Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder associated with impaired social behavior and communication, repetitive behaviors, and restricted interests. In addition to genetic factors, environmental factors such as prenatal drug exposure contribute to the development of ASD. However, how those prenatal factors induce behavioral deficits in the adult stage is not clear. To elucidate ASD pathogenesis at the molecular level, we performed a high-resolution mass spectrometry-based quantitative proteomic analysis on the prefrontal cortex (PFC) of mice exposed to valproic acid (VPA) in utero, a widely used animal model of ASD. Differentially expressed proteins (DEPs) in VPA-exposed mice showed significant overlap with ASD risk genes, including differentially expressed genes from the postmortem cortex of ASD patients. Functional annotations of the DEPs revealed significant enrichment in the Wnt/β-catenin signaling pathway, which is dysregulated by the upregulation of Rnf146 in VPA-exposed mice. Consistently, overexpressing Rnf146 in the PFC impaired social behaviors and altered the Wnt signaling pathway in adult mice. Furthermore, Rnf146-overexpressing PFC neurons showed increased excitatory synaptic transmission, which may underlie impaired social behavior. These results demonstrate that Rnf146 is critical for social behavior and that dysregulation of Rnf146 underlies social deficits in VPA-exposed mice.
Subject terms: Autism spectrum disorders, Proteomics
Autism spectrum disorder: Improving understanding at the molecular level
Increased levels of a protein that regulates neuronal signaling pathway, called Rnf146, are associated with social behavior deficits consistent with autism spectrum disorder (ASD) in a mouse model. Individuals with ASD show decreased sociability, repetitive behaviors, and restricted interests. Although both genetic and environmental factors are involved, the underlying molecular mechanisms are unclear. Yong-Seok Lee at Seoul National University in South Korea and co-workers investigated the mechanisms underlying ASD using a drug-induced mouse model. They found that the ASD-model mice showed deficits in social behaviors and had higher levels of Rnf146 compared to control mice. Boosting levels of Rnf146 in control mice produced ASD-like behavior. Further investigation showed that the ASD-model mice had increased excitatory synaptic transmission in the prefrontal cortex, which regulates social behavior. These results indicate a promising target for ASD research.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with a range of behavioral manifestations, such as impaired social interaction and communication, repetitive behaviors, and restricted interests1. Studies have been carried out to map risk genes, the alterations of which are associated with ASD development, and those results have been accumulated in the SFARI gene database (https://gene.sfari.org)2. Aside from genetic factors, pre- and postnatal environmental factors also play crucial roles in the generation of ASD phenotypes, especially impaired social behavior3,4.
Valproic acid (VPA), or 2-propyl pentanoic acid, is a short-chain fatty acid clinically used as a mood stabilizer and antiepileptic drug5. VPA is a well-known inducer of neural tube defects and its prenatal exposure highly likely causes teratogenic effects that often lead to autistic-like behavior and developmental disorders6. A distinct linkage between prenatal exposure to VPA and autism has been confirmed7–11. Clinical studies have reported that in utero exposure to VPA is associated with an increased risk of impaired embryonic neurodevelopment and cognitive development and a higher incidence of ASD in offspring12,13. Rodents exposed to prenatal VPA (hereafter, VPA-exposed rodents) are widely considered animal models of ASD associated with prenatal environmental risk factors that allow investigation of its underlying neurobiological mechanism11. For example, VPA-exposed rodents showed autistic-like behaviors, such as decreased social interaction, repetitive behaviors, high pain sensitivity, and increased basal anxiety14–16. VPA-exposed rodents also exhibit alterations seen in other autism mouse models, including changes in brain connectivity, an excitatory/inhibitory imbalance, aberrant dendritic spine development in the hippocampus and cerebellar cortical formation, and deformation of Purkinje cells16–21. These findings provide further evidence that the model of prenatal VPA exposure has face validity at the cellular level.
Previous studies have proposed that the wingless (Wnt)/β-catenin signaling pathway, which is critical for brain development and synaptic functions, is one of the convergent cellular processes altered in ASD22–24. Exposure to VPA has also been shown to activate the Wnt and mTOR pathways25. Correspondingly, downregulating canonical Wnt/β-catenin signaling ameliorated ASD-like behavioral phenotypes in VPA-exposed rats26. VPA was also shown to increase the proliferation of neural progenitor cells by activating Wnt/β-catenin signaling during development9. However, the molecular mechanism underlying VPA-induced activation of the Wnt/β-catenin signaling pathway in the adult brain and the causality between Wnt/β-catenin signaling dysregulation and behavioral deficits are unclear.
Recent advances in omics technologies have enabled us not only to identify dysregulated molecules but also to gain a comprehensive understanding of the molecular mechanisms underlying ASD27,28. Beyond global gene expression profiling, single-cell genomics techniques have been employed to investigate the molecular pathophysiology of ASD in a cell type-specific manner29. Recently, mass spectrometry (MS)-based proteomic techniques, which allow us to directly study molecular networks in tissues or cells at the protein level30,31, have emerged as a valuable tool for identifying molecular changes associated with ASD in various model systems, including mice, humans, and forebrain organoids28,32–34. However, limited research has focused on identifying dysregulated proteins and signaling networks in VPA-exposed mice.
While the prefrontal cortex (PFC) is critical for regulating social behaviors and dysregulation of the PFC is often observed in humans with ASD as well as mouse models, including VPA-exposed rodents35–37, it is still unknown how prenatal VPA exposure affects the mouse PFC proteome, which may underlie ASD-like behavioral manifestations. Therefore, in this study, we employed a high-resolution mass spectrometry (MS)-based quantitative proteomic approach to identify differentially expressed proteins (DEPs) and altered signaling pathways in the PFC of VPA-exposed mice. We found that the expression of the ring-type E3 ubiquitin transferase Rnf146, which is a key regulator of the Wnt/β-catenin signaling pathway, is increased in the PFC of VPA-exposed mice. Furthermore, overexpression of Rnf146 in the PFC impairs excitatory synaptic transmission and social behavior in adult mice, demonstrating that Rnf146 upregulation contributes to social behavior deficits in a VPA-induced mouse model of ASD.
Materials and methods
Mice
CD1 (ICR) mice (CriOri; OrientBio, Korea) were used for VPA-exposed mouse preparation as previously described38. VPA (350 mg/kg, 10 ml/kg) or vehicle (0.9% saline) was subcutaneously injected into pregnant mice over the interscapular region at E10. Social behavior tests were performed 6-7 weeks after birth. Six-week-old C57BL/6N male mice (C57BL/6NCrljOri; OrientBio, Korea) were used for the prefrontal Rnf146 overexpression experiments. Rnf146- or eGFP-overexpressing mice were subjected to behavioral tests 4 weeks after viral injection. Animals were reared with a standard 12 h light/12 h dark cycle. All animal experiments were approved by the Institutional Animal Care and Use Committee of KonKuk University (KU18054) and Seoul National University (SNU-180521-3-6).
Behavioral tests
Social behaviors were examined using the 3-chamber test as previously described38. Please see the supplementary methods for details regarding the behavioral tests.
Tissue sampling
PFC samples were collected from VPA-treated or control mice one week after the behavioral tests and used for proteomics and western blot analyses. Brain tissues for western blotting were dissected from Rnf146- or eGFP-overexpressing mice 2–3 weeks after the behavioral tests. For RNA-seq, the PFC from Rnf146- or eGFP-overexpressing mice was dissected 4 weeks after viral injection. Mice were sacrificed under 5% isoflurane anesthesia. Brain tissues from mice were immediately dissected and frozen on dry ice and then stored at −80 °C until further use.
Proteomic data generation and analysis
Quantitative tandem mass spectrometry data were acquired as previously described28,39. Briefly, PFC proteins were extracted from VPA-exposed (n = 4) and control mice (n = 4) and were enzymatically digested by sequencing-grade trypsin. Peptides were chemically labeled using TMT reagents, and pooled TMT-barcoded peptides were fractionated by high pH reversed-phase liquid chromatography. A peptide mixture in each fraction was analyzed with a high-resolution Orbitrap mass spectrometer (Thermo Scientific, USA) in data-dependent acquisition mode.
The tandem mass spectra obtained were analyzed with MaxQuant software as previously described40. Identified peptides and proteins were normalized to carry out principal component analysis, multiscatter plot analysis, and hierarchical clustering analysis. Please see the supplementary methods and Supplementary Fig. 2 for more details regarding proteomic data generation and analysis.
Enrichment analyses of DEPs
Proteins with significant differences in expression in VPA-exposed mice compared to control mice (p value ≤ 0.05) were considered VPA-DEPs. Using the homolog database from Mouse Genome Informatics (http://www.informatics.jax.org/homology.shtml), we converted the mouse protein list into a list of human homologs; mouse proteins without human homologs were excluded from the analysis. Please see the supplementary methods for a detailed description of the enrichment analyses.
Functional mapping and protein‒protein interaction (PPI) network
We evaluated the biological functions of the VPA-DEPs in terms of the pathways from the MsigDB Gene Ontology and Reactome Database41 using the EnrichR tool42. Please see the supplementary methods for a detailed description of the functional mapping and PPI network.
