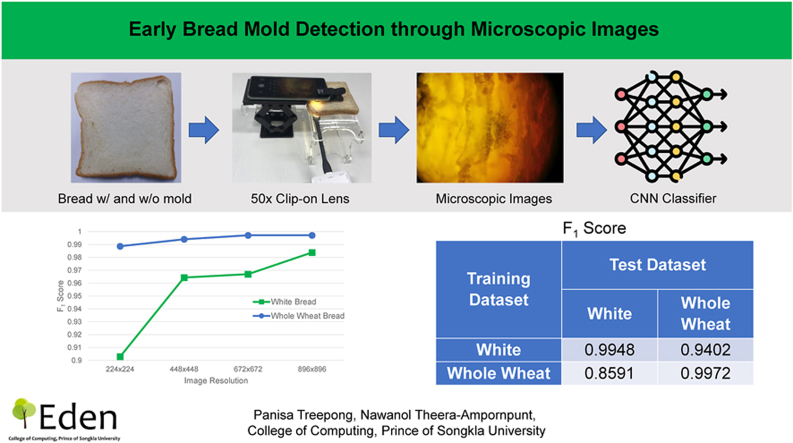
Abstract
Mold on bread in the early stages of growth is difficult to discern with the naked eye. Visual inspection and expiration dates are imprecise approaches that consumers rely on to detect bread spoilage. Existing methods for detecting microbial contamination, such as inspection through a microscope and hyperspectral imaging, are unsuitable for consumer use. This paper proposes a novel early bread mold detection method through microscopic images taken using clip-on lenses. These low-cost lenses are used together with a smartphone to capture images of bread at 50× magnification. The microscopic images are automatically classified using state-of-the-art convolutional neural networks (CNNs) with transfer learning. We extensively compared image preprocessing methods, CNN models, and data augmentation methods to determine the best configuration in terms of classification accuracy. The top models achieved near-perfect scores of 0.9948 for white sandwich bread and 0.9972 for whole wheat bread.
Keywords: Image classification, Microbiology, Food safety, Food computing
Graphical abstract
Highlights
-
•
A method for consumer to detect mold on bread in early stage.
-
•
Images of bread are taken using 50x clip-on lens together with a smartphone.
-
•
22 Convolutional Neural Networks are extensively compared.
-
•
Near-perfect F1 score of 0.99 is achieved.
-
•
Datasets are publicly available.
1. Introduction
Bread is one of the most commonly consumed foods worldwide. Depending on the type, moisture content, room temperature, and amount of preservative used, bread can be stored at room temperature for a few days or up to three or more weeks (Degirmencioglu et al., 2011). If stored for too long, bread can develop fungi or mold, making it unsafe for consumption. In the early growth stage, mold can be difficult to discern with the naked eye. Moreover, some types of bread have non-uniform textures or colors that make it harder to detect the mold. Consumers often rely on the expiration date or best-before date printed on the product’s packaging to determine whether the bread is safe for consumption. However, manufacturers assume typical storage conditions when setting these dates, and storage conditions can vary significantly across households, seasons, and regions. For example, many consumers store bread in a refrigerator or freezer, and expiration dates cannot be relied on in these cases. Visual and odor inspection is also not always reliable, especially in the early stage of mold growth. Therefore, in this work, we propose a novel method for regular consumers to detect bread mold. This method focuses on the early stages where signs of mold are not apparent to the naked eye, and it thus reduces the risk of food poisoning as well as food waste.
Since bread mold in the early stages of growth can be difficult to notice with the naked eye or even in high-resolution photographs, using magnified images of the bread is necessary for this task. Microscopes are the best tool for taking microscopic images, but they require expertise and are too cumbersome for regular consumers to use. Smartphone clip-on lenses have recently been developed (CU SmartLens, 2023; Apexel, 2023; SmartMicroOptics, 2023; knowthystore, 2023). These lenses provide 20–100× magnification at a fraction of the cost of a typical microscope and, more importantly, can be conveniently used by a layperson. Since enough light can pass through a slice of bread, the sample does not need to be cut to capture a well-lit photograph. However, having a light source behind the object is still necessary, and an adjustable microscope stage or camera stand will aid in obtaining a well-focused and clear image. Although 50× magnification is enough to see the mold clearly, regular consumers are unfamiliar with microscopic images of bread textures and mold. Therefore, in this work, we develop computer models that can automatically perform the task of mold detection in images for them.
