Unusual Argonaute (Ago) proteins have been recently identified in prokaryotic immune systems, which upon binding to foreign nucleic acids, activate their associated NADase effectors. Recent cryo-EM structures of such unique Ago-associated TIR and SIR2 NADases revealed an unconventional nucleic acid binding-mediated NADase activation, adding to the repertoire of canonical cyclic second messenger-driven TIR and SIR2 activation mechanisms.
Sharing their name with the ancient warriors of Greek mythology, Argonaute (Ago) proteins are critical defenders of the immune system, playing key roles in DNA/RNA silencing pathways spanning the three domains of life.1 Ago association with small interfering RNAs (siRNAs), microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs), guides their interaction with target nucleic acids through sequence complementarity, positioning the target for cleavage. Prokaryotic and eukaryotic Agos were originally described as composed of N-terminal domain (N), PIWI/Argonaute/Zwille domain (PAZ), middle domain (MID) and P-element induced wimpy testis domain (PIWI) with robust nuclease activities.2 However, their shorter prokaryotic counterparts, termed short pAgos, lost their nuclease activities, whereby their PAZ domains were replaced with an analog or APAZ domain, and gained novel enzymatic functions through association with a variety of effectors, including NADases, Toll/interleukin-1 receptor/resistance protein (TIR) and sirtuins (SIR2).3,4 This potential for recognition of target nucleic acids through guide RNA (gRNA)-mediated complementary target pairing by pAgos has been harnessed by the prokaryotic immune system as a sensor, which upon recognizing a target ssDNA, triggers conformational changes in the Ago PIWI domains. This in turn induces oligomerization-dependent or -independent NADase activity of the associated TIR and SIR2 effectors, ensuing NAD+ depletion and unleashing the potential of these systems in cell death signaling pathways of bacteria in their fight against invading bacteriophages and foreign DNA. A recent study published in Cell Research explores two such systems, namely TIR-APAZ/Ago and SIR2-APAZ/Ago, both of which are composed of short pAgos acting as sensors and TIR and SIR2 as their respective NADase effectors.5 Four other groups have also reported structural studies on the TIR-APAZ/Ago system,6–9 with a consensus between groups related to ssDNA-activated NADase activity.
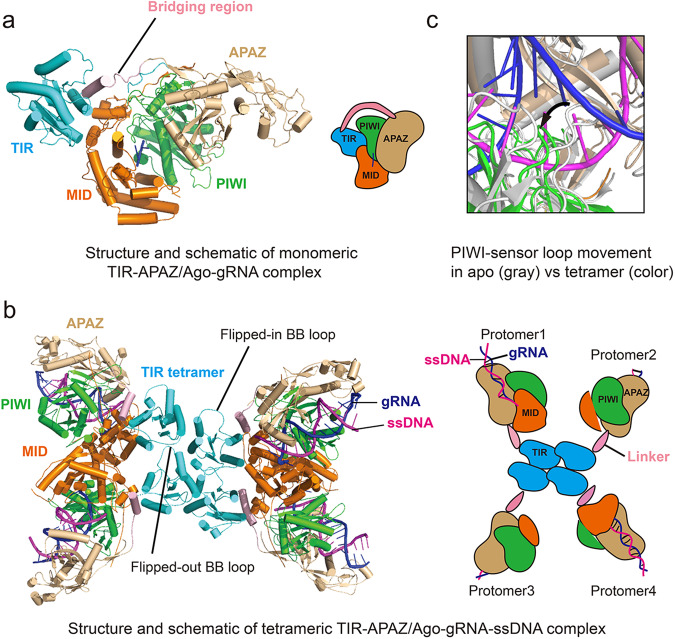
Cryo-EM-guided structural studies of the TIR/APAZ/Ago–gRNA ribonucleoprotein (RNP) system, also termed as short prokaryotic Ago and the associated TIR-APAZ (SPARTA), revealed separated TIR and APAZ domains held together by a bridging helix, which encompass the Ago MID and PIWI domains, with the TIR domain interacting with the MID domain and the APAZ domain interacting with the PIWI domain (Fig. 1a). The positioning of the 5′-phosphate-containing gRNA–target DNA heteroduplex within the positively charged channel created by the MID, PIWI and APAZ domains of this RNP complex, is manifested through RNP tetramerization into an assembly resembling a butterfly scaffold (Fig. 1b). The TIR domains are positioned through a parallel, two-stranded and head-to-head tetramerization alignment stabilized through key inter- and intrastrand interactions, thereby forming the central body, which in turn aligns MID, PIWI and APAZ domains toward the edges, thereby forming the wings of the butterfly scaffold (Fig. 1b). Upon closer inspection, these wings exhibit asymmetric alignments relative to each other, with the TIR-APAZ/Ago monomers of the same wing adopting different conformations owing to the clockwise conformational rotation of adjacent TIR-APAZ/Ago monomers relative to each other, a process vital for active site formation and achievement of a typical head-to-tail orientation of the TIR tetramer (Fig. 1b). Unlike other systems which employ associated oligomerization domains and filament formation, TIR-APAZ/Ago tetrameric higher-order complex formation is primarily mediated by TIR–TIR interactions.10,11 In addition, the TIR oligomer is not open-ended owing to the characteristic internalization of TIR monomers of the tetrameric TIR-APAZ/Ago complex, thereby prohibiting further oligomerization (Fig. 1b). Notably, two disordered/flexible or “flipped-out” BB-loops potentially restrict substrate binding, leaving two ordered or “flipped-in” BB-loops for NAD+ recognition, thereby creating two out of four active subunits (Fig. 1b). In support of this hypothesis, mutations leading to abrogation of interstrand and BB-loop-mediated intrastrand contacts within the TIR tetramer greatly diminished NADase activity. However, in the longer term, this will require further validation through determination of the structure of the NAD+-bound active complex.
