Abstract
Anorexia nervosa (AN) is a heritable eating disorder (50–60%) with an array of commonly comorbid psychiatric disorders and related traits. Although significant genetic correlations between AN and psychiatric disorders and related traits have been reported, their shared genetic architecture is largely understudied. We investigated the shared genetic architecture of AN and schizophrenia (SCZ), bipolar disorder (BIP), major depression (MD), mood instability (Mood), neuroticism (NEUR), and intelligence (INT). We applied the conditional false discovery rate (FDR) method to identify novel risk loci for AN, and conjunctional FDR to identify loci shared between AN and related phenotypes, to summarize statistics from relevant genome-wide association studies (GWAS). Individual GWAS samples varied from 72,517 to 420,879 participants. Using conditional FDR we identified 58 novel AN loci. Furthermore, we identified 38 unique loci shared between AN and major psychiatric disorders (SCZ, BIP, and MD) and 45 between AN and psychological traits (Mood, NEUR, and INT). In line with genetic correlations, the majority of shared loci showed concordant effect directions. Functional analyses revealed that the shared loci are involved in 65 unique pathways, several of which overlapped across analyses, including the “signal by MST1” pathway involved in Hippo signaling. In conclusion, we demonstrated genetic overlap between AN and major psychiatric disorders and related traits, and identified novel risk loci for AN by leveraging this overlap. Our results indicate that some shared characteristics between AN and related disorders and traits may have genetic underpinnings.
Subject terms: Medical genetics, Psychiatric disorders
Introduction
Anorexia nervosa (AN) is an eating disorder characterized by restricted energy intake, abnormally low body weight, intense fear of weight-gain, and body image disturbances [1]. The age of onset is typically in early adolescence and the disorder disproportionally affects females. The estimated lifetime prevalence is 0.62% among females and 0.04% among males [2], but prevalence has increased in recent decades [3], particularly during the Covid-19 pandemic [4, 5]. Medical complications due to malnutrition and low body weight are common [6] and mortality rates are high [7]. Evidence of effective psychological and pharmacological treatments is limited [8], and treatment outcomes are often unsatisfactory [8, 9].
Although its core clinical characteristics relate to eating and body image disturbances, a variety of comorbid psychopathologies and related traits are commonly associated with AN. Patients tend to be characterized by neuroticism and perfectionism, and commonly present with anxious personality disorders [10, 11]. AN also often co-occurs with alexithymia, which involves difficulties in identifying and describing emotions [12]. Other commonly comorbid psychiatric conditions include mood and anxiety disorders [9, 13, 14], most notably major depression (MD). Evidence of psychotic features has also been reported in AN, with patients at increased risk of schizophrenia (SCZ) [15] and exhibiting delusional thoughts in relation to body image [15–17]. Moreover, AN is associated with cognitive traits such as increased intelligence [18]. Understanding the genetic underpinnings of the association between AN and the abovementioned major psychiatric disorders and related traits may provide insights into the genetic architecture of AN and its comorbidities.
Genetic variation substantially influences risk for AN, with twin-based heritability estimates of 50–60% [19]. Recent genome-wide association studies (GWAS) have identified the first genetic risk loci for AN [20, 21] and highlighted significant positive genetic correlations between AN and other psychiatric disorders, including obsessive-compulsive disorder, SCZ, MD, and bipolar disorder (BIP) [21–23]. Furthermore, studies have documented familial associations between AN and MD [24–26], SCZ [15, 26], and related disorders [27]; indicating shared familial liability factors. These findings align with converging evidence of shared genetic risk across multiple distinct psychiatric disorders [22], underscoring the complexity of genetic relationships among psychiatric phenotypes, including AN. Despite these advances, the genetic architecture of the disorder is largely unknown, and few studies have identified the shared risk loci between AN and other mental disorders. Characterizing the shared genetic architecture between AN and related phenotypes is a promising avenue for insight into genetic liability for AN itself, to inform nosology, guide research into pathophysiological mechanisms and suggest novel pharmacological treatments [28].
In the current study, we sought to identify novel risk loci for AN and investigate the shared genetic architecture of AN and related phenotypes including SCZ, BIP, MD, mood instability (Mood), neuroticism (NEUR), and intelligence (INT). Specifically, we leveraged the boost in statistical power by combining two GWAS to identify AN loci conditioned on their association with other phenotypes, using pleiotropy-based conditional false discovery rate (FDR) statistics [29–31]. Further, we applied conjunctional FDR to identify shared genetic loci, thereby revealing additional shared polygenic architecture between AN and related psychiatric disorders and traits.
Methods
Genome-wide association samples
We used GWAS summary statistics for AN [21], SCZ [32], BIP [33], and MD [34]; from the Psychiatric Genomics Consortium. The AN sample included 16,992 individuals with AN and 55,525 controls [21]. The SCZ sample included 40,675 individuals with SCZ and 64,643 controls [32]. The BIP sample consisted of 40,463 individuals with BIP and 313,436 controls [33]. The MD sample consisted of 105,718 individuals with MD and 344,901 controls [34]. To avoid sample overlap, we excluded the UK Biobank cohort from our MD and BIP samples. Additionally, we used GWAS summary statistic results for the following psychological traits: Mood [35], NEUR [36], and INT [37]. The Mood sample included 363,705 individuals; the NEUR sample included 372,903 individuals; and the INT sample included 269,867 individuals. The Supplementary information provides a brief account of the phenotype definitions for each GWAS sample, and additional details are provided in the original GWAS studies [21, 32–37].
Statistical analyses
We estimated genetic correlations between AN and each disorder and psychological trait using linkage disequilibrium (LD) score regression [38, 39]. To assess the presence of cross-phenotype polygenic enrichment, we generated conditional quantile-quantile (Q-Q) plots [29], conditioning AN on each disorder (SCZ, BIP, MD) and psychological trait (Mood, NEUR, and INT), and vice versa. We compared the enrichment of associations of all single nucleotide polymorphisms (SNPs) with SNPs associated with the conditional trait (i.e., the major psychiatric disorders and psychological traits), at increasingly significant p-value thresholds (0.1, 0.01, and 0.001). Successive deflections of the Q-Q plot away from the null line with increasing strength of association with the conditional trait is indicative of cross-phenotype polygenic enrichment.
