Dear Editor,
2′-O-methylation (Nm) (Fig. 1a) is a prevalent post-transcriptional RNA modification present in many cellular RNAs and plays critical roles in modulating both physical properties and functions of eukaryotic RNA. Studies of Nm modifications in RNA have long been hampered by a lack of effective mapping methods. Previously reported approaches can work well for detecting Nm modifications on abundant RNAs,1–7 but face challenges when applied to less abundant RNAs such as mRNA, lack stoichiometric information, and suffer from RNA sample degradation due to chemical treatment. Here, we present Nm-Mut-seq, a mutation signature-based Nm mapping method, which uses a custom reverse transcriptase (RT) that installs mutations at Am-, Cm-, and Gm-modified sites (Um is undetectable by this method). Our work provides a much-needed approach to detect Nm at base resolution in low abundant RNAs and to estimate the stoichiometry of each modified site transcriptome-widely.
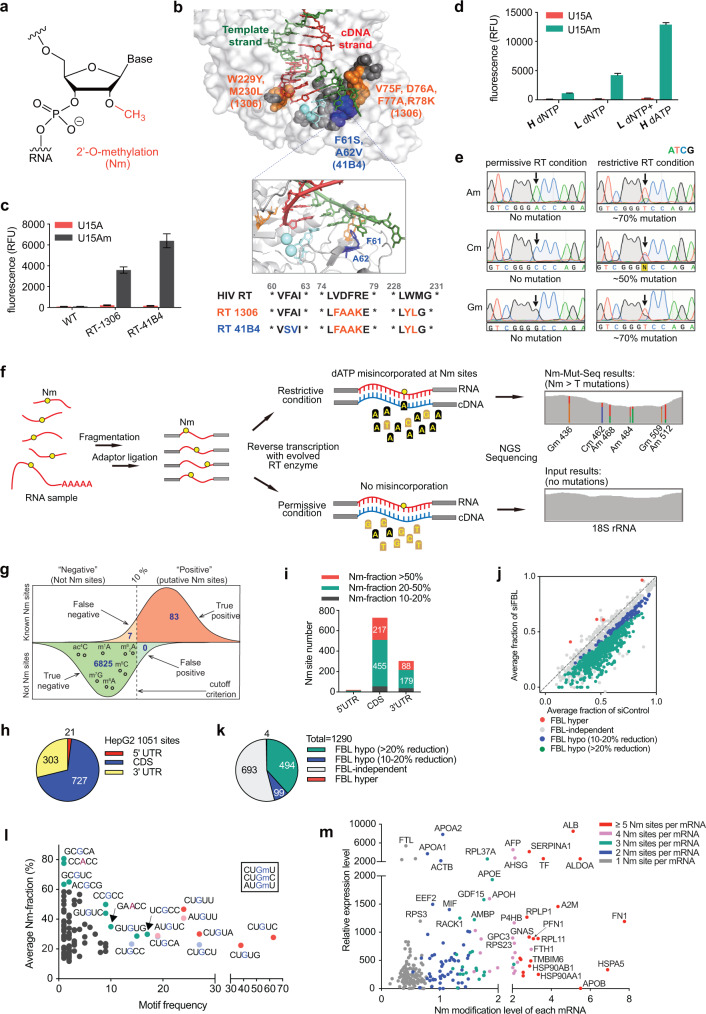
Fig. 1. Base-resolution quantitative mapping of transcriptome-wide 2′-O-methylation using Nm-Mut-seq.