RNA sequencing of DEGs in Rnf146-overexpressing mice
Raw reads were processed with Cutadapt43 for Illumina adapter trimming and removal of low-quality reads. We removed 15 bp of each read for low quality (q < 30) and discarded reads shorter than 100 base pairs. After a quality check with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), we quantified the abundance of transcripts using Salmon software (v1.9.0)44 in quasi-mapping-based mode referring to the mouse reference transcriptome (GENCODE vM30, GRCm39). Please see the supplementary methods for details.
WGCNA network construction and module identification
To identify the functional topology in samples, weighted gene coexpression network analysis (WGCNA, version 1.71) was applied to transcriptomics data. Please see the supplementary methods for details.
Viral vector construction and AAV packaging
An adeno-associated viral (AAV) vector for neuronal expression of Rnf146 (pAAV-hsyn-mRnf146-T2A-eGFP-WPRE) was constructed by recombinant PCR. The AAV was prepared as previously described45. Please see the supplementary methods for a detailed description of vector construction and AAV packaging.
Surgical procedures
Surgical procedures were performed as previously described46. After anesthetization with a mixture of Zoletil (30 mg/kg) and Rompun (10 mg/kg) via i.p. injection, mice were placed on the rodent stereotaxic apparatus. AAV (5 × 1012 GC/mL, 200 nL in ACSF) was bilaterally injected into the PFC (AP + 2.4 from bregma, ML ± 0.5 from midline, DV −2.4 and −1.8 from skull) using a Nanolitre 2010 (WPI, USA) at a speed of 20 μL/min. Animals were given at least 3 weeks of rest before further experiments.
Western blot analysis
Total protein lysates (10 µg) were separated on a 10% gel by SDS‒PAGE and transferred to a nitrocellulose membrane (Bio-Rad, USA). After blocking in 5% skim milk or BSA, membranes were incubated with primary antibodies (phospho-β-catenin, #9561, Cell Signaling; β-catenin, #9582, Cell Signaling; RNF146, NBP1-85318, NOVUS; α-Tubulin, sc-8035, Santa Cruz) overnight at 4 °C. After incubation with appropriate HRP-conjugated secondary antibodies, chemiluminescent signals were detected by AI600 (Amersham™, USA) using ECL solution (32209; Pierce, USA).
Statistical analysis
A two-tailed unpaired t test was used for the statistical comparisons of the preference index (PI) and protein expression levels of VPA- and vehicle-exposed mice. Statistical analyses for behavioral tests and western blot analyses were carried out using GraphPad Prism v7 software. All data are shown as the mean ± SEM.
Results
Social behavior deficits in VPA-exposed mice
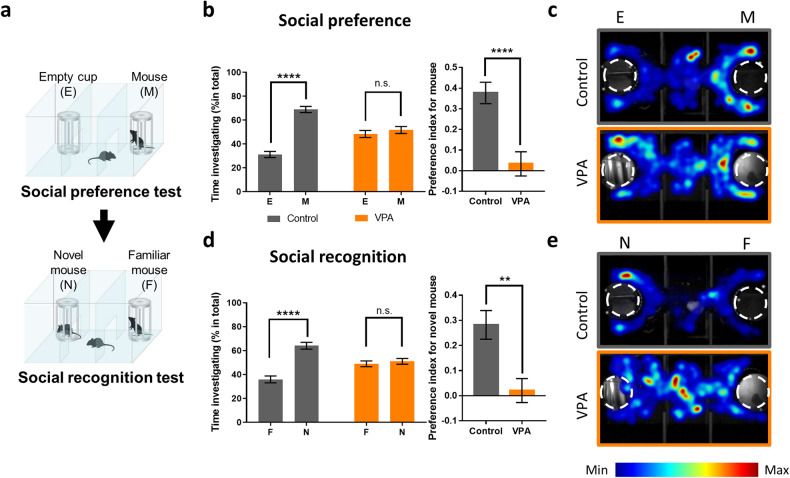
Studies have established that in utero exposure to VPA induces social behavior deficits in offspring14,38. The three-chamber test has been widely used to examine social behaviors in mice47. Since mice are social animals, they prefer to spend more time interacting with a conspecific than exploring an object or empty cup in the three-chamber test (Fig. 1a)47. We confirmed that VPA-exposed mice did not show a preference for the conspecific over the empty cup and subsequently showed a significantly lower social preference index than control mice (Fig. 1b, c). In addition, VPA-exposed mice did not show a preference for a novel conspecific over a familiar conspecific, while control mice showed a significant preference for the novel conspecific (Fig. 1d, e). In addition, we confirmed that in utero exposure to VPA induces teratogenic effects. We observed that VPA-exposed dams gave birth to significantly smaller litters than control dams (Supplementary Fig. 1a), and most VPA-exposed offspring exhibited a crooked tail phenotype resulting from neural tube defects (Supplementary Fig. 1b, c)6. Notably, we found a significant correlation between the social preference index and the angles of the crook point in VPA-exposed mouse tails (Supplementary Fig. 1d), providing further evidence that prenatal VPA exposure induces developmental deficits in offspring. After evaluating these behavioral and morphological phenotypes, the prefrontal cortex (PFC) of VPA- and vehicle-exposed mice was dissected for proteomic analysis.
Fig. 1. Social deficits in VPA-exposed mice.
a Schematic diagram of the 3-chamber social behavior test. The social preference and recognition of vehicle (control)- or VPA-exposed mice were evaluated. b Left: in the social preference test, VPA-exposed mice did not show a significant preference for a novel conspecific (M) over an empty cup (E), while control mice showed a significant preference for the novel conspecific (n = 19 mice for control, n = 20 mice for VPA, two-way ANOVA: VPA, F1, 74 = 2.290e−014, P > 0.9999; target, F1, 74 = 54.32, ****P < 0.0001; interaction, F1, 74 = 38.2, ****P < 0.0001; Sidak’s multiple comparisons test: control, mouse versus object t74 = 9.461, ****P < 0.0001; VPA, mouse versus object t74 = 0.8525, P = 0.6360). Right: preference index (PI) of control and VPA-exposed mice. VPA-exposed mice showed significantly lower PI values than control mice (n = 19 for control, n = 20 for VPA, two-tailed unpaired t test; t37 = 4.37, ****P < 0.0001). c Representative heatmap images of the social preference test of control (upper) and VPA-exposed mice (lower). d Left: in the social recognition test, VPA-exposed mice did not show a significant preference for a novel conspecific (N) versus a familiar conspecific (F), while control mice showed a significant preference for the novel conspecific (n = 19 mice for control, n = 20 mice for VPA, two-way ANOVA: VPA, F1, 74 = 0.000, P > 0.9999; target, F1, 74 = 33.36, ****P < 0.0001; interaction, F1, 74 = 24.82, ****P < 0.0001; Sidak’s multiple comparisons test: control, novel versus familiar t74 = 7.511, ****P < 0.0001; VPA, novel versus familiar t74 = 0.5683, P = 0.8164). Right: PI of control and VPA-exposed mice. VPA-exposed mice showed significantly lower PI values than control mice (n = 19 for control, n = 20 for VPA, two-tailed unpaired t test; t37 = 4.523, **P = 0.0012). e Representative heatmap images of the social recognition test of control mice (upper) and VPA-exposed mice (lower).
Proteomic analysis in VPA-exposed mice
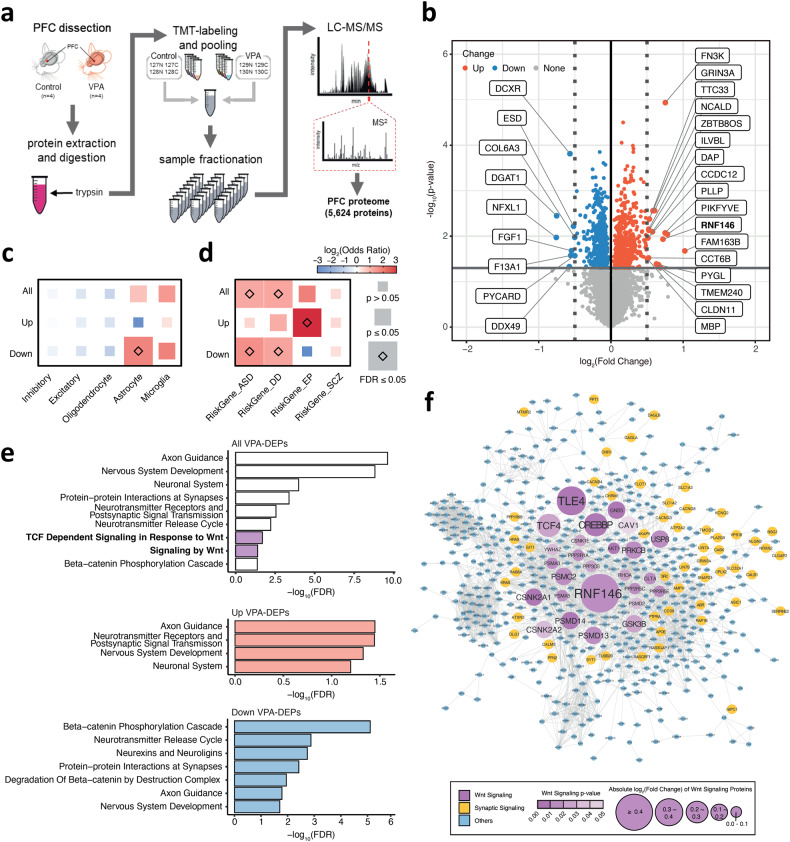
To characterize the effects of VPA on molecular signaling, we conducted a TMT-based high-throughput quantitative proteomic analysis using a high-resolution Orbitrap mass spectrometer (Fig. 2a). A total of 5624 proteins were quantitatively identified with ~17.5% median sequence coverage (Fig. 2a and Supplementary Fig. 2a). After quantile normalization (Supplementary Fig. 2b), we carried out principal component analysis, which showed a distinct separation between VPA-exposed and control brain tissues (Supplementary Fig. 2c). Pearson correlation coefficients between these samples had the lowest value of 98.7%, demonstrating good reproducibility between biological replicates (Supplementary Fig. 2d). The hierarchical clustering of DEPs was distinct, as shown in a heatmap visualizing the differences in protein expression (Supplementary Fig. 2e).
