Abstract
Background:
Patient-reported outcomes (PRO) allow clinicians to measure health-related quality of life (HRQOL) and understand patients’ treatment priorities, but obtaining PRO requires surveys which are not part of routine care. We aimed to develop a preliminary natural language processing (NLP) pipeline to extract HRQOL trajectory based on deep learning models using patient language.
Materials and methods:
Our data consisted of transcribed interviews of 100 patients undergoing surgical intervention for low-risk thyroid cancer, paired with HRQOL assessments completed during the same visits. Our outcome measure was HRQOL trajectory measured by the SF-12 physical and mental component scores (PCS and MCS), and average THYCA-QoL score.
We constructed an NLP pipeline based on BERT, a modern deep language model that captures context semantics, to predict HRQOL trajectory as measured by the above endpoints. We compared this to baseline models using logistic regression and support vector machines trained on bag-of-words representations of transcripts obtained using Linguistic Inquiry and Word Count (LIWC). Finally, given the modest dataset size, we implemented two data augmentation methods to improve performance: first by generating synthetic samples via GPT-2, and second by changing the representation of available data via sequence-by-sequence pairing, which is a novel approach.
Results:
A BERT-based deep learning model, with GPT-2 synthetic sample augmentation, demonstrated an area-under-curve of 76.3% in the classification of HRQOL accuracy as measured by PCS, compared to the baseline logistic regression and bag-of-words model, which had an AUC of 59.9%. The sequence-by-sequence pairing method for augmentation had an AUC of 71.2% when used with the BERT model.
Conclusions:
NLP methods show promise in extracting PRO from unstructured narrative data, and in the future may aid in assessing and forecasting patients’ HRQOL in response to medical treatments. Our experiments with optimization methods suggest larger amounts of novel data would further improve performance of the classification model.
Keywords: Machine learning, Artificial neural networks, Medical information systems, Natural language processing
1. Background
Patient-reported outcomes (PRO) allow clinicians to assess patient well-being directly without relying on interpretation by a clinician. They are traditionally ascertained using surveys. Despite the important role of PRO in making patient-centered treatment decisions, barriers remain to routine use of PRO outside of research contexts [1,2]. PRO instruments can be time-consuming and expensive to administer. In addition, they are not part of the routine clinical workflow, requiring patients to complete an extra task.
Health related quality of life (HRQOL) is an example of PRO representing a patient’s perceived physical and mental health [3]. Algorithms to determine psychosocial well-being from narrative data sources, such as patient interviews, hold potential in providing a streamlined way to gauge a patient’s well-being. Narrative data are increasingly available from patient communications and the electronic health record (EHR), and may provide insight into HRQOL [4,5]. However, lack of structure in natural language can complicate automated interpretation. Natural language processing (NLP) can aid in extracting meaning from narrative data.
Thyroid cancer is relatively common in the United States. Because mortality rates are low, and a variety of treatment options are available, each with its own risk profile and potential for lifelong morbidity, ascertaining HRQOL is essential in thyroid cancer. No tools currently exist to extract HRQOL from patient language. Extraction of HRQOL outcomes after surgery can help personalize treatment in the face of a myriad of options now available to patients [6]. Patients at risk for long-term decrements in HRQOL may need active referral to resources such as counseling and support groups. Furthermore, if we are able to identify these patients preoperatively, they may opt for less invasive treatment options.
Previous NLP work in the medical field largely draws from data available in the EHR [7,8]. Furthermore, many existing NLP approaches train algorithms using the written documentation of doctors and other clinicians [9], which are likely to be structured or include jargon [10] By contrast, we use a novel and unique dataset of prospectively collected and transcribed patient interviews paired with numeric scores on several validated Likert-like PRO measures for HRQOL, which serves as an annotated machine learning dataset. We also analyze speech from transcripts of patients, who have little to no medical background and are likely to be less domain-specific than that of medical professionals.
We iteratively implemented and evaluated several methods for inferring HRQOL trajectory from narrative data. In addition, we have developed a way to assess whether the size of the data set is large enough for training, which points to a distinction between the density of the data and its diversity.
2. Materials and methods
Fig. 1 gives an overview of the experiments described in this paper. After pre-processing, Linguistic Inquiry and Word Count (LIWC) was used with two different classifiers (logistic regression (LR) and a support vector machine (SVM)) to provide an initial baseline model. We then leveraged modern NLP models. Fine-tuned BERT-based models were trained and compared to the LIWC-based models. Finally, two data augmentation techniques (sequence generation using GPT-2, and sequence pairing) were used to optimize performance.
Fig. 1. Data Analysis Workflow.