Next, we employed the conditional FDR (condFDR) method [29, 31] which leverages cross-phenotype enrichment observed on the conditional Q-Q plots to improve the discovery of genetic loci associated with AN. This method builds on an empirical Bayesian statistical framework, using GWAS summary statistics from a primary trait of interest (e.g., AN) together with those of a conditional trait (e.g., SCZ) to estimate the posterior probability that a SNP has no association with the primary trait, provided that the p values for that SNP in both the primary and conditional traits are as small as or smaller than the observed p value. The condFDR method increases the power to identify genetic variants associated with the primary trait of interest by reranking the test statistics of the primary trait based on the strength of association with the conditional trait.
To determine shared genetic loci between AN and related psychiatric disorders and psychological traits we employed the conjunctional FDR (conjFDR) method [28–30]. This method is defined as the maximum of the two condFDR statistics for a specific SNP, i.e. for trait A conditional on trait B and trait B conditional on trait A. This represents an estimate for the posterior probability that a SNP is null for either or both traits, provided that the p values for both phenotypes are as small as or smaller than the p values for each trait individually. More details are found in the original [29] and subsequent publications [31].
In the current study, we used an FDR level of 0.05 per pairwise comparison for cond/conjFDR. Manhattan plots based on the conjFDR were generated to depict the genomic location of shared genetic loci [29]. All analyses were performed after excluding SNPs in the extended major histocompatibility complex (MHC; hg19 location chr 6: 25119106-33854733) and the 8p23.1 (hg19 location chr 8: 7242715-12483982) regions to avoid potential biases due to complex LD.
Genomic loci definition
We used the FUMA protocol to define independent genomic loci jointly associated with AN and each psychiatric disorder and psychological trait [40]. The genomic loci were defined based on the lead SNPs and candidate SNPs within each locus as significant SNPs within a LD r2 >= 0.6 and cond/conjFDR <0.1 with at least one related independent significant SNP. Independent significant SNPs were considered as cond/conjFDR < 0.05 and r2 < 0.6 while lead SNPs were defined if they were in approximate linkage equilibrium with each other (r2 < 0.1). If two or more lead SNPs located within one LD block (in 250 kb), we merged them into one genomic risk locus. LD information from the European population was derived from the 1000 Genomes Project reference panel [41]. We used Bedtools [42] to identify shared loci across pair-wise analyses and to identify unique genetic loci across all analyses. Novel loci were determined by cross-referencing identified loci with previous GWASs and other relevant studies.
Functional annotation
We used FUMA to annotate identified shared variants according to function, the predicted deleteriousness (Combined Annotation Dependent Depletion score, [43], regulatory effect (RegulomeDB scores, [44] and chromatin states. We next mapped genes to candidate SNPs using three gene-mapping methodologies using FUMA: 1) positional mapping which matches SNPs by physical position to all genes within a 10 kb window 2) expression quantitative trait loci (eQTL) mapping that identifies genes whose expression is associated with the SNPs’ allelic variation, 3) chromatin interaction mapping that matches SNPs to genes with which they are predicted to interact based on chromatin structure [40]. We applied gene-set analyses (on all genes) using FUMA [45, 46], pathway analyses using Consensus PathDB [47] and spatiotemporal gene expression analysis of mapped genes in the R package “cerebroViz” using BrainSpan RNA sequencing data [48, 49]. All analyses were corrected for multiple comparisons using Bonferroni correction.
Results
Genetic overlap and correlation between AN and related psychiatric disorders and psychological traits
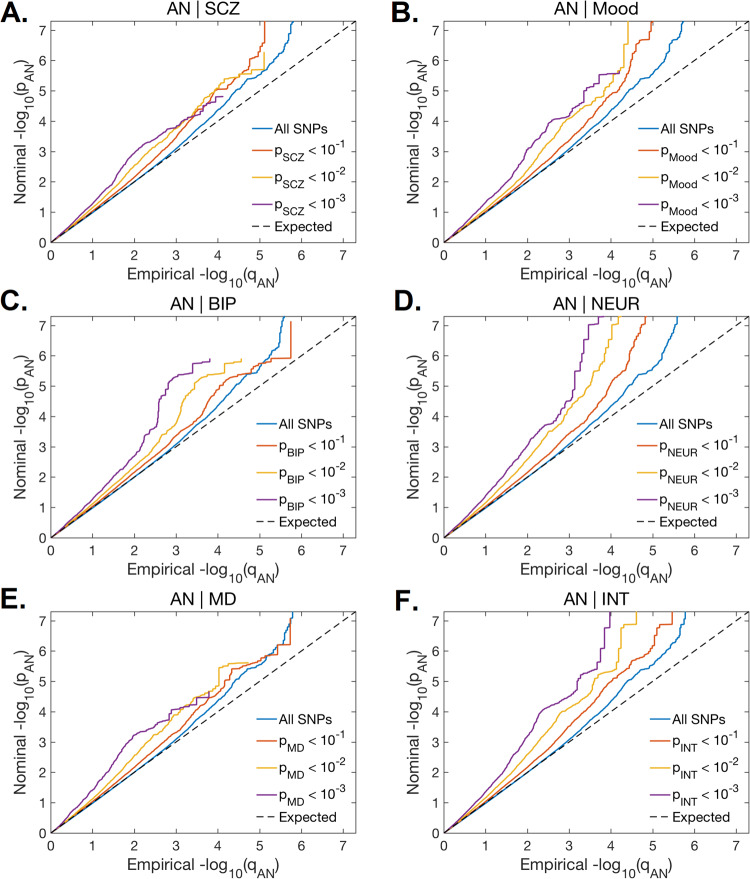
The conditional Q-Q plots showed successive increments of SNP enrichment for AN conditioned on association p-values for each psychiatric disorder and psychological trait (Fig. 1). The consistently increasing leftward deflection for subsets of variants with higher significance in the conditional trait in both directions indicates substantial polygenic overlap between AN and the other psychiatric disorders and related traits. See Supplemental Figure S1 for the reverse Q-Q plots.
Fig. 1. Panels A-F show conditional quantile-quantile plots of nominal vs.
empirical anorexia nervosa (AN) -log10 p-values as a function of the significance of the association with schizophrenia (SCZ), bipolar disorder (BIP), major depression (MD), mood instability (Mood), neuroticism (NEUR), and intelligence (INT) at the level of p < .10, p < .01 and p < .001. These plots show the quantiles of the observed p-values on the y-axis against the theoretical quantiles under no association on the x-axis. Deflections from the null line indicate systematic association.