Fig. 2. VPA-exposed mice show upregulation of the Wnt signaling pathway.
a The overall design of the proteomic experiment. The dissected PFC tissues were homogenized and sonicated, and tryptic digestion was performed to break proteins down into peptides, which were further labeled with TMT isobaric labeling of 8 samples for 24 fractionations, followed by mass spectrometry quantitative analysis. b Volcano plot showing differentially expressed proteins between VPA-exposed mice and control mice (VPA-DEPs). The x-axis represents the log2 scale of the fold change in differential expression, while the y-axis represents the −log10 scale of the significance in changes. VPA-DEPs exhibiting expression changes with an absolute log2-fold change equal to or greater than 0.05 are labeled in the plot. c Enrichment analysis of VPA-DEPs with genes specific to five distinct cell types, as defined by single-cell RNA-seq data from the mouse nervous system. The log2(odds ratio) values that exceeded the range of −3 to 3 were adjusted to fit within this range. The blue and red colors represent the log2(odds ratio) values, and the size of the box corresponds to the significance in enrichment. d Enrichment analysis of VPA-DEPs with disorder risk genes. The log2(odds ratio) values that exceeded the range of −3 to 3 were adjusted to fit within this range. The blue and red colors represent the log2(odds ratio) values, and the size of the box corresponds to the significance in enrichment. Autism spectrum disorder (ASD), developmental disorder (DD), epilepsy (EP), and schizophrenia (SCZ). e Functional annotations for VPA-DEPs showing significantly enriched biological pathways (FDR ≤ 0.05). The x-axis represents the −log10 scale of significance in enrichment. The purple highlighted bars indicate the pathway associated with Wnt signaling enriched with all VPA-DEPs. The enriched pathways for upregulated and downregulated VPA-DEPs are highlighted in red and blue, respectively. f Protein–protein interaction (PPI) network of VPA-DEPs revealing proteins related to Wnt signaling and neuronal development.
Among the 5624 identified proteins, 941 proteins were identified as DEPs in VPA-exposed mice (VPA-DEPs) compared to controls, including 460 upregulated and 481 downregulated DEPs (Fig. 2b, Supplementary Table 1a). To determine the cellular specificity of VPA-DEPs for the mouse nervous system, we conducted an enrichment test using cell-type marker genes across the central nervous system (Fig. 2c, Supplementary Table 1b)48. We found that downregulated VPA-DEPs were significantly enriched in astrocyte marker genes (odds ratio [OR] = 3.6, false discovery rate [FDR] = 1.9 × 10−4, one-sided Fisher’s exact test [FET]). Furthermore, these downregulated VPA-DEPs showed significant overlap with astrocytes in the human brain when compared with cell-type marker genes for the developing human brain cortex (OR = 3.7, FDR = 9.0 × 10−6, one-sided FET; Supplementary Fig. 3, Supplementary Table 1c)49. Taken together, these findings suggest potential decreases in the expression of astrocyte-specific proteins in the PFC by in utero VPA exposure.
We further evaluated the enrichment of VPA-DEPs with differentially expressed genes (DEGs) in the postmortem cortex of ASD patients. From Parikshak et al.50, we used two gene sets: (1) up- or downregulated DEGs enriched in ASD brains compared to control brains and (2) coexpressed network genes of 25 modules. We found a significant enrichment of downregulated VPA-DEPs in the cortex. M9 (OR = 2.4, FDR = 2.6 × 10−3, one-sided FET) (Supplementary Fig. 4, Supplementary Table 1d), which was an astrocyte-specific module associated with ASD brains. We further assessed whether VPA exposure alters the expression of known ASD or neurodevelopmental disorder (NDD) risk genes. To do this, we collated a list of high-confidence risk genes for ASD, developmental delay (DD), epilepsy (EP), and schizophrenia (SCZ), prioritized based on large-scale exome studies51–54. Notably, we found that downregulated VPA-DEPs were significantly enriched in ASD risk genes (OR = 3.0, FDR = 8.4 × 10−3, one-sided FET) and DD risk genes (OR = 2.4, FDR = 2.7 × 10−2, one-sided FET) (Fig. 2d, Supplementary Table 1e). Among them, several genes have been implicated in the Wnt/β-catenin signaling pathway. Crebbp is part of a transcriptional regulatory complex inhibiting the Wnt/β-catenin signaling pathway55. Synaptotagmin-1 (Syt1) was reported to be the Wnt target molecule for neurotransmitter release at excitatory synapses56. In addition, ubiquitin-specific peptidase 9 X-linked (USP9X) was shown to regulate the canonical and noncanonical Wnt pathways in cancer development via the deubiquitylation of DVL257. We also compared our findings with previous proteomics studies using other ASD mouse models, such as Cntnap2, Elp2, and Pten mutant mice (Supplementary Fig. 5, Supplementary Table 1f)28,33,34. The downregulated VPA-DEPs in our study showed significant overlap with upregulated DEPs in other ASD proteomics studies, primarily involving proteins associated with the neurodegeneration pathway (KEGG: hsa05022, OR = 15.5, p-value = 1.9 × 10−3).
We performed functional annotations and identified significant enrichment of VPA-DEPs in Wnt/β-catenin signaling-related pathways (Fig. 2e, Supplementary Table 1g–i). VPA-DEPs were significantly enriched for signaling by WNT (R-HSA-195721, OR = 1.8, FDR = 4.0 × 10−2) and TCF-dependent signaling in response to WNT (R-HSA-201681, OR = 2.2, FDR = 2.0 × 10−2). In particular, downregulated VPA-DEPs were significantly enriched for β-catenin-related pathways, including the beta-catenin phosphorylation cascade (R-HSA-196299, OR = 2.4, FDR = 7.8 × 10−6) and degradation of beta-catenin by the destruction complex (R-HSA-195253, OR = 7.9, FDR = 1.1 × 10−2). Among these pathway genes, the largest perturbation of protein expression was observed for RNF146 (log2(fold change) = 0.72; p-value = 1.2 × 10−2), which has been suggested to be a key regulator of Wnt/β-catenin signaling (Fig. 2f). This result implies that the upregulation of RNF146 mediates the VPA-induced modulation of the Wnt signaling pathway.
Verification of RNF146 upregulation in the PFC of VPA-exposed mice
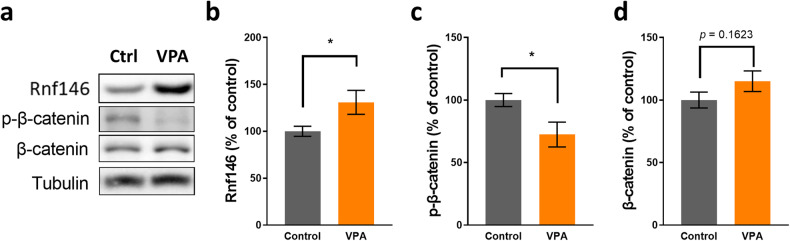
To verify the proteomics results of alterations in Wnt/β-catenin signaling, we examined the protein expression level of a selected target protein, Rnf146. Consistent with the proteomics data, the protein expression level of Rnf146 was significantly increased in the PFC of VPA-exposed mice compared to that in the vehicle-exposed control mice (Fig. 3a, b). Rnf146 positively regulates the Wnt/β-catenin pathway by inducing the degradation of Axin, which is the concentration-limiting component of β-catenin58. Axin forms a multiprotein β-catenin destruction complex with GSK3 and adenomatous polyposis coli (APC), which induces the phosphorylation and subsequent degradation of β-catenin59. Consistent with these findings, it has been shown that the activity of Wnt/β-catenin signaling is elevated in VPA-exposed mice9. Moreover, in the present experiment, phosphorylated β-catenin (p-β-catenin) was significantly decreased in the PFC of VPA-exposed mice compared with the control group, demonstrating alterations in the Rnf146-Wnt/β-catenin signaling pathway in VPA-exposed mice (Fig. 3c). However, while there was a mild trend of increase in β-catenin levels in VPA-exposed mice compared to control mice, the total amount of β-catenin was not significantly different between the two groups (Fig. 3d).