a Chemical structure of 2′-O-methylation (Nm). b Top, the space-filling model of HIV-1 RT structure (PDB ID: 1RTD) with mutations identified in RT-1306 (orange), RT-41B4 (blue), and randomized mutation libraries (gray). Middle, the ribbon model of HIV-1 RT active site with key amino acid residues (F61, A62). Bottom, the sequence alignment of the three RTs. c Comparison of fluorescence intensity generated by HIV RT wildtype (WT), RT-1306, and RT-41B4 pure protein with a 33-mer RNA probe containing U15A (in red) or U15Am (in gray) as substrate. d Comparison of fluorescence intensity generated by RT-41B4 under different dNTP concentrations in the reverse transcription reaction step (“H” = 1 mM, “L” = 40 µM). e Sanger sequencing results of the cDNA products generated by RT-41B4 with Am15, Cm15, and Gm15 RNA probes under restrictive and permissive conditions. Mutations at Nm sites are indicated with arrows. f Scheme of Nm-Mut-seq pipeline based on NGS platform. g The sensitivity and specificity of Nm-Mut-seq in mapping Nm sites in human rRNA. Small black circles refer to other types of RNA modifications in rRNA. h Distribution of 1051 Nm candidates in HepG2 mRNA. i Distribution of Nm site count vs Nm fraction of three segments of HepG2 mRNA. j Scatter plot of Nm-Mut-seq data showing changes in Nm fraction at Nm sites in FBL-knockdown cells. k Pie chart of FBL hypo-regulated, hyper-regulated and independent Nm sites. l Correlation between the frequency of Nm motifs (x-axis) and the fraction of Nm motifs (y-axis) for 494 FBL hypo-regulated Nm sites. The three most frequent Gm motifs (CU(Gm)U, CU(Gm)U, and AU(Gm)U) are highlighted in red, light blue, and pink, respectively. Other motifs of a high frequency or a high average Nm fraction are marked in green. m Relative gene expression level vs Nm modification strength per mRNA at FBL hypo-regulated Nm sites in siControl HepG2 cells (Nm modification strength = the sum of Nm stoichiometric fractions at all Nm-modified sites on each mRNA).
To develop an RT that is mutagenic at Nm sites, we employed the fluorescence-based RT selection platform we previously developed (RT-PCR-IVT assay) (Supplementary information, Fig. S1a and Data S1).8 Briefly, we utilized a synthetic RNA oligo,9 a 33-mer fluorogenic Broccoli aptamer with an Am15 modification, to select the RT variant capable of incorporating a mutation at Nm site during the reverse transcription reaction. We began with the previously developed RT-1306,8 an evolved variant of the p66 subunit of HIV RT with six mutations (V75F, D76A, F77A, R78K, W229Y and M230L) due to its improved basal Am detection compared to commercially available RTs (Supplementary information, Fig. S1b and Data S2). Iterative rounds of selection yielded our lead RT, the variant RT-41B4, which contains F61S and A62V substitutions in addition to the mutagenic background of RT-1306 (Fig. 1b; Supplementary information, Fig. S1c, d and Data S3). Further in vitro biochemical characterization of purified RT confirmed that RT-41B4 produces higher fluorescence intensity as well as improved mutagenic efficiency than RT-1306 (Fig. 1c; Supplementary information, Fig. S1e, f). Moreover, steady-state kinetics analyses and Sanger sequencing results of RT-41B4 revealed that dATP, rather than dCTP and dGTP, is the optimal nucleotide substrate for misincorporation at Nm sites during reverse transcription reaction (Supplementary information, Fig. S1g, h). After screening a suite of dNTP concentrations (Supplementary information, Fig. S1i), we found that a “restrictive RT condition” — consisting of a low concentration of dNTP (10 μM) and a high concentration of dATP (1 mM) — significantly increases the fluorescence response (Fig. 1d) and generates an over 50% Nm-to-T mutation rate at the modified sites, with no detectable mutation at any of the Nm sites under the “permissive RT condition” (1 mM dNTP) (Fig. 1e). Furthermore, the readthrough efficiency of RT-41B4 under the restrictive condition is considerably higher than that of RT-1306 (Supplementary information, Fig. S1j), indicating the potential application of the evolved RT in mapping Nm modifications.
Encouraged by these results, we employed RT-41B4 and the optimized next-generation sequencing (NGS) conditions to build the Nm-Mut-seq pipeline (Fig. 1f) and validated Nm modifications in ribosomal RNA (rRNA) from HeLa cells. We analyzed 90 annotated Nm sites (Am, Cm and Gm) of the human 80S ribosome that were confirmed by several studies (Um sites are not included in the list),10–13 of which 83 were identified by our method (Fig. 1g; Supplementary information, Table S1). Notably, the mutation signatures at Nm sites displayed internal A-to-T mutations within the reads, indicating that the mutations were generated during RT readthrough at the modified site and ruling out the mutation signals arising from other factors such as random RT tails (Supplementary information, Fig. S2a, b). Other known modified types, such as m6A, m7G, ac4C, m5C, etc., showed very low mutation rates (< 5%), readily distinguishable from the mutation signatures at Nm sites (Fig. 1g), indicating that Nm-Mut-seq enables a clean detection of Nm sites in rRNA (Supplementary information, Table S2).
