrawal of the paw was considered as a positive response. The number of withdrawal responses within 10 applications was recorded as percentage withdrawal frequency.
Heat nociceptive hypersensitivity was determined by measuring paw withdrawal latencies upon heat stimulation, as described.14,24,26,27 Briefly, mice were placed on a glass surface in individual Plexiglas cages. A beam of light from a Model 336 Analgesia Meter (IITC Inc. Life Science Instruments) was applied to the middle of the plantar surface of each hind paw. A quick lift of the hind paw was considered as the paw withdrawal latency. A cut-off time of 20 s was used to avoid tissue damage to the hind paw.
Cold nociceptive hypersensitivity was detected by measuring paw withdrawal latencies to noxious cold (0°C), as previously described.14,24,26,27 Briefly, mice were placed in a Plexiglas chamber on the cold aluminium plate. The paw withdrawal latency was recorded as the length of time between placement and the first sign of the mouse jumping and/or flinching. This procedure was repeated three times with 10-min intervals. A cut-off time of 20 s was used.
Spontaneous ongoing pain was examined by carrying out the CPP test, as described.14,24,26,27 Briefly, the apparatus consisted of a pair of chambers with distinct stripes (horizontal versus vertical) on the walls and different texture (rough versus smooth) on the floor connected with a door in the middle (Med Associates Inc). Following preconditioning, each animal was monitored for 15 min through photobeam detectors installed along the chamber walls to automatically record basal time spent in each chamber, via MED-PC IV CPP software. Conditional training for lidocaine- or saline-paired chamber was conducted for the following 3 days with the door closed. The mice first received an intrathecal injection of saline (5 μl) specifically paired with one conditioning chamber, in the morning for 15 min. Six hours later, lidocaine (0.8% in 5 μl saline) was given intrathecally paired with another opposite conditioning chamber, in the afternoon, for 15 min. The injection order of saline and lidocaine every day was alternated. On the test day, the mice were placed in two chambers with the door open. The time spent in each chamber was recorded for 15 min for analysis of chamber preference. Place preference was indexed by the difference between the post-test time and preconditioning time spent in the lidocaine-paired chamber.
Locomotor activity tests including placing, grasping and righting reflexes were measured as previously described.14,24,26,27 For the placing reflex, the placed positions of the hind limbs were slightly lower than those of the forelimbs, and the dorsal surfaces of the hind paws were brought into contact with the edge of a table. Whether the hind paws were placed on the table surface reflexively was recorded. For the grasping reflex, after the mouse was placed on a wire grid, whether the hind paws grasped the wire on contact was recorded. For the righting reflex, when the mouse was placed on its back on a flat surface, whether it immediately assumed the normal upright position was recorded. Each trial was repeated five times at 5-min intervals and the scores for each reflex were recorded based on counts of each normal reflex.
Dorsal root ganglion microinjection
DRG microinjection was performed as described previously, with minor modification.14,24,26,27 Briefly, a 3-cm long skin incision was made aseptically at the midline of the lower back after mice were anaesthetized with 2% isoflurane. The unilateral L4 and/or L3 articular processes were exposed and then removed. The glass micropipette was carefully inserted into the exposed ipsilateral L4 or L3/4 DRGs and 1 µl of either siRNA solution (40–80 µM) or viral solution (4–9 × 1012 tu/µl) was injected into each DRG at a rate of 0.1 µl/min. The micropipette was removed 10 min after the injection. The wound was copiously irrigated with sterile saline and the skin was closed by suturing.
Cell culture and transfection
CAD cell cultures and DRG neuronal cultures were prepared as described previously.11,14,26,32 Briefly, CAD cells were cultured in Dulbecco’s modified Eagle medium (DMEM)/F-12 HEPES (Gibco/Thermo Fisher Scientific) supplemented with 8% foetal bovine serum and 1% antibiotics. Dissociated DRG neurons were obtained from 4-week-old CD1 mice and collected in cold neurobasal medium (Gibco/Thermo Fisher Scientific) supplemented with 10% foetal bovine serum, 5 ml L-glutamine (200 mM) (Gibco/ThermoFisher Scientific), 10 ml B-27® Supplement (50×) (Gibco/Thermo Fisher Scientific), 100 units/ml penicillin and 100 µg/ml streptomycin (Quality Biological). Dissociated cells were centrifuged and resuspended in mixed neurobasal medium, then cultured in a six-well plate coated with 50 µg/ml poly-D-lysine (Sigma) at 1.5–4 × 105 cells. The cells were incubated at 5% CO2 and 37°C. On Day 2, 4–10 µl of virus (titre ≥ 6–9 × 1012/µl), 300–500 ng of vector or siRNA (100 nM; transfected with Lipofectamine 3000) was added directly to neurons cultured in 6-well plates (2 ml media per well), and the cells were collected after 3 days.
Real-time PCR
Total RNA from mouse tissues was extracted and purified using the RNeasy mini kit (Qiagen). Total DNase-treated RNA from human DRGs were purchased from Clontech Laboratories, Inc. cDNA was synthesized by using the Omniscript RT Kit (Qiagen) with specific RT-primers or oligo(dT) primers. Quantitative real-time PCR assays were conducted using SYBR Green real-time PCR Master Mix. The primers used are listed in Supplementary Table 4.
Single-cell RT-PCR
The single-cell RT-PCR procedure was performed according to the manufacturer’s instructions with the Single-Cell RT-PCR Assay Kit (Signosis). In brief, the freshly dissociated mouse DRG neurons from adult mice were first prepared as described.11,14,26,32 Four hours after plating, single living large (>35 μm), medium (25–35 μm) and small (<25 μm) DRG neurons were sorted using a micromanipulator device (Leica Microsystems) under an inverted microscope and collected in a PCR tube containing 6–8 μl of cell lysis buffer (Signosis). Following centrifugation, the supernatants were collected and divided into PCR tubes for different genes. All nest-PCR primers used are listed in Supplementary Table 4. The PCR products were analysed on ethidium bromide-stained 2% agarose gels.
Plasmid constructs and virus production
Full-length cDNAs encoding Ebf1, Hnrnpm, SS-lncRNA and Kcnn1 were amplified by RT-PCR from mouse DRG RNA with the Platinum Taq High Fidelity Kit (Invitrogen/Thermo Fisher Scientific) and primers with restriction enzymes (Supplementary Table 4), respectively. Double enzyme-digested PCR products were ligated into the pHpa-tra-SK plasmids (University of North Carolina, Chapel Hill) to replace enhanced green fluorescent protein (GFP) sequence or the multiple cloning site of the pAAV-MCS vector (Cell Biolabs). The resulting vectors expressed the genes under the control of the cytomegalovirus promoter. The sequences for targeting gene shRNAs and control scrambled shRNA were annealed and ligated into the pAAV-shRNA-EF1a-EYFP using the BamHI and XbaI cut sites. To produce viral particles, the recombinant viral vectors and packaging vectors were co-transfected into HEK-293T cells. Adeno-associated virus (AAV) particles were harvested and purified by using AAVpro Purification Kit (Takara). Negative control siRNA and Ebf1 siRNA (s201270), Hnrnpm siRNA (s94804) and Kcnn1 siRNA (s96527) were purchased from Thermo Fisher Scientific, Inc.
Rapid amplification of cDNA ends
The rapid amplification of cDNA ends (RACE) was performed using the second generation 5′/3′ RACE Kit (Roche Diagnostics) following the manufacturer’s instructions to determine the 5′ and 3′ end of SS-lncRNA. Briefly, total RNA from mouse DRG was extracted with RNeasy Mini Kit (QIAGEN) and treated with overdose of DNase I (RNase-free) (Invitrogen). For the 5′ RACE, first-strand cDNA synthesis was performed by strand-specific primers followed by poly(A) tailing. The 3′ RACE analysis was performed by reverse transcription of cDNA using oligo dT-anchor primer followed by gene-specific and anchor primer amplification. All primers are listed in Supplementary Table 4. All PCR products were purified from an agarose gel and cloned to pCR-Blunt II-TOPO vector (Thermo Fisher) for DNA sequencing. The obtained sequences were assembled to get the full-length cDNA sequence.
RNA fractionation
The separation of nuclear and cytoplasmic fractions from primary cultured mouse DRG neurons was carried out by using the PARIS Kit (Invitrogen). Briefly, cultured neurons were rinsed with ice-cold 1× PBS and lysed in ice-cold cell fractionation buffer. After incubation for 30 min on ice, the lysate was centrifuged at 15 000g for 15 min at 4°C to separate the nuclear and cytoplasmic fractions. Total RNA of each fraction was extracted by following the kit instructions. Various gene/transcript expression levels in both nuclear and cytoplasmic fractions of all samples were quantified by quantitative real-time RT-PCR as described above.
Western blotting
The fresh tissues were homogenized and lysed in ice-cold lysis buffer containing proteinase inhibitor and phosphatase inhibitor. The samples were loaded onto a 4–15% stacking/7.5% separating SDS-PAGE (Bio-Rad Laboratories), and then electrophoretically transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories). The membranes were blocked with 5% non-fat milk in Tris-buffered saline/0.1% Tween-20 for 1 h and incubated overnight at 4°C with the following primary antibodies including goat anti-EBF1 (1:500, R&D systems), mouse anti-hnRNPM (1:500, Santa Cruz), rabbit anti-KCNN1 (1:,000, LifeSpan BioSciences, Inc), rabbit anti-phospho-ERK1/2 (Thr202/Tyr204, 1:800, Cell Signaling), rabbit anti-ERK1/2 (1:800, Cell Signaling), mouse anti-GFAP (1:800, Cell Signaling), rabbit anti-GAPDH (1:2000, Santa Cruz), and rabbit anti-histone H3 (1:1000, Cell Signaling). The proteins were detected by western peroxide reagent and luminol/enhancer reagent (Clarity Western ECL Substrate, Bio-Rad) and exposed using the ChemiDoc XRS System with Image Lab software (Bio-Rad). The intensity of blots was quantified with densitometry using Image Lab software (Bio-Rad). GAPDH and histone H3 were used as loading controls for cytoplasmic and nuclear protein, respectively.
Northern blotting
Northern blot analysis was performed as previously described.11,33 Briefly, digoxigenin-dUTP labelled complementary RNA (cRNA) probes of mouse SS-lncRNA were made by PCR using genomic DNA from mouse as the template. Total extracted RNA (10 μg) was separated on agarose-formaldehyde gel, transferred to BrightStar-plus positively charged nylon membrane (AM10100, Ambition) and was exposed to UV irradiation (150 mJ/cm2) for crosslinking into the membrane. The membrane was hybridized with the probe at 68°C overnight. The next day, the membranes were washed once with 2× SSC, and twice with 0.1× SSC, followed by detection with the CDP-Star chemiluminescent substrate (Roche).
RNAscope in situ hybridization and co-immunohistochemistry
The RNAscope in situ hybridization (ISH) was carried out by using RNAscope® 2.5 HD Detection reagent-red kit (REF:322360, ACD) following the manufacturers’ instructions. The positive control probe for the housekeeping gene Ppib (REF: 320881, ACD) and the negative control probe for the bacterial gene DapB were used to validate the ISH procedure. The signals were detected by using the probe for SS-lncRNA (REF:573241, ACD) and revealed by using BASEscope™ Fast RED assay following the manufacturer’s instructions. Following ISH, the immunohistochemistry staining was performed as described previously.14,26,34,35 After being blocked for 1 h at room temperature in 0.01 M PBS/0.3% Triton X-100 containing 4% goat serum, the sections were incubated with chicken anti-β-tubulin III (1:200, EMD Millipore), rabbit anti-glutamine synthetase (1:500, Sigma-Aldrich), rabbit anti-NF200 (1:100, Sigma), biotinylated IB4 (1:200, Sigma), rabbit anti-P2X3 (1:500, Neuromics), mouse anti-TH (1:1000, EMD Millipore) and rabbit anti-TRPV1 (1:400, Abcam), respectively, at 4°C overnight. The fluorescent signals were developed with appropriate fluorescence-conjugated secondary antibodies. Images were captured with an inverted microscope (DMI4000, Leica Microsystems). Immunoreactive positive neurons containing three or more particles of SS-lncRNA were considered to be ‘co-expressed’ cells, according to preceding studies.36,37
Bioinformatics prediction of transcription factors
The 2000-bp promoter sequence of SS-lncRNA gene was achieved based on the University of Santa Cruz (UCSC) genomic database (https://genome.ucsc.edu). The motifs of EBF1 in this promoter region were predicted according to JASPAR database (http://jaspar.genereg.net/). To avoid false-positive scoring, the relative profile score threshold was set at 90%. Through analysing the predicted score from the potential EBF1-binding regions, the region with the highest score was used to clone and to investigate the binding between EBF1 and SS-lncRNA.
In vitro protein translation
The full-length sequence of SS-lncRNA containing the T7 promoter was construed by RT-PCR with the primers in Supplementary Table 4 and was in vitro transcribed and translated by the Transcend™ Non-Radioactive Translation Detection System procedures described in the manufacturer's manual (Promega). In this procedure, biotinylated lysine residues are incorporated into nascent proteins during translation, allowing for in vitro non-radioactively labelled protein synthesis. Proteins were detected with horseradish peroxidase-conjugated streptavidin. The signals were visualized by using SuperSignal western blotting kit detection reagents (Thermo Pierce). Luciferase was used as a coding gene control.