Fig. 3. VPA-exposed mice show increased Rnf146 expression.
a Representative western blot results. Ctrl, vehicle-exposed control mice; VPA, VPA-exposed mice. b VPA-exposed mice showed significantly increased Rnf146 levels compared to control mice (n = 14 mice for control, n = 13 mice for VPA, two-tailed unpaired t test; t25 = 2.283, *P = 0.0312). Rnf146 expression levels were normalized to tubulin levels. c VPA-exposed mice showed significantly decreased p-β-catenin levels, which were normalized to β-catenin levels, compared to control mice (n = 14 mice for control, n = 13 mice for VPA, two-tailed unpaired t test; t25 = 2.519, *P = 0.0185). d VPA-exposed mice showed comparable β-catenin levels, which were normalized to tubulin levels, when compared to control mice (n = 14 mice for control, n = 13 mice for VPA, two-tailed unpaired t test; t25 = 1.456, P = 0.1577).
Social behavior deficits in RNF146-overexpressing mice
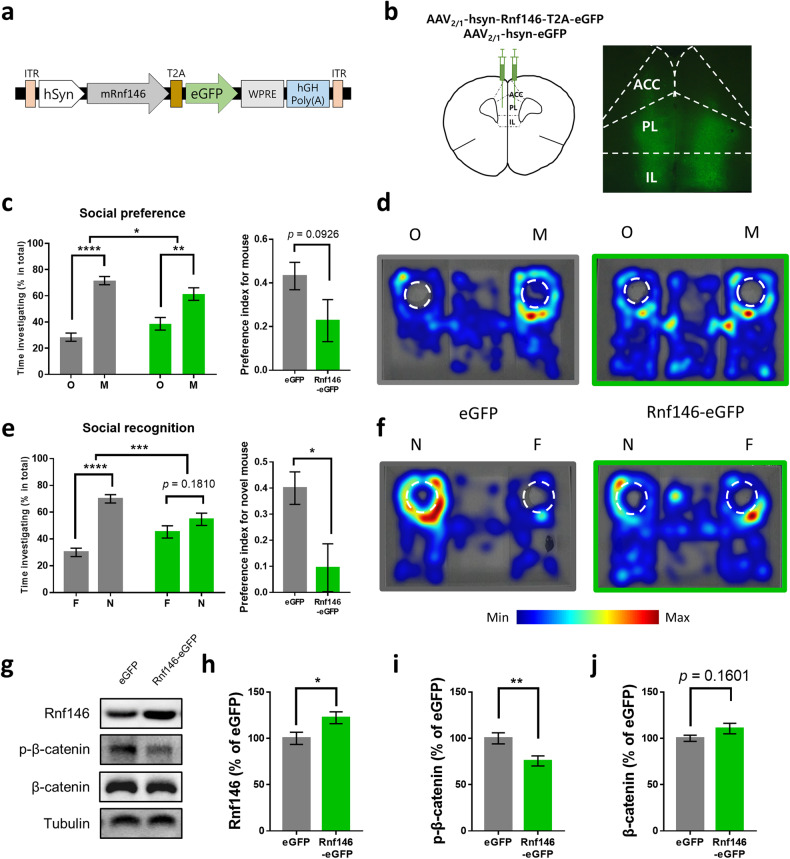
To examine whether the elevated expression of Rnf146 is associated with social behavior deficits in VPA-exposed mice, we overexpressed Rnf146 in the PFC of naïve C57BL/6 mice using an AAV (Fig. 4a, b). In the social preference test, both Rnf146- and eGFP-overexpressing mice spent significantly more time exploring a conspecific than an object, while the Rnf146-expressing mice showed a weaker social preference than GFP-expressing mice (Fig. 4c, d). Interestingly, however, Rnf146-overexpressing mice showed a deficit in the social recognition test, as they did not show a preference for a novel conspecific over a familiar conspecific, while control eGFP-overexpressing mice showed a significant preference for a novel conspecific (Figs. 4e, f). These results demonstrate that Rnf146 overexpression caused a severe deficit in social recognition with a mild impact on social preference in mice. Consistent with the results from VPA-exposed mice, the level of p-β-catenin was significantly decreased in the PFC of Rnf146-overexpressing mice compared with the control group, and the amount of total β-catenin tended to increase in Rnf146-overexpressing mice compared to that in eGFP-overexpressing mice (Fig. 4g–j). Rnf146-overexpressing mice showed basal locomotor activity and anxiety levels comparable to those of eGFP-overexpressing control mice (Supplementary Fig. 6).
Fig. 4. Rnf146 overexpression in the PFC causes social deficits in mice.
a Diagram for AAV constructs encoding mouse Rnf146 (mRNF146) and eGFP. ITR, inverted terminal repeat sequence; WPRE, Woodchuck hepatitis virus (WHV) posttranscriptional regulatory element. b Schematic diagram of bilateral AAV virus injection in the PFC (left) and an example of a brain slice expressing Rnf146 in the PFC (right). ACC, anterior cingulate cortex; PL, prelimbic cortex; IL, infralimbic cortex. c Left: In the social preference test, Rnf146-overexpressing mice showed a significant preference for a novel conspecific (M) versus an object (O) compared to eGFP-overexpressing control mice (n = 11 mice for eGFP, n = 11 mice for Rnf146, two-way ANOVA: Rnf146, F1, 40 = 0.000, P > 0.9999; target, F1, 40 = 65.87, ****P < 0.0001; interaction, F1, 20 = 6.025, *P = 0.0234; Sidak’s multiple comparisons test: eGFP, mouse versus object t40 = 7.52, ****P < 0.0001; Rnf146, mouse versus object t40 = 3.958, ***P = 0.0006). Right: preference index (PI) of eGFP- or Rnf146-overexpressing mice. Rnf146-overexpressing mice showed PI values comparable to those of control mice (n = 11 mice for eGFP, n = 11 mice for Rnf146, two-tailed unpaired t test; t20 = 1.781, P = 0.0901). d Representative heatmap images from eGFP-overexpressing mice (left) and Rnf146-overexpressing mice (right) of social behavior during the social preference test. e Left: in the social recognition test, Rnf146-overexpressing mice did not show a significant preference for a novel conspecific (N) versus a familiar conspecific (F), while eGFP-overexpressing control mice showed a significant preference for the novel conspecific (n = 11 mice for eGFP, n = 11 mice for Rnf146, two-way ANOVA: Rnf146, F1, 40 = 0, P > 0.9999; target, F1, 40 = 39.94, ****P < 0.0001; interaction, F1, 40 = 15.22, ***P = 0.0004; Sidak’s multiple comparisons test: eGFP, novel versus familiar t40 = 7.227, ****P < 0.0001; Rnf146, novel versus familiar t40 = 1.71, P = 0.1810). Right: PI of eGFP- or Rnf146-overexpressing mice. Rnf146-overexpressing mice showed significantly lower PI values than control mice (n = 11 mice for eGFP, n = 11 mice for Rnf146 group, two-tailed unpaired t test; t20 = 2.759, *P = 0.0121). f Representative heatmap images from eGFP-overexpressing mice (left) and Rnf146-overexpressing mice (right) of social behavior during the social recognition test. g Western blot analysis of the Wnt signaling pathway in the PFC of Rnf146-overexpressing mice. Samples from the PFC of GFP-overexpressing mice were used as controls. h Rnf146-overexpressing mice showed significantly increased Rnf146 levels (normalized to tubulin levels) compared to eGFP-overexpressing mice (n = 7 mice for eGFP, n = 10 mice for Rnf146, two-tailed unpaired t test; t14 = 2.381, *P = 0.0320). i Rnf146-overexpressing mice showed significantly decreased p-β-catenin levels (normalized to β-catenin levels) compared to eGFP-overexpressing mice (n = 7 mice for eGFP, n = 10 mice for Rnf146, two-tailed unpaired t test; t14 = 2.995, **P = 0.0096). j Rnf146-overexpressing mice showed a comparable β-catenin level (normalized to tubulin levels) compared to eGFP-overexpressing mice (n = 7 mice for eGFP, n = 10 mice for Rnf146, two-tailed unpaired t test; t14 = 1.483, P = 0.1601).