Genome-wide LD score regression showed significant positive genetic correlations between AN and the following disorders and traits: SCZ (rg = 0.26, SE = 0.03, p = 6.64 × 10−14), BIP (rg = 0.19, SE = 0.04, p = 1.18 × 10−6), MD (rg = 0.35, SE = 0.06, p = 1.59 ×10−9), NEUR (rg = 0.26, SE = 0.04, p = 1.52 × 10−12), and INT (rg = 0.10, SE = 0.04, p = 3.80 × 10−3). Genetic correlation between AN and Mood were not significant (rg = 0.06, SE = 0.05, p = 0.22). Please note that the statistical significance of certain genetic correlations (e.g., AN and SCZ) is higher than others (e.g., MD and SCZ) despite a lower point estimate because the precision of the estimate is greater. This is likely due to the higher heritability and discoverability of certain phenotypes (e.g., higher for SCZ compared to MD).
AN-associated loci identified with condFDR
Using condFDR analysis (FDR < 0.05), we identified 40, 37, and 49 loci associated with AN conditionally on SCZ, BIP, and MD, respectively (Supplementary Tables 1–3). For the psychological traits, we identified 48, 55, and 60 loci associated with AN conditionally on Mood, NEUR, and INT, respectively (Supplementary Tables 4–6). Across all conditional analyses, there were 58 unique loci associated with AN, of which all were novel.
Loci shared between AN and related psychiatric disorders and psychological traits
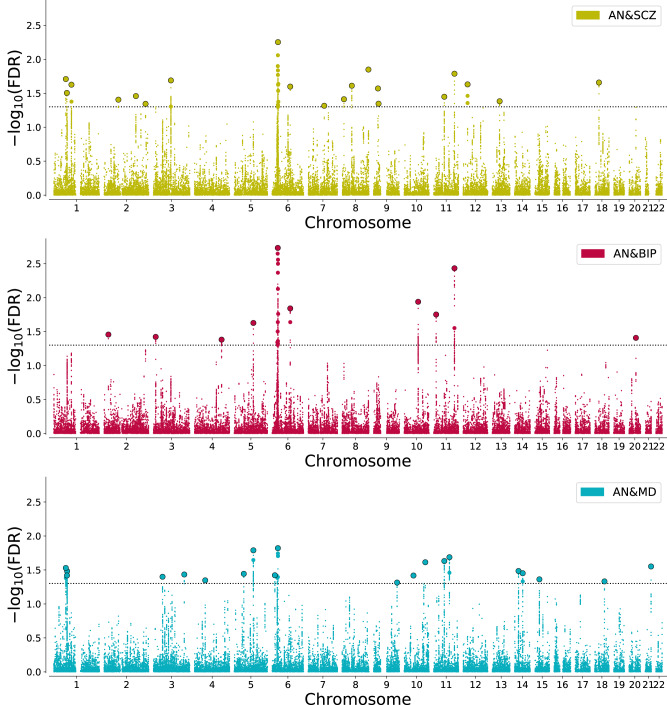
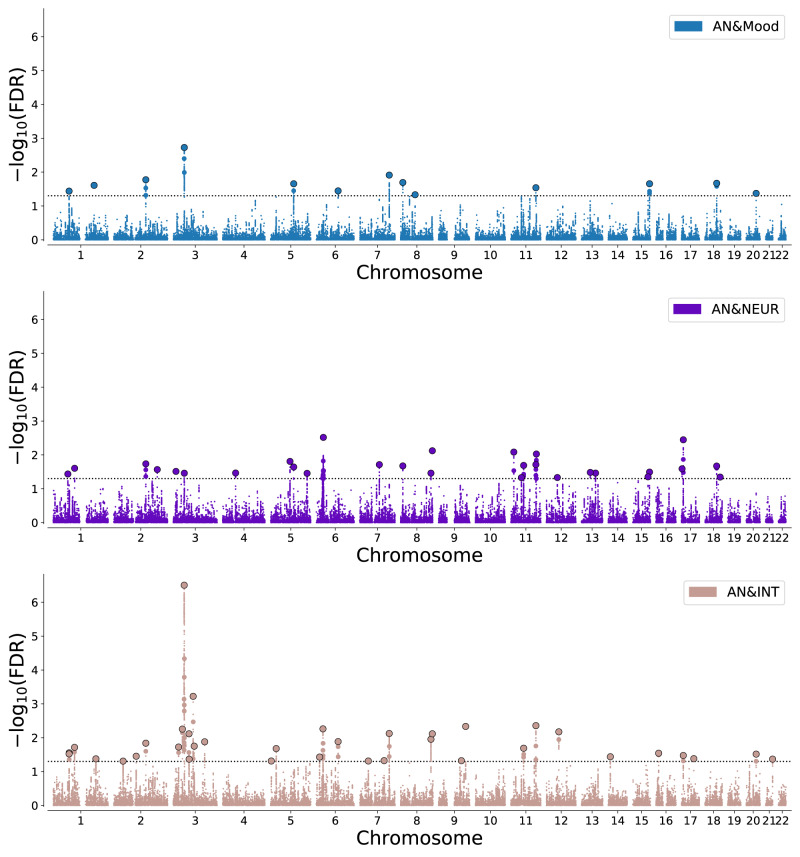
ConjFDR analysis revealed numerous loci shared between AN and the related disorders and traits; see Table 1 and Supplementary Tables 7–12. Specifically, 20, 10, and 20 loci were jointly associated with SCZ, BIP, and MD, respectively. In total, we identified 38 unique shared loci between AN and related psychiatric disorders. Furthermore, 13, 29, and 36 loci were jointly associated with Mood, NEUR, and INT, respectively. In total, 45 loci were uniquely shared between AN and the psychological traits; of which 9 overlapped with the 38 uniquely shared between AN and psychiatric disorders. After merging physically overlapping condFDR loci across all analyses, there were 56 “unique” loci associated with AN. See Figs. 2–3 for Manhattan plots of all shared loci.
Table 1.
Results of conjunctional FDR showing number of shared loci between AN and related disorders and psychological traits.
| Secondary phenotype | Secondary GWAS (n/neffective for case-control GWASs)1 | Shared loci with AN at conjFDR < 0.05 | Novel loci in AN (n) | Novel loci in comparison phenotype (n) |
|---|---|---|---|---|
| SCZ | 126,282 | 20 | 18 | 4 |
| BIP | 150,670 | 10 | 10 | 3 |
| MD | 389,039 | 20 | 19 | 9 |
| Mood | 356,933 | 13 | 12 | 4 |
| NEUR | 372,903 | 29 | 26 | 7 |
| INT | 269,867 | 36 | 31 | 16 |
Note: AN Anorexia nervosa, BIP Bipolar disorder, conjFDR Conjunctional False Discovery Rate, INT Intelligence, Mood Mood instability, NEUR Neuroticism, SCZ Schizophrenia. 1neffective = 4/(1/ncases + 1/ncontrols).