RNA pull-down and mass spectrometry
The RNA pull-down assay was performed by using a Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher) according to the manufacturer’s instructions. The SS-lncRNA sense and SS-lncRNA antisense RNA (Supplementary Table 4) were synthesized in vitro by PCR using the Biotin RNA Labeling Mix (Roche) and purified by the Thermo GeneJET RNA Purification Kit (Thermo Fisher). The sense RNA was used as a negative control. The cultured DRG neurons were homogenized and cross-linked by 37% formaldehyde for 10 min at room temperature, followed sheared by sonication. Hybridization was performed at 37°C overnight. Biotinylated RNA was captured by Dynabeads MyOne Streptavidin T1 beads (Invitrogen, Cat. No. 65601). Proteins were separated by the 4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) followed by mass spectrometry analysis at the Center for Advanced Proteomics Research at Rutgers New Jersey Medical School. To verify the binding of SS-lncRNA to hnRNPM, western blot analysis was carried out, as described above.
Luciferase reporter assay
A 600-bp fragment from the SS-lncRNA promoter region (including the EBF1-binding motif) and a 2000-bp fragment from the Kcnn1 promoter region (including hnRNPM-binding sites) were amplified by PCR from genomic DNA using appropriate primers (Supplementary Table 4) to construct the reporter plasmids, respectively, as previously described.11 Luciferase activity was assayed by the Dual-Luciferase Reporter Assay System (Promega). Analyses of the test samples were performed in duplicate and were repeated three independent times. Relative luciferase was calculated by normalizing firefly luciferase activity (reporter) to Renilla luciferase activity (an internal control).
RNA sequencing
The ipsilateral L4 DRGs from eight SNL or sham mice with microinjection of AAV5-Gfp or AAV5-SS-lncRNA 4 weeks before surgery were collected on Day 7 post-surgery and pooled together to achieve enough RNA. Total RNA (1.2 µg/sample) extracted as described above was subjected to rRNA depletion by Ribo-Zero rRNA Removal Kit (Illumina). Stranded-specific RNA sequencing libraries were prepared by using the Illumina TruSeq Stranded Total RNA Kit without poly-A selection. All assays were performed in triplicate according to each manufacturer’s instructions. Sequencing was carried out on an Illumina HiSeq2500 platform High Output Mode, in a 2 × 150 bp paired-end configuration, with a total of more than 190 M reads per lane (at least 60 M reads per sample).38
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed by using the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Briefly, DRG homogenates were crosslinked at room temperature for 10 min with 1% formaldehyde. The crosslinking reaction was quenched by adding glycine (0.25 M final concentration). After centrifugation, the pellet was resuspended and lysed in SDS lysis buffer with protease inhibitor cocktail. Cross-linked chromatin was sonicated to obtain DNA fragments of 200–1000 bp. Lysate was precleaned with 20 μl Pierce Protein A/G Agarose beads. Immunoprecipitation was carried out at 4°C overnight using rabbit anti-EBF1 (2 μg, Millipore), mouse anti-hnRNPM (2 μg, Santa Cruz Biotechnology, Inc) or purified rabbit or mouse IgG. Input (2% of the sample for immunoprecipitation) was used as a positive control. DNA fragments were purified with a QIAquick DNA purification Kit (Qiagen) and analysed by using SYBR green real-time PCR with the primers listed in Supplementary Table 4.
RNA-binding protein immunoprecipitation
The RNA-binding protein immunoprecipitation (RIP) assay was carried out by using a Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Briefly, DRGs were collected and homogenized in ice-cold PBS. After centrifugation (1500 rpm, for 5 min at room temperature), the pellet was resuspended in an equal volume of RIP lysis buffer containing protease and RNase inhibitors. Magnetic beads protein A/G were incubated with 2 μg of mouse anti-hnRNPM (Santa Cruz Biotechnology, Inc) or of purified mouse IgG (Millipore) at room temperature for 30 min. The RIP lysates were then incubated with the bead-antibody complexes overnight at 4°C. The samples were washed by using a magnetic separator six times with 500 μl of ice-cold RIP wash buffer. After treatment with proteinase K, the immunoprecipitated RNAs were purified by phenol/chloroform extraction and eluted in 15 μl of RNase–DNase free water. About 10–20% of RIP lysate was used as input. The relative abundance of SS-lncRNA transcripts was then quantified by RT-qPCR. All primers used are listed in Supplementary Table 4.
Chromatin isolation by RNA purification
Chromatin isolation by RNA purification (ChIRP) was carried out as reported.39,40 Briefly, the cultured neurons were rinsed with chilled PBS and crosslinked by 1% formaldehyde for 10 min. The reactions were quenched by adding 0.125 M glycine for 5 min. After cell pellets were dissolved in nuclear lysis buffer, the lysates were sonicated to break DNA to 100–500 bp fragments. A total of 14 different biotinylated antisense DNA probes that were complementary to the sequence of SS-lncRNA were designed using an online tool (Singlemoleculefish.com) and numbered. The negative control probes that were not complementary to the sequence of SS-lncRNA. Seven odd-numbered probes, seven even-numbered probes and negative control probes were hybridized, respectively, with the cell lysates overnight. The complex of beads/probes/DNA was pulled down by using streptavidin magnetic C1 beads (Invitrogen). The potential binding DNA was ready for analysis by quantitative PCR assay as described above.
Whole-cell patch-clamp recording
The acute disassociated DRG neurons were cultured on laminin-coated coverslip, as previously described.11,12,14,35 Whole-cell patch clamp recording was carried out 4 to 6 h after plating. To increase the recording efficiency, only AAV9-GFP (green)-labelled small-sized neurons (<25 µm) were recorded at the temperature of 36°C. Kv currents were determined using a method described in our previous studies.11,12,14 In brief, for Kv current isolation, bath solution contained the following regents (in mM): choline-Cl 130, KCl 5, CdCl2 1, CaCl2 2, MgCl2 1, HEPES 10, glucose 10 (pH 7.4 with Tris-base, 320 mOsm). The intracellular pipette solution contained (in mM): potassium gluconate 120, KCl 20, MgCl2 2, EGTA 10, HEPES 10, Mg-ATP 4 (pH 7.3 with KOH, 310 mOsm). Kv currents were recorded under voltage clamp with a 500 ms depolarizing pulse (command voltages to potentials ranging between −30 to +50 mV in 10 mV increments, holding at −80 mV). The resting membrane potential was taken 3 min after a stable recording was first obtained. Action potentials (APs) were recorded under current clamp. The rheobase current was defined as the first step current that induced one AP by 50 ms depolarizing step current. APs were evoked by using a 1-s depolarizing current pulse (80 pA, 1.5 × of average rheobase current). The neurons showing single spike or no firing after 80 pA current injection were excluded. The AP amplitude was measured between the peak and the baseline. The AP overshoot was measured between the AP peak and 0 mV. AHP currents were recorded under voltage clamp with a 500 ms depolarizing pulse (from −60 to 0 mV, holding at −60 mV). The AHP amplitude was measured between the maximum hyperpolarization and the final plateau voltage. The artificial CSF consisted of (in mM): NaCl 140, KCl 5, CaCl2 2, MgCl2 1, HEPES 10 and glucose 10, with pH adjusted to 7.3 by NaOH. The electrode resistances of micropipettes ranged from 4 to 6 MΩ. The patch pipette solution contained (mM): KCl 135, MgATP 3, Na2ATP 0.5, CaCl2 1.1, EGTA 2, and glucose 5; pH was adjusted to 7.38 with KOH and osmolarity adjusted to 300 mOsm with sucrose. The number of spontaneous APs was calculated as the number of APs per second in 3 min during extracellular buffer application. Recording signals were amplified with Axopatch 700B (Molecular Devices), filtered at 2 kHz, and sampled at 5 kHz using pCLAMP 10 (Molecular Devices).
Statistical analysis
For in vivo experiments, mice were randomly distributed across experimental cohorts. For in vitro experiments, the cells were evenly suspended and then randomly distributed into each well tested. The sample sizes were determined based on our pilot studies, previous reports in the field11,31,41–43 and power analyses (power of 0.90 at P < 0.05). All results are shown as the means ± standard error of the mean (SEM) of at least three independent experiments. Data distribution was assumed to be normal, but this was not formally tested. The data were statistically analysed using two-tailed, paired Student’s t-test and a one-way, two-way or three-way ANOVA. When ANOVA showed a significant difference, pairwise comparison between means was performed using the post hoc Tukey method (SigmaPlot 12.5, San Jose, CA). Significance was set at P < 0.05.
Data availability
All information necessary to evaluate the reported findings are included in the main text or the Supplementary material. Additional data or further methodological details will be provided by the authors upon request.
Results
Identification of SS-lncRNA in the neurons of dorsal root ganglion
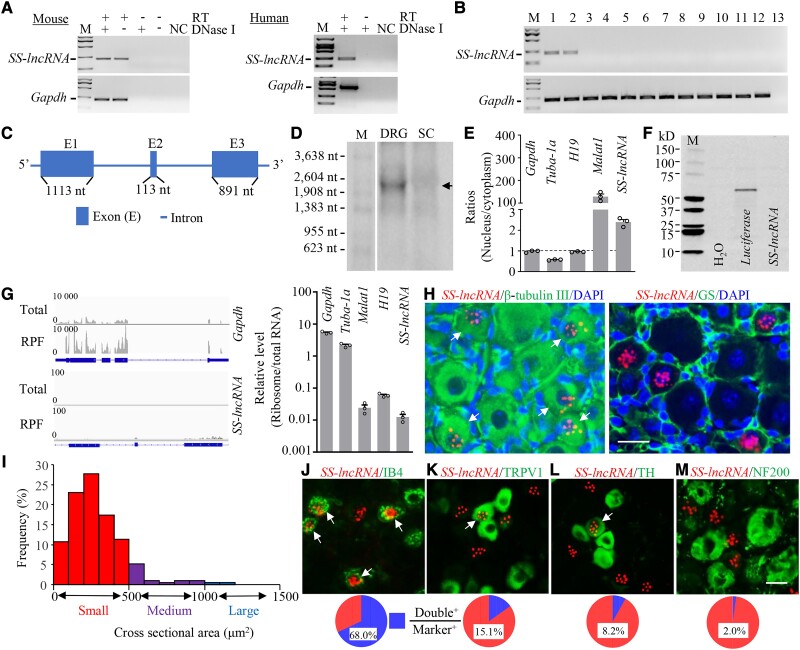
To detect SS-lncRNA, we first searched our RNA-sequencing database38 and found a significant decrease of the stacked reads in the genomic region (158 205 157–158 209 430) of chromosome 1 from the ipsilateral L4 DRG on Day 7 following SNL as compared to sham mice (Supplementary Fig. 1). We then designed strand-specific primers for reverse transcription and identified the SS-lncRNA transcripts in the DRGs of mouse and human (Fig. 1A), although the sequences were not completely identical between two species. In addition to DRG, this transcript was also detected in the trigeminal ganglion, but not in the spinal cord, various brain regions and other body organs of mice (Fig. 1B). We carried out RACE for directional sequencing of 5′ and 3′ ends plus RT-PCR assay and further identified a 2.118-kb full-length SS-lncRNA (containing three exons and poly-A tail) in mouse DRG (Fig. 1C and Supplementary Fig. 2) and a 2.454-kb full-length SS-lncRNA (containing three exons and poly-A tail) in human DRG (Supplementary Fig. 3). Northern blot analysis confirmed the existence of SS-lncRNA at the expected size in the DRG, but not in the spinal cord, of adult mice (Fig. 1D). Quantitative analysis of nuclear/cytoplasmic RNA extracted from the DRG revealed that SS-lncRNA was more enriched in the nucleus than in the cytoplasm (Fig. 1E). Coding substitution frequency analysis of SS-lncRNA showed no apparent open reading frame (Supplementary Figs 2 and 3). Consistently, in vitro translation of SS-lncRNA did not produce any peptides/proteins (Fig. 1F). Furthermore, ribosome profiling analysis proved no/minimal ribosomes on SS-lncRNA (Fig. 1G). Together, these results indicate that SS-lncRNA is a non-coding RNA.
Figure 1.
Identification and characterization of sensory neuron-specific long non-coding RNA (SS-lncRNA) in DRG. (A) SS-lncRNA transcript was detected in naïve mouse and human DRG by using reverse transcription (RT)-PCR with strand-specific primers. The extracted RNA samples were treated with excess DNase I to exclude genomic DNA contamination. Without RT primer, the PCR products of neither SS-lncRNA nor Gapdh (control) were detected in the DNase I-treated samples. n = 3 biological repeats/species. NC = H2O; M = DNA ladder marker. (B) SS-lncRNA transcript is expressed in the DRG (1) and trigeminal ganglion (2), but not in spinal cord (3), frontal cortex (4), brainstem (5), hippocampus (6), thalamus (7), cerebellum (8), lung (9), heart (10), liver (11) and kidney (12), from adult male mice. Lane 13 = H2O. Gapdh = a control; n = 3 mice. (C) Schematic diagram of full-length SS-lncRNA analysed by 5′ and 3′ RACE assay. Blue boxes and thick lines indicate exons and introns, respectively. (D) Northern blot analysis showing that SS-lncRNA (arrow) is detected in the DRG, but not in the spinal cord (SC), from naïve mice. n = 3 biological repeats (five mice/repeat). M = RNA marker. (E) Ratios of nucleus to cytoplasm for Gapdh mRNA, Tuba-1a mRNA, H19, Malat1 and SS-lncRNA in mouse DRG. n = 3 mice. (F) In vitro translation of SS-lncRNA using the transcend non-radioactive translation detection systems. Luciferase was used as a control for coding RNA. n = 3 biological repeats. (G) Ribosome profiling of SS-lncRNA and Gapdh. The blue rectangles represent exons. RFP = ribosome-protected fragments. Signal ratios of ribosome profiling to RNA sequencing (total) for mRNAs (Gapdh and Tuba-1a) and long non-coding RNAs (SS-lncRNA, H19 and Malat1). n = 3 mice. (H) Representative photomicrographs showing that SS-lncRNA (Red; signal particles ≥3) is co-expressed with β-tubulin III (green, left) in individual DRG cells (arrows) and undetected in glutamine synthetase (GS, green, right)-labelled DRG cells. 4′, 6-diamidino-2-phenylindole (DAPI, blue) is a marker of cellular nucleus. Scale bar = 25 µm. (I) Histogram showing the distribution pattern of SS-lncRNA-positive somata in normal DRG. Small, 77%; medium, 18%; large, 5%. n = 3 mice. (J–M) Co-expression of SS-lncRNA (red) with isolectin B4 (IB4, green, J), transient receptor potential vanilloid type 1 (TRPV1, green, K), tyrosine hydroxylase (TH, green, L) and neurofilament-200 (NF200, green, M) in individual DRG neurons (arrows). n = 8–10 sections from 4 to 5 mice. Scale bar = 25 µm.