Altered Wnt/β-catenin signaling pathway in RNF146-overexpressing PFC
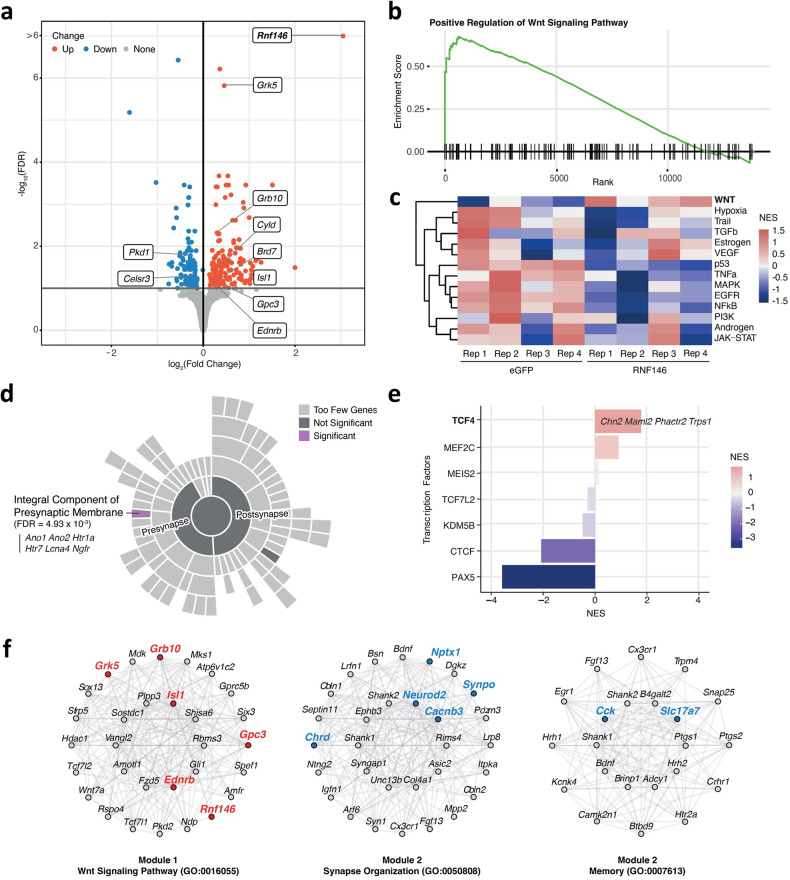
To investigate whether Rnf146 overexpression induces transcriptional changes in the PFC through Wnt/β-catenin signaling activation and to elucidate the molecular mechanism underlying the social deficits induced by Rnf146 overexpression, we conducted RNA sequencing analysis using mice that were confirmed to overexpress Rnf146 and exhibit social deficits. When compared with eGFP-overexpressing mice, Rnf146-overexpressing mice exhibited a total of 294 DEGs (Rnf146-DEGs) (FDR ≤ 0.1), some of which overlapped with VPA-DEPs (n = 11, Adgrb2, Chrm1, Fmnl1, Islr2, Mtmr2, Plppr1, Ripor2, Rnf146, Smpd4, Tmem132a, and Trir) (Fig. 5a and Supplementary Table 2a). Importantly, Rnf146 overexpression resulted in a significant enrichment with positive regulation of Wnt signaling pathways (GO:0030177; normalized enrichment score (NES) = 1.7, FDR = 4.9 × 10−2) (Fig. 5b and Supplementary Table 2b). Along with Rnf146, the Rnf146-DEGs that were mainly enriched in this pathway included Gpc3, which regulates the interaction between Wnt and Frizzled and leads to the activation of downstream signaling pathways60. We further calculated the pathway activity scores of 14 signaling processes in each sample. Rnf146-overexpressing mice showed elevated Wnt signaling pathway (NES = 1.5, p-value = 1.2 × 10−2) (Fig. 5c and Supplementary Table 2c). These results indicate that Rnf146 plays a pivotal role in modulating Wnt/β-catenin signaling in the mouse PFC.
Fig. 5. The Wnt signaling pathway is promoted by Rnf146 overexpression.
a Volcano plot showing differentially expressed genes between Rnf146-overexpressing mice and eGFP-overexpressing mice (Rnf146-DEGs). The x-axis represents the log2 scale of the fold change in differential expression, while the y-axis represents the -log10 scale of the significance changes. Rnf146-DEGs associated with the Wnt signaling pathway are labeled in the plot. b Gene set enrichment plot depicting the elevated positive regulation of the Wnt signaling pathway in Rnf146-overexpressing mice. c Heatmap of the pathway activity for each sample inferred by PROGENy. The levels of pathway activity reflect the extent of pathway deregulation. d Enrichment analysis of the upregulated Rnf146-DEGs with pathway terms describing synaptic locations in the brain. Pathway analysis was performed using one-sided Fisher’s exact test to identify significantly overrepresented terms. The upregulated Rnf146-DEGs significantly overlapped (FDR < 0.01) with the integral component of the presynaptic membrane pathway (purple). Pathway terms with two or fewer overlapping genes were excluded and indicated as too few genes. e Enrichment analysis of Rnf146-DEGs with the inferred activity of transcription factors associated with ASD. Upregulated target genes of the transcription factor TCF4 are annotated in the plot. NES, normalized enrichment score. f Network of genes in WGCNA module 1 enriched in the Wnt signaling pathway and genes in module 2 enriched in synapse organization and memory. In each network, the top 30 hub genes (22 for memory) with the highest levels of intramodular connectivity were depicted. Rnf146-DEGs are highlighted in bold, with red and blue colors indicating up- and downregulation, respectively.
To further explore whether Rnf146 overexpression alters the expression of synaptic molecules that may be associated with behavioral deficits, we analyzed 266 pathway terms of synaptic locations and processes from the SynGO database61. The upregulated Rnf146-DEGs (Ano1, Ano2, Htr1a, Htr7, Kcna4, and Ngfr) significantly overlapped with an integral component of the presynaptic membrane (GO:0099056, FDR = 4.9 × 10−3) (Fig. 5d, Supplementary Table 2d). As Wnt is known to regulate presynaptic assembly and neurotransmitter release across synapses, this finding supports the role of Rnf146 in Wnt/β-catenin signaling62. Moreover, regarding the transcription factors linked to ASD, Rnf146 overexpression significantly increased the activity of the transcription factor TCF4, which was accompanied by an upregulation of TCF4 target genes such as Chn2, Maml2, Phactr2 and Trps1 (NES = 1.8, p-value = 3.66 × 10−3) (Fig. 5e, Supplementary Table 2e). TCF4 binds to β-catenin and transactivates Wnt target genes, indicating that TCF4 could be the primary target of Rnf146 overexpression for activating the Wnt/β-catenin signaling pathway63. Overall, these findings suggest that Rnf146 plays a crucial role in regulating the Wnt/β-catenin signaling pathway in the PFC of mice.
To delineate the core gene network of Rnf146-overexpressing mice, we analyzed gene coexpression using Rnf146- and eGFP-overexpressing transcriptomics datasets. We found 51 coexpression modules with ~200 to 1200 genes (Fig. 5f and Supplementary Fig. 7, Supplementary Tables 2f, 2g, and 2h), representing biological pathways such as nervous system development (Module 1), synapse organization, and memory (Module 2). Module 1 was characterized by the Wnt signaling pathway (GO:0016055, FDR = 4.8 × 10−3), in which Rnf146 and its interacting genes (n = 36) were coexpressed. This module was significantly enriched in upregulated Rnf146-DEGs, implicating the core genes perturbed by Rnf146 overexpression (OR = 47.3, FDR = 9.2 × 10−108, one-sided FET). In contrast, Module 2, which was characterized by pathways associated with synapse organization (GO:0050808, FDR = 4.0 × 10−8) and memory (GO:0007613, FDR = 3.1 × 10−5), was significantly enriched in downregulated Rnf146-DEGs, including Cacnb3, Cck, Chrd, Neurod2, Nptx1, Slc17a7, and Synpo. Coexpressed genes of Module 2 also included Ube3b, which was significantly perturbed by Rnf146 overexpression and highly associated with intellectual disability (eigengene-based connectivity = 0.96) (Supplementary Table 2f)64,65.
Increased excitatory synaptic transmission in RNF146-overexpressing neurons
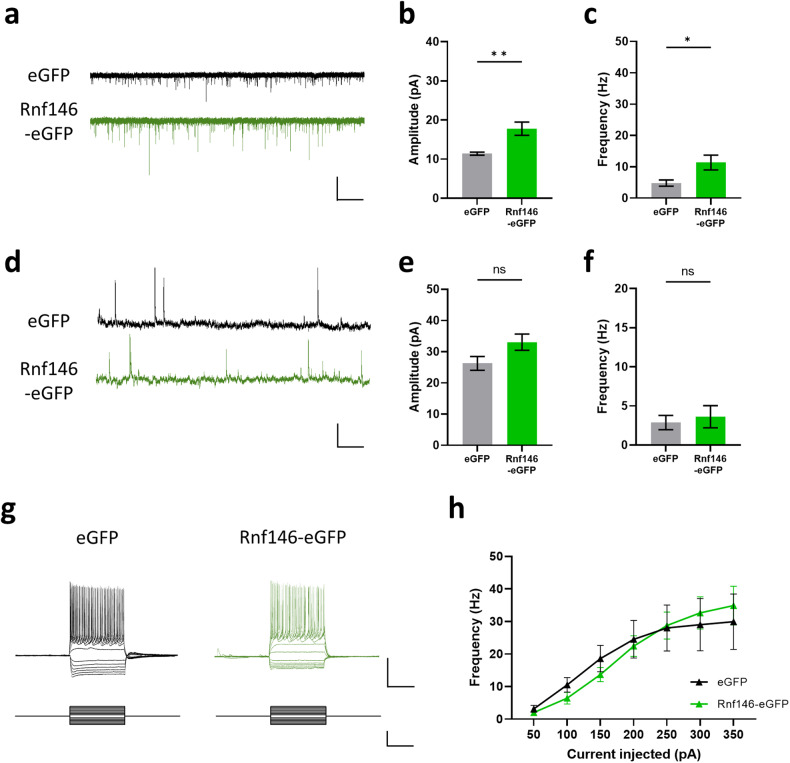
To further investigate the neuronal mechanism underlying impaired social behaviors in Rnf146-overexpressing mice, we performed whole-cell patch-clamp recordings. Previous studies have shown that electrophysiological properties are altered in the PFC of VPA-exposed mice38,66,67. We found that both the amplitude and frequency of spontaneous excitatory postsynaptic currents (sEPSCs) were significantly increased in Rnf146-expressing neurons compared to eGFP-expressing control neurons (Fig. 6a–c), whereas there was no significant change in spontaneous inhibitory postsynaptic currents (sIPSCs) (Fig. 6d–f). Finally, Rnf146 overexpression did not affect neuronal excitability (Fig. 6g, h) or other electrophysiological properties (Supplementary Table 3). These data suggest that Rnf146 overexpression may cause social behavior deficits by altering the excitation/inhibition balance in neurons of the PFC.
