Abstract
The Dutch Pharmacogenetics Working Group (DPWG) aims to facilitate PGx implementation by developing evidence-based pharmacogenetics guidelines to optimize pharmacotherapy. This guideline describes the starting dose optimization of the anti-cancer drug irinotecan to decrease the risk of severe toxicity, such as (febrile) neutropenia or diarrhoea. Uridine diphosphate glucuronosyl transferase 1A1 (UGT1A1 encoded by the UGT1A1 gene) enzyme deficiency increases risk of irinotecan-induced toxicity. Gene variants leading to UGT1A1 enzyme deficiency (e.g. UGT1A1*6, *28 and *37) can be used to optimize an individual’s starting dose thereby preventing carriers from toxicity. Homozygous or compound heterozygous carriers of these allele variants are defined as UGT1A1 poor metabolisers (PM). DPWG recommends a 70% starting dose in PM patients and no dose reduction in IM patients who start treatment with irinotecan. Based on the DPWG clinical implication score, UGT1A1 genotyping is considered “essential”, indicating that UGT1A1 testing must be performed prior to initiating irinotecan treatment.
Subject terms: Predictive markers, Genetic testing
Introduction
The role of heritable genetic variation on drug response is referred to as pharmacogenetics (PGx). Personalized medicine, also known as precision medicine, can be achieved with the use of PGx information. Knowledge of an individual’s genetic composition for drug metabolizing enzymes, drug transporters, receptors or effector proteins may be used to guide pharmacological treatment. To implement the use of PGx in a clinical setting, guidelines informing physicians are essential. In order to accommodate, the Royal Dutch Pharmacists Association (KNMP) has appointed the Dutch Pharmacogenetics Working Group (DPWG) in 2005, a group of 15 professionals consisting of (clinical) pharmacists, physicians, a general practitioner, clinical pharmacologists, clinical chemists and epidemiologists [1]. The role of the DPWG is to develop evidence-based PGx-guided therapeutic recommendations based on systematic literature review and to implement these into computerized systems used nationwide in The Netherlands for medication prescription, dispensing and monitoring. In order to meet the public request for this information also outside the Dutch pharmacist and physician systems, the DPWG guidelines and future updates are published [2–5].
The current guideline presents the gene-drug interaction between UGT1A1 and the anti-cancer drug irinotecan. The pharmacotherapeutic rationale for use of irinotecan as well as the cost-effectiveness of PGx-guided dosing is outside the scope of this guideline. This manuscript provides information on the development of this guideline and presents an overview of the PGx therapeutic recommendations. Background information of irinotecan and of the UGT1A1 gene and its genetic variation is provided. This genetic information is followed by the evidence from literature on the gene-drug interaction between UGT1A1 and irinotecan. Finally, therapeutic recommendations for the clinic and clinical decision support systems are provided. These DPWG PGx-guided recommendations are also compared to other international guidelines. The goal of this DPWG recommendation is to individualize the starting dose of irinotecan thereby decreasing the risk of severe and potentially fatal toxicity.
Drug: irinotecan
Irinotecan is a commonly applied anticancer drug and is registered for first-line treatment of pancreatic cancer, the second-line treatment of advanced and metastatic colorectal cancer and several other cancer types, including lung cancer and Ewing sarcoma. Of all treated patients, up to 40% experience common Toxicity Criteria grade ≥ III delayed diarrhoea, and up to 50% of the patients experience grade ≥ III neutropenia [6, 7].
Irinotecan is a prodrug that is converted predominantly by carboxylesterases (CES) in the liver and intestines to the active metabolite SN-38, which has 100 to 1000-fold higher activity compared to irinotecan. Irinotecan is, besides by CES, also metabolised by CYP3A4/5 in the liver to inactive metabolites. SN-38 is predominantly glucoronidated by UGT1A1 and also by UGT1A6, UGT1A7, UGT1A9 and UGT1A10 to the inactive metabolite SN-38-glucuronide. A schematic overview of the metabolism of irinotecan and its active metabolite SN-38 is depicted in Fig. 1.
Fig. 1. Irinotecan metabolism. Irinotecan and its metabolites are presented in rectangles.