The cellular distribution pattern of SS-lncRNA in mouse DRG was also examined by using the RNAscope ISH assay followed by immunohistochemical staining of various cell markers. SS-lncRNA-labelled signal particles (≥3) were more enriched in the nucleus than in the cytoplasm within the β-tubulin III (a specific neuronal marker)-positive cells from naïve DRG (Fig. 1H and Supplementary Fig. 4A). In contrast, no signal particles were detected in the glutamine synthetase (a marker for satellite glial cells)-positive cells of naïve DRG (Fig. 1H and Supplementary Fig. 4B). Approximately 10.6% (253/2397) of the β-tubulin III-positive neurons were labelled by SS-lncRNA, of which about 77% were small (<25 µm in diameter), 18% medium (25–35 µm in diameter), and 5% large (> 35 µm in diameter) (Fig. 1I). Consistently, ∼68% of isolectin B4 (IB4, a marker for small non-peptidergic neurons)-labelled neurons (Fig. 1J and Supplementary Fig. 5A) and 67.7% of P2X purinoceptor 3 (P2X3, another marker for small non-peptidergic neurons)-labelled neurons (Supplementary Fig. 5B) were positive for SS-lncRNA. In addition, ∼15.1% of transient receptor potential vanilloid type 1 (TRPV1, a marker for small peptidergic neurons)-labelled neurons, 8.2% of tyrosine hydroxylase (TH, a marker for small low-threshold neurons)-labelled neurons, and 2% of neurofilament-200 (NF200, a marker for medium/large cells and myelinated amyloid-β fibres)-labelled neurons were SS-lncRNA-positive (Fig. 1K–M and Supplementary Fig. 6). Distribution of SS-lncRNA mainly in small DRG neurons suggests its involvement in the transmission and modulation of nociceptive information.
Peripheral nerve injury downregulates SS-lncRNA in injured DRG
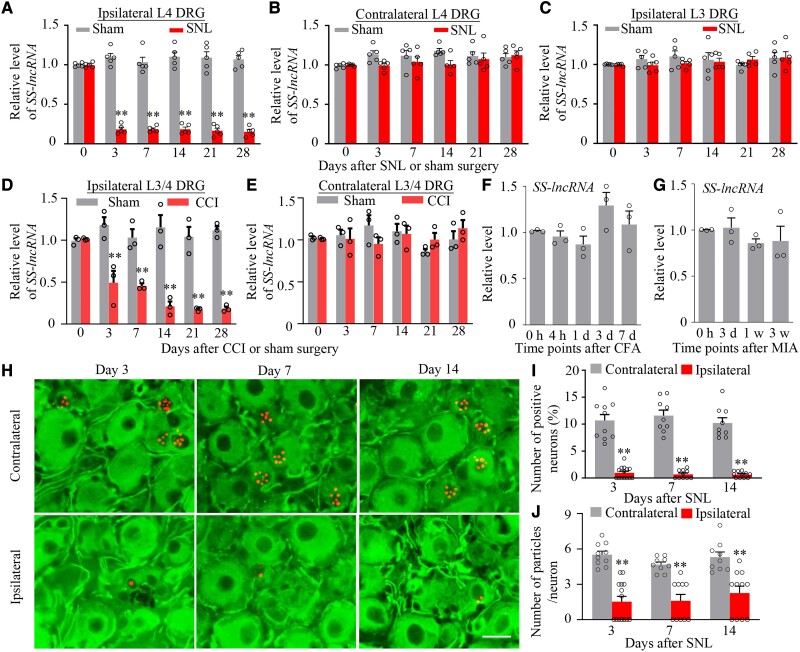
We next examined whether the expression of SS-lncRNA was altered in mouse DRG after peripheral nerve injury. The level of SS-lncRNA was dramatically reduced in the ipsilateral L4 DRG on Days 3, 7, 14, 21 and 28 after unilateral L4 SNL, but not sham surgery (Fig. 2A). Neither SNL nor sham surgery changed basal expression of SS-lncRNA in the contralateral L4 DRG and ipsilateral L3 DRG (Fig. 2B and C). Results were similar in injured DRG following CCI (Fig. 2D and E). Interestingly, basal level of SS-lncRNA was not substantially altered in the ipsilateral L3/4 DRGs during the observation periods in animal models of chronic inflammatory pain caused by plantar injection of CFA into unilateral hind paw (Fig. 2F) or intra-articular injection of MIA into unilateral knee joint (Fig. 2G). Consistently, number of SS-lncRNA-positive neurons, number of total particles per positive neuron, number of nuclear particles per positive neurons, and number of cytoplastic particles per positive neurons in the ipsilateral L4 DRG were significantly less than those in the contralateral L4 DRG on Days 3, 7 and 14 post-SNL (Fig. 2H–J and Supplementary Fig.7A–D). These reductions occurred in all three types of DRG neurons, but predominantly in small DRG neurons (Supplementary Fig. 8). The evidence indicates the downregulation of SS-lncRNA in injured DRG neurons after peripheral nerve injury.
Figure 2.
SS-lncRNA downregulation in injured DRG of male mice after peripheral nerve injury. (A–C) Level of SS-lncRNA in the ipsilateral (A) and contralateral (B) L4 DRG and ipsilateral L3 DRG (C) after unilateral L4 SNL or sham surgery. n = 20 mice/time point/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (D and E) Level of SS-lncRNA in the ipsilateral (D) and contralateral (E) L3/4 DRGs after unilateral CCI or sham surgery. n = 6 mice/time point/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (F and G) Level of SS-lncRNA in the ipsilateral L3/4 DRGs at different time points after unilateral hind paw injection of the complete Freund’s adjuvant (CFA, F) or unilateral intra-articular injection of sodium mono-iodoacetate (MIA, G). n = 6 mice/time point/group. One-way ANOVA with post hoc Tukey test. (H–J) Representative photomicrographs (H) and corresponding statistical analysis (I and J) showing number of SS-lncRNA-labelled neurons (signal particles ≥3) and number of particles per labelled neuron in the ipsilateral and contralateral L4 DRG on Days 3, 7 and 14 after SNL. n = 9–14 sections/time point from four to five mice. **P < 0.01 by two-way ANOVA with post hoc Tukey test. Scale bar = 25 µm.
EBF1 participates in nerve injury-induced downregulation of DRG SS-lncRNA
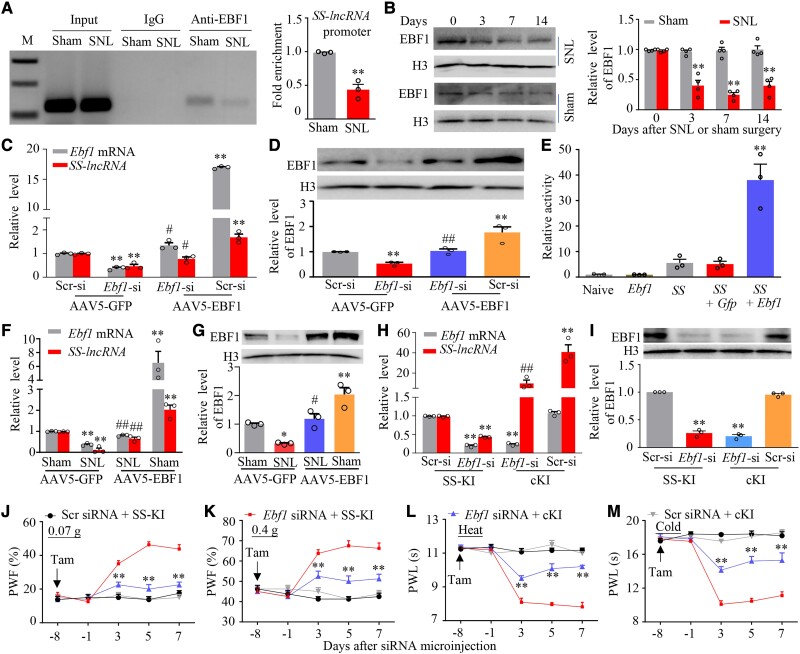
We further explored how peripheral nerve injury downregulated SS-lncRNA in injured DRG. Considering that transcription factors commonly regulate cellular gene expression, we used the online JASPAR software and found a consensus binding site (−179GTCCCAAGGGA−169) for early B cell transcription factor 1 (EBF1) within mouse SS-lncRNA promotor. In addition, Ebf1 mRNA expression in our RNA sequencing database was reduced in injured DRG post-SNL.38 We speculated that a reduction of EBF1 expression might be responsible for the SS-lncRNA downregulation in injured DRG after nerve injury. Indeed, the binding of EBF1 to SS-lncRNA promoter was verified by the evidence demonstrating that the fragment of SS-lncRNA promoter containing this binding site could be amplified from the complex immunoprecipitated with the anti-EBF1 antibody in nuclear fraction from the sham DRG (Fig. 3A). SNL decreased this binding, as evidenced by a 56% reduction in binding activity in the ipsilateral L4 DRG on Day 7 after SNL mice as compared to that after sham surgery (Fig. 3A). This reduction could be explained by a time-dependent decrease of EBF1 expression in the ipsilateral L4 DRG after SNL, not sham surgery (Fig. 3B).
Figure 3.
SS-lncRNA downregulation caused by EBF1 decrease in injured DRG of male mice after peripheral nerve injury. (A) SS-lncRNA gene promoter fragment immunoprecipitated by rabbit anti-EBF1 antibody in the ipsilateral L4 DRG on Day 7 after SNL or sham surgery. Input = total purified fragments; IgG = purified rabbit IgG; M = DNA ladder marker. n = 36 mice (three repeats)/group. **P < 0.01, by unpaired Student’s t-test. (B) Level of EBF1 protein in the ipsilateral L4 DRG at different days after SNL or sham surgery. n = 16 mice (four repeats)/time point/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (C and D) Levels of Ebf1 mRNA (C), SS-lncRNA (C) and EBF1 protein (D) in cultured DRG neurons transfected/transduced as indicated. Ebf1-si = Ebf1 siRNA; Scr-si = control scrambled siRNA. n = 3 biological repeats/treatment/assay. **P < 0.01, by one-way ANOVA with post hoc Tukey test. (E) SS-lncRNA promoter activities in CAD cells transfected as shown. Naïve = no treatment; Ebf1 = proviral vector expressing full-length Ebf1 mRNA; Gfp = proviral vector expressing Gfp mRNA; SS = pGL3 reporter vector containing SS-lncRNA promoter. n = 3 biological repeats/treatment. **P < 0.01, by one-way ANOVA followed by post hoc Tukey test. (F and G) Levels of Ebf1 mRNA (F), SS-lncRNA (F) and EBF1 protein (G) in the ipsilateral L4 DRG on Day 14 after SNL or sham surgery in mice with microinjection of AAV5-GFP or AAV5-EBF1 into the ipsilateral L4 DRG 35 days before surgery. n = 12 mice/group/assay. *P < 0.05, **P < 0.01, by two-way ANOVA with post hoc Tukey test. (H and I) Levels of Ebf1 mRNA (H), SS-lncRNA (H) and EBF1 protein (I) in the ipsilateral L3/4 DRGs on Day 7 after microinjection of Scr siRNA or Ebf1 siRNA into the ipsilateral L3/4 DRGs from SS-KI mice and cKI mice with intraperitoneal injection of tamoxifen daily for seven consecutive days before siRNA microinjection. n = 6 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (J–M) Paw withdrawal frequency (PWF) to 0.07 g (J) and 0.4 g (K) von Frey filaments and paw withdrawal latency (PWL) to heat (L) and cold (M) stimuli on the ipsilateral side at the different days as indicated before and after Ebf1 siRNA or scrambled (Scr) siRNA microinjection into unilateral L3/4 DRGs in SS-KI mice and cKI mice with intraperitoneal injection of tamoxifen (Tam) daily for seven consecutive days before siRNA microinjection. n = 10 mice/group. **P < 0.01 versus the Ebf1 siRNA plus SS-KI mice at the corresponding time points by three-way ANOVA with repeated measures followed by post hoc Tukey test.