Fig. 2. Manhattan plots based on the -log10 transformed conjFDR values for each single-nucleotide polymorphism, depicting common genetic variants shared between anorexia nervosa (AN), schizophrenia (SCZ), bipolar disorder (BIP), and major depression (MD).
The dotted horizontal line represents the threshold chosen for reporting shared associations (conjFDR < 0.05). Circles indicate lead SNPs.
Fig. 3. Manhattan plots based on the -log10 transformed conjFDR values for each single-nucleotide polymorphism, depicting common genetic variants shared between anorexia nervosa (AN), mood instability (Mood), neuroticism (NEUR), and intelligence (INT).
The dotted horizontal line represents the threshold chosen for reporting shared associations (conjFDR < 0.05). Circles indicate lead SNPs.
A pattern of concordant genetic effects was evident when comparing the effect directions of lead SNPs. Concordant effect directions were observed for 19 of 20 (95%) shared loci in AN and SCZ, 7 of 10 (70%) shared loci in AN and BIP, and 18 of 20 (90%) shared loci in AN and MD (Supplementary Tables 7–9). For the psychological traits, concordant association directions were observed for 8 of 13 (62%) shared loci in AN and Mood, 28 of 29 (96%) shared loci in AN and NEUR, and 21 of 36 (58%) shared loci in AN and INT (Supplementary Tables 10–12).
Annotation of loci shared between AN and related psychiatric disorders and psychological traits
We functionally annotated all SNPs in LD (r2 ≥ 0.6) with a significant independent SNP and with conjFDR <0.1 within the shared loci associated with AN and related disorders and traits using FUMA. The majority of SNPs jointly associated with AN and SCZ, BIP, Mood and INT were intronic; while the majority of SNPs jointly associated with AN and MD and NEUR were intergenic. For more details see Supplementary information, Figure S2 and Supplementary Tables 7–12).
We mapped candidate SNPs to genes by three independent gene-mapping strategies. In total 187 unique protein coding genes, including 125, 115, and 109 genes were mapped between AN and SCZ, BIP and MD, respectively. Eighty of these genes were mapped by at least two out of the three gene mapping strategies. Gene set analysis of these genes did not show any overrepresented gene-ontology (GO) terms after excluding MHC region from the analysis. Similarly, 294 unique protein coding genes were mapped to the candidate SNPs in the shared loci across AN and psychological traits, including 165, 204 and 53 mapped across AN and INT, NEUR and Mood, respectively. The gene set analysis showed that the mapped genes were overrepresented in 223 GO terms for AN and psychological traits. See Supplementary Table 37 for all GO gene sets. The most significantly overrepresented gene-set was “synapse organization” (p = 2.87 × 10−7), followed by “negative regulation of intracellular steroid hormone receptor signaling pathway” (p = 1.16 × 10−6), “regulation of transport” and “neuron development”.
Among genes mapped to loci shared between AN and psychiatric disorders, several immune-related genes were implicated, including the C4A, C4B, NCAM1, HLA-B, HLA-C, and HLA-E genes. Both the C4A [50, 51] and C4B [51] genes have previously been associated with the etiology of SCZ and BIP. The C4A, NACM1, HLA-B, HLA-C, and HLA-E genes were also mapped to loci shared between AN and NEUR, and AN and INT. This suggests genes associated with AN risk are also associated with a range of other disorder- and trait-related phenotypes. However, genes mapped to the MHC region must be interpreted with caution due to the complex LD in this genomic region. See Table 2 for an overview of genes mapped to lead SNPs within loci jointly associated with anorexia nervosa and at least two related phenotypes (see Supplementary Tables 25–30 for full list of genes).
Table 2.
Genes mapped to lead single nucleotide polymorphisms within loci jointly associated with anorexia nervosa and at least two related phenotypes.
| Phenotypes | Symbol | Chr | Phenotypes | Symbol | Chr |
|---|---|---|---|---|---|
| MDD, INT | ADARB1 | 21 | Mood, NEUR | LINC00923 | 15 |
| MDD, INT | AMT | 3 | Mood, INT | LRGUK | 7 |
| Mood, NEUR, INT | ARHGAP15 | 2 | SCZ, BIP, MDD, NEUR | MICB | 6 |
| BIP, NEUR | ARNTL | 11 | SCZ, NEUR | MSH5 | 6 |
| BIP, MDD | BAG6 | 6 | SCZ, Mood, NEUR | MSRA | 8 |
| SCZ, NEUR | C2 | 6 | BIP, Mood, NEUR, INT | NCAM1 | 11 |
| SCZ, BIP, MDD, NEUR | C4A | 6 | MDD, INT | NCKIPSD | 3 |
| MDD, INT | CCDC71 | 3 | SCZ, MDD, Mood, INT | NEGR1 | 1 |
| SCZ, BIP, MDD, NEUR | CCHCR1 | 6 | Mood, INT | NEURL2 | 20 |
| SCZ, NEUR | CFB | 6 | MDD, NEUR, INT | OR5AK2 | 11 |
| SCZ, NEUR | CSNK2B | 6 | MDD, NEUR | P4HTM | 3 |
| Mood, INT | CTSA | 20 | Mood, INT | PCIF1 | 20 |
| SCZ, BIP, MDD, NEUR | DDAH2 | 6 | SCZ, MDD, NEUR | PDE4B | 1 |
| MDD, INT | DTNBP1 | 6 | Mood, NEUR | RBM6 | 3 |
| Mood, NEUR | ERI1 | 8 | Mood, NEUR | RNF123 | 3 |
| Mood, INT | EXOC4 | 7 | Mood, INT | SPATA25 | 20 |
| Mood, NEUR | FAM212A | 3 | MDD, Mood, NEUR | TCF4 | 18 |
| BIP, MDD | FLOT1 | 6 | BIP, NEUR | TIMP4 | 3 |
| MDD, Mood, NEUR | GPX1 | 3 | BIP, INT | TRIM26 | 6 |
| BIP, MDD | HLA-B | 6 | SCZ, NEUR, INT | TSNARE1 | 8 |
| SCZ, BIP, MDD, NEUR | HLA-C | 6 | SCZ, BIP, MDD, NEUR, INT | VARS2 | 6 |
| SCZ, NEUR | HLA-DQA1 | 6 | MDD, INT | WDR6 | 3 |
| SCZ, NEUR | HLA-DQB1 | 6 | Mood, INT | ZSWIM1 | 20 |
Note: Genes mapped to lead SNPs within loci jointly associated with AN and at least two related phenotypes (SCZ, BIP, MD, Mood, NEUR, and INT) at conjFDR < 0.05. Genes shared between AN and related phenotypes mapped to genes using positional mapping, eQTL mapping, and chromatin interaction strategies. AN Anorexia nervosa, BIP Bipolar disorder, conjFDR Conjunctional False Discovery Rate, INT Intelligence, Mood Mood instability, NEUR Neuroticism, SCZ Schizophrenia.