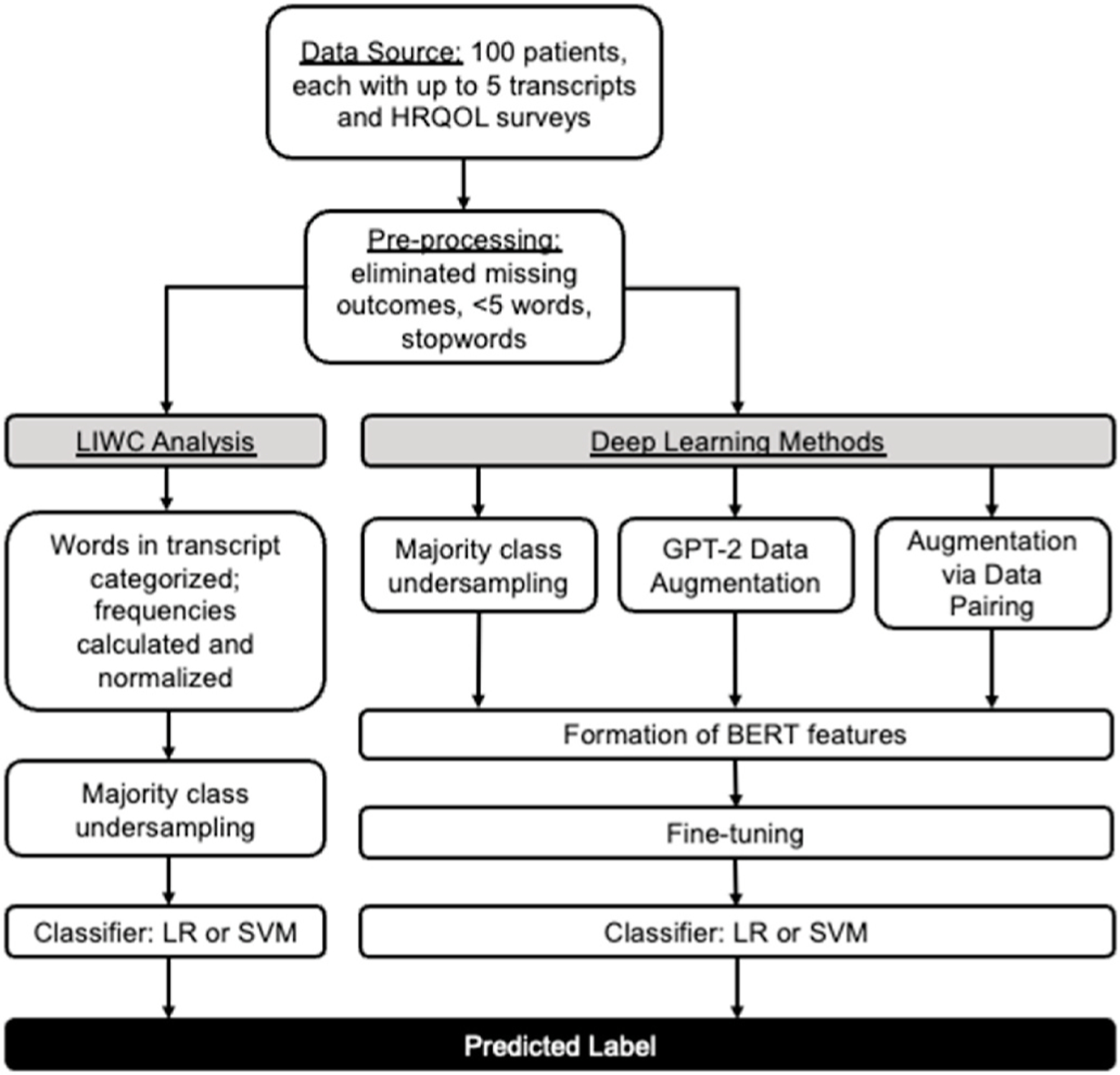
Abbreviations: LIWC, Logistic Inquiry and Word Count; LR, logistic regression; SVM, support vector machine
A diagram of the experiments described in this paper. After basic pre-processing of transcripts, baseline classification models based on logistic regression as well as SVM using LIWC features were performed as baselines. This was then compared to fine-tuning BERT, and using two data augmentation models to further improve performance.
2.1. Dataset
We used a novel dataset from a prospective randomized controlled clinical trial assessing surgical interventions for low-risk thyroid cancer (see Declarations for details of ethical approval and consent to participate). Each patient was evaluated at up to five time-points: preoperative (6 weeks–24 h before operation), and 2 weeks, 6 weeks, 6 months, and 1 year postoperative. At each visit, the patient participated in a semistructured interview and completed several assessments of HRQOL, including the 12-item short form survey (SF-12), and the THYCA-QoL. The SF-12 is a 12-item questionnaire measuring the impact of health on an individual’s everyday life, and is validated in Americans with cancer [11,12]. The THYCA-QoL is a thyroid-cancer specific questionnaire that focuses on common symptoms of thyroid cancer or its treatment and has also been extensively validated [13,14].
Each patient had up to 5 transcripts (one from each time point). Each transcript consisted of alternating utterances (sequences) between patient and interviewer. Each sequence was assigned the HRQOL scores of the transcript from which it originated; thus all sequences taken from the same transcript had the same HRQOL scores. We selected the trajectories of three HRQOL scores as primary outcome measures: the SF-12 Physical and Mental Component Scores (PCS and MCS) and the average THYCA-QoL score (Average THYCA). A random sample of sequences from the data is shown in Supplemental Table 4a. Many of the sequences do not obviously relate to health issues.