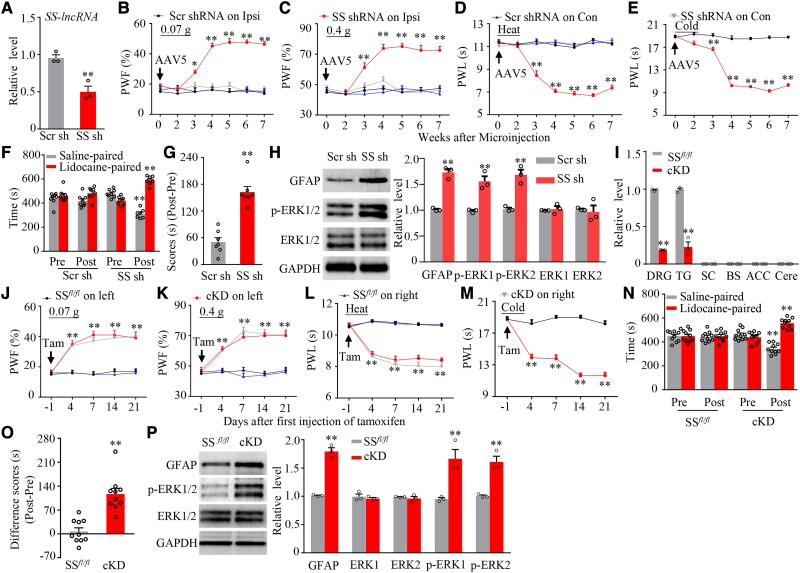
To define whether EBF1 directly regulates SS-lncRNA expression, we first knocked down EBF1 through transfecting a specific Ebf1 siRNA in cultured DRG neurons. The Ebf1 siRNA, but not control scrambled siRNA, markedly reduced basal levels of not only Ebf1 mRNA and EBF1 protein, but also SS-lncRNA in AAV5-GFP (as a control)-transduced neurons (Fig. 3C and D). These reductions could be reversed by co-transduction of AAV5 expressing full-length Ebf1 mRNA (AAV5-EBF1; Fig. 3C and D), suggesting that SS-lncRNA reduction was in specific response to EBF1 knockdown. As expected, AAV5-EBF1 transduction alone significantly increased basal amount of SS-lncRNA in the scrambled siRNA-treated neurons (Fig. 3C). Dual-luciferase reporter assays showed that co-transfection of full-length Ebf1 (not control Gfp) vector with SS-lncRNA reporter vector increased the activity of SS-lncRNA gene promoter (Fig. 3E). In in vivo experiments, rescuing the SNL-induced reduction of DRG EBF1 through microinjection of AAV5-EBF1 (not AAV5-GFP) into the ipsilateral L4 DRG of male mice 35 days before surgery attenuated the SNL-induced mechanical, heat and cold nociceptive hypersensitivities on the ipsilateral (not contralateral) side and stimulation-independent spontaneous ongoing pain during the development period (Supplementary Fig. 9A–I). Given that DRG neuronal hyperexcitability triggers the hyperactivation of spinal cord dorsal horn neurons and astrocytes through enhancing the release of neurotransmitters/neuromodulators in primary afferents under neuropathic pain conditions,7 DRG microinjection of AAV5-EBF1 (not AAV5-GFP) also blocked the SNL-induced neuronal/astrocyte hyperactivities in the ipsilateral L4 dorsal horn 14 days after SNL (Supplementary Fig. 9J). This microinjection also reversed the SNL-induced reduction in the amount of SS-lncRNA in the ipsilateral L4 DRG (Fig. 3F and G). In addition, mimicking the SNL-induced reduction of DRG EBF1 through microinjection of Ebf1 siRNA (but not scrambled siRNA) into unilateral L3/4 DRGs of naïve male mice produced a marked decrease in the level of SS-lncRNA in the microinjected DRGs, enhanced responses to mechanical, heat and cold stimuli on the ipsilateral (not contralateral) side and the increases in neuronal/astrocyte activities in the ipsilateral L3/4 dorsal horn (Supplementary Fig. 10A–G). To further demonstrate whether these enhanced behavioural responses are mediated by DRG downregulated SS-lncRNA, we generated Rosa26SS-lncRNA knock-in (SS-KI) mice (Supplementary Fig. 11) and crossbred them with sensory neuron-specific advillin-CreERT2 mice to achieve cKI mice, in which intraperitoneal (i.p.) injection of tamoxifen at 1 mg daily for 7 days induces SS-lncRNA overexpression only in sensory neurons (Supplementary Fig. 12A). Ebf1 siRNA-induced reduction of DRG SS-lncRNA and enhanced responses to mechanical, heat and cold stimuli on the ipsilateral side were greatly reversed and attenuated, respectively, in the tamoxifen-treated cKI mice (Fig. 3H–M). Expectedly, basal behavioural responses on the contralateral side in these mice were not altered during the observation period (Supplementary Fig. 12B–D). Neither siRNA nor virus affected locomotor function in the mice used (Supplementary Table 1). Given that SS-lncRNA co-expressed with Ebf1 mRNA mainly in small DRG neurons (Supplementary Fig. 13A–C), our results suggest that nerve injury-induced downregulation of SS-lncRNA is attributed, at least in part, to the silence of Ebf1 expression in injured DRG.
Rescuing downregulated DRG SS-lncRNA relieves chronic neuropathic pain
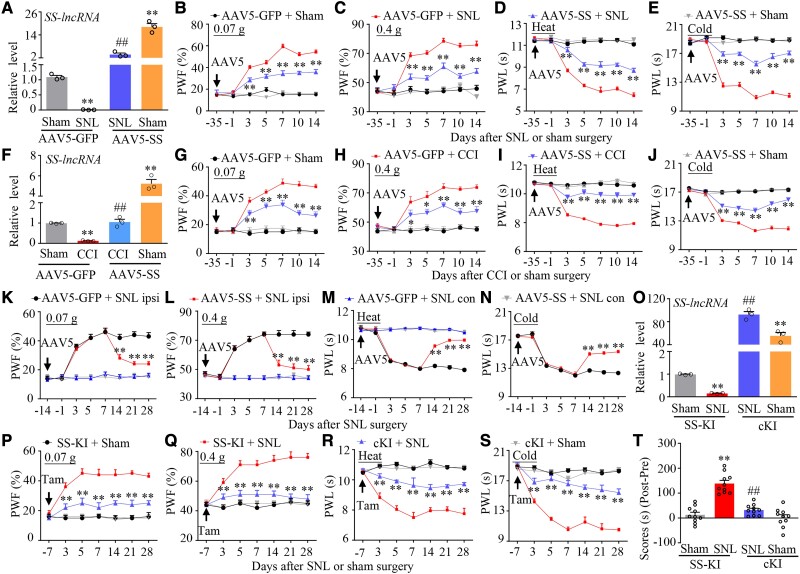
To investigate whether downregulated SS-lncRNA in injured DRG contributes to neuropathic pain development, we rescued this downregulation through microinjection of AAV5 expressing full-length SS-lncRNA (AAV5-SS) into the ipsilateral L4 DRGs 35 days before surgery in male mice (Fig. 4A). Consistent with previous reports,14,24,26 the mice microinjected with AAV5-GFP exhibited mechanical, heat and cold nociceptive hypersensitivities on the ipsilateral side from Days 3 to 14 post-SNL (Fig. 4B–E) and the increases of neuronal/astrocyte activities in the ipsilateral L4 dorsal horn on Day 14 post-SNL (Supplementary Fig. 14A). These hypersensitivities/increases were ameliorated in the mice microinjected with AAV5-SS during the observation period (Fig. 4B–E and Supplementary Fig. 14A). As expected, microinjection of neither AAV5-GFP nor AAV5-SS altered basal paw withdrawal responses on the contralateral side of SNL mice and on either side of sham-operated mice (Fig. 4B–E and Supplementary Fig. 14B–D). Similar results were observed after DRG microinjection of AAV5-SS or AAV5-GFP in male CCI mice (Fig. 4F–J and Supplementary Fig. 14E–G). The role of DRG downregulated SS-lncRNA in the maintenance of neuropathic pain was also investigated. Because AAV5 requires 3–4 weeks to become expressed,11,12,14,24 male mice were subjected to SNL surgery 2 weeks after DRG viral microinjection. Mechanical, heat and cold nociceptive hypersensitivities were fully developed on the ipsilateral (not contralateral) side in both AAV5-GFP- and AAV5-SS-microinjected mice on Day 7 post-SNL (Fig. 4K–N). These hypersensitivities were markedly alleviated from Days 14 to 28 post-SNL in the AAV5-SS-microinjected mice (Fig. 4K–N). Moreover, SNL-induced increases of neuronal/astrocyte activities in the ipsilateral L4 dorsal horn on Day 28 post-SNL were abolished in the AAV5-SS-microinjected mice (Supplementary Fig. 14H).
Figure 4.
Rescuing nerve injury-induced downregulation of DRG SS-lncRNA attenuated the development and maintenance of nerve injury-induced nociceptive hypersensitivity in male mice. (A) Level of SS-lncRNA in the ipsilateral L4 DRG on Day 14 after SNL or sham surgery in male mice with microinjection of AAV5-GFP or AAV5-SS into unilateral L4 DRGs 35 days before surgery. n = 12 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (B–E) Effect of microinjection of AAV5-SS or AAV5-GFP into the ipsilateral L4 DRG on the development of SNL-induced nociceptive hypersensitivity. Paw withdrawal frequency (PWF) to 0.07 g (B) and 0.4 g (C) von Frey filament and paw withdrawal latency (PWL) to heat (D) and cold (E) stimuli on the ipsilateral side at different days indicated before and after surgery. n = 12 mice/group. **P < 0.01 versus the AAV5-GFP plus SNL group at the corresponding time points by three-way ANOVA with repeated measures followed by post hoc Tukey test. (F) Level of SS-lncRNA in the ipsilateral L3/4 DRGs on Day 14 after CCI or sham surgery in male mice with microinjection of AAV5-GFP or AAV5-SS into unilateral L3/4 DRGs 35 days before surgery. n = 6 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (G–J) Effect of microinjection of AAV5-SS or AAV5-GFP into the ipsilateral L3/4 DRGs on the development of CCI-induced nociceptive hypersensitivity. Paw withdrawal frequency (PWF) to 0.07 g (G) and 0.4 g (H) von Frey filament and paw withdrawal latency (PWL) to heat (I) and cold (J) stimuli on the ipsilateral side at different days indicated before and after surgery. n = 12 mice/group. *P < 0.05, **P < 0.01 versus the AAV5-GFP plus CCI group at the corresponding time points by three-way ANOVA with repeated measures followed by post hoc Tukey test. (K–N) Effect of microinjection of AAV5-SS or AAV5-GFP into unilateral L4 DRG on the maintenance of SNL-induced nociceptive hypersensitivity. Paw withdrawal frequency (PWF) to 0.07 g (K) and 0.4 g (L) von Frey filament and paw withdrawal latency (PWL) to heat (M) and cold (N) stimuli on the ipsilateral (ipsi) and contralateral (con) sides at different days indicated before and after surgery. n = 12 mice/group. **P < 0.01 versus the AAV5-GFP plus SNL group at the corresponding time points by three-way ANOVA with repeated measures followed by post hoc Tukey test. (O) Level of SS-lncRNA in the ipsilateral L4 DRGs on Day 28 after SNL or sham surgery in SS-KI mice and cKI mice with intraperitoneal injection of tamoxifen daily for seven consecutive days before surgery. n = 12 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (P–S) Paw withdrawal frequency (PWF) to 0.07 g (P) and 0.4 g (Q) von Frey filaments and paw withdrawal latency (PWL) to heat (R) and cold (S) stimuli on the ipsilateral side at the different days as indicated before and after SNL or sham surgery in SS-KI mice and cKI mice with intraperitoneal injection of tamoxifen (Tam) daily for seven consecutive days before surgery. n = 12 mice/group. **P < 0.01 versus the SNL SS-KI mice at the corresponding time points by three-way ANOVA with repeated measures followed by post hoc Tukey test. (T) Spontaneous ongoing pain as assessed by the CPP paradigm 3 weeks after SNL or sham surgery in SS-KI mice and cKI mice with intraperitoneal injection of tamoxifen daily for seven consecutive days before surgery. n = 10 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test.
To exclude the possibility that the observed phenotypic changes above may be caused by microinjection-induced tissue damage, we performed SNL or sham surgery in male SS-KI mice and cKI mice. Rescuing the SNL-induced reduction of SS-lncRNA in the ipsilateral L4 DRG of the cKI mice (not SNL SS-KI mice) following tamoxifen injection (Fig. 4O) impaired the SNL-induced mechanical, heat and cold nociceptive hypersensitivities on the ipsilateral side from Days 3 to 28 post-SNL (Fig. 4P–S). This rescue also abolished the SNL-induced stimulation-independent spontaneous ongoing pain and increases of neuronal/astrocyte activities in the ipsilateral L4 dorsal horn on Day 28 post-SNL (Fig. 4T and Supplementary Fig. 15 A and B). Basal paw responses on the contralateral side of SNL cKI mice or SNL SS-KI mice and on both sides of the corresponding sham mice were not altered, although a significant increase of SS-lncRNA in the ipsilateral L4 DRG of the tamoxifen-treated sham cKI mice (Fig. 4O–S and Supplementary Fig. 15C–E). Similar findings were seen in SNL female SS-KI mice and cKI mice following tamoxifen injection (Supplementary Fig. 16A–K). All AAV5-microinjected mice and tamoxifen-treated SS-KI mice or cKI mice displayed normal locomotor activity (Supplementary Table 1).
Collectively, these findings suggest that rescuing DRG SS-lncRNA downregulation relieves nerve injury-induced nociceptive hypersensitivity during both development and maintenance periods.