Fig. 6. Rnf146 overexpression increases excitatory synaptic transmission in prefrontal pyramidal neurons.
a Spontaneous excitatory postsynaptic current (sEPSC) traces of prefrontal neurons in Rnf146- and eGFP-overexpressing mice. Scale bar: 50 pA, 500 ms. b The sEPSC amplitude of prefrontal neurons in Rnf146-overexpressing mice was significantly larger than that in eGFP-overexpressing mice (n = 13 cells from 5 mice for eGFP, n = 16 cells from 6 mice for Rnf146; two-tailed unpaired t test, t27 = 3.302, **P = 0.0027). c The sEPSC frequency of prefrontal neurons in Rnf146-overexpressing mice was significantly larger than that in eGFP-overexpressing mice (n = 13 cells from 5 mice for eGFP, n = 16 cells from 6 mice for Rnf146; two-tailed unpaired t test, t27 = 2.373, *P = 0.0250). d Spontaneous inhibitory postsynaptic current (sIPSC) traces of prefrontal neurons in Rnf146- and eGFP-overexpressing mice. Scale bar: 100 pA, 500 ms. e The sIPSC amplitude of prefrontal neurons in Rnf146-overexpressing mice is comparable to that in eGFP-overexpressing mice (n = 11 cells from 4 mice for eGFP, n = 12 cells from 4 mice for Rnf146; two-tailed unpaired t test, t21 = 1.980, P = 0.0609). f The sIPSC frequency of prefrontal neurons in Rnf146-overexpressing mice is comparable to that in eGFP-overexpressing mice (n = 11 cells from 4 mice for eGFP, n = 12 cells from 4 mice for Rnf146; two-tailed unpaired t test, t21 = 0.4323, P = 0.6699). g Representative traces of voltage responses of prefrontal neurons in Rnf146- or eGFP-overexpressing mice evoked by current step stimuli (−300-300 pA, 50 pA increment, 1 s). Scale bar: upper, 50 mV, 500 ms; lower, 500 pA, 500 ms. h Summary data of the number of action potentials evoked in response to 300 pA current steps. The number of AP spikes was comparable in the prefrontal neurons of Rnf146-overexpressing mice relative to those of eGFP-overexpressing mice (n = 13 cells from 5 mice for eGFP, n = 16 cells from 5 mice for Rnf146; two-way ANOVA with repeated measures: effect of injected current, F6, 162 = 28.24, ****P < 0.0001; effect of Rnf146 expression, F1, 27 = 0.006249, P = 0.9376; interaction between current injection and Rnf146 expression, F6, 162 = 0.7368, P = 0.6207, Sidak’s multiple comparison test for eGFP versus Rnf146. 50 pA, t189 = 0.1724, P > 0.9999; 100 pA, t189 = 0.6127, P = 0.9957; 150 pA, t189 = 0.7362, P = 0.9870; 200 pA, t189 = 0.3139, P > 0.9999; 250 pA, t189 = 0.1120, P > 0.9999; 300 pA, t189 = 0.5416, P = 0.9980; 350 pA, t189 = 0.7398, P = 0.9867).
Discussion
ASD can be caused by environmental factors, including prenatal drug exposure. One of the most widely used mouse models of ASD is prenatal VPA exposure; these mice exhibit an ASD-like phenotype, including social deficits. Despite several proteomic studies in a similar context, no study has reported quantitative proteomic analysis of the PFC of the VPA-exposed mouse model of ASD. Here, we utilized a TMT-based quantitative proteomic approach to identify novel molecular pathways underlying chemical-induced ASD pathophysiology. We identified Rnf146 as a hub protein responsible for the activation of the Wnt/β-catenin signaling pathway in VPA-exposed mice. Rnf146 overexpression in naïve adult mice impaired social behaviors, which may be caused by increased excitatory synaptic transmission. Consistent with these findings, our transcriptome analysis of the PFC of Rnf146-overexpressing mice revealed alterations in the Wnt signaling pathway. This study showed, for the first time, that Rnf146 plays a key role in modulating social behavior.
We found a significant overlap between DEPs in the PFC of VPA-exposed mice (VPA-DEPs) and DEGs in the postmortem cortex of patients with ASD. Additionally, the downregulated VPA-DEPs were significantly enriched in previously identified ASD and DD risk alleles. Moreover, when comparing our proteomics data with other ASD mouse models, such as Cntnap2, Elp2, and Pten mutant mice, we observed significant overlaps among DEPs. However, it is interesting that downregulated VPA-DEPs in our study showed significant overlap with upregulated DEPs in other ASD models28,33,34. This suggests an opposite direction of pathophysiology among ASD mouse models and a potentially distinct pathway in VPA-exposed mice. Importantly, only VPA-exposed mice showed Rnf146 as a DEP.
Rnf146 is an E3 ubiquitin-protein ligase that prevents the formation of the β-catenin destruction complex by inducing Axin degradation58,68. Research has mainly focused on its role in the pathogenesis of cancer or other neurodegenerative disorders, such as Parkinson’s disease69,70. Other proteins engaged in the Wnt signaling pathway, such as BTRC and DIXDC1, have been reported as ASD candidate genes71–73; however, RNF146 was suggested as an ASD candidate gene only recently74. Our proteomic analyses of VPA-exposed mice and behavioral data from Rnf146-overexpressing mice suggest that Rnf146 is a strong candidate molecule responsible for social deficits associated with ASD.
The Wnt signaling pathway is critically involved in early vertebrate development, such as embryonic patterning and synaptic formation75–77. Thus, dysregulation of the Wnt/β-catenin signaling pathway is associated with many neurodevelopmental disorders, including autism22,24,75. Consistent with our finding that Wnt/β-catenin signaling is activated in VPA-exposed mice, upregulation of the Wnt signaling pathway has been reported to cause ASD-like behavioral phenotypes in rodents. For example, heterozygous GSK3β-knockout mice showed altered sociability and anxiety levels, paralleling the changes associated with chronic lithium administration, which activates Wnt/β-catenin signaling78,79. In utero injection of XAV939, a tankyrase inhibitor that subsequently activates β-catenin signaling, induced autism-like behaviors such as impaired sociability in mice80. A recent study reported that exposing a human organoid to VPA led to decreased expression of Lhx9, which has been suggested to negatively regulate Wnt signaling81,82. However, downregulation of the Wnt/β-catenin signaling pathway was also proposed as a candidate etiology of ASD in mouse models such as Fmr1-/y mice83,84. These results suggest that bidirectional dysregulation of Wnt signaling is responsible for the etiology of ASD.
In addition to its role in development, Wnt signaling is also implicated in the adult brain in functions ranging from adult neurogenesis to synaptic plasticity85,86. Our finding that overexpressing Rnf146 in the adult PFC is sufficient to induce deficits in social behavior in mice strongly demonstrates the critical role of Wnt/β-catenin signaling in the adult PFC in regulating synaptic functions and social behaviors. We found that PFC neurons overexpressing Rnf146 displayed increased excitatory synaptic transmission, which is consistent with previous findings in VPA-exposed mice38. This increase in the amplitude and frequency of EPSCs without changes in IPSCs may lead to an E/I imbalance in the PFC of Rnf146-overexpressing mice, which in turn causes social deficits. Changes in excitatory synaptic transmission are highly correlated with the activated Wnt/β-catenin signaling pathway since this pathway contributes to the formation and functional regulation of central synapses87. Consistently, the AMPA-mediated synaptic current was shown to be regulated by postsynaptic β-catenin88.
Furthermore, our transcriptome analysis of the Rnf146-overexpressing PFC suggests that the expression of genes involved in synaptic organization and memory is significantly changed, which may underlie the observed increased excitatory transmission and social deficits in these mice. Interestingly, the expression of a component of the bone morphogenic protein (BMP) signaling pathway, Chrd, for which null mice showed an increased frequency of excitatory synaptic transmission, was significantly decreased in the Rnf146-overexpressing PFC (Supplementary Table 2a)89. Moreover, the expression of Ube3b, which is associated with intellectual disability64,65, was significantly decreased by Rnf146 overexpression (Supplementary Table 2a).