Convolutional neural networks (CNNs) are a class of bio-inspired computer models that have emerged as one of the top-performing models for image classification. CNNs have been extensively studied and used in many domains, including in the detection and classification of microorganisms. Sun et al. (2016) explored the problem of detecting and identifying mold colonies in images of unhulled rice. The models used include the support vector machine, artificial neural network, convolutional neural network, and deep belief network. Mishra et al. (2019) and Jubayer et al. (2021) used enhanced image segmentation and object detection algorithms, respectively, to detect mold in various foods, including bread. While these approaches do not require specialized equipment, they rely on non-magnified images. Hence, when used for mold detection, mold can only be detected once it has grown significantly enough to be seen with the naked eye; hence, a significant risk of food poisoning remains for the consumer even when mold is not detected.
CNNs developed in the recent years tend to have a large number of layers. These “deep” CNNs can capture and analyze complicated patterns but require large training datasets. In the absence of such datasets, transfer learning can be used to train accurate models. Deep CNNs are commonly used for classifying microscopic images. Wahid et al. (2019) used the Xception deep learning model to classify seven varieties of bacteria that cause human diseases. Mahbod et al. (2021) identified three categories of pollen grains and non-pollen (debris) based on their microscopic images. The authors used four CNN models together, and the final determination was based on the average output of the four models. Other studies have utilized deep learning for the classification of human cells, usually for clinical diagnosis (Liu et al., 2022; Nguyen et al., 2018; Iqbal et al., 2019; Alzubaidi et al., 2020; Qin et al., 2018; Meng et al., 2018). Our work proposes another application of deep learning to classify microscopic images, for a layperson to use as an expert system.
Hyperspectral and multispectral imaging, paired with machine vision or machine learning techniques, have been successfully used to analyze food quality and detect microbial, chemical, and physical contaminants. Both types of imaging work by capturing images of an object based on how different wavebands are altered by the object. Hyperspectral imaging utilizes wide, continuous wavebands that provide more complete data, while multispectral imaging utilizes only a few non-continuous wavebands to speed up image acquisition (Qin et al., 2013). Chu et al. (2020) used near-infrared hyperspectral imaging, principal component analysis, the successive projections algorithm, and a support vector machine to detect fungal infection in maize kernels. The top method achieved 99–100% accuracy in three maize hybrids. Bonah et al. (2020) used visible near-infrared hyperspectral imaging to detect, quantify, and visualize bacteria on the surface of pork. The most accurate approaches used only eight to 21 wavebands out of 618 available and achieved very low error rates. Xie et al. (2017) used hyperspectral imaging to detect tomato leaves infected with gray mold. Feature selection combined with the k-nearest neighbor algorithm produced the best classification accuracy of 97.22%. Meng et al. (2020) detected southern corn rust, caused by a fungal pathogen, using reflectance spectra of the leaf. Amodio et al. (2017), Teerachaichayut and Ho (2017), and Pissard et al. (2021) used near-infrared spectroscopy to evaluate the quality of strawberries, limes, and apples, respectively.
While the imaging techniques reviewed above provide excellent detection capability, they require expensive, sophisticated hardware unsuitable for consumer use. Food safety research generally focuses on food safety during production, processing, and distribution. In this work, we instead focus on making improvements on the consumers’ side. The objective of this study is to propose and evaluate a method for early bread mold detection that is appropriate for consumer use. Our method utilizes microscopic clip-on lenses to capture images of bread and CNNs to detect mold automatically in the images. To the best of our knowledge, this is the first proposal to use microscopic clip-on lenses to detect early microbial growth in food. A mobile application can be developed using our method and would allow consumers to conveniently check and ensure that their bread is mold-free.
The major contributions of this work can be summarized as follows.
-
1.
We propose a novel, low-cost method for consumers to detect early mold growth on bread through microscopic clip-on lenses.
-
2.
For untrained consumers, we propose using CNN models to automatically determine whether a microscopic image of bread contains mold.
-
3.
We extensively evaluate many aspects of CNN model training configurations, including image preprocessing methods, CNN model, image resolution, and data augmentation, to achieve optimum classification accuracy.
-
4.
We evaluate the proposed method on white and whole wheat sandwich bread stored in realistic conditions with natural mold growth. The top approaches achieve near-perfect classification accuracy, with scores of 0.9948 for white bread and 0.9972 for whole wheat bread. However, white bread is more difficult to classify, requiring higher image resolution and data augmentation to achieve comparable results to those for whole wheat bread.
The rest of the paper is organized as follows: Section 2 describes the data collection method, image preprocessing method, and CNN models. Section 3 presents the evaluation method and discusses the results, and conclusions are drawn in Section 4.