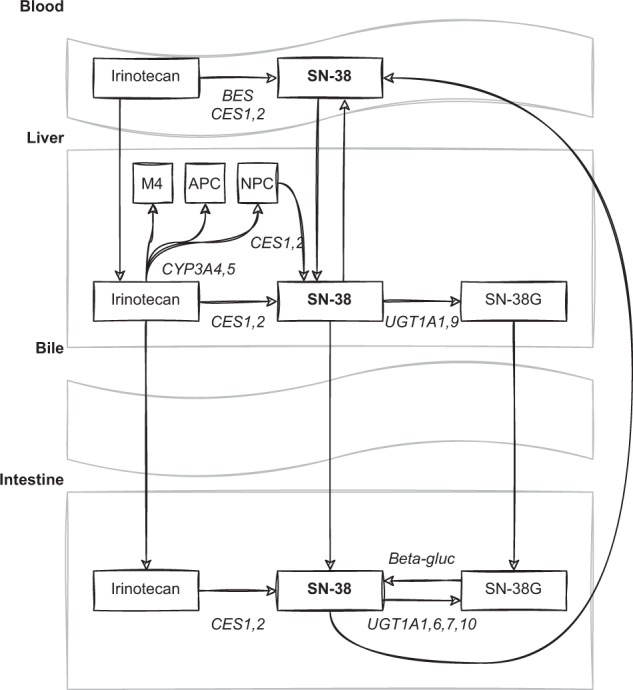
The active metabolite, SN-38, is presented in bold letters. Abbreviations: APC = 7-ethyl-10-[4-N-(5-aminopentanoic acid)-1-piperidino] carbonyloxycamptothecin, BES butyrylcholinesterase, CES carboxylesterase, CYP cytochrome P450 enzymes, NPC = 7-ethyl-10-[4-(1-piperidino)-1-amino] carbonyloxycamptothecin, UGT uridine diphosphate glucuronosyltransferase, SN-38G SN-38 glucuronide, beta-gluc beta-glucuronidase.
Gene: uridine diphosphate glucuronosyl transferase 1A1 (UGT1A1)
The UGT1A1 gene coding for the UGT1A1 enzyme is located on chromosome 2 (2q37.1) and consists of 5 exons, of which the first exon at the 5’ terminus is unique and exons 2 to 5 are shared with the genes UGT1A6 and UGT1A9.
Many variants exist for UGT1A1; more than 100 different alleles have been identified/described in the literature and are often associated with Gilbert syndrome or Crigler-Najjar syndrome. A number of these alleles and their functionality are listed in Supplementary Table 1. The most studied variation in UGT1A1 involves a repeat in the promoter region of the UGT1A1 gene. The number of “TA” tandem repeats in the TATA box of the promoter region varies. The UGT1A1 activity decreases with an increasing number of TA repeats. For example, the *28 variant contains 7 TA repeats instead of 6 TA repeats before the last TA in the TATA box and results in a 67 to 82 percent lower gene expression [8, 9].
The frequency of the various UGT1A1 alleles shows considerable inter-ethnic variability. The variant allele *28 is abundant in inhabitants in South Asia (41%) and much less frequently in the rest of Asia (10–12%) [10]. The prevalence in Europeans ranges from 22 to 39% [10]. The variant allele *6 is common in several Asian populations and also strongly associated with reduced enzyme activity [11–13]. UGT1A1*6 has allele frequencies in Japanese, Korean and Chinese populations of 13, 23 and 23%, respectively [14]. An overview of UGT1A1 allele and genotype frequencies in different populations based on these references and the gnomAD database, is provided in Supplementary Table 2.
Translation of genotype to predicted phenotype
The DPWG has concluded that variants resulting in decreased UGT1A1 metabolic capacity have sufficient evidence to be implemented into clinical care. In the case of the *36 variant, an allele that results in increased UGT1A1 metabolic capacity, there are currently no data to suggest that this results in clinically relevant effects. Therefore, for the time being, this is considered an allele with normal function. Notwithstanding, higher doses of irinotecan could potentially be indicated, but this requires further research. For UGT1A1, three different phenotypes are distinguished: normal metaboliser (NM), intermediate metaboliser (IM) and poor metaboliser (PM). The two phenotypes with reduced metabolic capacity (IM and PM) are further subdivided based on whether or not *28 is the only gene variant that results in decreased metabolic capacity. The genotype-phenotype translation is presented in Table 1. In addition, an extensive genotype-phenotype translation table that can be used to programme the translation of genotype results into predicted phenotypes in laboratory information systems is provided in Supplementary Table 3.