Fig. 1. Structural analysis of inactive and active states of TIR-APAZ/Ago–gRNA complexes.
a Structure and schematics of the inactive monomeric TIR-APAZ/Ago–gRNA complex depicting the encircling of Ago MID and PIWI domains by the TIR and APAZ domains. b Structure and schematics of the active tetrameric TIR-APAZ/Ago–gRNA–ssDNA complex depicting the asymmetric butterfly scaffold with MID, PIWI and APAZ domains toward the edges and the central tetrameric alignment of TIR domains highlighting flipped-in and flipped-out BB-loops. c Ago PIWI sensor loop displacement on the active tetrameric TIR-APAZ/Ago–gRNA–ssDNA complex (color) with respect to the sensor loop in the apo monomeric TIR-APAZ/Ago–gRNA complex (gray), enabling removal of the central gRNA–ssDNA base pairing block and faithful propagation of the heteroduplex.
Zeroing in on the Ago molecules of the TIR-APAZ/Ago tetramer revealed that Ago dimerization was induced upon metal-dependent target binding, a novel feature essential for NAD+ hydrolysis. Of particular note, the Ago PIWI domains, upon target ssDNA binding, shifted away from the MID domains, allowing space for Ago dimerization. This shift enables a key “sensor loop” occluding the gRNA:ssDNA binding channel to swing away, potentially removing the steric barrier for gRNA–DNA heteroduplex formation, especially at the central region (bp positions 13–15 in the duplex), thereby acting as a mechanism for inspection and faithful propagation of the heteroduplex (Fig. 1c). This feature was corroborated by the loss of NADase activity observed upon deletion of the sensor loop and mutation of a key tryptophan residue that stabilizes the loop.
Contrary to gRNA–DNA duplex-mediated oligomerization of TIR monomers in the TIR-APAZ/Ago complex, the SIR2-APAZ/Ago complex did not reveal higher-order oligomerization upon target ssDNA binding.5 Instead, it retained a typical bilobed Ago architecture, with SIR2 and APAZ encircling Ago. In addition, unlike active sites created by two TIR monomers in the tetrameric TIR-APAZ/Ago complex bound to target nucleic acid, SIR2-APAZ/Ago complex is likely to employ a single active site contributed by only the SIR2 domain. However, similar to the TIR-APAZ/Ago system, the SIR2-APAZ/Ago system also possesses a functionally equivalent “sensor loop”.5 The role of the sensor loop needs further validation by analysis of the sensor loop positioning in the SIR2-APAZ/Ago structure in its inactive state. Notably, the SIR2 domain was missing from the cryo-EM density of the target ssDNA-bound state, which is indicative of its destabilization, a phenomenon also observed in ThsA effector of the Thoeris antiphage defense system, where it was suggested to be required for its NADase activity.12
It is interesting to speculate that Ago-associated TIR NADases possess a close-ended parallel assembly which is opposed to all other reported antiparallel TIR NADase orientations, perhaps hinting at it being a reminiscent of parallel scaffold TIR assemblies which lack NADase activity.10,11,13 Adding to this uniqueness, unlike other TIR-containing defense systems which are activated by signaling cyclic nucleotide second messengers, TIR-APAZ/Ago and SIR2-APAZ/Ago systems are activated by ssDNA with a robust regulatory mechanism enabled by the functionally conserved “sensor loops”, akin to molecular checkpoints, in place.
Author contributions
A.C wrote the first draft with subsequent edits together with D.J.P.
Contributor Information
Arpita Chakravarti, Email: chakraa2@mskcc.org.
Dinshaw J. Patel, Email: pateld@mskcc.org
References
- 1.Hutvagner G, Simard MJ. Nat. Rev. Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 2.Meister G. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 3.Swarts DC, et al. Nat. Struct. Mol. Biol. 2014;21:743–753. doi: 10.1038/nsmb.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryazansky S, Kulbachinskiy A, Aravin AA. mBio. 2018;9:e01935–18. doi: 10.1128/mBio.01935-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. Cell Res. 2023 doi: 10.1038/s41422-023-00839-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Z, et al. Nature. 2023 doi: 10.1038/s41586-023-06456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, et al. bioRxiv. 2023 doi: 10.1101/2023.07.12.548734. [DOI] [Google Scholar]
- 8.Zhang J-T, et al. bioRxiv. 2023 doi: 10.1101/2023.07.14.549122. [DOI] [Google Scholar]
- 9.Guo M, et al. Cell Res. 2023 doi: 10.1038/s41422-023-00850-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morehouse BR, et al. Nature. 2022;608:803–807. doi: 10.1038/s41586-022-04999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogrel G, et al. Nature. 2022;608:808–812. doi: 10.1038/s41586-022-05070-9. [DOI] [PubMed] [Google Scholar]
- 12.Manik MK, et al. Science. 2022;377:eadc8969. doi: 10.1126/science.adc8969. [DOI] [PubMed] [Google Scholar]
- 13.Essuman K, et al. Curr. Biol. 2018;28:421–430. doi: 10.1016/j.cub.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]