In this study, we used only male mice for proteomics and Rnf146 overexpression experiments because previous studies have shown that VPA exposure does not affect social behaviors in females90,91. In addition, Rnf146 was listed as one of the male-specific ASD candidate genes in a previous study74. Nevertheless, it would be intriguing to test whether Rnf146 overexpression in the PFC affects social behaviors in female mice in future studies.
Overall, our results provide novel insight into the molecular mechanism of prenatal drug exposure-associated ASD and suggest that the Rnf146-Wnt-β-catenin signaling pathway is a promising target to alleviate social deficits in ASD, even in adults.
Supplementary information
Acknowledgements
This study was supported by grants from the National Research Foundation of Korea (NRF-2017M3C7A1026942 to Y.-S.L. and M.-S.K.; NRF-2018R1A5A2025964 to Y.-S.L.; NRF-2019M3E5D3073568 and NRF-2020R1C1C1003426 to J.-Y.A.; NRF-2022R1A2C2013377 to M.-S.K.; and NRF-2018H1A2A1061381 to G.P.) and Korea University (to J.-Y.A.) and Korea Basic Science Institute research program (C270100 to J.Y.K.).
Author contributions
Y.-S.L., M.-S.K. C.Y.S., and J.-Y.A. designed and supervised the research. G.P., S.J.J., E.L.G., M.K., and S.K. prepared the VPA mice and performed behavioral analyses. G.P. was responsible for cloning, packaging the AAVs, stereotaxic injections, western blotting, and patch clamp recording. W.E.J., G.B., and J.Y.K. performed proteomic sample preparation and data acquisition. W.E.J., S.K., J.H.P., Y.K., K.P.K., and M.-S.K. analyzed the proteomic data and interpreted the proteomic results. S.K. and J.J. analyzed the transcriptomic data and performed the functional enrichment analysis. S.K. and S.C. conducted coexpression network and module enrichment analyses. W.E.J., J.H.P. and G.P. created the figures. W.E.J., G.P., S.K., Y.-S.L., M.-S.K., and J.-Y.A. wrote the manuscript with the assistance of the others.
Data availability
All the data necessary to reach the conclusions of this paper are presented in the paper and/or the Supplementary Materials. The raw proteome MS data and MaxQuant search results have been deposited in the PRIDE database (project accession: PXD036413). Sequencing data have been deposited in GEO under accession code GSE215871.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gaeun Park, Wooyoung Eric Jang, Seoyeon Kim.
Contributor Information
Chan Young Shin, Email: chanyshin@kku.ac.kr.
Joon-Yong An, Email: joonan30@korea.ac.kr.
Min-Sik Kim, Email: mkim@dgist.ac.kr.
Yong-Seok Lee, Email: yongseok7@snu.ac.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s12276-023-01065-2.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5™ 5th ed. (American Psychiatric Publishing, Inc., 2013).
- 2.Fischbach GD, Lord C. The Simons simplex collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Ehninger D, et al. Gestational immune activation and Tsc2 haploinsufficiency cooperate to disrupt fetal survival and may perturb social behavior in adult mice. Mol. Psychiatry. 2012;17:62–70. doi: 10.1038/mp.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed MD, et al. IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature. 2020;577:249–253. doi: 10.1038/s41586-019-1843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perucca E. Pharmacological and therapeutic properties of valproate: a summary after 35 years of clinical experience. CNS Drugs. 2002;16:695–714. doi: 10.2165/00023210-200216100-00004. [DOI] [PubMed] [Google Scholar]
- 6.Vorhees CV. Teratogenicity and developmental toxicity of valproic acid in rats. Teratology. 1987;35:195–202. doi: 10.1002/tera.1420350205. [DOI] [PubMed] [Google Scholar]
- 7.Bromley RL, Mawer G, Clayton-Smith J, Baker GA. Autism spectrum disorders following in utero exposure to antiepileptic drugs. Neurology. 2008;71:1923–1924. doi: 10.1212/01.wnl.0000339399.64213.1a. [DOI] [PubMed] [Google Scholar]
- 8.Rasalam AD, et al. Characteristics of fetal anticonvulsant syndrome associated autistic disorder. Dev. Med. Child Neurol. 2005;47:551–555. doi: 10.1017/S0012162205001076. [DOI] [PubMed] [Google Scholar]
- 9.Go HS, et al. Prenatal exposure to valproic acid increases the neural progenitor cell pool and induces macrocephaly in rat brain via a mechanism involving the GSK-3beta/beta-catenin pathway. Neuropharmacology. 2012;63:1028–1041. doi: 10.1016/j.neuropharm.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 10.Kim KC, et al. The critical period of valproate exposure to induce autistic symptoms in Sprague-Dawley rats. Toxicol. Lett. 2011;201:137–142. doi: 10.1016/j.toxlet.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Mabunga DF, Gonzales EL, Kim JW, Kim KC, Shin CY. Exploring the validity of valproic acid animal model of autism. Exp. Neurobiol. 2015;24:285–300. doi: 10.5607/en.2015.24.4.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roullet FI, Lai JK, Foster JA. In utero exposure to valproic acid and autism–a current review of clinical and animal studies. Neurotoxicol. Teratol. 2013;36:47–56. doi: 10.1016/j.ntt.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Chomiak T, Turner N, Hu B. What we have learned about autism spectrum disorder from valproic acid. Pathol. Res. Int. 2013;2013:712758. doi: 10.1155/2013/712758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolini C, Fahnestock M. The valproic acid-induced rodent model of autism. Exp. Neurol. 2018;299:217–227. doi: 10.1016/j.expneurol.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Schneider T, Przewłocki R. Behavioral alterations in rats prenatally exposed to valproic acid: animal model of autism. Neuropsychopharmacology. 2005;30:80–89. doi: 10.1038/sj.npp.1300518. [DOI] [PubMed] [Google Scholar]
- 16.Markram K, Rinaldi T, Mendola DL, Sandi C, Markram H. Abnormal fear conditioning and amygdala processing in an animal model of autism. Neuropsychopharmacology. 2008;33:901–912. doi: 10.1038/sj.npp.1301453. [DOI] [PubMed] [Google Scholar]
- 17.Wang, R. et al. Aberrant development and synaptic transmission of cerebellar cortex in a VPA induced mouse autism model. Front. Cell. Neurosci.12 (2018). [DOI] [PMC free article] [PubMed]
- 18.Yamaguchi H, et al. Environmental enrichment attenuates behavioral abnormalities in valproic acid-exposed autism model mice. Behav. Brain Res. 2017;333:67–73. doi: 10.1016/j.bbr.2017.06.035. [DOI] [PubMed] [Google Scholar]
- 19.Lazaro MT, et al. Reduced prefrontal synaptic connectivity and disturbed oscillatory population dynamics in the CNTNAP2 model of autism. Cell Rep. 2019;27:2567–2578.e2566. doi: 10.1016/j.celrep.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu S, Anderson CT, Levitt P, Shepherd GMG. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated met receptor tyrosine kinase. J. Neurosci. 2011;31:5855. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobbi G, Janiri L. Sodium- and magnesium-valproate in vivo modulate glutamatergic and GABAergic synapses in the medial prefrontal cortex. Psychopharmacology. 2006;185:255–262. doi: 10.1007/s00213-006-0317-3. [DOI] [PubMed] [Google Scholar]
- 22.Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol. Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwan V, Unda BK, Singh KK. Wnt signaling networks in autism spectrum disorder and intellectual disability. J. Neurodev. Disord. 2016;8:45. doi: 10.1186/s11689-016-9176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin L, Dai X, Yin Y. Valproic acid exposure sequentially activates Wnt and mTOR pathways in rats. Mol. Cell. Neurosci. 2016;75:27–35. doi: 10.1016/j.mcn.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, et al. Downregulating the canonical Wnt/beta-catenin signaling pathway attenuates the susceptibility to autism-like phenotypes by decreasing oxidative stress. Neurochem. Res. 2012;37:1409–1419. doi: 10.1007/s11064-012-0724-2. [DOI] [PubMed] [Google Scholar]
- 27.An JY, et al. Towards a molecular characterization of autism spectrum disorders: an exome sequencing and systems approach. Transl. Psychiatry. 2014;4:e394. doi: 10.1038/tp.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang WE, et al. Cntnap2-dependent molecular networks in autism spectrum disorder revealed through an integrative multi-omics analysis. Mol. Psychiatry. 2023;28:810–821. doi: 10.1038/s41380-022-01822-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velmeshev D, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364:685–689. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 31.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebat J, et al. Strong association of De Novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kojic M, et al. Elp2 mutations perturb the epitranscriptome and lead to a complex neurodevelopmental phenotype. Nat. Commun. 2021;12:2678. doi: 10.1038/s41467-021-22888-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thacker S, Eng C. Transcriptome-(phospho)proteome characterization of brain of a germline model of cytoplasmic-predominant Pten expression with autism-like phenotypes. Npj. Genom. Med. 2021;6:42. doi: 10.1038/s41525-021-00201-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Courchesne E, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 36.Rinaldi, T., Perrodin, C. & Markram, H. Hyper-connectivity and hyper-plasticity in the medial prefrontal cortex in the valproic acid animal model of autism. Front. Neural Circuit2 (2008). [DOI] [PMC free article] [PubMed]