2. Methodology
The overall steps in our method are as follows. First, we took microscopic photographs of the bread using 50x lenses clipped onto a smartphone. Both images with no mold and images with mold present were necessary. The images underwent preprocessing steps before they were used to train the CNN classifier using transfer learning. The output of the CNN classifier was the class of image, indicating whether mold was present in the image.
2.1. Datasets1
We used the two most popular types of sliced bread to train and evaluate our models: 1) white sandwich bread and 2) whole wheat sandwich bread, locally obtained in Phuket, Thailand. Both types of bread contained preservatives, and the expiration date was five days after the day the bread was purchased. We collected images of bread without mold within the first three days to ensure that no mold was present. To collect images of bread with mold, we stored the bread in its original packaging at room temperature (25–30 °C) and away from sunlight until a small amount of mold was present upon careful inspection. We then took photographs of the bread, focusing on the moldy parts. The photographs were taken using a Huawei P30 Pro smartphone and a 50x CU SmartLens microscopic clip-on lens, as shown in Fig. 1 (CU SmartLens, 2023). We chose this model of clip-on lenses because it has a smaller footprint and lower cost, and it does not require batteries/charging, making it suitable for consumer use. This model’s maximum magnification is 50x, which is sufficient for mold detection. The effect of using a lower magnification can be approximated through lower image resolution. The following camera settings were used: 40-megapixel image resolution, automatic sensor sensitivity (ISO), automatic shutter time, automatic white balance, and exposure value adjusted manually for each shot to produce well-illuminated images. The photography setup is depicted in Fig. 2. We placed the slice of bread directly on a transparent stage with an LED (light-emitting diode) light source underneath, and the smartphone was placed on an adjustable stand. We took the microscopic photograph of the bread directly, without cutting the bread and without using glass slides. A domain expert then classified the images into the following two classes.
-
1.
No mold (negative class): The magnified image contained no evidence of mold and was taken during the first three days after obtaining the bread, before the expiration date.
-
2.
Mold present (positive class): Mold was visible in the magnified image but was not immediately noticeable to the naked eye.
Fig. 1.
Clip-on microscopic lens used in the study.
Fig. 2.
Photography setup for bread image collection.
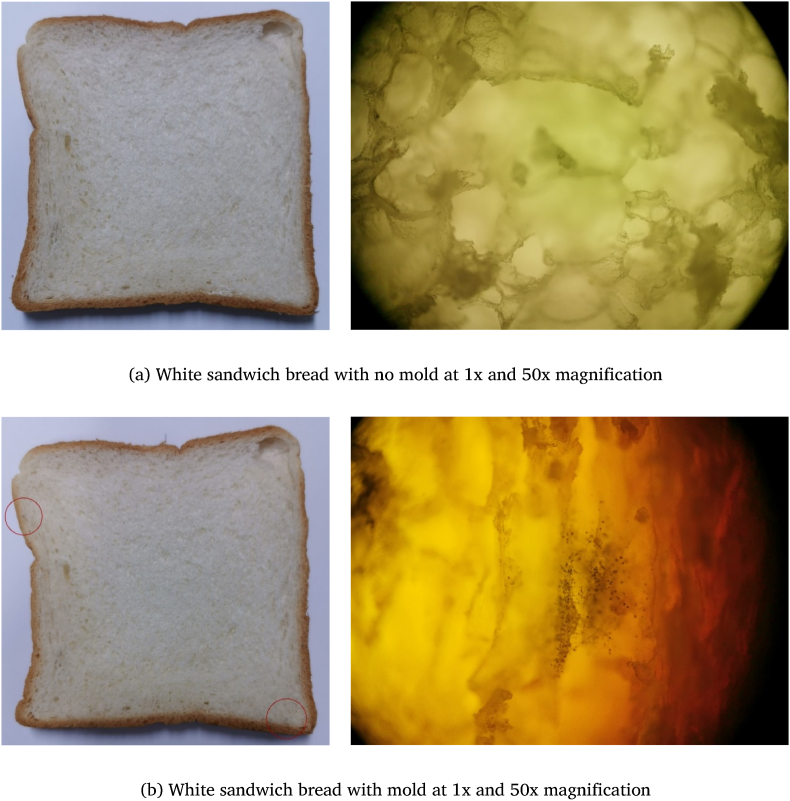
Examples of magnified and non-magnified images of both classes of white bread are presented in Fig. 3. Images of whole wheat bread also have similar patterns. Images that did not fit either of the two classes above, such as images of moldy bread where mold was not visible in the magnified image, were excluded. Blurry images and images that were too dark or too bright were also excluded.