2.2. Pre-processing
First, standard data pre-processing steps were undertaken (Supplemental Methods).
Next we calculated HRQOL score trajectory for each sequence. The original HRQOL scores were numeric in all cases. Rather than attempt to infer these values directly, we instead focused on the HRQOL trajectory, defined as the direction of difference in the HRQOL score (PCS, MCS, or average THYCA) between two time points. Sequences associated with a transcript with a positive HRQOL trajectory slope were labeled “class 1,” and negative HRQOL trajectory slope, “class 0.” Thus a positive slope or “class 1” an improvement in patient HRQOL, and vice versa. The problem thus became one of binary classification.
Of up to five transcripts from pre-defined time points at which the HRQOL surveys and interviews were administered, two trajectories were computed: the 1–2 trajectory (direction of HRQOL change between transcript 1 and transcript 2) and the 2-last trajectory (between transcript 2 and the last available transcript).1 Transcript 2 represents the first postoperative interview. These trajectories were selected, respec tively, to reflect the HRQOL change before and after surgery, and the subsequent recovery or lack thereof. We noted that 1) the total number of sequences was modest, on the order of a few thousand of samples for each class, and 2) the distribution of trajectories was imbalanced (Fig. 2 a–c).
Fig. 2. Distribution of HRQOL trajectories.
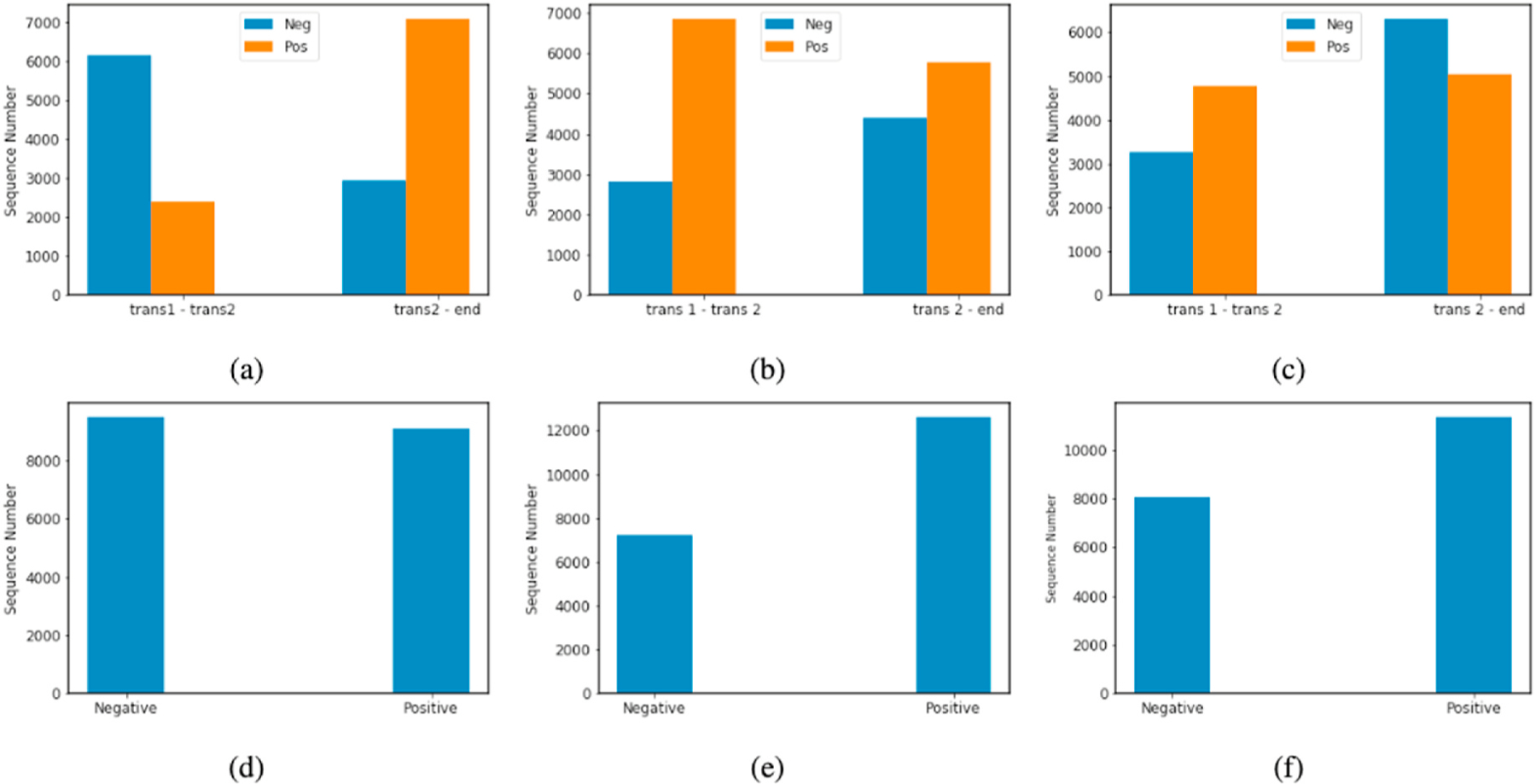
The distributions before combination of two trajectories for each outcome measurement are shown for (a) PCS, (b) MCS, and (c) average THYCA. The distributions after combining the trajectories are shown for (d) PCS, (e) MCS, and (f) Average THYCA.