Spatio-temporal analysis indicated that the expression of mapped genes to the shared loci between AN and psychiatric disorders (SCZ, BIP, and MD) decreased after middle childhood (Figure S2). The mapped genes to the shared loci between AN and Mood, and AN and NEUR were weakly expressed after late childhood, while the mapped genes to the shared loci between AN and INT were highly expressed from late childhood to late adolescence (Figure S3). A more detailed description of functional annotation analyses for all analyses and for each individual analysis is presented in Supplementary information.
Pathway analysis of genes mapped to loci shared between AN and related psychiatric disorders and psychological traits
After excluding MHC regions; 1, 2, and 10 pathways were enriched for genes linked to the candidate SNPs shared between AN and SCZ, BIP and MD, respectively. None of these pathways were identified across all three analyses. Furthermore, 15, 14, and 35 pathways were enriched for genes linked to the candidate SNPs shared between AN and Mood, NEUR, and INT, respectively. Several of these were shared across multiple analyses, including the “Signaling by MST1”, “amb2 integrin signaling”, “pathways regulating Hippo signaling”, and “rho-selective guanine exchange factor akap13 mediates stress fiber formation” pathways. See Supplemental Tables 31–36 for all overrepresented pathways.
Based on unique genes mapped to the shared loci between AN and the psychiatric disorders (SCZ, BIP, and MD) we identified 7 significantly overrepresented pathways (see Supplemental Table 38). No new pathways emerged in this analysis; all pathways were also enriched for genes linked to the candidate SNPs shared between AN and MD. For the unique genes mapped to the shared loci between AN and the psychological traits (Mood, NEUR, and INT) we identified 36 significantly overrepresented pathways (see Supplemental Table 39). Most of these were also identified in individual analyses of Mood, NEUR, and INT. However, some additional pathways emerged, including “transcriptional regulation of granulopoiesis”, “EPHA forward signaling”, and “signaling by TGF-β receptor complex”.
Discussion
In the current study, we demonstrated genetic overlap between AN and major psychiatric disorders and psychological traits, and identified novel risk loci for AN by leveraging this overlap. Our findings suggest a large degree of genetic overlap between AN and related psychiatric disorders and traits, possibly indicating overlapping genetic mechanisms [31]. Our findings shed light on the genetic etiology of AN, and could have clinical utility in developing risk prediction models and psychopharmacological treatments.
We identified 38 unique loci jointly associated with AN and psychiatric disorders, while 45 unique loci were jointly associated with AN and psychological traits. Nine of these loci overlapped. The effect direction of the lead SNPs on AN and each psychiatric disorder and related psychological traits revealed that the majority of lead SNPs had concordant effects, but to different degrees. Most lead SNPs had concordant effects on AN and each psychiatric disorder (70–95%), including 95% of loci shared with SCZ. Effect directions were also mostly concordant for the psychological traits (58–96%), the highest of which was for NEUR. These findings are in line with the positive genetic correlations between AN and MD, SCZ, BIP, and NEUR identified in the current and previous studies [20–22, 38]. Of note, the genetic correlation between AN and Mood was non-significant in our study, and our observed significant correlation between AN and BIP contrasts with findings from an earlier study based on smaller GWAS samples [38].
The genetic overlap between AN and MD aligns with epidemiological evidence of associations between these two disorders. MD is among the disorders most frequently comorbid with AN [9, 13, 14]. Both AN and MD are characterized by mood disturbances, and studies have shown that such symptoms are central features among patients with AN [52, 53]. Family studies have shown that AN and MD tend to co-aggregate in families [24–26], suggesting the disorders may share familial liability factors. Our findings suggests that at least part of the co-occurrence of these disorders is due to shared genetic liabilities, mirroring results from twin studies [54, 55].
Our finding that AN shows genetic overlap with BIP and SCZ is intriguing and may reflect shared clinical features or vulnerabilities. There is evidence of psychotic features in AN [56], including the pervasive body image disturbances that may be delusional by nature [15–17]. Also, difficulty recognizing and describing feelings (alexithymia) is frequent in eating disorders [12] and is also reported in MD, BIP and SCZ [57, 58], possibly representing a shared vulnerability factor. Previous findings indicate familial co-aggregation of AN and SCZ [15, 26],—implicating a genetic basis for the association. A recent systematic review also highlighted that BIP and eating disorders are frequently comorbid [59]. It is possible that AN, SCZ, and BIP share genetic vulnerabilities that are only partly reflective of the clinical features of each disorder. Instead, the genetic overlap between these disorders may reflect general liabilities to multiple psychiatric disorders, akin to the suggested “p” factor [60]. These overlapping vulnerabilities may be reflected in the underlying genetic architecture.
Our study also demonstrated genetic overlap between AN and the psychological traits Mood, NEUR, and INT. The overlap with Mood and NEUR aligns with epidemiological and genetic evidence supporting an association between AN and psychiatric disorders characterized by mood disturbances and high anxiety [9, 13, 14]. Previous studies have shown that patients with AN are characterized by high levels of depressive and anxious features even following recovery [61, 62]. In support of this, one study found that NEUR was prospectively related to development of AN [63]. Additionally, we documented genetic overlap between AN and INT, which could be related to the increased intelligence associated with AN [18]. In line with this, a previous GWAS found significant positive genetic correlations between AN and years of education [21]. They also found positive genetic correlations between INT and AN when specific AN subtypes were considered. Importantly, our findings show that the genetic overlap between AN and related phenotypes is not limited to diagnostically defined psychopathology, but extends to trait phenotypes (e.g., neuroticism) that are shared among common psychiatric disorders and may underlie the high comorbidity.