Table 1.
Genotype-phenotype translation.
| Genotype | Phenotype predicted based on genotype | |
|---|---|---|
| Description | Examples | |
| two alleles with normal (or increased) enzyme activity | *1/*1, *1/*36 | NM |
| *28 and one allele with normal (or increased) enzyme activity | *1/*28, *28/*36 | IM (*1/*28) |
| one allele with decreased enzyme activity other than *28 and one allele with normal (or increased) enzyme activity | *1/*6, *1/*37, *36/*37 | IM other |
| two *28 alleles | *28/*28 | PM (*28/*28) |
| two alleles with decreased enzyme activity, of which at least one is not *28 | *6/*6, *6/*28, *28/*37 | PM other |
NM normal metaboliser, IM intermediate metaboliser, PM poor metaboliser.
Gene-drug interaction
Pharmacological mechanism
UGT1A1 is mainly present in the liver and intestines and is the most important enzyme to inactivate irinotecan’s active metabolite SN-38. Decreased UGT1A1 activity leads to increased concentrations of SN-38, which in turn could lead to an increased risk of severe toxicities, such as (febrile) neutropenia and diarrhoea [15]. Variations in the UGT1A1 gene can result in reduced, or even absent enzyme activity. For example, the UGT1A1*28/*28 genotype leads to an 18-159% increased systemic exposure of SN-38, and SN-38 metabolic clearance decreases by 61% [16–21].
Supporting body of evidence
A detailed description of the methods used for literature collection, assessment and preparation of the gene-drug monograph has previously been published [1]. In brief, a systematic review of literature was performed, relevant articles were summarized, and therapeutic recommendations were proposed by a scientist of the Royal Dutch Association for the Advancement of Pharmacy (MN). The performed search strategy can be found in Supplementary Material 1 and was conducted until March 19, 2021. The quality of evidence was scored on a 5-point scale ranging from 0 (lowest) to 4 (highest) and the impact of the clinical effect was scored on a 7-point scale ranging from AA# (positive effect) to F (highest negative effect). This clinical impact scale (AA#-F) runs parallel to the Common Terminology Criteria for Adverse Events (CTCAE); where CTCAE grade 5 severity is equal to clinical relevance score F (death) and CTCAE grade 1 severity is equal to clinical relevance score B. The clinical relevance score additionally includes the scores AA#, AA and A, since these do not exist in the CTCAE. These regard AA#: “Positive clinical effect”, AA: “No significant clinical and/or kinetic effect”, and A: “Significant kinetic effect or not clinically relevant negative effect”. The summaries and scores of the articles reviewed to devise this guideline are described in Supplementary Table 4. The summary and scores of each article were checked by two independent DPWG members. The DPWG made the final decision on the therapeutic recommendations. DPWG guidelines are checked for agreement with current evidence every 5 years in general. An updated version of the guideline will be published if recommendations are altered.
The initial literature search was performed on September 18, 2006, followed by searches on October 27, 2008, March 19, 2014, July 20, 2017 and March 19, 2021. Given the large number of articles, the only articles included after July 2006 were those that included at least 25 subjects with one or more *28 alleles. The only clinical studies included for the period 2008–2017 were meta-analyses, as large individual studies (n > 200) were already included in the meta-analyses. From 2008 to 2014 only meta-analyses with mainly White patients were included. Three Asian meta-analyses investigating the effect of *6 and *28 were not included as these are insufficiently relevant to the situation in the Netherlands. For the period after 2014, meta-analyses were included if the effect of *28 was analysed, either alone or in combination with *6. For the period after 2017, clinical studies were only included if they investigated more than 500 patients with the additional requirements of more than 150 cases for case-control studies, and analysis of the effect of *28 in the case of meta-analyses. Pharmacokinetic studies were only included if exposure to or clearance of SN-38 was determined for the *1/*1, *1/*28 and *28/*28 genotypes and if these were the most important genotypes investigated within the population (i.e. studies among Whites) (for the period from 2008 to 2014) or for the *1/*1, *1/*28 and/or *1/*6, and *28/*28 and/or *6/*28 and/or *28/*28 genotypes (for the period from 2014). For the periods from 2008 to 2014 and after 2017, there were no relevant studies investigating the effect of dose adjustments. This means that there were no studies that investigated the effect of approximately 30% lower initial doses for PM compared to the standard dose for NM and IM in this period.