To determine the stoichiometry of Nm modifications, we prepared Nm-Mut-seq libraries using synthetic oligos containing -NNNmNN- with different percentages of stoichiometric amounts of Nm (0%, 25%, 50%, 75%, 100%). Results on probes containing 100% Nm revealed that RT-41B4 produces variable mutation rates at the Nm sites across different sequence contexts, indicating that the calibration of sequence context-dependent mutation rates could promote a more accurate estimation of modification stoichiometry (Supplementary information, Fig. S2c). Results on probes containing 0% Nm revealed very low mutation rates occurring at unmodified oligos (Supplementary information, Fig. S2d). We then obtained sequence context-dependent calibration curves (Supplementary information, Fig. S2e and Table S3) and converted the mutational frequencies of rRNA into Nm stoichiometry. As a result, 69 Nm sites in rRNA were almost fully modified (Nm fraction > 80%), while the rest were partially modified (Supplementary information, Fig. S3a and Table S1). The estimated stoichiometry was mostly consistent with previous results reported by Erales et al.11 and Taoka et al.13 (Supplementary information, Fig. S3b). To further investigate whether Nm-Mut-seq can monitor the changes in Nm modification levels generated by genetic alterations, we performed Nm-Mut-seq on rRNA from fibrillarin (FBL)-depleted HepG2 cells (Supplementary information, Fig. S3c). Upon FBL depletion, several sites showed significant decreases in Nm levels, which is mostly consistent with the previous results reported by Sharma et al.12 and Erales et al.11 (Supplementary information, Fig. S3d). As the duration of siRNA-mediated knockdown increased, the Nm fraction continuously trended downward (Supplementary information, Table S4).
We next applied this method to map Nm modifications in polyA+ RNA from HeLa and HepG2 cells. LC-MS/MS confirmed the presence of Nm modification in purified mRNA, with an Nm/N ratio ranging from 0.09% to 0.15% (Supplementary information, Fig. S4a). We employed Nm-Mut-seq in both HeLa and HepG2 cell lines, which yielded 961 and 1051 mRNA Nm candidate sites overlapped from three biologically replicates, respectively (Fig. 1h; Supplementary information, Fig. S4b–d and Tables S5 and S6). Additionally, we prepared sequencing libraries with modification-free HepG2 mRNA generated from in vitro transcription (IVT), which were used as a negative control. 75 sites (out of 1051 sites) identified as putative Nm sites in HepG2 cellular mRNA were observed with similar mutation signatures in the IVT RNA library, indicating that these sites are not 2′-O-methylated while the remaining sites identified are likely Nm sites (Supplementary information, Fig. S4e, f). In both cell lines, most of the putative Nm sites displayed a stoichiometric fraction of 20%–50%, mainly located in coding sequence (CDS) (Fig. 1i; Supplementary information, Fig. S4g). The distribution of Nm stoichiometry and site number across modification types revealed that Gm and Am are in a relatively higher modification level (Supplementary information, Fig. S4h, i). The Nm candidate sites were mapped to hundreds of adequately expressed genes, including many genes of low abundance with an RPKM < 10, indicating that our method can detect Nm modifications on the less abundant transcripts (Supplementary information, Fig. 4j). As expected, the mutation signatures detected at mRNA Nm sites were located at internal positions of reads, without biased mutation signals from read end (Supplementary information, Figs. S2a and S4k, l).
To investigate the potential “writer” proteins for mRNA Nm methylomes, we next knocked down two known Nm methyltransferases in human cells, FBL and FTSJ3, respectively. LC-MS/MS quantification with purified polyA+ RNA revealed a significant decrease in Cm/C and Gm/G ratios after FBL loss, suggesting that FBL might affect Nm methylation in mRNA (Supplementary information, Fig. S5a). We performed Nm-Mut-seq following FBL knockdown in HepG2 cells. Compared with siControl, 494 sites showed a significantly decreased Nm fraction (> 20% reduction, P < 0.05) in FBL-depleted cells, while only 6 sites showed an increased Nm stoichiometry, suggesting that FBL may install or affect 2′-O-methylation in mRNA (Fig. 1j, k). In parallel experiments with FTSJ3 depletion, we only identified 31 hypo-methylated Nm sites, indicating that FTSJ3 may not be a major “writer” protein for Am, Cm and Gm installation in HepG2 cells (Supplementary information, Fig. S5b).