As imbalanced data sets can bias classification results, we balanced the classes by combining the negative-slope 1–2 trajectories with the negative-slope 2-last trajectories (and the same with the positive-slope trajectories) into a single dataset. This improved the overall balance (Fig. 2 d–f). During training, we under-sampled the majority class while keeping the original distribution of classes in the validation/test set. We repeated this process for MCS and average THYCA. After this, there were 18194 sequences for PCS, 14450 for MCS, and 16056 for average THYCA.
In all experiments, we used 70% of the data as a training set, 20% as a test set, and the remaining 10% as a validation set for each of the variables.
2.3. Experiments
Our objective was to infer HRQOL trajectory from sequences. The input of each classifier consisted of features extracted from sequences using different encoding methods, discussed in the following sub-sections. The output was the class label.
2.3.1. Analysis with LIWC features
We used LIWC as an initial benchmark encoding method. LIWC is a dictionary-based text analysis tool widely used to ascertain the linguistic characteristics of text [15]. It assigns every word in the text to one or more categories in an internal dictionary and produces an output specifying the percentage of the total words in the text that fall into each linguistic category. LIWC does not consider the underlying structure or sequence of speech, but only its lexical contents. Though LIWC is limited by the accuracy and comprehensiveness of the dictionary they are based upon, it has been used in many studies evaluating indicators of psychosocial well-being [16] and has also served as a comparison point for deep learning methods [17–19].
For each sequence, frequencies were calculated for 9 LIWC categories of interest (Table 1), selected by an expert in communications research and medical sentiment analysis (JH) based on the research question and dataset properties. The LIWC frequencies became input features for the training of the logistic regression (LR) and support vector machine (SVM) classifiers.
Table 1.
Linguistic features of patient interview transcripts.
| LIWC CATEGORY | EXAMPLES |
|---|---|
| POSITIVE EMOTIONS | Love, nice, sweet |
| NEGATIVE EMOTION | Hurt, ugly, nasty |
| FEELING | Feels, touch |
| BIOLOGIC PROCESSES | Eat, blood, pain |
| BODY | Cheek, hands, spit |
| HEALTH | Clinic, flu, pill |
| PAST-FOCUSED | Ago, did, talked |
| PRESENT-FOCUSED | Today, is, now |
| FUTURE-FOCUSED | May, will, soon |
Abbreviations: LIWC, Linguistic Inquiry and Word Count.
The nine LIWC categories used for construction of the baseline predictive model. The right column gives examples of words falling into each category.
2.3.2. Analysis using BERT
BERT is a transformer-based language model which has achieved state-of-the-art performance on many natural language processing tasks [20]. BERT works by parsing large quantities of data and representing words and their statistical correlations with other words in a high dimensional feature space. BERT is then fine-tuned on a domain-specific corpus of text and applied to downstream tasks. In our experiments, we used the fine-tuned BERT as an encoder to extract sequence embeddings, which were then input into the classifier. Specifically, we added a linear layer followed by a sigmoid function on top of the pre-trained BERT, and they were fine-tuned together for classification.
We used the BERT implementation of Wolf et al. [21]. The first token of the BERT output ([CLS]) was used as the sequence embedding, a vector with dimension 768 (Fig. 3a). An independent BERT model was fine-tuned separately for the analysis of each HRQOL measure (PCS, MCS, and average THYCA).
Fig. 3. Structure of BERT-based classification model.