Mimicking nerve injury-induced DRG SS-lncRNA downregulation produces nociceptive hypersensitivity
Is the downregulated SS-lncRNA in injured DRG sufficient for neuropathic pain? To this end, we mimicked the nerve injury-induced downregulation of DRG SS-lncRNA through microinjection of AAV5 expressing a specific SS-lncRNA shRNA (AAV5-SS shRNA) into unilateral L3/4 DRGs of naïve male mice. AAV5 expressing a scrambled shRNA (AAV5-Scr shRNA) was used as a control. As expected, the amount of SS-lncRNA was markedly reduced in the microinjected DRGs 7 weeks after DRG microinjection of AAV5-SS shRNA, but not AAV5-Scr shRNA (Fig. 5A). More importantly, this microinjection produced substantial increases in paw withdrawal frequencies to 0.07 and 0.4 g von Frey filaments and decreases in paw withdrawal latencies to heat and cold stimuli on the ipsilateral (not contralateral) side (Fig. 5B–E). These augmented responses occurred 3 weeks post-microinjection and persisted for at least 7 weeks (Fig. 5B–E). This microinjection also led to stimulation-independent spontaneous ongoing pain and increases of neuronal/astrocyte activities in the ipsilateral L3/4 dorsal horn 7 weeks post-microinjection (Fig. 5F–H).
Figure 5.
DRG knockdown of SS-lncRNA produced neuropathic pain-like symptoms in male naïve mice. (A) Level of SS-lncRNA in the ipsilateral L3/4 DRGs 7 weeks after microinjection of AAV5-scrambled shRNA (Scr sh) or AAV5-SS-lncRNA shRNA (SS sh) into unilateral L3/4 DRGs. n = 8 mice/group. **P < 0.01, by unpaired Student’s t-test. (B–E) Effect of microinjection of Scr sh or SS sh into the ipsilateral L3/4 DRGs on paw withdrawal frequency (PWF) to 0.07 g (B) and 0.4 g (C) von Frey filament stimuli and paw withdrawal latency (PWL) to heat (D) and cold (E) stimuli on the ipsilateral (Ipsi) and contralateral (Con) at time points shown after viral microinjection. n = 8 mice/group. *P < 0.05, **P < 0.01 versus the Scr sh-treated mice at the corresponding time points by two-way ANOVA with repeated measures followed by post hoc Tukey test. (F and G) Effect of microinjection of Scr sh or SS sh into the ipsilateral L3/4 DRGs on spontaneous ongoing pain as assessed by the CPP paradigm 7 weeks after viral microinjection. Pre= preconditioning; Post = post-conditioning. n = 8 mice/group. **P < 0.01, by two-way ANOVA with repeated measures followed by post hoc Tukey test (F) or by unpaired Student’s t-test (G). (H) Neuronal and astrocyte activities in the ipsilateral L3/4 dorsal horn as assessed by levels of p-ERK1/2 and GFAP, respectively, 7 weeks after DRG microinjection of Scr sh or SS sh into unilateral L3/4 DRGs. Total ERK1/2 were used as a control. n = 3 mice/group. **P < 0.01, by unpaired Student’s t-test. (I) Level of SS-lncRNA in DRG, TG, spinal cord (SC), brainstem (BS), anterior cingulate cortex (ACC) and cerebellum (Cere) from SSfl/fl mice and cKD mice 21 days after first injection of tamoxifen. n = 6 mice, **P < 0.01, by unpaired Student’s t-test. (J–M) Paw withdrawal frequency (PWF) to 0.07 g (J) and 0.4 g (K) von Frey filaments and paw withdrawal latency (PWL) to heat (L) and cold (M) stimuli on the left and right sides at the different days as indicated in SSfl.fl mice and cKD mice with intraperitoneal injection of tamoxifen (Tam) daily for seven consecutive days. n = 10 mice, **P < 0.01 versus the SSfl/fl group by three-way ANOVA with repeated measures followed by post hoc Tukey test. (N and O) Spontaneous ongoing pain as assessed by the CPP paradigm 21 days after first tamoxifen injection in SSfl.fl mice and cKD mice. Pre = preconditioning; Post = post-conditioning. n = 10 mice/group. **P < 0.01, by two-way ANOVA with repeated measures followed by post hoc Tukey test (N) or by unpaired Student’s t-test (O). (P) Neuronal and astrocyte activities in the L3/4 dorsal horn from SSfl.fl mice and cKD mice as assessed by levels of p-ERK1/2 and GFAP, respectively, 21 days after first tamoxifen injection. Total ERK1/2 were used as a control. n = 9 mice/group. **P < 0.01, by unpaired Student’s t-test.
Given that shRNA may have potential off-targets, we generated SS-lncRNAfl/fl (SSfl/fl) mice (Supplementary Fig. 17) and cross-bred them with sensory neuron-specific advillin-CreERT2 mice to obtain cKD mice, in which intraperitoneal injection of tamoxifen at 1 mg daily for 7 days knocked down SS-lncRNA expression only in sensory neurons (Fig. 5I). Like the AAV5-SS shRNA-microinjected mice, the cKD mice (but not SSfl/fl mice) displayed mechanical, heat and cold hypersensitivities from Days 4 to 21 after the first injection of tamoxifen and stimulation-independent spontaneous ongoing pain and increases of neuronal/astrocyte activities in L3/4 dorsal horn on Day 21 after the first injection of tamoxifen on the bilateral sides (Fig. 5J–P). Similar results were observed in female SSfl/fl mice and female cKD mice after tamoxifen injection (Supplementary Fig. 18A–H). Expectedly, no changes in locomotor activity were found in either AAV5-microinjected mice or tamoxifen-injected SSfl/fl mice or cKD mice (Supplementary Table 1).
Taken together, these data suggest that, in the absence of nerve injury, DRG SS-lncRNA downregulation leads to neuropathic pain-like symptoms.
Downregulated SS-lncRNA participates in the nerve injury-induced reduction of KCNN1 expression in injured DRG
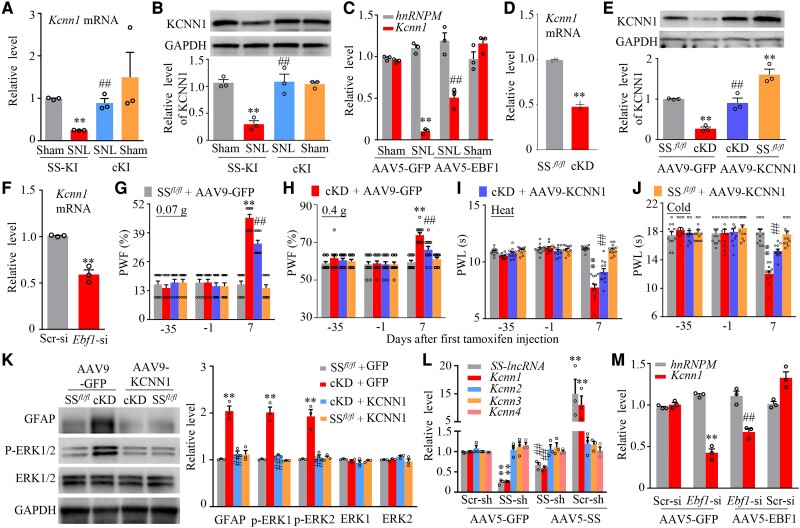
To elucidate how DRG downregulated SS-lncRNA contributes to neuropathic pain, we carried out high-throughput RNA sequencing to search the downstream genes of SS-lncRNA in injured DRG after SNL. The unbiased gene expression analyses revealed that approximately 1574 genes out of a total of 18 865 identified genes were markedly changed in the ipsilateral L4 DRG from the AAV5-GFP-treated mice 7 days after SNL as compared to those after sham surgery (Supplementary Fig. 19A). About 27.9% of these changed genes were upregulated and 72.1% downregulated (Supplementary Fig. 19A). Among these changed genes, 44 upregulated genes and 483 downregulated genes were reversed in the ipsilateral L4 DRG from the AAV5-SS-treated mice 7 days after SNL (Supplementary Fig. 19B). These reversed genes-encoding proteins are particularly enriched on the plasma membrane (Supplementary Fig. 19C).
Among these reversed genes, one of the most striking genes is Kcnn1 mRNA (encoding KCNN1). Like SS-lncRNA, the level of Kcnn1 mRNA was considerably reduced in the ipsilateral L4 DRG from Days 3 to 28 after unilateral L4 SNL, but not sham surgery (Supplementary Fig. 20A). Consistently, the amount of KCNN1 protein was also significantly reduced in the ipsilateral L4 DRG from days 3 to 14 post-SNL (Supplementary Fig. 20B). To determine whether its reduction participated in neuropathic pain genesis, we rescued the SNL-induced KCNN1 reduction in injured DRG through microinjection of AAV9 expressing full-length Kcnn1 mRNA (AAV9-KCNN1) into the ipsilateral L4 DRG 35 days before surgery (Supplementary Fig. 20C). Microinjection of AAV9-KCNN1, but not AAV9-GFP, diminished the SNL-induced mechanical, heat and cold nociceptive hypersensitivities from Days 3 to 28 post-SNL on the ipsilateral side, stimulation-independent spontaneous ongoing pain and increases of neuronal/astrocyte activities in the ipsilateral L4 dorsal horn on Day 28 post-SNL (Supplementary Figs 20D–K and 21). Neither AAV9 microinjection changed basal paw responses on the contralateral side of SNL mice and on both sides of sham mice (Supplementary Fig. 20D–J) and locomotor function (Supplementary Table 1), although there was a substantial increase of KCNN1 in the ipsilateral L4 DRG of sham mice microinjected with AAV9-KCNN1 (Supplementary Fig. 20C). Furthermore, we mimicked the SNL-induced reduction of DRG KCNN1 through microinjection of a specific Kcnn1 siRNA into unilateral L3/4 DRGs of naïve mice (Supplementary Fig. 22A and B). The mice microinjected with Kcnn1 siRNA, but not a control scrambled siRNA, displayed enhanced responses to mechanical, heat and cold stimuli on the ipsilateral (not contralateral) side from Days 3 to 7 post-microinjection (Supplementary Fig. 22C–F) and increases of neuronal/astrocyte activities in the ipsilateral L3/4 dorsal horn on Day 7 post-microinjection (Supplementary Fig. 22G). These findings strongly suggest that DRG KCNN1 reduction is required for nerve injury-induced nociceptive hypersensitivity.
To demonstrate whether KCNN1 reduction is due to SS-lncRNA downregulation in injured DRG under neuropathic pain conditions, we first examined the effect of rescuing SS-lncRNA downregulation on the SNL-indued reduction in KCNN1 expression in injured DRG. The tamoxifen-treated cKI mice exhibited significant reversion of the SNL-induced reductions in the levels of Kcnn1 mRNA and KCNN1 protein in the ipsilateral L4 DRG on Day 28 post-SNL (Fig. 6A and B). A similar change of Kcnn1 mRNA was observed in the AAV5-EBF1-microinjected mice 14 days post-SNL (Fig. 6C). Conversely, the cKD mice displayed the reductions in the amounts of Kcnn1 mRNA and KCNN1 protein in the DRGs on Day 21 after first injection of tamoxifen (Fig. 6D and E). Microinjection of Ebf1 siRNA into unilateral L3/4 DRGs of naïve mice produced a similar change of Kcnn1 mRNA in the microinjected DRGs on Day 7 post-microinjection (Fig. 6F). More importantly, mechanical, heat and cold nociceptive hypersensitivities and increases of neuronal/astrocyte activities in the L3/4 dorsal horn on the ipsilateral (but not contralateral) side in the AAV9-GFP-microinjected cKD mice were not seen in the AAV9-KCNN1-microinjected cKD mice on Day 7 after first injection of tamoxifen (Fig. 6E, G–K and Supplementary Fig. 23A–C). This suggests that DRG KCNN1 reduction mediates the downregulated SS-lncRNA-induced nociceptive hypersensitivity. Neither virus nor tamoxifen injection altered basal responses on both sides of SSfl/fl mice (Fig. 6G–J and Supplementary Fig. 23A–C) and locomotor activity in both SSfl/fl mice and cKD mice (Supplementary Table1). Consistently, the amount of Kcnn1 mRNA (but not Kcnn2-4 mRNAs) was reduced in cultured DRG neurons transduced with AAV5-SS shRNA plus AAV5-GFP and increased in cultured DRG neurons transduced with AAV5-SS plus AAV5 Scr shRNA (Fig. 6L). More importantly, the AAV5-SS shRNA-induced Kcnn1 mRNA reduction was markedly rescued by co-transduction of AAV5-SS (Fig. 6L). Additionally, the Ebf1 siRNA-induced Kcnn1 mRNA reduction could be reversed by AAV5-EBF1 transduction in cultured DRG neurons (Fig. 6M). Collectively, these data suggest that SS-lncRNA positively regulates the transcriptional activity of Kcnn1 gene and alleviates nerve injury-induced nociceptive hypersensitivity through rescuing Kcnn1 reduction in injured DRG.
Figure 6.