By leveraging the genetic overlap between AN and the included psychiatric disorders and psychological traits, we identified risk loci associated with AN conditional on each of the phenotypes. Specifically, we identified 40, 37, and 49 loci associated with AN conditional on SCZ, BIP, and MD respectively. We also identified 60, 55, and 48 loci associated with AN conditional on Mood, NEUR, and INT respectively. Among these risk loci were the eight loci previously reported in a recent GWAS study of AN [21]. Across all analyses, 58 novel risk loci for AN were identified. These findings highlight the benefit of leveraging the polygenic overlap between psychiatric disorders to boost discovery of novel risk loci. Extending our analyses to other phenotypes holds potential in revealing more risk loci for AN.
The mapped genes for the shared loci included a number of genes related to the immune system, for example the C4A gene, which was implicated across multiple analyses. While no pathways were identified for loci shared between AN and psychiatric disorders, several overlapping pathways were identified for loci shared between AN and the psychological traits. In particular, the “Pathways Regulating Hippo Signaling”, “Wnt Signaling Pathway“ and “signaling by MST1” pathways were implicated for shared loci with both Mood and INT. Hippo signaling regulates stemness, cell proliferation and apoptosis and disruption of these systems has been associated with metabolic and neurodegenerative diseases, mirroring findings from the primary AN GWAS which highlighted associations with genes involved in metabolic dysregulation [21]. Further, recent research showed that genes within the Hippo pathway modulate key molecular and cellular processes that are involved in the pathophysiology of stress-related-psychiatric disorders [64]. In addition to this, various proteins of the Hippo signaling pathway are linked via Wnt-signaling and other pathways to stress-regulated signaling cascades [65], while overexpression of MST1 induces memory impairment via disturbances in the patterns of neural activities. This mechanism may therefore contribute to the genetic overlap observed between AN and INT [66].
Our study has limitations. As with all GWAS findings, it is challenging to identify the true ‘causal SNP’ from correlated SNPs due to LD. Therefore, experimental studies and improved fine-mapping strategies are needed to determine the true causal variants underlying the shared associations reported here and whether the same causal variants are involved in AN and the included phenotypes. We also note that sample size for the AN GWAS is still small, and future studies utilizing larger samples may elucidate more shared genetic loci. Furthermore, the AN GWAS sample lacks ethnic diversity, with samples confined to North-America, Europe, Australia and New Zealand. Finally, we note that a small proportion of our AN sample (4.5%; 768 AN cases and 3065 controls) was derived from the UK Biobank, which also contributed to the MOOD, NEUR and INT samples (see Supplementary information). Sample overlap may inflate the cond/conjFDR statistics resulting in an increase in false positives, although since UKB cases and controls represented <5% of the total AN, this is unlikely to significantly change our results.
In conclusion, we used conditional and conjunctional FDR approach to identify novel risk loci for AN and demonstrate genetic overlap between AN and related psychiatric disorders and psychological traits. This expands on previous findings of shared genetic risk between AN and related phenotypes, advancing our understanding of the genetic underpinnings of AN. This increases the understanding the genetic architecture of AN, which can form a platform for developing risk prediction models and novel psychopharmacological treatments.
Supplementary information
Acknowledgements
We gratefully acknowledge support from the National Institutes of Health (NS057198, EB00790); the Research Council of Norway (229129, 213837, 223273, 283798); the South-East Norway Regional Health Authority (2017-112, 2020-034); KG Jebsen Stiftelsen; and the EU’s Horizon2020 (grant #847776 CoMorMent). CMB is supported by NIMH (R01MH120170; R01MH124871; R01MH119084; R01MH118278; R01MH124871); Brain and Behavior Research Foundation Distinguished Investigator Grant; Swedish Research Council (Vetenskapsrådet, award: 538-2013-8864); Lundbeck Foundation (Grant no. R276-2018-4581). The funding bodies had no role in the design or conduct of the study; the collection, management, analysis or interpretation of the data; the preparation, review, or approval of the manuscript; nor the decision to submit the manuscript for publication.
We want to thank all the research participants taking part in the GWASs included in this study, including Complex Trait Genetics lab, the GWASs of intelligence; Mood and Neuroticism from UKB and the GWAS of anorexia nervosa; schizophrenia; bipolar and major depressive disorder from PGC. In addition, we want to thank the consortia for making their GWAS summary statistics available, and the people who provided DNA samples.
The computations were performed on resources provided by UNINETT Sigma 2 – the National Infrastructure for High-Performance Computing and Data Storage in Norway.
Author contributions
LB, SB, and OAA conceived the study. SB analyzed the data. LB, SB, and GH interpreted the results and spearheaded the writing. SB drafted the online methods. All authors gave conceptual input on the methods and/or results and all authors contributed to and approved the final manuscript.
Data availability
Data supporting the findings of this study are openly available from an online repository or are available on request from study authors. All code is freely available at https://github.com/precimed and https://github.com/bulik/ldsc. Analyses were conducted in Python v3.5, Matlab R2020b, and R v3.6.3. Locus definition, functional annotation, and gene-set analysis were performed using FUMA (https://fuma.ctglab.nl/).
Competing interests
OAA has received speaker’s honorarium from Sunovion, and Lundbeck and is a consultant for Healthlytix. CMB reports: Shire (grant recipient, Scientific Advisory Board member); Idorsia (consultant); Pearson (author, royalty recipient); Equip Health Inc. (Clinical Advisory Board). The remaining authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lasse Bang, Shahram Bahrami.
Contributor Information
Shahram Bahrami, Email: shahram.bahrami@medisin.uio.no.
Ole A. Andreassen, Email: o.a.andreassen@medisin.uio.no
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-023-02585-1.
References
- 1.APA. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 5th ed. Arlington, VA: American Psychiatric Association; 2013.