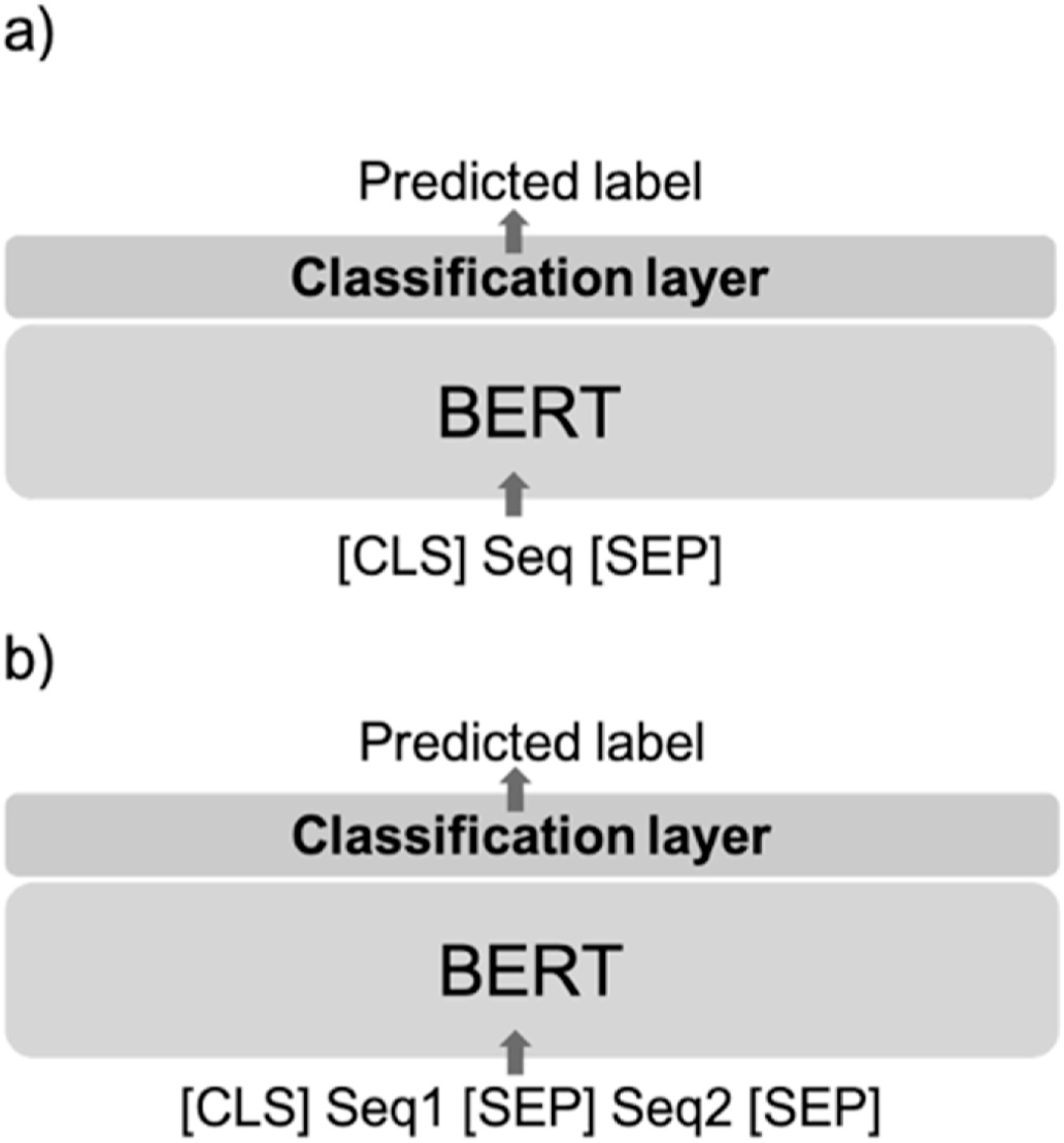
In (a), a natural language sequence Seq is sandwiched by two special tokens [CLS] and [SEP], indicating the start and end of the sequence. This is input into BERT, which transforms the text into a feature vector used to carry out the classification, i.e., the prediction of the label. In (b), two sequences Seq1 and Seq2 are concatenated and separated (again using [CLS] and [SEP] tokens) and then jointly input into BERT.
As previously noted, the total number of sequences in our dataset was relatively small. We then implemented two methods of data augmentation to further improve classification performance.
2.3.3. Training set augmentation via GPT-2
GPT-2 is a large transformer-based auto-regressive decoder pretrained on large bodies of text to generate representative sequences. We used GPT-2 to augment the training set (and not the test set) by synthesizing new sequences semantically similar to the original data.
The pre-trained GPT-2 was first fine-tuned on our dataset D, and then the fine-tuned model Gtuned was used to generate synthetic sequences of each class (Supplemental Methods). The synthesized sequences Dgenerated were considered to be additional examples of the class corresponding to the prompt used. In this way, an arbitrary number of label-invariant sequences can be generated.
A BERT was fine-tuned on D, and the trained model was used as a classifier h to evaluate the quality of the generated sequences. Each generated sequence was input into h, and output was the inferred label alongside a probability score (the softmax of the output). Sequences were discarded if their inferred labels did not match the initial prompt and if their probability scores were less than 0.9. Thus the filtering attempts to retain only the most salient of the synthetic utterances. The effectiveness of this filtering step was tested using an ablation study (Supplemental Methods, Supplemental Table 5)
We repeated the BERT fine-tuning trained on D∪Dgenerated. The training set was augmented by 2, 3, and 4 times its original size and the model was evaluated on the unchanged test set. The hyperparameters were the same as for the non-augmented model.
2.3.4. Training set augmentation via pairwise sequences
Since we attempted to classify changes in the trajectories (and not specific numerical values), the input to the classifier could be augmented to use data from more than one sequence. We approached this by using a novel method that pairs sequences from two transcripts, both from the same patient. This is motivated by the fact that transcripts from the same patient are likely to have shared semantics. We paired sequences from transcript 1 and transcript 2 (called text a) with sequences from transcript 2 and the final transcript (called text b). Thus we created augmented feature vectors from two transcripts of the same patient where each pair shared one class label (Supplemental Methods).