General conclusion of evidence
For *28/*28 and “PM other”, there is strong evidence that these genotypes are associated with an increased risk of grade ≥3 toxicity such as neutropenia or diarrhoea. All nine meta-analyses investigating adverse events and 16 of the 23 included studies reported this increased risk. In addition, all seven meta-analyses and three studies investigating the effect of *28/*28 and/or *6/*6 and/or *6/*28 compared with all other genotypes, found that this toxicity risk was also increased for *28/*28 and/or PM patients compared to all other patients. With regard to efficacy, four of the five meta-analyses and eight of the ten studies did not show the *28 and/or *6 variants to be associated with increased effectiveness of treatment. See Supplementary Table 4 and 5 for a detailed description of the literature and the rationale of the therapeutic recommendations. In addition, recently the results of a prospective implementation study of UGT1A1 genotype-guided dosing of irinotecan in PM patients were published showing that UGT1A1 genotype-guided dosing of irinotecan in PM patients with applying a 30% dose reduction significantly improved safety while maintaining therapeutic drug exposure [22].
In summary, for *28/*28 and “PM other” there is ample evidence for an increased risk of serious adverse events such as neutropenia or diarrhoea at normal doses (also when compared to all other genotypes/phenotypes), while convincing evidence for an increased efficacy has not been demonstrated. Therefore, the DPWG concludes that a UGT1A1 gene-drug interaction is present and that it necessitates a dose adjustment of irinotecan. Ongoing debate persists on whether or not there is a clinically relevant higher risk of toxicity in PM patients treated with lower dosages of irinotecan (<150 mg/m2). However, two meta-analyses [23, 24] indicate that the risk of grade 3–4 neutropenia is also elevated at lower doses of irinotecan and therefore the DPWG recommend dose adjustment of irinotecan in all dosing categories.
For *1/*28 and “IM other”, a similar amount of evidence is present as for *28/*28 and “PM other”. See Supplementary Table 4 and 5 for a detailed description of the literature and the rationale of the therapeutic recommendations. However, *1/*28 is the major group among White populations. The initial standard irinotecan dose derived in earlier phase I studies was therefore mainly driven by the *1/*28 genotype. This is confirmed by Lu et al. 2015 [25], showing that most *1/*28 + *1/*1 patients tolerate the standard dose, whereas *28/*28 patients did not. Furthermore, there were negligible dose adjustments calculated for *1/*28 compared to all genotypes (a weighted mean calculated dose adjustment to 95% of the dose for all patients based on 6 studies with a total of 112 patients with the *1/*28 genotype) (Supplementary Table 5). This means that a priori dose reduction for patients with *1/*28 would lead to subtherapeutic doses for this patient group. Because the kinetic and clinical effects of *28 and *6 are comparable, the same holds true for IM predicted phenotype as a whole. Therefore, the DPWG concludes that a gene-drug interaction is present, but that therapy adjustment is neither required nor advisable in UGT1A1 IM patients.
Based on the above, the dose for *1/*1 may be increased. As three meta-analyses did not identify a difference in effectiveness of therapy between *1/*28 and *1/*1, an increase for *1/*1 patients has not yet proven to be useful. Therefore, the DPWG decided to refrain from a recommendation for *1/*1.
Pharmacotherapeutic recommendations
The DPWG therapeutic recommendations to optimize the starting dose of irinotecan in patients known to have a variant UGT1A1 predicted phenotype is summarized in Table 2. In brief, UGT1A1 PM patients, including *28/*28, should receive a 70% starting dose of irinotecan, with the number of 70% primarily based on kinetic data and early dose-finding studies as described below. Further dose titration is possibly guided on neutrophil count and clinical tolerance. For UGT1A1*1/*28 and UGT1A1 “IM other” patients no dose reduction is recommended.
Table 2.