To validate the Nm candidate sites detected by Nm-Mut-seq, we treated the HepG2 cell RNA as previously described in the Nm-seq method1 and calculated the fold changes at Nm sites between “untreated” and “treated” groups by performing RT-qPCR (Supplementary information, Fig. S5c). Using known Nm sites in 18S rRNA as a control, we determined that a fold change > 1.5 is appropriate for Nm site validation in RT-qPCR assay of Nm-seq. We selected 115 sites for further analysis (including the top 100 Nm sites and 15 additional FBL-regulated sites in HepG2 mRNA); 69 out of 115 sites displayed a fold change > 1.5, with 32 of them > 2 in three biologically independent replicates (Supplementary information, Fig. S5d and Table S7). We next validated the same subset of putative Nm sites based on MeTH-seq;7 90 out of 115 Nm candidates showed a fold change > 1.5 using MeTH-seq assay in three biologically independent replicates (Supplementary information, Fig. S5e). Altogether, 69 out of 115 highly confident Nm sites were verified by at least three orthogonal methods (Nm-Mut-seq, Nm-seq, and MeTH-seq) (Supplementary information, Fig. S5f). Notably, most of the FBL-regulated sites were verified by all three orthogonal methods, indicating that the sites with genetic dependence on Nm methyltransferase are more reliable.
We next focused on the 494 FBL hypo-methylated Nm sites as confident sites for downstream investigation (Supplementary information, Table S8). These sites consist primarily of Gm (Supplementary information, Fig. S6a), which is consistent with our LC-MS/MS results. The 494 FBL-regulated sites are mainly located in CDS, and over half of them showed above 50% Nm fraction in siControl (Supplementary information, Fig. S6b–d). Further analysis of the sequence context around the Nm sites revealed diverse motifs, with CU(Gm)U, CU(Gm)C, and AU(Gm)U as the most frequently modified ones (Fig. 1l). Metagene profiling revealed that Val and Cys codons in the CDS are most frequently modified by Nm methylation (Supplementary information, Fig. S6e). The 494 FBL-regulated sites are primarily located in the first position of the codon and mapped to 266 adequately expressed genes, exhibiting enriched gene ontology clusters in ribosome, translation, and RNA processing (Supplementary information, Fig. S6f, g). Our observations revealed the presence of transcripts containing multiple Nm sites per mRNA; these mRNAs bearing a higher Nm methylation strength tend to exhibit higher expression levels (Supplementary information, Fig. S6h, i). Specifically, we have identified a set of highly expressed mRNAs that are heavily modified by Nm (Fig. 1m). Upon FBL depletion, we observed a significant decrease in Nm modification levels in several abundant mRNAs containing multiple Nm sites (Supplementary information, Fig. S7a). We evaluated the expression levels of the 266 mRNA and found that 147 of them were decreased upon FBL depletion (Supplementary information, Fig. S7b), indicating a correlation between Nm modification levels and abundance of its modified transcripts, consistent with a previous report.14
Human FBL is a C/D box snoRNA-dependent methyltransferase, which allows computational prediction of snoRNA-targeted Nm sites in human mRNA. Among the 494 Nm sites, 109 sites were predicted to potentially bind snoRNA (Supplementary information, Table S9). Specifically, 95 sites containing Gm and Cm could be bound by seven snoRNAs (Supplementary information, Fig. S8a), of which SNORD21, SNORD96B, and SNORD88C have previously been characterized as snoRNAs that function in rRNA Nm methylation (Supplementary information, Fig. S8b–d). To further investigate the direct dependence of snoRNAs on their targeted Nm sites, we performed Antisense Oligonucleotide-mediated SNORD21 knockdown in WT HepG2 cells. The knockdown efficiency was confirmed by RT-qPCR (Supplementary information, Fig. S8e). We observed that one Nm site showed a significantly decreased Nm fraction in SNORD21-depleted cells (P < 0.05), indicating SNORD21-directed Nm installation in HepG2 mRNA (Supplementary information, Fig. S8f, g).