Fig. 3.
Examples of images of the same slice of white sandwich bread.
The two datasets, one for white sandwich bread and another for whole wheat sandwich bread, were kept separated and never combined. Each dataset was divided into a training set, a validation set, and a test set with a rough ratio of 60:15:25, separated by the slice of bread from which the images were taken, to ensure that different subsets never contained images from the same slice of bread. The training set was used to train the CNN model, while the validation set was used for learning rate optimization, and the test set was used to evaluate the models. The number of images in each subset is listed in Table 1, Table 2.
Table 1.
Size of white sandwich bread dataset.
| Subset | Number of Images |
||
|---|---|---|---|
| No Mold | Mold Present | Total | |
| Training | 632 | 245 | 877 |
| Validation | 153 | 75 | 228 |
| Test | 303 | 87 | 390 |
| Total | 1,088 | 407 | 1,495 |
Table 2.
Size of whole wheat sandwich bread dataset.
| Subset | Number of Images |
||
|---|---|---|---|
| No Mold | Mold Present | Total | |
| Training | 471 | 460 | 931 |
| Validation | 105 | 113 | 218 |
| Test | 160 | 179 | 339 |
| Total | 736 | 752 | 1,488 |
2.2. Image preprocessing
Microscopic images taken using a 50x CU SmartLens have black corners due to vignetting. However, the presence of these black corners does not affect CNN models’ prediction performance, so it is unnecessary to remove them from the images. Following the convention of image classification research, the images were cropped to a square size. However, we skipped center cropping (zooming in) because in some of our images, the mold was only present near the edge of the image, and center cropping would remove it. Significant portions of dark corners were unintentionally removed in the process of square cropping.
The most noticeable sign of mold was the presence of its spores, seen in 50x microscopic images as clusters of tiny black dots. In some images, the mycelium, the root-like structure of a fungus, was also visible. However, some images of bread with no mold also contained isolated small black dots, which could confuse untrained humans. In addition, the overall colors of microscopic images of bread varied between shots, ranging from red to yellow to green, due to the different densities of bread, independent of whether mold was present. Since these patterns did not contain apparent color information, converting the image to grayscale could improve the models’ prediction performance. Furthermore, the brightness of the images also varied between shots. Therefore, we compared the following image representations.
-
1.
RGB (red-green-blue): Images are represented in red-green-blue format and undergo no color or brightness change.
-
2.
RGB w/normalized mean: The brightness of each image is adjusted so that its brightness mean is equal to the brightness mean of the entire dataset.
-
3.
RGB w/normalized mean and SD (standard deviation): The brightness of each image is adjusted so that its brightness mean and standard deviation are equal to the brightness mean and standard deviation of the entire dataset.
-
4.
Grayscale: Images are converted to grayscale.
-
5.
Grayscale w/normalized mean: Images are converted to grayscale, and the brightness of each image is adjusted so that its brightness mean is equal to the brightness mean of the entire dataset.
-
6.
Grayscale w/normalized mean and SD: Images are converted to grayscale, and the brightness of each image is adjusted so that its brightness mean and standard deviation are equal to the brightness mean and standard deviation of the entire dataset.
Conversion from RGB to grayscale accounts for differences in humans’ perceived brightness of red, green, and blue light using the following weighted average:
| (1) |
where is the grayscale value and , , and are the values of the red, green, and blue components, respectively.
2.3. Data augmentation
Because our datasets were small, techniques that could help expand the datasets were potentially beneficial. A commonly used technique is data augmentation, where data points are modified realistically and are either added to the dataset or used in place of the original data points. Data augmentation can reduce overfitting and allow the model to recognize more patterns in data. Many operations are commonly used for image data, such as flipping, rotation, zooming, cropping, translation, and adjusting brightness and contrast. As some of our images contained bread mold near the edges of the images, we excluded operations that were likely to remove the images' edges. Therefore, we used and compared the following three image operations for data augmentation.
-
1.
Brightness adjustment, where brightness is modified by a random factor in the range of [-0.2, 0.1] relative to the entire range of the pixel value, which is 255
-
2.
Flipping, both horizontally and vertically, with a probability of 0.5 each, and performed independently
-
3.
Rotation, from 0 to 360°, with points outside the boundary of the original image filled by the reflection of the original image
Instead of producing separate datasets with augmented data before model training, we implemented data augmentation operations as the first layers of the CNN model. With this setup, every image is randomly transformed just before it is processed during training. Each image may be transformed differently in each training pass (epoch). Compared with generating a fixed dataset, this approach has the benefit that many different transformations of each image can be generated without increasing the size of the dataset, which would increase the computational requirements.