- 2.Qian J, Wu Y, Liu F, Zhu Y, Jin H, Zhang H, et al. An update on the prevalence of eating disorders in the general population: a systematic review and meta-analysis. Eat Weight Disord - Stud Anorex Bulim Obes [Internet]. 2021 Apr 8 [cited 2021 Dec; Available from: 10.1007/s40519-021-01162-z [DOI] [PMC free article] [PubMed]
- 3.Galmiche M, Déchelotte P, Lambert G, Tavolacci MP. Prevalence of eating disorders over the 2000-18 period: a systematic literature review. Am J Clin Nutr. 2019;109:1402–13. doi: 10.1093/ajcn/nqy342. [DOI] [PubMed] [Google Scholar]
- 4.Taquet M, Geddes JR, Luciano S, Harrison PJ. Incidence and outcomes of eating disorders during the COVID-19 pandemic. Br J Psychiatry. 2021;220:262–4. doi: 10.1192/bjp.2021.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toulany A, Kurdyak P, Guttmann A, Stukel TA, Fu L, Strauss R, et al. Acute care visits for eating disorders among children and adolescents after the onset of the COVID-19 pandemic. J Adolesc Health. 2022;70:42–7. doi: 10.1016/j.jadohealth.2021.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell JE, Crow S. Medical complications of anorexia nervosa and bulimia nervosa. Curr Opin Psychiatry. 2006;19:438–43. doi: 10.1097/01.yco.0000228768.79097.3e. [DOI] [PubMed] [Google Scholar]
- 7.Keshaviah A, Edkins K, Hastings ER, Krishna M, Franko DL, Herzog DB, et al. Re-examining premature mortality in anorexia nervosa: a meta-analysis redux. Compr Psychiatry. 2014;55:1773–84. doi: 10.1016/j.comppsych.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 8.Watson HJ, Bulik CM. Update on the treatment of anorexia nervosa: review of clinical trials, practice guidelines and emerging interventions. Psychol Med. 2013;43:2477–500. doi: 10.1017/S0033291712002620. [DOI] [PubMed] [Google Scholar]
- 9.Steinhausen HC. The outcome of anorexia nervosa in the 20th century. Am J Psychiatry. 2002;159:1284–93. doi: 10.1176/appi.ajp.159.8.1284. [DOI] [PubMed] [Google Scholar]
- 10.Cassin SE, von Ranson KM. Personality and eating disorders: a decade in review. Clin Psychol Rev. 2005;25:895–916. doi: 10.1016/j.cpr.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Farstad SM, McGeown LM, von Ranson KM. Eating disorders and personality, 2004-16: a systematic review and meta-analysis. Clin Psychol Rev. 2016;46:91–105. [Google Scholar]
- 12.Westwood H, Kerr-Gaffney J, Stahl D, Tchanturia K. Alexithymia in eating disorders: systematic review and meta-analyses of studies using the Toronto Alexithymia Scale. J Psychosom Res. 2017;99:66–81. doi: 10.1016/j.jpsychores.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godart NT, Perdereau F, Rein Z, Berthoz S, Wallier J, Jeammet P, et al. Comorbidity studies of eating disorders and mood disorders. Critical review of the literature. J Affect Disord. 2007;97:37–49. doi: 10.1016/j.jad.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Marucci S, Ragione LD, De Iaco G, Mococci T, Vicini M, Guastamacchia E, et al. Anorexia nervosa and comorbid psychopathology. Endocr Metab Immune Disord Drug Targets. 2018;18:316–24. doi: 10.2174/1871530318666180213111637. [DOI] [PubMed] [Google Scholar]
- 15.Zhang R, Larsen JT, Kuja-Halkola R, Thornton L, Yao S, Larsson H, et al. Familial co-aggregation of schizophrenia and eating disorders in Sweden and Denmark. Mol Psychiatry. 2020;26:5389–97. doi: 10.1038/s41380-020-0749-x. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann AS, Thomas JJ, Wilson AC, Wilhelm S. Insight impairment in body image disorders: Delusionality and overvalued ideas in anorexia nervosa versus body dysmorphic disorder. Psychiatry Res. 2013;210:1129–35. doi: 10.1016/j.psychres.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Phillips KA, Kim JM, Hudson JI. Body image disturbance in body dysmorphic disorder and eating disorders: obsessions or delusions? Psychiatr Clin North Am. 1995;18:317–34. [PubMed] [Google Scholar]
- 18.Lopez C, Stahl D, Tchanturia K. Estimated intelligence quotient in anorexia nervosa: a systematic review and meta-analysis of the literature. Ann Gen Psychiatry. 2010;9:40. doi: 10.1186/1744-859X-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz Z, Hardaway JA, Bulik CM. Genetics and epigenetics of eating disorders. Adv Genomics Genet. 2015;5:131–50. doi: 10.2147/AGG.S55776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of Anorexia Nervosa. Am J Psychiatry. 2017;174:850–8. doi: 10.1176/appi.ajp.2017.16121402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson HJ, Yilmaz Z, Thornton LM, Hübel C, Coleman JRI, Gaspar HA, et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019;51:1207–14. doi: 10.1038/s41588-019-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell. 2019;179:1469-.e11. [DOI] [PMC free article] [PubMed]
- 23.Yilmaz Z, Halvorsen M, Bryois J, Yu D, Thornton LM, Zerwas S, et al. Examination of the shared genetic basis of anorexia nervosa and obsessive–compulsive disorder. Mol Psychiatry. 2020;25:2036–46. doi: 10.1038/s41380-018-0115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lilenfeld LR, Kaye WH, Greeno CG, Merikangas KR, Plotnicov K, Pollice C, et al. A controlled family study of anorexia nervosa and bulimia nervosa: psychiatric disorders in first-degree relatives and effects of proband comorbidity. Arch Gen Psychiatry. 1998;55:603–10. doi: 10.1001/archpsyc.55.7.603. [DOI] [PubMed] [Google Scholar]
- 25.Logue CM, Crowe RR, Bean JA. A family study of anorexia nervosa and bulimia. Compr Psychiatry. 1989;30:179–88. doi: 10.1016/0010-440x(89)90071-0. [DOI] [PubMed] [Google Scholar]
- 26.Grigoroiu-Serbanescu M, Magureanu S, Milea S, Dobrescu I, Marinescu E. Modest familial aggregation of eating disorders in restrictive anorexia nervosa with adolescent onset in a Romanian sample. Eur Child Adolesc Psychiatry. 2003;12:i47–53. doi: 10.1007/s00787-003-1107-7. [DOI] [PubMed] [Google Scholar]
- 27.Strober M, Freeman R, Lampert C, Diamond J. The association of anxiety disorders and obsessive compulsive personality disorder with anorexia nervosa: evidence from a family study with discussion of nosological and neurodevelopmental implications. Int J Eat Disord. 2007;40:S46–51. doi: 10.1002/eat.20429. [DOI] [PubMed] [Google Scholar]