The input to BERT then takes the following form: [CLS] sequence 1 [SEP] sequence 2 [SEP] where the special separating token [SEP] indicates the end of sequence 1 and the start of sequence 2. The model structure is shown in Fig. 3b. The two sequences in each pair were truncated in turns to fit the maximum length requirement (256). Similar to the GPT-2 methods, the training set was augmented by 2, 3, and 4 times its original size and the model was evaluated on the unchanged test set. Other hyperparameters were the same as previously stated.
3. Results
3.1. Baseline models
We began with LR and SVM models trained on LIWC features. These had some classification power, but were only slightly better than chance (Table 2; for succinctness, only results of evaluation on the variable of PCS are shown; results for MCS and average THYCA are given in the Supplement). Though the modest performance may have been due to the low dimensionality of LIWC features, which failed to capture features correlated with the class labels, more importantly, LIWC does not consider the underlying structure or sequence of speech, but only its lexical contents.
Table 2.
Baseline model characteristics.
| MODEL | LR | SVM | BERT-BASE | CLINICAL BERT |
|---|---|---|---|---|
| ACCURACY | 57.24 | 57.54 | 63.95 | 63.00 |
| AUC | 59.90 | 60.90 | 69.73 | 69.00 |
| F-1 (MACRO) | 57.20 | 57.48 | 63.89 | 61.00 |
| PRECISION | 57.22 | 57.52 | 63.97 | 64.00 |
| RECALL | 57.21 | 57.50 | 63.90 | 59.00 |
Abbreviations: LR, logistic regression; SVM, support vector machine.
Baseline model metrics for inference of PCS trajectory are shown (metrics for baseline models inferring MCS, average THYCA trajectory are given in the Supplement). Numbers are percentages.
We then turned to modern deep learning methods to improve upon these results by fine-tuning BERT on the down-stream binary classification. We used both the pretrained BERT-base and clinical BERT, which are pretrained on the Wikipedia dataset and massive clinical domain data, respectively. Fine-tuning BERT-base on the classification problem improved the accuracy for inferring PCS trajectory by 11.72% and 11.14% compared to LR and SVM, respectively. This is expected because BERT considers context semantics, and better feature representations can be generated, hence improving performance. Overall, clinical BERT performed slightly worse than BERT-base.
3.2. Augmentation via GPT-2
The overall model performance for inferring HRQOL trajectory improved with the GPT-2-augmented training set (Table 3). We show only results with LR. With a training set four times of the original dataset size, the accuracy and AUC improved by 9.60% and 9.44% compared to the baseline, respectively. This demonstrates that, up to a point, adding synthetic samples can boost the classification accuracy of the model. As GPT-2 captures the semantics of the original sequences during finetuning, it can generate sequences semantically similar to the original data. However, although GPT-2 increases the training set size, it does not expand the scope of vocabulary or syntax as evidenced by a plateau effect in performance at four times the size of the original training set. Further increases in size did not improve the performance. A random selection of utterances created by GPT-2 augmentation and filtering can be found in Supplemental Table 4b.
Table 3.
Augmentation via GPT-2 on PCS.
| TRAINING SET SIZE (AS MULTIPLE OF ORIGINAL) | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| ACCURACY | 63.95 | 68.16 | 69.21 | 70.09 |
| AUC | 69.73 | 75.06 | 75.97 | 76.31 |
| F-1 (MACRO) | 63.89 | 67.94 | 69.08 | 70.00 |
| PRECISION | 63.97 | 68.40 | 69.37 | 70.20 |
| RECALL | 63.90 | 68.03 | 69.12 | 70.02 |
Model metrics for inference of PCS trajectory after augmentation using GPT-2, with augmentation of the training dataset to a 2, 3 and 4 times the original size compared to the baseline (metrics for models inferring MCS, average THYCA trajectory are given in the Supplement). Numbers are percentages.
3.4. Augmentation via pairwise sequences
Using the pairwise sequences technique, with a training set augmented to 4 times the original dataset size (equal to the total number of sequences present in both transcripts associated with each trajectory), the LR accuracy and AUC for inferring PCS improved by 9.50% and 8.37% compared to using the non-augmented dataset, respectively (Table 4). In contrast to the GPT-2 augmentation method, this pairing-based data augmentation technique does not add new samples; rather, it changes the representation of the data to a format that results in more data points, and improves model performance by enhancing the sentiment semantics via variations of pairing available samples. In this case, we also observed limits to the diversity of the data points that this technique could generate, given the plateau in increasing model performance with a certain size of the augmented training set.
Table 4.