2.4. Models
Many CNN models designed for image classification have been proposed. The best model depends on the task at hand, among other things. However, based on previous studies, a large dataset is necessary to train an accurate CNN model that can recognize complex patterns. Our datasets contain only 1,488–1,495 images, which is insufficient to train an accurate model from scratch. Fortunately, a technique called transfer learning allows us to use complex CNN models with our datasets. First, the model is trained on a large dataset for a separate task. Then, the last few layers of the model are replaced with new layers for our task, and the weights of only the new layers are trained using our smaller dataset. This approach can be viewed as using the original CNN model as the feature extractor for the new task. Application of this approach in many domains has shown that it produces robust and highly accurate classifiers, even when the images in both datasets are dissimilar. The CNN models we used are summarized in Table 3.
Table 3.
CNN models included in the comparison.
| Model | Number of Parameters |
|---|---|
| DenseNet121 (Huang et al., 2017) | 7.0M |
| EfficientNetV2B0 (Tan and Le, 2021) | 5.9M |
| EfficientNetV2B1 (Tan and Le, 2021) | 6.9M |
| EfficientNetV2B2 (Tan and Le, 2021) | 8.8M |
| EfficientNetV2B3 (Tan and Le, 2021) | 12.9M |
| EfficientNetV2L (Tan and Le, 2021) | 117.7M |
| EfficientNetV2M (Tan and Le, 2021) | 53.2M |
| EfficientNetV2S (Tan and Le, 2021) | 20.3M |
| InceptionResNetV2 (Szegedy et al., 2017) | 51.8M |
| InceptionV3 (Szegedy et al., 2016) | 21.8M |
| MobileNet (Howard et al., 2017) | 3.2M |
| MobileNetV2 (Sandler et al., 2018) | 2.3M |
| MobileNetV3Large (Howard et al., 2019) | 3.0M |
| MobileNetV3Small (Howard et al., 2019) | 0.9M |
| NASNetMobile (Zoph et al., 2018) | 4.3M |
| RegNetX016 (Radosavovic et al., 2020) | 7.9M |
| RegNetX032 (Radosavovic et al., 2020) | 14.4M |
| RegNetY016 (Radosavovic et al., 2020) | 9.9M |
| RegNetY032 (Radosavovic et al., 2020) | 18.0M |
| ResNet50 (He et al., 2016a, He et al., 2016ba) | 23.6M |
| ResNet50V2 (He et al., 2016a, He et al., 2016bb) | 23.6M |
| Xception (Chollet, 2017) | 20.9M |
For our work, all CNN models were pretrained using the ImageNet dataset (Russakovsky et al., 2015). The original output layer was replaced by a two-dimensional global average pooling layer, a dropout layer with a dropout rate of 0.2, and an output layer with a single node. The pretrained weights were fixed, and the binary cross-entropy loss function was used. Important hyperparameters of the models that needed to be carefully chosen or optimized included the following.
-
1.
Optimizer and its parameters, such as learning rate
-
2
Batch size
-
3
Number of epochs or steps for which to train each model
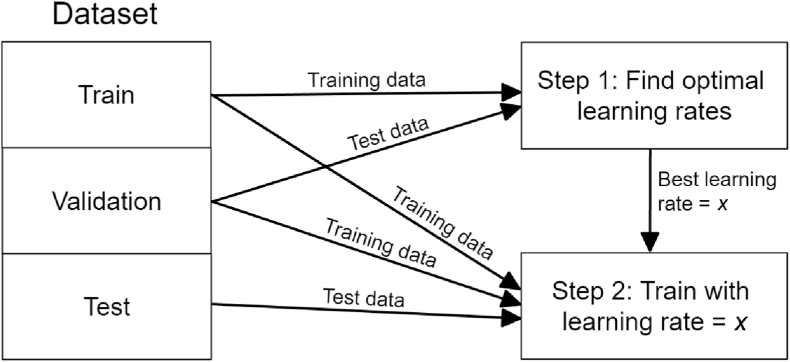
We used Adam as the optimizer, as it has demonstrated superior performance and is often provided by libraries as the default optimizer (Kingma and Ba, 2014). We fixed the batch size at 64, as it was the maximum value that allowed all CNN models to run on our testbed without out-of-memory errors. The number of epochs refers to the number of complete passes the training process iterates over the training dataset. To keep the amount of computation reasonable and bounded, we fixed the number of epochs to train each model at 100. The Adam optimizer has many parameters, the most important being the learning rate (Godbole et al., 2023). The optimal learning rate depends on the CNN model, among other things. Therefore, each time a model was trained and evaluated, we optimized the learning rate using the following two-step process, illustrated in Fig. 4.