- 28.Smeland OB, Frei O, Fan CC, Shadrin A, Dale AM, Andreassen OA. The emerging pattern of shared polygenic architecture of psychiatric disorders, conceptual and methodological challenges. Psychiatr Genet. 2019;29:152–9. doi: 10.1097/YPG.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreassen OA, Djurovic S, Thompson WK, Schork AJ, Kendler KS, O’Donovan MC, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92:197–209. doi: 10.1016/j.ajhg.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeland OB, Frei O, Shadrin A, O’Connell K, Fan CC, Bahrami S, et al. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 2020;139:85–94. doi: 10.1007/s00439-019-02060-2. [DOI] [PubMed] [Google Scholar]
- 32.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29. doi: 10.1038/s41588-021-00857-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward J, Tunbridge EM, Sandor C, Lyall LM, Ferguson A, Strawbridge RJ, et al. The genomic basis of mood instability: identification of 46 loci in 363,705 UK Biobank participants, genetic correlation with psychiatric disorders, and association with gene expression and function. Mol Psychiatry. 2020;25:3091–9. doi: 10.1038/s41380-019-0439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7. doi: 10.1038/s41588-018-0151-7. [DOI] [PubMed] [Google Scholar]
- 37.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B, Loh PR, Finucane HK, Ripke S, Yang J. Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015 Oct;526:68–74. [DOI] [PMC free article] [PubMed]
- 42.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma Oxf Engl. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–94. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–34. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11:1889–907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
- 48.Bahl E, Koomar T, Michaelson JJ. cerebroViz: an R package for anatomical visualization of spatiotemporal brain data. Bioinforma Oxf Engl. 2017;33:762–3. doi: 10.1093/bioinformatics/btw726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melbourne JK, Rosen C, Feiner B, Sharma RP. C4A mRNA expression in PBMCs predicts the presence and severity of delusions in schizophrenia and bipolar disorder with psychosis. Schizophr Res. 2018;197:321–7. doi: 10.1016/j.schres.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monteleone AM, Mereu A, Cascino G, Criscuolo M, Castiglioni MC, Pellegrino F, et al. Re-conceptualization of anorexia nervosa psychopathology: a network analysis study in adolescents with short duration of the illness. Int J Eat Disord. 2019;52:1263–73. doi: 10.1002/eat.23137. [DOI] [PubMed] [Google Scholar]
- 53.Solmi M, Collantoni E, Meneguzzo P, Tenconi E, Favaro A. Network analysis of specific psychopathology and psychiatric symptoms in patients with anorexia nervosa. Eur Eat Disord Rev J Eat Disord Assoc. 2019;27:24–33. doi: 10.1002/erv.2633. [DOI] [PubMed] [Google Scholar]
- 54.Thornton LM, Welch E, Munn-Chernoff MA, Lichtenstein P, Bulik CM. Anorexia nervosa, major depression, and suicide attempts: shared genetic factors. Suicide Life Threat Behav. 2016;46:525–34. doi: 10.1111/sltb.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wade TD, Bulik CM, Neale M, Kendler KS. Anorexia nervosa and major depression: shared genetic and environmental risk factors. Am J Psychiatry. 2000;157:469–71. doi: 10.1176/appi.ajp.157.3.469. [DOI] [PubMed] [Google Scholar]
- 56.Miotto P, Pollini B, Restaneo A, Favaretto G, Sisti D, Rocchi MBL, et al. Symptoms of psychosis in anorexia and bulimia nervosa. Psychiatry Res. 2010;175:237–43. doi: 10.1016/j.psychres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 57.Ospina LH, Shanahan M, Perez-Rodriguez MM, Chan CC, Clari R, Burdick KE. Alexithymia predicts poorer social and everyday functioning in schizophrenia and bipolar disorder. Psychiatry Res. 2019;273:218–26. doi: 10.1016/j.psychres.2019.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li S, Zhang B, Guo Y, Zhang J. The association between alexithymia as assessed by the 20-item Toronto Alexithymia Scale and depression: a meta-analysis. Psychiatry Res. 2015;227:1–9. doi: 10.1016/j.psychres.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Thiebaut S, Godart N, Radon L, Courtet P, Guillaume S. Crossed prevalence results between subtypes of eating disorder and bipolar disorder: a systematic review of the literature. L’Encephale. 2019;45:60–73. doi: 10.1016/j.encep.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Caspi A, Houts RM, Belsky DW, Goldman-Mellor SJ, Harrington H, Israel S, et al. The p factor: one general psychopathology factor in the structure of psychiatric disorders? Clin Psychol Sci J Assoc. 2014;2:119–37. doi: 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holtkamp K, Müller B, Heussen N, Remschmidt H, Herpertz-Dahlmann B. Depression, anxiety, and obsessionality in long-term recovered patients with adolescent-onset anorexia nervosa. Eur Child Adolesc Psychiatry. 2005;14:106–10. doi: 10.1007/s00787-005-0431-5. [DOI] [PubMed] [Google Scholar]
- 62.Pollice C, Kaye WH, Greeno CG, Weltzin TE. Relationship of depression, anxiety, and obsessionality to state of illness in anorexia nervosa. Int J Eat Disord. 1997;21:367–76. doi: 10.1002/(sici)1098-108x(1997)21:4<367::aid-eat10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 63.Bulik CM, Sullivan PF, Tozzi F, Furberg H, Lichtenstein P, Pedersen NL. Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch Gen Psychiatry. 2006;63:305–12. doi: 10.1001/archpsyc.63.3.305. [DOI] [PubMed] [Google Scholar]
- 64.Stepan J, Anderzhanova E, Gassen NC. Hippo signaling: emerging pathway in stress-related psychiatric disorders? Front Psychiatry. 2018;9:715. doi: 10.3389/fpsyt.2018.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, Jho E-hoon. Merlin, a regulator of Hippo signaling, regulates Wnt/β-catenin signaling. BMB Rep. 2016;49:357–8. doi: 10.5483/BMBRep.2016.49.7.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shang Y, Yan Y, Chen B, Zhang J, Zhang T. Over-expressed MST1 impaired spatial memory via disturbing neural oscillation patterns in mice. Genes Brain Behav. 2020;19:e12678. doi: 10.1111/gbb.12678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are openly available from an online repository or are available on request from study authors. All code is freely available at https://github.com/precimed and https://github.com/bulik/ldsc. Analyses were conducted in Python v3.5, Matlab R2020b, and R v3.6.3. Locus definition, functional annotation, and gene-set analysis were performed using FUMA (https://fuma.ctglab.nl/).