SS-lncRNA downregulation participated in the SNL-induced KCNN1 decrease in injured DRG of male mice. (A and B) Levels of Kcnn1 mRNA (A) and KCNN1 protein (B) in the ipsilateral L4 DRGs on Day 28 after SNL or sham surgery in the SS-KI mice and cKI mice with intraperitoneal injection of tamoxifen daily for seven consecutive days before surgery. n = 12 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (C) Levels of hnRNPM and Kcnn1 mRNAs in the ipsilateral L4 DRGs on Day 14 after SNL or sham surgery in the mice with microinjection of AAV5-GFP or AAV5-EBF1 into ipsilateral L4 DRG 35 days before surgery. n = 12 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (D) Level of Kcnn1 mRNA in L3/4 DRGs from SSfl/fl mice and cKD mice 21 days after first intraperitoneal injection of tamoxifen. n = 8 mice. **P < 0.01, by unpaired Student’s t-test. (E) Level of KCNN1 protein in the ipsilateral L3/4 DRGs on Day 7 after first intraperitoneal injection of tamoxifen in SSfl/fl mice and cKD mice with microinjection of AAV9-GFP or AAV9-KCNN1 into unilateral L3/4 DRGs 35 days before tamoxifen injection. n = 12 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (F) Level of Kcnn1 mRNA in the ipsilateral L3/4 DRGs on Day 7 after microinjection of Scr si or Ebf1 siRNA into unilateral L3/4 DRGs. n = 6 mice/group. **P < 0.01, by unpaired Student’s t-test. (G–J) Paw withdrawal frequency (PWF) to 0.07 g (G) and 0.4 g (H) von Frey filament stimuli and paw withdrawal latency (PWL) to heat (I) and cold (J) stimuli on the ipsilateral side on Day 7 after first intraperitoneal injection of tamoxifen in SSfl/fl mice and cKD mice with microinjection of AAV9-GFP or AAV9-KCNN1 into the unilateral L3/4 DRGs 35 days before tamoxifen injection. n = 12 mice/group. **P < 0.01, by three-way ANOVA with repeated measures followed by post hoc Tukey test. (K) Neuronal and astrocyte activities in the L3/4 dorsal horn as assessed by levels of p-ERK1/2 and GFAP, respectively, on Day 7 after first intraperitoneal injection of tamoxifen in SSfl/fl mice and cKD mice with microinjection of AAV9-GFP or AAV9-KCNN1 into the unilateral L3/4 DRGs 35 days before tamoxifen injection. Total ERK1/2 were used as a control. n = 6 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (L) Levels of SS-lncRNA, Kcnn1 mRNA, Kcnn2 mRNA, Kcnn3 mRNA and Kcnn4 mRNA in the cultured DRG neurons transduced/transfected as indicated. Scr-sh = AAV5 expressing control scrambled shRNA; SS-sh = AAV5 expressing SS-lncRNA shRNA; AAV5-SS = AAV5 expressing SS-lncRNA; AAV5-EFP = AAV5 expressing Gfp mRNA. n = 3 biological repeats/group. **P < 0.01, by one-way ANOVA followed by post hoc Tukey test.
Mimicking nerve injury-induced SS-lncRNA downregulation results in the KCNN1-controlled increase in small DRG neuronal excitability
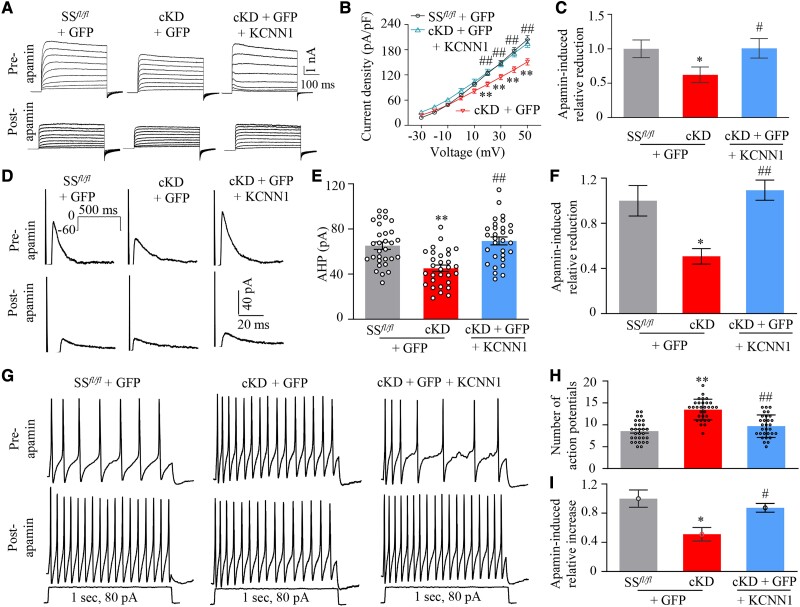
Given that KCNN1 mediates the AHP of action potential and contributes to neuronal excitability,15 we investigated the impact of mimicking the SNL-induced DRG SS-lncRNA downregulation on the KCNN1-controlled AHP, Kv currents and excitability of DRG neurons. To increase the recording efficiency, we microinjected AAV9-GFP alone or a mixed viral solution of AAV9-KCNN1 and AAV9-GFP into unilateral L3/4 DRGs of SSfl/fl mice or cKD mice 35 days before first tamoxifen injection. Whole-cell patch-clamp recordings were performed only in acutely disassociated GFP-labelled small DRG neurons from the ipsilateral L3/4 DRGs 7 days after first intraperitoneal injection of tamoxifen, as KCNN1 and SS-lncRNA were expressed predominantly in small DRG neurons.16 Compared to AAV9-GFP-microinjected SSfl/fl mice, total Kv current density and AHP currents were significantly reduced in small DRG neurons from AAV9-GFP-microinjected cKD mice following tamoxifen injection (Fig. 7A, B, D and E). These reductions were completely reversed in AAV9-KCNN1-microinjected cKD mice (Fig. 7A, B, D and E). Bath application of 100 nM apamin, a selective KCNN blocker,44 led to less reductions of Kv currents and AHP currents in the DRG neurons from AAV9-GFP-microinjected cKD mice than those from AAV9-GFP-microinjected SSfl/fl mice or AAV9-KCNN1-microinjected cKD mice (Fig. 7C and F), strongly indicating that reductions of total Kv current density and AHP currents in small DRG neurons of tamoxifen-treated cKD mice are likely due to KCNN1 downregulation.
Figure 7.
Downregulated SS-lncRNA produced the KCNN1-controlled increase in the excitability of small DRG neurons. All recordings were carried out on Days 7–10 after first intraperitoneal injection of tamoxifen into SSfl/fl mice and cKD mice with microinjection of AAV9-GFP (GFP) or a mixed solution of AAV9-KCNN1 (KCNN1) and AAV9-GFP into unilateral L3/4 DRGs 4 weeks before tamoxifen injection. (A) Representative traces of total Kv current before and after apamin application in small DRG neurons from three treated groups as indicated. (B) I–V curves in small DRG neurons before apamin application from AAV9-GFP plus SSfl/fl group (n = 30 neurons, 18 mice), AAV9-GFP plus cKD (n = 30 neurons, 15 mice) and AAV9-KCNN1/AAV5-GFP plus cKD (n = 30 neurons, 16 mice). **P < 0.01 versus the AAV9-GFP-microinjected SSfl/fl group at the corresponding voltage and ##P < 0.01 versus the AAV9-GFP-microinjected cKD group at the corresponding voltage by three-way ANOVA followed by post hoc Tukey test. (C) At 50 mV, relative reductions in total Kv currents in small DRG neurons after apamin application from three treated groups as indicated. *P < 0.05, by two-way ANOVA with post hoc Tukey test. (D) Representative traces of afterhyperpolarization (AHP) currents elicited by depolarizing pulse injection (from −60 to 0 mV, holding at −60 mV, 500 ms) in small DRG neurons before and after apamin application from three treated groups as indicated. (E and F) Amplitudes of AHP currents before apamin application (E) and relative reductions in the amplitudes of AHP currents after apamin treatment (F) in small DRG neurons from AAV9-GFP-microinjected SSfl/fl mice (n = 30 neurons, 11 mice), AAV9-GFP-microinjected cKD mice (n = 30 neurons, 14 mice) or AAV9-KCNN1-microinjected cKD mice (n = 30 neurons, 15 mice). **P < 0.01, by two-way ANOVA with post hoc Tukey test. (G) Representative traces of action potentials elicited by current injection (1 s, 80 pA) in small DRG neurons before and after apamin application from three treated mice as indicated. (H and I) Frequency of action potentials before apamin application (H) and relative increases in the frequency of action potentials after apamin treatment in small DRG neurons from three treated groups as indicated. Number of cells recorded was same as in E. **P < 0.01, by two-way ANOVA with post hoc Tukey test.
In addition, DRG neuronal excitability was also examined. Compared to AAV9-GFP-microinejcted SSfl/fl mice, the number of action potentials evoked by stimulation of 80 pA (1 s) was substantially increased in small DRG neurons from AAV9-GFP-microinejcted cKD mice following tamoxifen injection (Fig. 7G–I). This increase was not seen in AAV9-KCNN1-microinjected cKD mice (Fig. 7G–I). Bath application of 100 nM apamin produced less increase in number of action potentials in the DRG neurons from AAV9-GFP-microinjected cKD mice than that from AAV9-GFP- microinjected SSfl/fl mice or AAV9-KCNN1-microinjected cKD mice (Fig. 7I). No significant changes were seen in the resting membrane potentials, current thresholds, membrane input resistances, number of spontaneous action potentials and other action potential parameters including threshold, amplitude and overshoot among three treated groups (Supplementary Table 2). Together, SS-lncRNA downregulation increases neuronal excitability likely through silencing KCNN1 expression in small DRG neurons.
SS-lncRNA downregulation reduces the recruitment of hnRNPM to Kcnn1 promoter to lead to Kcnn1 silence in injured DRG
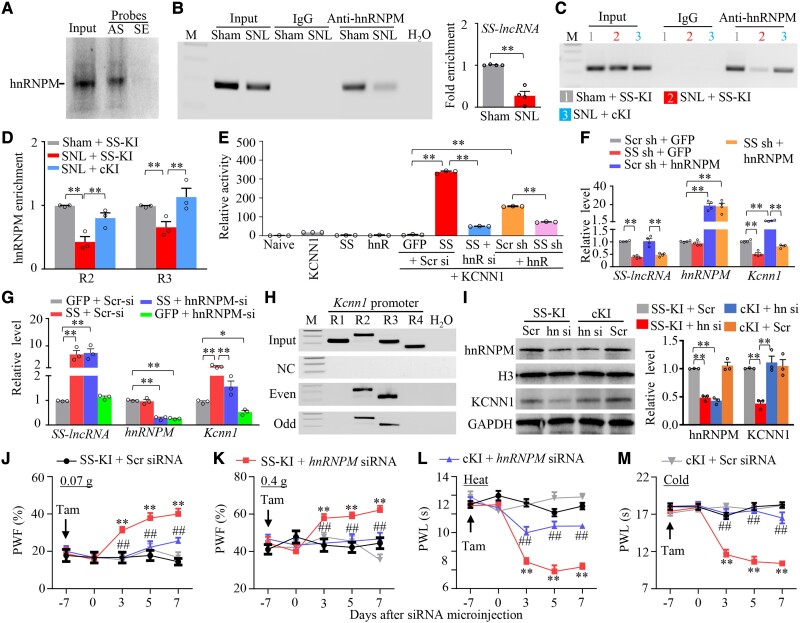
Finally, we asked how Kcnn1 mRNA was reduced by downregulated SS-lncRNA in injured DRG neurons. Given that RNA-binding proteins regulate gene transcription,24,45 we searched for SS-lncRNA binding proteins by designing antisense RNA probes to capture SS-lncRNA and then carrying out the mass spectrometry (MS) assay. We found that 16 proteins bound to SS-lncRNA in cultured DRG neurons (Supplementary Table 3). hnRNPM (heterogeneous nuclear ribonucleoprotein M) appeared to be the most likely binding partner among these binding proteins (Supplementary Table 3). The binding of hnRNPM to SS-lncRNA was verified in cultured DRG neurons (Fig. 8A). Furthermore, a SS-lncRNA fragment was immunoprecipitated by anti-hnRNPM antibody, but not normal purified IgG, in the ipsilateral L4 DRG of sham mice (Fig. 8B). SNL significantly reduced this immunoprecipitating activity by 73% of the value of sham group in the ipsilateral L4 DRG on Day 7 post-surgery (Fig. 8B). The reduction in the binding ability of SS-lincRNA to hnRNPM was mainly due to the SNL-induced DRG SS-lncRNA downregulation in injured DRG, as the level of hnRNPM protein was not altered in the ipsilateral L4 DRG from Days 3 to14 post-SNL (Supplementary Fig. 24A).
Figure 8.