Summary therapeutic recommendations based on UGT1A1 predicted phenotype for irinotecan.
| UGT1A1 predicted phenotype | Therapeutic recommendation |
|---|---|
|
PM (*28/*28) |
Start with 70% of the normal dosea. If the patient tolerates this initial dose, the dose can be increased, guided by the neutrophil count. |
| PM other |
Start with 70% of the normal dosea. If the patient tolerates this initial dose, the dose can be increased, guided by the neutrophil count. |
|
IM (*1/*28) |
No action required. |
| IM other | No action required. |
IM intermediate metaboliser, PM poor metaboliser.
aThe normal dose is defined as the dose the patient would receive if he/she would not have a gene variant.
The dose calculation for *28/*28 was based on the SN-38 exposure (area under the curve (AUC)) or clearance in 6 studies with a total of 28 patients with *28/*28. The weighted mean of the calculated dose adjustment is a dose of 58% (range 39–85%, median 53%) of the dose for *1/*1 and a dose of 69% (range 48–92%, median 64%) of the dose for all patients. As the frequency of *1/*1 in Europe is less than 50%, and as caution should be exercised to prevent subtherapeutic doses, the calculated dose compared to all patients was chosen. This is translated into a starting dose of 70% which is more achievable in clinical practice (Supplementary Table 5). The SN-38 glucuronide/SN-38 area under the curve (AUC) ratios are comparable for *28/*28 and *6/*6, suggestive of a similar effect size on irinotecan metabolism [26]. Therefore, the recommendations for the “PM other” predicted phenotype (for example caused by *6), is the same as the recommendations for the *28/*28 genotype, respectively.
More information on the rationale, kinetic and clinical consequences of these therapeutic recommendations are depicted in Supplementary Table 5.
Supplementary Table 6 provides an overview of suggested pop-up (or look-up) texts for electronic prescribing systems for pharmacists and physicians. These can be used to program alerts into the clinical decision support system (CDSS).
Implications for clinical practice
Ongoing debate persists whether and which single gene-drug pairs should be implemented into routine care. Points of debate include the amount of evidence that is necessary supporting effectiveness of genotyping prior to initiating therapy, cost-effectiveness of PGx testing in the pre-therapeutic setting and its reimbursement [27]. As a consequence, gene-drug pairs which are ready for implementation are hampered in application in clinical practice [28]. In an effort to overcome this inconclusiveness and to direct clinicians on whether or not to order relevant PGx genotyping tests before initiating therapy, the DPWG has developed the Clinical Implication Score. The DPWG Clinical Implication Score for a gene-drug pair can be scored as: essential, beneficial or potentially beneficial. These categories are clarified in Supplementary Table 7. The development of these categories and the systematic scoring criteria are discussed elsewhere [29]. In brief, the implications for clinical practice are based on four criteria: the clinical effect associated with gene–drug interaction; the level of evidence supporting the associated clinical effect; the number needed to genotype (NNG) in the Dutch population; and the availability of and type of PGx information in the drug label issued by the Dutch drug agency CBG-MEB. The scores provided for each of these criteria by the DPWG can be found in Supplementary Table 7. Only gene-drug interactions which are actionable are subject to receiving a Clinical Implication Score.
The Clinical Implication Score of the gene-drug interaction between UGT1A1 and irinotecan is 8 out of the maximum of 10 points. This indicates that genotyping before starting irinotecan is considered “essential” for drug safety. Genotyping must be performed before drug therapy has been initiated to guide dose selection. The feasibility and clinical benefit of such an approach has also recently been demonstrated. A recent prospective implementation study on UGT1A1 genotype-guided dosing of irinotecan in PM patients showed that genotype-guided dosing in PM patients increases safety, provides therapeutic drug exposure, and is cost-effective, and supports the recommendation of a 70% starting dose in UGT1A1 PM patients [22].
Differences between available pharmacogenetic guidelines
To the best of our knowledge there are two other pharmacogenetic guidelines available on the gene-drug interaction of irinotecan and UGT1A1. First, a guideline by the French joint working group comprising the National Pharmacogenetics Network (RNPGx) and the Group of Clinical Onco-pharmacology (GPCO-Unicancer) [30]. Second, an Italian guideline by the Italian association of medical oncologists (AIOM) and the Italian Society of Pharmacology (SIF) [31]. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has no guideline available, but indicates this gene-drug interaction as an actionable PGx [32]. Both guidelines are shortly discussed below.