Fig. 4.
Two-step model training process for learning rate optimization.
Step 1: Find the optimal learning rate. The model is trained on the training set using multiple learning rate values. The value that produces the highest prediction performance on the validation set is then used in step 2. The learning rate values used are 0.0001, 0.0002, 0.0005, 0.001, 0.002, 0.005, 0.01, 0.02, and 0.05. When the learning rate is too low, the accuracy will be low. By contrast, when the learning rate is too high, the accuracy could be low or high, but it is unstable across epochs, as it jumps back and forth between the local minimum and other points away from it. To avoid learning rates that are too high, the maximum (i.e., worst) validation loss over the last 10 epochs is used to compare the learning rates.
Step 2: Train and evaluate the model. The best learning rate from step 1 is used to train the model from scratch again using the training set and validation set combined, and the prediction performance on the test set is reported. We trained each model five times and reported the average performance to reduce variance across runs.
Other parameters of the Adam optimizer are left at their default values: 0.9 for , 0.999 for , for ε, and no weight decay.
3. Evaluation
3.1. Experiment setup
We evaluated the classification accuracy of the proposed method using two datasets containing 1,495 and 1,488 microscopic images of white and whole wheat bread, respectively. These datasets were used separately and never combined. Each dataset was divided into a non-overlapping training set, validation set, and test set. Section 2 details the datasets, image preprocessing methods, and selection of model hyperparameters.
Since our image classification task focuses on the positive class (mold present), we used precision and recall instead of accuracy as the primary metrics to evaluate and compare the models. Precision is the fraction of positive predictions that are correct. That is, when the model detects mold in an image, precision is the probability that the image actually contains mold. Recall is the fraction of data points in the positive class that are correctly classified. That is, of all images that actually contain mold, recall is the fraction that the model detects. Both metrics range from 0 to 1, with higher values indicating a more accurate classification. To compute an accurate and representative precision value, it is necessary to use the test set with the same class distribution as the distribution in actual use (i.e., by consumers). This distribution differs from our datasets’ distributions because ours depend solely on the number of images of each class we managed or chose to collect. To avoid bias toward either class, we assumed that both classes are evenly distributed. Instead of modifying our test datasets, we assigned the appropriate weight to data points in each class such that both classes had equal overall importance. That is, we calculated precision as
| (2) |
where is the number of true positives; is the number of false positives; and is the relative weight of each negative data point compared to each positive data point, defined as
| (3) |
where and are the numbers of data points in the positive and negative classes, respectively, in the test set. This allowed us to use all data points in our test set while ensuring a representative value of precision. The weight of each positive data point was fixed at 1. Recall is not affected by class distribution, so no adjustment is necessary.
To enable a simple comparison of prediction performance, precision and recall are summarized into score, which is the harmonic mean of precision and recall, defined as
| (4) |
The range of scores is 0–1, with higher values indicating a more accurate classification. A value of 1 means all data points are correctly classified.
We designed five experiments, each aimed to evaluate or optimize certain aspects of the proposed method, as follows.
Experiment 1: Comparing image representations
Experiment 2: Comparing CNN models for each image resolution
Experiment 3: Comparing data augmentation methods
Experiment 4: Effect of transfer learning
Experiment 5: Cross-dataset prediction
In experiments 1–3, we aimed to find the best configuration/model in terms of classification accuracy. Experiment 2 would additionally inform us how the magnification power of the lens affects classification accuracy. Experiment 4 investigated the degree to which transfer learning improves classification accuracy. Experiment 5 evaluated the models’ accuracy when the test images were of a slightly different type of bread than the types in the training images.
To keep the amount of computation reasonable, we started by finding the best combination of image representation and CNN model using greedy search, where only one hyperparameter/aspect is changed at a time, rather than a complete grid search where all configurations are evaluated. The values from the best configuration(s) were then used as fixed hyperparameters in each experiment. The batch size was always fixed at 64. The Adam optimizer was used, with of 0.9, of 0.999, ε of , and no weight decay. The learning rate was optimized using the two-step process specified in Section 2.4.