Augmentation via pairing sequences for PCS. Numbers are percentages.
| TRAINING SET SIZE (AS MULTIPLE OF ORIGINAL) | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| ACCURACY | 61.54 | 64.11 | 65.35 | 67.39 |
| AUC | 65.68 | 68.25 | 70.06 | 71.18 |
| F-1 (MACRO) | 61.49 | 63.93 | 65.35 | 66.26 |
| PRECISION | 61.49 | 64.60 | 65.41 | 66.16 |
| RECALL | 61.49 | 64.24 | 65.41 | 66.38 |
Model metrics for inference of PCS trajectory after augmentation using the sequence pairing method, with augmentation of the training dataset to a 2, 3 and 4 times the original size compared to the baseline (metrics for models inferring MCS, average THYCA trajectory are given in the Supplement). Numbers are percentages.
4. Discussion
We inferred the trajectory of HRQOL measurements in patient interviews using several techniques. While we progressively improved model performance on our classification task, overall accuracy remained modest, and at this stage is not ready for implementation in clinical care. However, this pilot application is an initial approach that serves as a starting point for further investigation.
In this work, we first performed logistic regression as well as SVM with LIWC features as a baseline. Then the performance was improved by fine-tuning BERT on the downstream task since BERT could capture the context semantics and thus generate better feature representation compared to dictionary-based LIWC features. Importantly, training with clinical BERT did not lead to any improvement over BERT base likely because the language used by patients was primarily lay language rather than professional medical language. Furthermore, we presented two data augmentation methods to enhance the diversity and size of the training set. Both approaches produced improvements over the baseline by generating a certain number of new data points, and they are novel for use in this application.
The size of the dataset was the primary limitation, and more real data should improve accuracy as supported by the improvements with synthetic data. The data contained only a small portion of the likely utterances of the patients. The process of adding synthetic data using GPT-2 can “fill in” missing regions in the space and thus increase the density of the data, but cannot expand the diversity of the utterances. Thus the success of the data augmentation schemes in improving the classification (even if only modestly) demonstrates that more data will improve performance, and motivates the gathering of larger data sets. The plateauing effect shows that the new data must be real, and must address the diversity of the information rather than just the density of the representation. The dataset utilized here is unique as it is rare to find transcribed patient language paired with HRQOL survey data. Given the cost of obtaining patient data, approaches to expand datasets will continue to prove worthwhile.
Classifying HRQOL trajectory is of great clinical relevance for assessing and forecasting a cancer patient’s response to treatment. In all cases, we found that classification of PCS and MCS trajectories was more accurate than average THYCA despite comparable levels of data quality. This is a somewhat surprising result, as the average THYCA is the only QOL measurement that assesses thyroid-specific outcomes [14]. This may suggest that other domains, aside from procedure specific symptoms, are most important in determining HRQOL for this cohort [22–24]. With more data, we anticipate increased accuracy of the NLP system, and more direct analysis of patient language for deriving HRQOL.
Based on these findings, we propose using the following elements of our methodology may be useful and expanded upon for the extraction of PRO using patient narrative data: first, retraining BERT models for feature extraction prior to classification; next, using sequence generation via GPT-2 and a novel pairing strategy that exploits the expected structure of the problem as improvements/deterioration of the QOL values.
5. Conclusion
We demonstrated several proof-of-concept NLP methods for inferring HRQOL trajectory from patient language. Leveraging modern machine-learning methods improved classification performance compared to logistic regression and bag-of-words type NLP methods. Future work with larger datasets pairing narrative data with patient-reported outcome measures will provide insights into determinants of HRQOL and facilitate improvements in communication between patients and clinicians. As more patient utterances and HRQOL measurements are digitally preserved, larger datasets will become available to train and perfect algorithms that glean HRQOL information directly from patient language.
Supplementary Material
Acknowledgements
The authors would like to thank Manasa Venkatesh in her assistance in preparing the dataset for analysis, and Sasha Rosser for her contributions to this project.
Funding
This work was supported by grant T32 DC009401 from the NIH/ NIDCD (VH) and by NIH/NCATS UL1TR002373, and in part from grant R01CA176911 from the National Cancer Institute and P30CA014520 from the National Cancer Institute to the University of Wisconsin Carbone Cancer Center.
Footnotes
Ethics approval and consent to participate
We used a novel dataset from a prospective randomized controlled clinical trial assessing surgical interventions for low-risk thyroid cancer (NCT02138214). The original study was approved by the University of Wisconsin-Madison Institutional Review Board (2014–0391) on 5/4/ 2015. The randomized controlled trial as well as this secondary use of the data was approved by the University of Wisconsin, Madison Institutional Review Board.
Declaration of competing interest
The authors have no conflicts of interest to disclose.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ibmed.2023.100097.
If transcript 2 was not available, transcript 3 was used in its place.