RNPGx and GPCO-Unicancer
The genotype-phenotype translation in the RNPGx/GPCO guideline is in line with the DPWG guideline; the *36 allele can be interpreted as a *1 allele and the *37 allele as a *28 allele. In addition, in both guidelines pre-treatment UGT1A1 genotyping is strongly recommended and the advised dose reduction at the first cycle for *28/*28 patients is similar, namely 25–30%.
However, the RNPGx/GPCO guideline does not recommend pre-therapeutic UGT1A1 genotyping for low irinotecan doses (<180 mg/m2) because haematological and gastrointestinal toxicities are quite similar regardless of the genotype for low irinotecan doses. In contrast, the DPWG concluded that the risk of grade 3–4 neutropenia is also elevated at lower doses of irinotecan based on two meta-analyses [23, 24] and therefore recommends to genotype all patients treated with irinotecan. Moreover, in the RNPGx/GPCO guideline it is recommended that *28/*28 patients must not receive high-dose irinotecan (≥240 mg/m2) because of a much higher risk of haematological toxicity (neutropenia) compared to other genotypes, whereas the DWPG guideline does not advocate a contra-indication for high-dose irinotecan in these patients.
AIOM and SIF
This Italian guideline only provides guidance on the *28 gene variant of UGT1A1. They recommend a dose reduction of 30% in *28/*28 patients which is in line with the current DPWG guideline.
Conclusion
In conclusion, the DPWG recommends a 70% starting dose in PM patients that start treatment with irinotecan. In IM patients, an a priori dose reduction is not recommended. Based on the DPWG clinical implication score, UGT1A1 genotyping is considered “essential”, therefore directing towards pre-therapeutic UGT1A1 testing in patients intended for treatment with irinotecan.
Disclaimer
The Pharmacogenetics Working Group of the KNMP (DPWG) formulates the optimal recommendations for each phenotype group based on the available evidence. If this optimal recommendation cannot be followed due to practical restrictions, e.g. therapeutic drug monitoring or a lower dose is not available, then the health care professional should consider the next best option.
Supplementary information
Supplementary Table 1 genetic variant and functionality
Supplementary Table 2 allele and genotype frequencies
Supplementary Table 3 genotype phenotype translation for LIS
Supplementary Table 5 therapeutic recommendation
Supplementary Table 6 clinical decision support text
Supplementary Table 7 clinical implication score
Author contributions
ECH and MJD drafted the manuscript. JJS and HJG supervised drafting of the manuscript and contributed to conceiving the work and interpretation of the results. MN performed most of the literature searches and article summaries, and suggested clinical decision support texts. BS had the clinical decision support texts translated in English and published them. NBV, AB, EJHF, AR, GAR, RHNS, DT, JW, and RW contributed to conceiving the work and interpretation of the results. VHMD led the meetings in which the DPWG decided about the article summaries and clinical decision supports texts and contributed to conceiving the work and interpretation of the results. In addition, all authors revised the manuscript and approved the final version.
Funding
The U-PGx consortium received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 668353. The DPWG received funding from the Royal Dutch Pharmacists Association.
Data availability
All data and material are either included in the supplementary information or publicly available (i.e. the published articles, PubMed). The guidelines and background information are available on the website of the Royal Dutch Pharmacists Association (KNMP) (Pharmacogenetic Recommendation Text. Available from: https://www.knmp.nl/).
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: Supplementary table 3 has been corrected.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Emma C. Hulshof, Maarten J. Deenen.
These authors jointly supervised this work: Henk-Jan Guchelaar, Jesse J. Swen.
Change history
2/9/2023
Supplementary table 3 has been corrected.
Change history
2/16/2023
A Correction to this paper has been published: 10.1038/s41431-023-01315-x
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-022-01243-2.