The experiments were run on a computer with an Intel Core i5-12400F processor, 64 gigabytes of memory, and an Nvidia GeForce RTX 3090 graphic processing unit (GPU), and running the Ubuntu 22.10 operating system. The TensorFlow library was used to preprocess images, process data, train the models, and evaluate the models.
3.2. Experiment 1: comparing image representations
The first experiment aimed to determine the best way to represent and normalize the images. Section 2.2 describes the six image representations/approaches that we compared. The image resolution was fixed at 896 × 896 pixels, and the top four to five CNN models for each type of bread were used. Table 4 lists the mean and SD values used to normalize images in each dataset. Note that the means of RGB and grayscale representations differ because we converted images from RGB to grayscale using a weighted average that accounts for differences in humans’ perceived brightness of different colors.
Table 4.
Mean and SD values used to normalize images in each dataset, 0–255 scale.
| Type of Bread | Image Representation | Mean | SD |
|---|---|---|---|
| White Bread | RGB | 127.62 | 73.10 |
| White Bread | Grayscale | 151.33 | 30.84 |
| Whole Wheat Bread | RGB | 111.10 | 86.43 |
| Whole Wheat Bread | Grayscale | 134.13 | 39.49 |
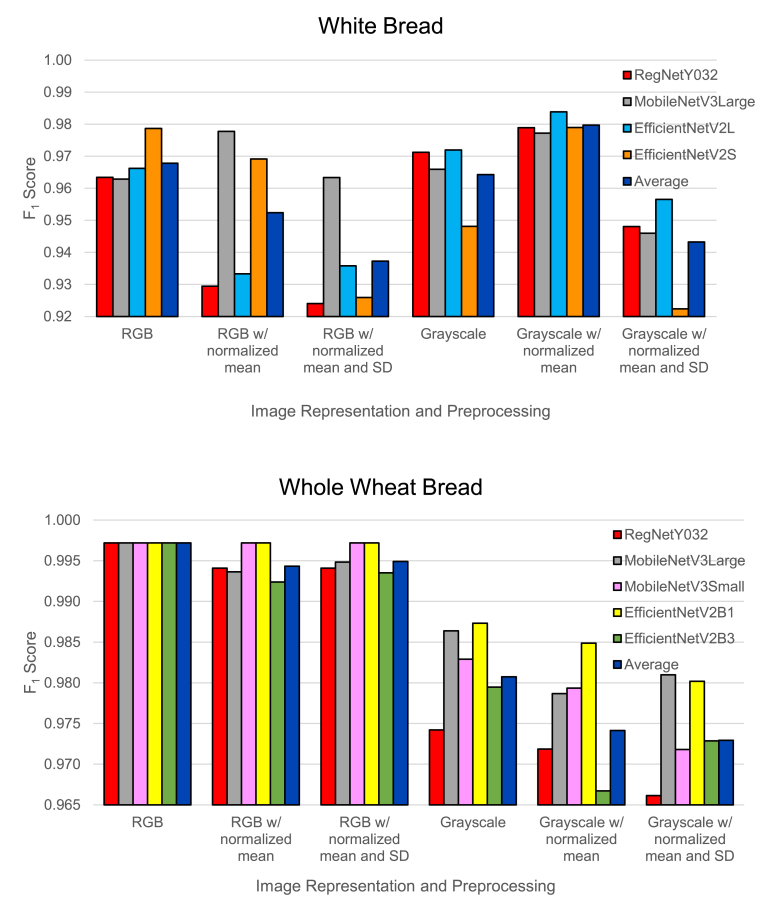
Fig. 5 illustrates the classification performance. For white bread, grayscale representations tended to produce slightly better results than RGB. Grayscale with normalized mean representation produced the best results, with an average score of 0.9797.
Fig. 5.
Top models’ classification performance for different image representations.
For whole wheat bread, however, RGB produced noticeably better results than grayscale. Normalizing the mean and SD slightly lowered the performance for both RGB and grayscale. The best results were obtained using RGB representation, with an score of 0.9972 for all top models.
These results suggest that the best image representation is different for white and whole wheat bread. We hypothesize that microscopic images of whole wheat bread contain color patterns or information that is helpful – although not necessarily apparent upon inspection – to the models for detecting mold. In subsequent experiments, we will use grayscale with normalized mean representation for white bread and RGB representation for whole wheat bread.