In summary, Nm-Mut-seq stoichiometrically quantified not only known Nm sites in rRNA, but also thousands of candidate Nm sites in human mRNA except for Um sites. Among those mRNA Nm sites, we revealed hundreds of FBL-dependent Nm sites. Our new approach provides an effective method to detect Nm at base resolution in low abundant RNAs such as mRNA and measure the stoichiometry of each modified site transcriptome-widely.
Supplementary information
Acknowledgements
This work was supported by the National Human Genome Research Institute (RM1 HG008935 to C.H. and B.C.D.) and the National Institute of Mental Health (R01 MH122142 to B.C.D.) of the National Institutes of Health (NIH), the University of Chicago Medicine Comprehensive Cancer Center (P30 CA14599) and the international postdoctoral exchange fellowship program (20190048) (L.C.). The evolved RT biochemistry work was partially supported by the National Key R&D Program of China (2018YFA0903200 to C.Z.). C.H. is an investigator at Howard Hughes Medical Institute. We thank Dr. Qing Dai for his help and suggestions on library construction. We thank Dr. Pieter W. Faber in Genomics Facility of the University of Chicago for help with high-throughput sequencing. We thank S. Ahmadiantehrani for editing the manuscript.
Author contributions
B.C.D., C.H. and L.C. conceived and planned the experiments. L.C. performed the RT screening experiments with suggestions from H.Z. L.C. evolved the RT enzyme, optimized RT conditions, carried out NGS library preparations, and characterized Nm methyltransferase. L.S.Z. performed bioinformatic analyses of Nm-Mut-seq data. C.Y. performed computational analyses on calibration curve and snoRNA prediction, with the suggestions from B.L. B.G. performed mass spectrometry measurement. L.C. performed RT enzyme kinetics and Nm site validation analyses under guidance from C.Z. and Z.D. L.C., C.H. and B.C.D. wrote the manuscript with input from all authors.
Data availability
Sequencing data listed in Supplementary information, Table S10 are available in the Gene Expression Omnibus database under the accession number of GSE174518.
Competing interests
C.H. is a scientific founder, a member of the scientific advisory board and equity holder of Aferna Bio, Inc. and AccuraDX Inc., a scientific cofounder and equity holder of Accent Therapeutics, Inc., and a member of the scientific advisory board of Rona Therapeutics. B.C.D. is a founder and holds equity in Tornado Bio, Inc.
Footnotes
These authors contributed equally: Li Chen, Li-Sheng Zhang, Chang Ye.
Contributor Information
Changming Zhao, Email: cmzhao03@whu.edu.cn.
Chuan He, Email: chuanhe@uchicago.edu.
Bryan C. Dickinson, Email: dickinson@uchicago.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00836-w.
References
- 1.Dai Q, et al. Nat. Methods. 2017;14:695–698. doi: 10.1038/nmeth.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkedal U, et al. Angew. Chem. Int. Ed. Engl. 2015;54:451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 3.Krogh N, et al. Nucleic Acids Res. 2016;44:7884–7895. doi: 10.1093/nar/gkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchand, V. et al. Biomolecules7, 13 (2017). [DOI] [PMC free article] [PubMed]
- 5.Zhu Y, Holley CL, Carmichael GG. Methods Mol. Biol. 2022;2404:393–407. doi: 10.1007/978-1-0716-1851-6_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y, Pirnie SP, Carmichael GG. RNA. 2017;23:1303–1314. doi: 10.1261/rna.061549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartoli, K. M., Schaening, C., Carlile, T. M. & Gilbert, W. V. bioRxiv10.1101/271916 (2018).
- 8.Zhou H, et al. Nat. Methods. 2019;16:1281–1288. doi: 10.1038/s41592-019-0550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filonov GS, Moon JD, Svensen N, Jaffrey SR. J. Am. Chem. Soc. 2014;136:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piekna-Przybylska D, Decatur WA, Fournier MJ. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erales J, et al. Proc. Natl. Acad. Sci. USA. 2017;114:12934–12939. doi: 10.1073/pnas.1707674114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Marchand V, Motorin Y, Lafontaine DLJ. Sci. Rep. 2017;7:11490. doi: 10.1038/s41598-017-09734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taoka M, et al. Nucleic Acids Res. 2018;46:9289–9298. doi: 10.1093/nar/gky811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott BA, et al. Nat. Commun. 2019;10:3401. doi: 10.1038/s41467-019-11375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data listed in Supplementary information, Table S10 are available in the Gene Expression Omnibus database under the accession number of GSE174518.