SS-lncRNA downregulation reduces the recruitment of hnRNPM to Kcnn1 promoter to produce Kcnn1 silence in injured DRG. (A) hnRNPM pulled down by SS-lncRNA antisense probe (AS), but not sense probe (SE), in cultured DRG neurons. Input = extracted total protein. n = 3 biological repeats. (B) SS-lncRNA fragment immunoprecipitated by mouse anti-hnRNPM antibody in the ipsilateral L4 DRGs on Day 7 after SNL or sham surgery. M = marker; Input = total purified RNA. IgG: purified mouse IgG. n = 40 mice (four repeats)/group. **P < 0.01, by unpaired Student’s t-test. (C and D) R2 and R3 regions of Kcnn1 promoter immunoprecipitated by mouse anti-hnRNPM antibody in the ipsilateral L4 DRGs on Day 7 after SNL or sham surgery from SS-KI mice or cKI mice with intraperitoneal injection of tamoxifen daily for seven consecutive days before surgery. Representative image showing the binding of hnRNPM to the R2 region of Kcnn1 promoter (C) and quantitative analysis of the bindings (D). M = marker; Input = total purified fragments; IgG = purified mouse IgG. n = 36 mice (three repeats)/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (E) Kcnn1 gene promoter activities in in vitro CAD cells transfected/transduced as shown. GFP = proviral vector expressing Gfp; SS = proviral vector expressing full-length SS-lncRNA; hnR = proviral vector expressing full-length hnRNPM; KCNN1 = pGL3 reporter vector with KCNN1 promoter; Scr si = control scrambled siRNA; hnR si = hnRNPM siRNA; SS sh = AAV5 expressing SS-lncRNA shRNA; Scr sh = AAV5 expressing control scrambled shRNA. n = 3 biological repeats/treatment. **P < 0.01, by one-way ANOVA post hoc Tukey test. (F and G) Levels of SS-lncRNA, hnRNPM mRNA and Kcnn1 mRNA in the cultured DRG neurons transduced/transfected as indicated. SS = AAV5 expressing full length SS-lncRNA; Scr sh = AAV5 expressing control scrambled shRNA; SS sh = AAV5 expressing SS-lncRNA shRNA; GFP = AAV5 expressing Gfp; Scr si = control scrambled siRNA; hnRNPM si = hnRNPM siRNA; hnRNPM = AAV5 expressing full length hnRNPM; n = 3–4 biological repeats/group. *P < 0.05, **P < 0.01, by one-way ANOVA with post hoc Tukey test. (H) Two regions (R2, −674/−362; R3, −381/−161), but not other two regions (R1, −900/−675; R4, −170/−16), from the Kcnn1 promoter were pulled down by SS-lncRNA probes in cultured DRG neurons. A total of 14 different biotinylated antisense DNA probes that were complementary to the sequence of full-length SS-lncRNA were designed and numbered. Seven old-number probes (Odd) and seven even-numbered probes (Even) were pooled together, respectively, for hybridization of cell lysate. M = marker; Input = extracted DNA; NC = negative control probes. n = 3 biological repeats. (I) Levels of hnRNPM and KCNN1 proteins in the ipsilateral L3/4 DRGs on Day 7 after microinjection of control scrambled siRNA (Scr) or hnRNPM siRNA (hn-si) into the unilateral L3/4 DRGs from male SS-KI mice or cKI mice with intraperitoneal injection of tamoxifen daily for 7 days before DRG siRNA microinjection. n = 6 mice/group. **P < 0.01, by two-way ANOVA with post hoc Tukey test. (J–M) Effect of microinjection of control scrambled (Scr) siRNA or hnRNPM siRNA into the ipsilateral L3/4 DRGs on paw withdrawal frequency (PWF) to 0.07 g (J) and 0.4 g (K) von Frey filament stimuli and paw withdrawal latency (PWL) to heat (L) and cold (M) stimuli on the ipsilateral side at time points as shown after siRNA microinjection in male SS-KI mice or cKI mice with intraperitoneal injection of tamoxifen (Tam) daily for 7 days before DRG siRNA microinjection. n = 8–9 mice/group. **P < 0.01 versus the Scr siRNA-treated SS-KI mice and ##P < 0.01 versus the hnRNPM siRNA-treated SS-KI mice at the corresponding time points by three-way ANOVA with repeated measures followed by post hoc Tukey test.
We further carried out the ChIP assay and found that hnRNPM bound to two regions (R2: −674/−362 and R3: −381/−161) of the Kcnn1 gene promoter, as documented by the amplification of only these two regions (out of five regions from −900 to +253) from the complexes immunoprecipitated with anti-hnRNPM antibody in nuclear fraction from the ipsilateral L4 DRG of tamoxifen-pretreated sham SS-KI mice (Fig. 8C and Supplementary Fig. 24B). These two binding activities were decreased by 57% and 34%, respectively, on Day 7 post-SNL in tamoxifen-pretreated SS-KI mice (Fig. 8C and D). The tamoxifen-pretreated SNL cKI mice did not exhibit these decreases (Fig. 8C and D), suggesting that rescuing SS-lncRNA downregulation can block the SNL-induced decrease in the binding activity between hnRNPM and the Kcnn1 promoter in injured DRG. The in vitro experiments showed that the promoter activity of Kcnn1 gene and the levels of Kcnn1 mRNA and its protein were dramatically increased in the CAD cells and cultured DRG neurons, respectively, with AAV9-hnRNPM transduction plus control scrambled siRNA transfection (Fig. 8E and F and Supplementary Fig. 24C). These increases could be diminished by co-transduction with AAV5-SS shRNA (Fig. 8E and F and Supplementary Fig. 24C). Additionally, SS-lncRNA overexpression in the CAD cells and cultured DRG neurons through transduction with AAV5-SS plus transfection with scrambled siRNA increased the promoter activity of Kcnn1 gene and amounts of Kcnn1 mRNA and its protein, respectively (Fig. 8E and G and Supplementary Fig. 24D). These increases were blocked by co-transfection with hnRNPM siRNA (Fig. 8E and G and Supplementary Fig. 24D). Given that SS-lncRNA also bound to R2 and R3 of the Kcnn1 gene promoter in cultured DRG neurons (Fig. 8H) and that SS-lncRNA co-expressed with Kcnn1 mRNA and hnRNPM mRNA in individual small DRG neurons (Supplementary Fig. 13), our findings indicate that SS-lncRNA downregulation results in a reduction in Kcnn1 transcriptional activity likely through decreasing the recruitment of hnRNPM to Kcnn1 gene promoter in injured DRG.
Although SNL did not alter basal expression of hnRNPM in injured DRG (Supplementary Fig. 21A), we asked whether hnRNPM expressed at normal/basal level was required for maintaining basal DRG KCNN1 expression and normal nociceptive information transmission and whether these effects were SS-lncRNA-dependent. The hnRNPM siRNA-microinjected SS-KI mice with intraperitoneal tamoxifen injection daily for 7 days before DRG siRNA microinjection displayed the decreases in the levels of Kcnn1 mRNA and KCNN1 protein in the L3/4 DRGs on Day 7 post-microinjection, mechanical, heat and cold nociceptive hypersensitivities from Days 3 to 7 post-microinjection and increases of neuronal/astrocyte activities in the L3/4 dorsal horn on Day 7 post-microinjection on the ipsilateral (but not contralateral) side (Fig. 8I–M and Supplementary Fig. 25A–E). These changes were not seen in the hnRNPM siRNA-microinjected cKI mice with intraperitoneal tamoxifen injection (Fig. 8I–M and Supplementary Fig. 25A–E). These findings suggests that normal expression of DRG hnRNPM is critical in maintaining basal DRG KCNN1 expression and basal/acute pain and that SS-lncRNA participates in these processes. As expected, neither hnRNPM siRNA nor control scrambled siRNA altered locomotor function in SS-KI mice or cKI mice (Supplementary Table 1).
Discussion
Previous work has attributed neuropathic pain initiation and progression at least in part to nerve injury-induced reduction of potassium channels in injured DRG.9 However, how potassium channels are reduced after peripheral nerve injury remains mysterious. Here we reported that the downregulation of a newly identified SS-lncRNA temporally correlated with KCNN1 reduction in injured DRG following peripheral nerve injury. The downregulated SS-lncRNA contributed to induction and maintenance of neuropathic pain through losing recruitment of hnRNPM to Kcnn1 promoter and subsequently leading to Kcnn1 transcriptional silence and neuronal hyperexcitability in injured DRG. SS-lncRNA likely plays an important role in nerve injury-induced nociceptive hypersensitivity.
SS-lncRNA, unlike other lncRNAs such as Kcna2 antisense RNA, NIS-lncRNA and H19,11,46,47 is transcriptionally silenced in injured DRG following peripheral nerve injury. Under normal conditions, SS-lncRNA was detected exclusively in DRG neurons, particularly in small DRG neurons. SS-lncRNA was downregulated in injured DRG neurons after SNL- or CCI-induced peripheral nerve injury, but not CFA- or MIA-induced peripheral inflammation. How this downregulation is in a specific response to peripheral nerve injury remains unknown, but the present study showed that nerve injury-induced transcriptional silence of DRG SS-lncRNA may be attributed to nerve injury-induced reduction in DRG EBF1. EBF1 could bind to and activate the SS-lncRNA promoter to trigger SS-lncRNA expression in DRG neurons. SNL-induced reduction of EBF1 resulted in a decrease in binding of EBF1 to SS-lncRNA promoter and then a transcriptional inactivation of SS-lncRNA gene in injured DRG. Whether DRG EBF1 reduction, like SS-lncRNA downregulation, also responds specifically to peripheral nerve injury remains to be determined. Admittedly, other transcription factors that may also be implicated in SS-lncRNA downregulation in injured DRG cannot be ruled out. Additionally, a decrease in RNA stability and/or epigenetic modifications48,49 may be involved in DRG SS-lncRNA downregulation after nerve injury.
DRG KCNN1 reduction is required for neuropathic pain genesis. Like SS-lncRNA, KCNN1 is expressed mainly in small DRG neurons.16 KCNN1 was decreased in the DRG avulsed from neuropathic pain patients.17 Our findings revealed that SNL reduced the expression of Kcnn1 mRNA and KCNN1 protein in injured mouse DRG neurons. Consistently, SNL decreased the apamin-sensitive Kv currents in injured rat DRG neurons.44 Interestingly, a previous study reported no change of KCNN1 protein in injured rat DRG 15 days after CCI.16 Given that this study did not mention behavioural data,16 unchanged expression of KCNN1 in injured DRG is likely due to an unsuccessful CCI model. More importantly, we found that rescuing KCNN1 reduction in injured DRG mitigated SNL-induced nociceptive hypersensitivity and dorsal horn neuronal/astrocyte hyperactivity on the ipsilateral side during the development period. In addition, knockdown of KCNN1 in the DRG of naïve mice produced enhanced responses to noxious stimuli and dorsal horn neuronal/astrocyte hyperactivity on the ipsilateral side. These findings strongly indicate an important role of DRG KCNN1 reduction in neuropathic pain genesis.
SS-lncRNA downregulation produces the hnRNPM-mediated reduction of Kcnn1 mRNA in injured DRG. HnRNPM is a member of the family of the (pre-) mRNA binding heterogeneous nuclear ribonucleoproteins (hnRNPs) that play major roles in the formation, packaging and processing of mRNA.50 We demonstrated that hnRNPM bound to the Kcnn1 gene promoter, triggered Kcnn1 transcriptional activation, and increased the expression of Kcnn1 mRNA and its protein in cultured DRG neurons. SNL did not significantly alter basal expression of hnRNPM in injured DRG, but hnRNPM expressed at normal level was required for maintaining basal KCNN1 expression in DRG neurons, because DRG knockdown of hnRNPM markedly reduced the levels of Kcnn1 mRNA and KCNN1 protein in in vivo and in vitro DRG and produced the enhanced responses to noxious stimuli. The impact of hnRNPM on Kcnn1 transcriptional activation may require the transcription factor TAF15, as interaction of hnRNPM with TAF15 promoted gene transcription.51SS-lncRNA knockdown blocked the hnRNPM-triggered activation of Kcnn1 promoter and expression of Kcnn1 mRNA and its protein in cultured DRG neurons. Moreover, SS-lncRNA overexpression attenuated the hnRNPM knockdown-induced the decreases of the expression of Kcnn1 mRNA and its protein in the DRG and the enhanced response to noxious stimuli. Given that SS-lncRNA bound to both hnRNPM and Kcnn1 promoter and that SS-lncRNA, hnRNPM mRNA and Kcnn1 mRNA co-expressed particularly in individual small DRG neurons, SS-lncRNA may function as a molecular scaffolder to recruit hnRNPM to the Kcnn1 promoter for maintaining normal KCNN1 expression in DRG neurons. Nerve injury-induced downregulation of SS-lncRNA may result in a decrease in its binding to hnRNPM, lost the recruitment of hnRNPM to the Kcnn1 promoter and silenced Kcnn1 expression in injured DRG neurons. This conclusion was further supported by the fact that SS-lncRNA-induced activation of the Kcnn1 promoter and increases in the levels of Kcnn1 mRNA and KCNN1 protein in cultured DRG neurons were prevented by hnRNPM knockdown. Besides hnRNPM, our MS assay revealed that the remaining 15 proteins also interacted with SS-lncRNA in DRG. Whether these proteins participate in the downregulated SS-lncRNA-induced reduction of KCNN1 in injured DRG remains to be determined.
Downregulated SS-lncRNA contributes to neuropathic pain likely through silencing KCNN1 expression in injured DRG. We demonstrated that rescuing nerve injury-induced downregulation of DRG SS-lncRNA reversed nerve injury-induced reductions of Kcnn1 mRNA and KCNN1 protein in injured DRG and attenuated the development and maintenance of nerve injury-induced nociceptive hypersensitivity. Conversely, mimicking nerve injury-induced DRG SS-lncRNA downregulation decreased the expression of DRG Kcnn1 mRNA and KCNN1 protein, reduced total Kv currents and AHP currents and increased excitability in small DRG neurons and augmented responses to noxious stimuli. Moreover, these electrophysiological changes and enhanced behavioural responses were significantly blocked after DRG overexpression of KCNN1. Considering that DRG KCNN1 reduction is required for neuropathic pain genesis, our findings suggest that SS-lncRNA alleviates nerve injury-induced nociceptive hypersensitivity likely through rescuing the hnRNPM-mediated DRG KCNN1 reduction, decreasing neuronal hyperexcitability in injured DRG, diminishing the release of transmitters/neuromodulators in primary afferents and consequently impairing dorsal horn central sensitization. In support of this conclusion, we found that rescuing SS-lncRNA downregulation in injured DRG prevented nerve injury-induced hyperactivation in dorsal horn neurons and astrocytes. It is worth noting that, besides Kcnn1 transcript, rescuing DRG SS-lncRNA downregulation also impacted the SNL-induced changes in other DRG transcripts. Thus, other potential mechanisms by which downregulated SS-lncRNA contributes to neuropathic pain cannot be excluded.