References
- [1].Nelson EC, Eftimovska E, Lind C, Hager A, Wasson JH, Lindblad S. Patient reported outcome measures in practice. BMJ 2015:350. [DOI] [PubMed]
- [2].Valderas JM, Kotzeva A, Espallargues M, Guyatt G, Ferrans CE, Halyard MY, et al. The impact of measuring patient-reported outcomes in clinical practice: a systematic review of the literature. Qual Life Res 2008;17(2):179–93. [DOI] [PubMed] [Google Scholar]
- [3].McKenna SP. The limitations of patient-reported outcome measurement in oncology. J Clin Pathways 2016;2(7):37–46. [Google Scholar]
- [4].Basch E, Deal AM, Dueck AC, Scher HI, Kris MG, Hudis C, et al. Overall survival results of a trial assessing patient-reported outcomes for symptom monitoring during routine cancer treatment. JAMA; 2017. [E1–E2 pp.]. [DOI] [PMC free article] [PubMed]
- [5].Shah DV, Cappella JN, Neuman WR. Big data, digital media, and computational social science: possibilities and perils. Ann Am Acad Polit Soc Sci 2015;659(1): 6–13. [Google Scholar]
- [6].Park JH, Jung YS, Kim JY, Jo Y, Bae SH. Trajectories of health-related quality of life in breast cancer patients. Support Care Cancer 2020;28(7):3381–9. [DOI] [PubMed] [Google Scholar]
- [7].Pakhomov S, Shah N, Hanson P, Balasubramaniam S, Smith SA. Automatic quality of life prediction using electronic medical records. AMIA Annu Symp Proc 2008: 545–9. [PMC free article] [PubMed]
- [8].Pakhomov SV, Shah ND, Van Houten HK, Hanson PL, Smith SA. The role of the electronic medical record in the assessment of health related quality of life. AMIA Annu Symp Proc 2011:1080–8. 2011. [PMC free article] [PubMed] [Google Scholar]
- [9].Clinical decision support systems: a survey of NLP-based approaches from unstructured data In: Reyes-Ortiz JA, González-Beltrán BA, Gallardo-López L, editors. 26th international workshop on database and expert systems applications (DEXA). IEEE; 2015. 2015. [Google Scholar]
- [10].Krishna K, Pavel A, Schloss B, Bigham JP, Lipton ZC. Extracting structured data from physician-patient conversations by predicting noteworthy utterances. In: Explainable AI in healthcare and medicine Springer; 2021. p. 155–69. [Google Scholar]
- [11].SF-12v2 Health survey. https://www.optum.com/optum-outcomes/what-we-do/health-surveys/sf-12v2-health-survey.html; 2016.
- [12].Bhandari NR, Kathe N, Hayes C, Payakachat N. Reliability and validity of SF-12v2 among adults with self-reported cancer. Res Soc Adm Pharm 2018;14(11):1080–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roth EM, Lubitz CC, Swan JS, James BC. Patient-reported quality-of-life outcome measures in the thyroid cancer population. Thyroid 2020;30(10):1414–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Husson O, Haak HR, Mols F, Nieuwenhuijzen GA, Nieuwlaat WA, Reemst PH, et al. Development of a disease-specific health-related quality of life questionnaire (THYCA-QoL) for thyroid cancer survivors. Acta Oncol 2013;52(2):447–54. [DOI] [PubMed] [Google Scholar]
- [15].Pennebaker JW, Booth RJ, Boyd RL, Francis ME. Linguistic Inquiry and word Count. LIWC; 2015.
- [16].Greaves F, Ramirez-Cano D, Millett C, Darzi A, Donaldson L. Use of sentiment analysis for capturing patient experience from free-text comments posted online. J Med Internet Res 2013;15(11):e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Biggiogera J, Boateng G, Hilpert P, Vowels M, Bodenmann G, Neysari M, et al. BERT meets LIWC: exploring state-of-the-art language models for predicting communication behavior in couples’ conflict interactions 2021. arXiv preprint arXiv:210601536.
- [18].Detection of mental health from Reddit via deep contextualized representations In: Jiang ZP, Levitan SI, Zomick J, Hirschberg J, editors. Proceedings of the 11th international workshop on health text mining and information analysis; 2020. [Google Scholar]
- [19].Tanana MJ, Soma CS, Kuo PB, Bertagnolli NM, Dembe A, Pace BT, et al. How do you feel? Using natural language processing to automatically rate emotion in psychotherapy. Behav Res Methods 2021:1–14. [DOI] [PMC free article] [PubMed]
- [20].Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. Comput Res Repos 2018. abs/1810.04805.
- [21].Wolf T, Debut L, Sanh V, Chaumond J, Delangue C, Moi A, et al. HuggingFace’s Transformers: state-of-the-art natural language processing 2019. arXiv preprint arXiv:191003771.
- [22].Sippel RS, Robbins SE, Poehls JL, Pitt SC, Chen H, Leverson G, et al. A randomized controlled clinical trial: no clear benefit to prophylactic central neck dissection in patients with clinically node negative papillary thyroid cancer. Ann Surg 2020;272 (3):496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pitt SC, Wendt E, Saucke MC, Voils CI, Orne J, Macdonald CL, et al. A qualitative analysis of the preoperative needs of patients with papillary thyroid cancer. J Surg Res 2019;244:324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Randle RW, Bushman NM, Orne J, Balentine CJ, Wendt E, Saucke M, et al. Papillary thyroid cancer: the good and bad of the “good cancer”. Thyroid 2017;27 (7):902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