References
- 1.Swen J, Wilting I, Goede A, de, Grandia L, Mulder H, Touw D, et al. Pharmacogenetics: From Bench to Byte. Clin Pharm Ther. 2008;83:781–7. doi: 10.1038/sj.clpt.6100507.. [DOI] [PubMed] [Google Scholar]
- 2.Guchelaar H-J. Pharmacogenomics, a novel section in the European Journal of Human Genetics. Eur J Hum Genet. 2018;26:1399–400. doi: 10.1038/s41431-018-0205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lunenburg CATC, van der Wouden CH, Nijenhuis M, Crommentuijn-van Rhenen MH, de Boer-Veger NJ, Buunk AM, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene–drug interaction of DPYD and fluoropyrimidines. Eur J Hum Genet. 2020;28:508–17. doi: 10.1038/s41431-019-0540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matic M, Nijenhuis M, Soree B, de Boer-Veger NJ, Buunk A-M, Houwink EJF, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2D6 and opioids (codeine, tramadol and oxycodone). Eur J Hum Genet. 2021. 10.1038/s41431-021-00920-y. [DOI] [PMC free article] [PubMed]
- 5.Brouwer JMJL, Nijenhuis M, Soree B, Guchelaar H-J, Swen JJ, van Schaik RHN, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2C19 and CYP2D6 and SSRIs. Eur J Hum Genet. 2021. 10.1038/s41431-021-01004-7. [DOI] [PMC free article] [PubMed]
- 6.Rougier P, Bugat R. CPT-11 in the treatment of colorectal cancer: clinical efficacy and safety profile. Semin Oncol. 1996;23:34–41. [PubMed] [Google Scholar]
- 7.Rothenberg ML. Efficacy and toxicity of irinotecan in patients with colorectal cancer. Semin Oncol. 1998;25:39–46. [PubMed] [Google Scholar]
- 8.Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s Syndrome. N. Engl J Med. 1995;333:1171–5. doi: 10.1056/nejm199511023331802.. [DOI] [PubMed] [Google Scholar]
- 9.UGT1A1 allele nomenclature n.d. https://www.pharmacogenomics.pha.ulaval.ca/wp-content/uploads/2015/04/UGT1A1-allele-nomenclature.html (accessed September 6, 2021).
- 10.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019:531210. 10.1101/531210.
- 11.Akiyama Y, Fujita K, Nagashima F, Yamamoto W, Endo H, Sunakawa Y, et al. Genetic testing for UGT1A1*28 and *6 in Japanese patients who receive irinotecan chemotherapy. Ann Oncol. 2008;19:2089–90. doi: 10.1093/annonc/mdn645.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teh LK, Hashim H, Zakaria ZA, Salleh MZ. Polymorphisms of UGT1A1*6, UGT1A1*27 & UGT1A1*28 in three major ethnic groups from Malaysia. Indian J Med Res. 2012;136:249–59. [PMC free article] [PubMed] [Google Scholar]
- 13.Sung C, Lee PL, Tan LL, Toh DSL. Pharmacogenetic risk for adverse reactions to irinotecan in the major ethnic populations of Singapore. Drug Saf. 2011;34:1167–75. doi: 10.2165/11594440-000000000-00000.. [DOI] [PubMed] [Google Scholar]
- 14.Akaba K, Kimura T, Sasaki A, Tanabe S, Ikegami T, Hashimoto M, et al. Neonatal hyperbilirubinemia and mutation of the bilirubin uridine diphosphate-glucuronosyltransferase gene: a common missense mutation among Japanese, Koreans and Chinese. Biochem Mol Biol Int. 1998;46:21–6. doi: 10.1080/15216549800203512.. [DOI] [PubMed] [Google Scholar]
- 15.de Man FM, Goey AKL, van Schaik RHN, Mathijssen RHJ, Bins S. Individualization of irinotecan treatment: a review of pharmacokinetics, pharmacodynamics, and pharmacogenetics. Clin Pharmacokinet. 2018;57:1229–54. doi: 10.1007/s40262-018-0644-7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goetz MP, McKean HA, Reid JM, Mandrekar SJ, Tan AD, Kuffel MA, et al. UGT1A1 genotype-guided phase i study of irinotecan, oxaliplatin, and capecitabine. Invest N. Drugs. 2013;31:1559–67. doi: 10.1007/s10637-013-0034-9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denlinger CS, Blanchard R, Xu L, Bernaards C, Litwin S, Spittle C, et al. Pharmacokinetic analysis of irinotecan plus bevacizumab in patients with advanced solid tumors. Cancer Chemother Pharm. 2009;65:97–105. doi: 10.1007/s00280-009-1008-7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer L, Das S, Janisch L, Wen M, Ramírez J, Karrison T, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–7. doi: 10.1038/sj.tpj.6500072.. [DOI] [PubMed] [Google Scholar]