Mechanisms underlying neuropathic pain are complicated and still incompletely understood. Several lines of evidence support the role of medium and large DRG neurons in neuropathic pain. An increase in spontaneous ectopic activity after nerve injury was found in injured myelinated afferents and the corresponding medium and large DRG neurons.52,53 Nerve injury-induced mechanical allodynia was mediated by amyloid-β fibres.54,55 After oxaliplatin administration, bursts of action potentials induced by the cooling applied on mouse and human peripheral axons were detected in myelinated A fibres.56 Moreover, DRG knockdown of Nav1.6,57Kcan2 antisense RNA,11 or NIS-lncRNA47 expressed mainly in medium and large DRG neurons significantly mitigated nerve injury-induced nociceptive hypersensitivities and prevented abnormal spontaneous activity in myelinated neurons of injured DRG. In contrast, if small nociceptive DRG neurons participate in neuropathic pain remains to be determined. The mice with knockdown or knockout of Nav1.7, Nav1.8, or Nav1.9 in small DRG neurons displayed intact neuropathic pain in early studies,58–60 but peripheral nerve injury increased the excitability and decreased membrane threshold and rheobase in injured small DRG neurons.61 A recent study revealed that blocking nerve injury-induced upregulation of GPR151 in small DRG neurons attenuated nerve injury-induced neuronal hyperexcitability and nociceptive hypersensitivities.62 Consistent with this observation, the present study showed that downregulated SS-lncRNA and KCNN1 expressed predominantly in small DRG neurons16 were required for DRG neuronal excitability and nociceptive hypersensitivities following nerve injury. The evidence strongly suggests that small DRG neurons, like medium and large DRG neurons, are also implicated in neuropathic pain genesis.
In conclusion, the present study demonstrated that SS-lncRNA overexpression in injured DRG alleviated nerve injury-induced nociceptive hypersensitivity without altering responses to acute/basal noxious stimuli and locomotor function. Given that SS-lncRNA is exclusively expressed in sensory neurons, targeted SS-lncRNA may produce an antinociceptive effect on neuropathic pain with no or fewer unwanted side effects.
Supplementary Material
Acknowledgements
We thank Biocytogen (Worcester, MA) for generating SS-lncRNAfl/fl mice and Rosa26SS-lncRNA knock-in mice, Dr Yun-Juan Chang for analysing ribosome profiling data and Dr Li Zhang for ribosome prefiling data analysis.
Contributor Information
Bing Wang, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Longfei Ma, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Xinying Guo, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Shibin Du, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Xiaozhou Feng, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Yingping Liang, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Gokulapriya Govindarajalu, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Shaogen Wu, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Tong Liu, Center for Advanced Proteomics Research, Departments of Biochemistry, Microbiology and Molecular Genetics, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Hong Li, Center for Advanced Proteomics Research, Departments of Biochemistry, Microbiology and Molecular Genetics, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Shivam Patel, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Alex Bekker, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Huijuan Hu, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA; Department of Physiology, Pharmacology and Neuroscience, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Yuan-Xiang Tao, Department of Anesthesiology, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA; Department of Physiology, Pharmacology and Neuroscience, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA; Department of Cell Biology and Molecular Medicine, New Jersey Medical School, Rutgers, The State University of New Jersey, Newark, NJ 07103, USA.
Funding
The mass spectrometry data were obtained from an Orbitrap mass spectrometer funded by NIH grants NS046593 and 1S10OD025047-01 to H.L. This work was supported by NIH grants R01NS111553 and RFNS113881 to Y.X.T. and NIH grant R01NS117484 to H.H. and Y.X.T.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Dieleman JP, Kerklaan J, Huygen FJPM, Bouma PAD, Sturkenboom MCJM. Incidence rates and treatment of neuropathic pain conditions in the general population. Pain. 2008;137:681–688. [DOI] [PubMed] [Google Scholar]
- 2. van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: A systematic review of epidemiological studies. Pain. 2014;155:654–662. [DOI] [PubMed] [Google Scholar]
- 3. O'Connor AB. Neuropathic pain: Quality-of-life impact, costs and cost effectiveness of therapy. Pharmacoeconomics. 2009;27:95–112. [DOI] [PubMed] [Google Scholar]
- 4. de Leon-Casasola OA. Opioids for chronic pain: New evidence, new strategies, safe prescribing. Am J Med. 2013;126:S3–11. [DOI] [PubMed] [Google Scholar]
- 5. Meyer R, Patel AM, Rattana SK, Quock TP, Mody SH. Prescription opioid abuse: A literature review of the clinical and economic burden in the United States. Popul Health Manag. 2014;17:372–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campbell JN, Meyer RA. Mechanisms of neuropathic pain. Neuron. 2006;52:77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costigan M, Scholz J, Woolf CJ. Neuropathic pain: A maladaptive response of the nervous system to damage. Annu Rev Neurosci. 2009;32:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith PA. K(+) channels in primary afferents and their role in nerve injury-induced pain. Front Cell Neurosci. 2020;14:566418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan L, Guan X, Wang W, et al. Impaired neuropathic pain and preserved acute pain in rats overexpressing voltage-gated potassium channel subunit Kv1.2 in primary afferent neurons. Mol Pain. 2014;10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao X, Tang Z, Zhang H, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang L, Gu X, Zhao JY, et al. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci Rep. 2016;6:37704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun L, Gu X, Pan Z, et al. Contribution of DNMT1 to neuropathic pain genesis partially through epigenetically repressing Kcna2 in primary afferent neurons. J Neurosci. 2019;39:6595–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao JY, Liang L, Gu X, et al. DNA Methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun. 2017;8:14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kshatri AS, Gonzalez-Hernandez A, Giraldez T. Physiological roles and therapeutic potential of Ca(2+) activated potassium channels in the nervous system. Front Mol Neurosci. 2018;11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mongan LC, Hill MJ, Chen MX, et al. The distribution of small and intermediate conductance calcium-activated potassium channels in the rat sensory nervous system. Neuroscience. 2005;131:161–175. [DOI] [PubMed] [Google Scholar]
- 17. Boettger MK, Till S, Chen MX, et al. Calcium-activated potassium channel SK1- and IK1-like immunoreactivity in injured human sensory neurones and its regulation by neurotrophic factors. Brain. 2002;125:252–263. [DOI] [PubMed] [Google Scholar]
- 18. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francescatto M, Vitezic M, Heutink P, Saxena A. Brain-specific noncoding RNAs are likely to originate in repeats and may play a role in up-regulating genes in cis. Int J Biochem Cell Biol. 2014;54:331–337. [DOI] [PubMed] [Google Scholar]
- 23. Lutz BM, Bekker A, Tao YX. Noncoding RNAs: New players in chronic pain. Anesthesiology. 2014;121:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan Z, Du S, Wang K, et al. Downregulation of a dorsal root ganglion-specifically enriched long noncoding RNA is required for neuropathic pain by negatively regulating RALY-triggered ehmt2 expression. Adv Sci (Weinh). 2021;8:e2004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu S, Bono J, Tao YX. Long noncoding RNA (lncRNA): A target in neuropathic pain. Expert Opin Ther Targets. 2019;23:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y, Guo X, Sun L, et al. N(6)-methyladenosine demethylase FTO contributes to neuropathic pain by stabilizing G9a expression in primary sensory neurons. Adv Sci (Weinh). 2020;7:1902402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma L, Yu L, Jiang BC, et al. ZNF382 Controls mouse neuropathic pain via silencer-based epigenetic inhibition of Cxcl13 in DRG neurons. J Exp Med. 2021;218:e20210920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Atianjoh FE, Yaster M, Zhao X, et al. Spinal cord protein interacting with C kinase 1 is required for the maintenance of complete Freund's adjuvant-induced inflammatory pain but not for incision-induced post-operative pain. Pain. 2010;151:226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chu YC, Guan Y, Skinner J, Raja SN, Johns RA, Tao YX. Effect of genetic knockout or pharmacologic inhibition of neuronal nitric oxide synthase on complete Freund's adjuvant-induced persistent pain. Pain. 2005;119:113–123. [DOI] [PubMed] [Google Scholar]
- 30. Mailhot B, Christin M, Tessandier N, et al. Neuronal interleukin-1 receptors mediate pain in chronic inflammatory diseases. J Exp Med. 2020;217:e20191430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tao YX, Rumbaugh G, Wang GD, et al. Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J Neurosci. 2003;23:6703–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rigaud M, Gemes G, Barabas ME, et al. Species and strain differences in rodent sciatic nerve anatomy: Implications for studies of neuropathic pain. Pain. 2008;136:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gallagher PE, Diz DI. Analysis of RNA by northern-blot hybridization. Methods Mol Med. 2001;51:205–213. [DOI] [PubMed] [Google Scholar]
- 34. Li Z, Mao Y, Liang L, et al. The transcription factor C/EBPbeta in the dorsal root ganglion contributes to peripheral nerve trauma-induced nociceptive hypersensitivity. Sci Signal. 2017;10:eaam5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mo K, Wu S, Gu X, et al. MBD1 Contributes to the genesis of acute pain and neuropathic pain by epigenetic silencing of Oprm1 and Kcna2 genes in primary sensory neurons. J Neurosci. 2018;38:9883–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Samineni VK, Grajales-Reyes JG, Sundaram SS, Yoo JJ, Gereau RW. Cell type-specific modulation of sensory and affective components of itch in the periaqueductal gray. Nat Commun. 2019;10:4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steinkellner T, Zell V, Farino ZJ, et al. Role for VGLUT2 in selective vulnerability of midbrain dopamine neurons. J Clin Invest. 2018;128:774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu S, Marie LB, Miao X, et al. Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Mol Pain. 2016;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp. 2012;(61):3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu C, Zhang QC, da Rocha ST, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Z, Gu X, Sun L, et al. Dorsal root ganglion myeloid zinc finger protein 1 contributes to neuropathic pain after peripheral nerve trauma. Pain. 2015;156:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park JS, Voitenko N, Petralia RS, et al. Persistent inflammation induces GluR2 internalization via NMDA receptor-triggered PKC activation in dorsal horn neurons. J Neurosci. 2009;29:3206–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu JT, Zhou X, Zhao X, et al. Opioid receptor-triggered spinal mTORC1 activation contributes to morphine tolerance and hyperalgesia. J Clin Invest. 2014;124:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sarantopoulos CD, McCallum JB, Rigaud M, Fuchs A, Kwok WM, Hogan QH. Opposing effects of spinal nerve ligation on calcium-activated potassium currents in axotomized and adjacent mammalian primary afferent neurons. Brain Res. 2007;1132:84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vanderweyde T, Youmans K, Liu-Yesucevitz L, Wolozin B. Role of stress granules and RNA-binding proteins in neurodegeneration: A mini-review. Gerontology. 2013;59:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wen J, Yang Y, Wu S, et al. Long noncoding RNA H19 in the injured dorsal root ganglion contributes to peripheral nerve injury-induced pain hypersensitivity. Transl Perioper Pain Med. 2020;7:176–184. [PMC free article] [PubMed] [Google Scholar]
- 47. Du S, Wu S, Feng X, et al. A nerve injury-specific long noncoding RNA promotes neuropathic pain by increasing Ccl2 expression. J Clin Invest. 2022;132:e153563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weskamp K, Barmada SJ. RNA Degradation in neurodegenerative disease. Adv Neurobiol. 2018;20:103–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Piñol-Roma S, Swanson MS, Gall JG, Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989;109:2575–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ainaoui N, Hantelys F, Renaud-Gabardos E, et al. Promoter-Dependent translation controlled by p54nrb and hnRNPM during myoblast differentiation. PLoS One. 2015;10:e0136466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu CN, Wall PD, Ben-Dor E, Michaelis M, Amir R, Devor M. Tactile allodynia in the absence of C-fiber activation: Altered firing properties of DRG neurons following spinal nerve injury. Pain. 2000;85:503–521. [DOI] [PubMed] [Google Scholar]
- 53. Tal M, Wall PD, Devor M. Myelinated afferent fiber types that become spontaneously active and mechanosensitive following nerve transection in the rat. Brain Res. 1999;824:218–223. [DOI] [PubMed] [Google Scholar]
- 54. Duan B, Cheng L, Bourane S, et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell. 2014;159:1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu ZZ, Kim YH, Bang S, et al. Inhibition of mechanical allodynia in neuropathic pain by TLR5-mediated A-fiber blockade. Nat Med. 2015;21:1326–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sittl R, Lampert A, Huth T, et al. Anticancer drug oxaliplatin induces acute cooling-aggravated neuropathy via sodium channel subtype Na(V)1.6-resurgent and persistent current. Proc Natl Acad Sci U S A. 2012;109:6704–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xie W, Strong JA, Zhang JM. Local knockdown of the NaV1.6 sodium channel reduces pain behaviors, sensory neuron excitability, and sympathetic sprouting in rat models of neuropathic pain. Neuroscience. 2015;291:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Amaya F, Wang H, Costigan M, et al. The voltage-gated sodium channel Na(v)1.9 is an effector of peripheral inflammatory pain hypersensitivity. J Neurosci. 2006;26:12852–12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kerr BJ, Souslova V, McMahon SB, Wood JN. A role for the TTX-resistant sodium channel Nav 1.8 in NGF-induced hyperalgesia, but not neuropathic pain. Neuroreport. 2001;12:3077–3080. [DOI] [PubMed] [Google Scholar]
- 60. Nassar MA, Levato A, Stirling LC, Wood JN. Neuropathic pain develops normally in mice lacking both Na(v)1.7 and Na(v)1.8. Mol Pain. 2005;1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gong K, Kung LH, Magni G, Bhargava A, Jasmin L. Increased response to glutamate in small diameter dorsal root ganglion neurons after sciatic nerve injury. PLoS One. 2014;9:e95491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xia LP, Luo H, Ma Q, et al. GPR151 In nociceptors modulates neuropathic pain via regulating P2X3 function and microglial activation. Brain. 2021;144:3405–3420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All information necessary to evaluate the reported findings are included in the main text or the Supplementary material. Additional data or further methodological details will be provided by the authors upon request.