- 37.Bicks LK, Koike H, Akbarian S, Morishita H. Prefrontal cortex and social cognition in mouse and man. Front. Psychol. 2015;6:1805. doi: 10.3389/fpsyg.2015.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J-W, et al. Pharmacological modulation of AMPA receptor rescues social impairments in animal models of autism. Neuropsychopharmacology. 2019;44:314–323. doi: 10.1038/s41386-018-0098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JH, et al. Disruption of nucleocytoplasmic trafficking as a cellular senescence driver. Exp. Mol. Med. 2021;53:1092–1108. doi: 10.1038/s12276-021-00643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Jassal B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- 44.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryu H-H, et al. Excitatory neuron-specific SHP2-ERK signaling network regulates synaptic plasticity and memory. Sci. Signal. 2019;12:eaau5755. doi: 10.1126/scisignal.aau5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, G. et al. Social isolation impairs the prefrontal-nucleus accumbens circuit subserving social recognition in mice. Cell Rep.35 (2021). [DOI] [PubMed]
- 47.Yang M, Silverman JL, Crawley JN. Automated three-chambered social approach task for mice. Curr. Protoc. Neurosci. 2011;Chapter 8:Unit 8 26. doi: 10.1002/0471142301.ns0826s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeisel A, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e1022. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowakowski TJ, et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parikshak NN, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu JM, et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54:1320–1331. doi: 10.1038/s41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplanis J, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heyne HO, et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018;50:1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]
- 54.Singh T, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–516. doi: 10.1038/s41586-022-04556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, et al. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 2007;26:2284–2294. doi: 10.1038/sj.emboj.7601667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciani L, et al. Wnt signalling tunes neurotransmitter release by directly targeting Synaptotagmin-1. Nat. Commun. 2015;6:8302. doi: 10.1038/ncomms9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen CP, Jernigan KK, Diggins NL, Webb DJ, MacGurn JA. USP9X deubiquitylates DVL2 to regulate WNT pathway specification. Cell. Rep. 2019;28:1074–1089.e1075. doi: 10.1016/j.celrep.2019.06.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 59.Lee E, Salic A, Krüger R, Heinrich R, Kirschner MW. The roles of APC and axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003;1:e10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao W, Ho M. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep. 2011;1:14–19. [PMC free article] [PubMed] [Google Scholar]
- 61.Koopmans F, et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron. 2019;103:217–234.e214. doi: 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmad-Annuar A, et al. Signaling across the synapse: a role for Wnt and Dishevelled in presynaptic assembly and neurotransmitter release. J. Cell Biol. 2006;174:127–139. doi: 10.1083/jcb.200511054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ravindranath A, et al. Wnt–β-catenin–Tcf-4 signalling-modulated invasiveness is dependent on osteopontin expression in breast cancer. Br. J. Cancer. 2011;105:542–551. doi: 10.1038/bjc.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheon S, et al. The ubiquitin ligase UBE3B, disrupted in intellectual disability and absent speech, regulates metabolic pathways by targeting BCKDK. Proc. Natl Acad. Sci. USA. 2019;116:3662–3667. doi: 10.1073/pnas.1818751116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Basel-Vanagaite L, et al. Deficiency for the ubiquitin ligase UBE3B in a blepharophimosis-ptosis-intellectual-disability syndrome. Am. J. Hum. Genet. 2012;91:998–1010. doi: 10.1016/j.ajhg.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brumback AC, et al. Identifying specific prefrontal neurons that contribute to autism-associated abnormalities in physiology and social behavior. Mol. Psychiatry. 2018;23:2078–2089. doi: 10.1038/mp.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rinaldi T, Kulangara K, Antoniello K, Markram H. Elevated NMDA receptor levels and enhanced postsynaptic long-term potentiation induced by prenatal exposure to valproic acid. Proc. Natl Acad. Sci. 2007;104:13501–13506. doi: 10.1073/pnas.0704391104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, Y. et al. RNF146 inhibits excessive autophagy by modulating the Wnt-β-catenin pathway in glutamate excitotoxicity injury. Front. Cell. Neurosci.11 (2017). [DOI] [PMC free article] [PubMed]
- 69.Kim, H. et al. Estrogen receptor activation contributes to RNF146 expression and neuroprotection in Parkinson’s disease models. Oncotarget8 (2017). [DOI] [PMC free article] [PubMed]
- 70.Shen J, Yu Z, Li N. The E3 ubiquitin ligase RNF146 promotes colorectal cancer by activating the Wnt/β-catenin pathway via ubiquitination of Axin1. Biochem. Biophys. Res. Commun. 2018;503:991–997. doi: 10.1016/j.bbrc.2018.06.107. [DOI] [PubMed] [Google Scholar]
- 71.Abrahams BS, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruzzo EK, et al. Inherited and De Novo genetic risk for autism impacts shared networks. Cell. 2019;178:850–866.e826. doi: 10.1016/j.cell.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin PM, et al. DIXDC1 contributes to psychiatric susceptibility by regulating dendritic spine and glutamatergic synapse density via GSK3 and Wnt/β-catenin signaling. Mol. Psychiatry. 2018;23:467–475. doi: 10.1038/mp.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, et al. Genetic evidence of gender difference in autism spectrum disorder supports the female-protective effect. Transl. Psychiatry. 2020;10:4. doi: 10.1038/s41398-020-0699-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freese JL, Pino D, Pleasure SJ. Wnt signaling in development and disease. Neurobiol. Dis. 2010;38:148–153. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang H, He X. Wnt/β-catenin signaling: new (and old) players and new insights. Curr. Opin. Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mulligan KA, Cheyette BNR. Wnt signaling in vertebrate neural development and function. J. Neuroimmune Pharmacol. 2012;7:774–787. doi: 10.1007/s11481-012-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brien WT, et al. Glycogen synthase kinase-3β haploinsufficiency mimics the behavioral and molecular effects of lithium. J. Neurosci. 2004;24:6791. doi: 10.1523/JNEUROSCI.4753-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Latapy C, Rioux V, Guitton MJ, Beaulieu JM. Selective deletion of forebrain glycogen synthase kinase 3β reveals a central role in serotonin-sensitive anxiety and social behaviour. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 2012;367:2460–2474. doi: 10.1098/rstb.2012.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fang W-Q, et al. Overproduction of upper-layer neurons in the neocortex leads to autism-like features in mice. Cell Rep. 2014;9:1635–1643. doi: 10.1016/j.celrep.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 81.Cui K, et al. Neurodevelopmental impairment induced by prenatal valproic acid exposure shown with the human cortical organoid-on-a-chip model. Microsyst. Nanoeng. 2020;6:49. doi: 10.1038/s41378-020-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peukert D, Weber S, Lumsden A, Scholpp S. Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 2011;9:e1001218. doi: 10.1371/journal.pbio.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mines MA, Yuskaitis CJ, King MK, Beurel E, Jope RS. GSK3 influences social preference and anxiety-related behaviors during social interaction in a mouse model of fragile X syndrome and autism. PLoS ONE. 2010;5:e9706. doi: 10.1371/journal.pone.0009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi CH, et al. Pharmacological reversal of synaptic plasticity deficits in the mouse model of Fragile X syndrome by group II mGluR antagonist or lithium treatment. Brain Res. 2011;1380:106–119. doi: 10.1016/j.brainres.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arredondo SB, Valenzuela-Bezanilla D, Mardones MD, Varela-Nallar L. Role of Wnt signaling in adult hippocampal neurogenesis in health and disease. Front. Cell Dev. Biol. 2020;8:860. doi: 10.3389/fcell.2020.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McLeod F, Salinas PC. Wnt proteins as modulators of synaptic plasticity. Curr. Opin. Neurobiol. 2018;53:90–95. doi: 10.1016/j.conb.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosso, S., Inestrosa, N. & Rosso, S. WNT signaling in neuronal maturation and synaptogenesis. Front. Cell. Neurosci.7 (2013). [DOI] [PMC free article] [PubMed]
- 88.Okuda T, Yu LMY, Cingolani LA, Kemler R, Goda Y. β-Catenin regulates excitatory postsynaptic strength at hippocampal synapses. Proc. Natl Acad. Sci. 2007;104:13479–13484. doi: 10.1073/pnas.0702334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun M, et al. Presynaptic contributions of chordin to hippocampal plasticity and spatial learning. J. Neurosci. 2007;27:7740. doi: 10.1523/JNEUROSCI.1604-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim KC, et al. Male-specific alteration in excitatory post-synaptic development and social interaction in pre-natal valproic acid exposure model of autism spectrum disorder. J. Neurochem. 2013;124:832–843. doi: 10.1111/jnc.12147. [DOI] [PubMed] [Google Scholar]
- 91.Kataoka S, et al. Autism-like behaviours with transient histone hyperacetylation in mice treated prenatally with valproic acid. Int. J. Neuropsychopharmacol. 2013;16:91–103. doi: 10.1017/S1461145711001714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data necessary to reach the conclusions of this paper are presented in the paper and/or the Supplementary Materials. The raw proteome MS data and MaxQuant search results have been deposited in the PRIDE database (project accession: PXD036413). Sequencing data have been deposited in GEO under accession code GSE215871.