- 19.de Jong FA, Kehrer DFS, Mathijssen RHJ, Creemers G, de Bruijn P, van Schaik RHN, et al. Prophylaxis of irinotecan‐induced diarrhea with neomycin and potential role for UGT1A1*28 genotype screening: a double‐blind, randomized, placebo‐controlled study. Oncologist. 2006;11:944–54. doi: 10.1634/theoncologist.11-8-944.. [DOI] [PubMed] [Google Scholar]
- 20.Paoluzzi L, Singh AS, Price DK, Danesi R, Mathijssen RHJ, Verweij J, et al. Influence of genetic variants in UGT1A1 and UGT1A9 on the in vivo glucuronidation of SN-38. J Clin Pharm. 2004;44:854–60. doi: 10.1177/0091270004267159.. [DOI] [PubMed] [Google Scholar]
- 21.Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382–8. doi: 10.1200/JCO.2004.07.173.. [DOI] [PubMed] [Google Scholar]
- 22.Hulshof EC, de With M, de Man FM, Creemers GJ, Deiman BALM, Swen JJ, et al. UGT1A1 genotype-guided dosing of irinotecan: a prospective safety and cost analysis in poor metaboliser patients. Eur J Cancer. 2022;162:148–57. doi: 10.1016/J.EJCA.2021.12.009.. [DOI] [PubMed] [Google Scholar]
- 23.Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: Low doses also increase risk. Clin Cancer Res. 2010;16:3832–42. doi: 10.1158/1078-0432.CCR-10-1122.. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association of UGT1A1*28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: a meta-analysis in Caucasians. Pharmacogenomics J. 2014;14:120–9. doi: 10.1038/tpj.2013.10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu CY, Huang CW, Wu IC, Tsai HL, Ma CJ, Yeh YS, et al. Clinical implication of UGT1A1 promoter polymorphism for irinotecan dose escalation in metastatic colorectal cancer patients treated with bevacizumab combined with FOLFIRI in the first-line setting. Transl Oncol. 2015;8:474–9. doi: 10.1016/j.tranon.2015.11.002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami H, Sai K, Saeki M, Saito Y, Ozawa S, Suzuki K, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17:497–504. doi: 10.1097/FPC.0b013e328014341f.. [DOI] [PubMed] [Google Scholar]
- 27.Pirmohamed M, Hughes DA. Pharmacogenetic tests: the need for a level playing field. Nat Rev Drug Discov. 2013;12:3–4. 10.1038/nrd3921. [DOI] [PubMed]
- 28.Swen JJ, Huizinga TW, Gelderblom H, de Vries EGE, Assendelft WJJ, Kirchheiner J, et al. Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med. 2007;4:e209. doi: 10.1371/journal.pmed.0040209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swen JJ, Nijenhuis M, van Rhenen M, de Boer-Veger NJ, Buunk AM, Houwink EJF, et al. Pharmacogenetic information in clinical guidelines: The European perspective. Clin Pharm Ther. 2018;103:795–801. doi: 10.1002/cpt.1049.. [DOI] [PubMed] [Google Scholar]
- 30.Etienne-Grimaldi MC, Boyer JC, Thomas F, Quaranta S, Picard N, Loriot MA, et al. UGT1A1 genotype and irinotecan therapy: general review and implementation in routine practice. Fundam Clin Pharm. 2015;29:219–37. doi: 10.1111/fcp.12117.. [DOI] [PubMed] [Google Scholar]
- 31.AIOM - SIF. Raccomandazioni par analisi farmacogenetiche n.d. https://www.aiom.it/raccomandazioni-2019-per-analisi-farmacogenetiche/ (accessed November 15, 2021).
- 32.Stanford University & St. Jude Children’s Research Hospital. Clinical Pharmacogenetics Implementation Consortium. PharmGKB PGRN 2020. https://cpicpgx.org/ (accessed May 8, 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 genetic variant and functionality
Supplementary Table 2 allele and genotype frequencies
Supplementary Table 3 genotype phenotype translation for LIS
Supplementary Table 5 therapeutic recommendation
Supplementary Table 6 clinical decision support text
Supplementary Table 7 clinical implication score
Data Availability Statement
All data and material are either included in the supplementary information or publicly available (i.e. the published articles, PubMed). The guidelines and background information are available on the website of the Royal Dutch Pharmacists Association (KNMP) (Pharmacogenetic Recommendation Text. Available from: https://www.knmp.nl/).