3.3. Experiment 2: comparing CNN models for each image resolution
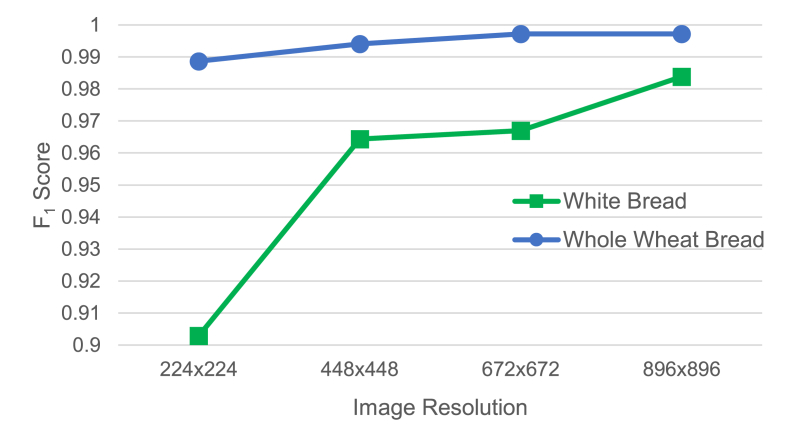
In this experiment, we aimed to identify the most accurate CNN model, measured in terms of score, and to investigate how image resolution affects classification accuracy. However, the most accurate model may be different for different image resolutions. Therefore, we performed and presented the comparison separately for each image resolution. We used image resolutions of 224 × 224, 448 × 448, 672 × 672, and 896 × 896 pixels. All 22 models listed in Section 2.4 were included in the comparison. Using a lower image resolution can approximate the effect of using a lens with lower magnification power, with the four resolutions corresponding to 12.5x, 25x, 37.5x, and 50× magnification, respectively.
Table 5, Table 6 present the classification performance of all models at different image resolutions. In Fig. 6, information in both tables is summarized to show how image resolution affects the classification performance of the top model. Higher image resolution produced better classification accuracy, as one would expect. However, the error rate increased more significantly as image resolution decreased for white bread compared with whole wheat bread. At the lowest resolution of 224 × 224 pixels, the score of the top model was 0.9029 for white bread and 0.9887 for whole wheat bread. We can conclude that detecting mold in whole wheat bread is easier than in white bread. A low resolution of 224 × 224 pixels was sufficient to produce high accuracy for whole wheat bread, while a high resolution of 896 × 896 pixels was necessary to achieve similarly high accuracy for white bread. We hypothesize that such a significant difference was an indirect consequence of mold being more noticeable on white bread than on whole wheat bread. As a result, when a small amount of mold was noticeable and images were taken, mold on whole wheat bread had grown more compared with mold growth on white bread.
Table 5.
Comparison of scores of CNN models at various image resolutions for white bread.
| CNN Model | Image Resolution (pixels) |
|||
|---|---|---|---|---|
| 224 224 | 448 448 | 672 672 | 896 896 | |
| DenseNet121 | 0.8263 | 0.9162 | 0.9526 | 0.9316 |
| EfficientNetV2B0 | 0.8538 | 0.9187 | 0.9609 | 0.9547 |
| EfficientNetV2B1 | 0.8308 | 0.9277 | 0.9457 | 0.9602 |
| EfficientNetV2B2 | 0.8490 | 0.9316 | 0.9477 | 0.9743 |
| EfficientNetV2B3 | 0.7726 | 0.9498 | 0.9583 | 0.9602 |
| EfficientNetV2L | 0.7501 | 0.9219 | 0.9633 | 0.9838 |
| EfficientNetV2M | 0.7123 | 0.8523 | 0.9511 | 0.9683 |
| EfficientNetV2S | 0.8351 | 0.8853 | 0.9598 | 0.9790 |
| InceptionResNetV2 | 0.8317 | 0.8347 | 0.9141 | 0.9347 |
| InceptionV3 | 0.7755 | 0.8978 | 0.9367 | 0.8909 |
| MobileNet | 0.7223 | 0.8824 | 0.9451 | 0.9589 |
| MobileNetV2 | 0.8414 | 0.8951 | 0.9567 | 0.9465 |
| MobileNetV3Large | 0.7902 | 0.9571 | 0.9669 | 0.9772 |
| MobileNetV3Small | 0.8339 | 0.8627 | 0.8741 | 0.9296 |
| NASNetMobile | 0.7982 | 0.8622 | 0.8719 | 0.8539 |
| RegNetX016 | 0.8988 | 0.9147 | 0.9415 | 0.9400 |
| RegNetX032 | 0.8532 | 0.9138 | 0.9318 | 0.9732 |
| RegNetY016 | 0.9029 | 0.9190 | 0.9564 | 0.9571 |
| RegNetY032 | 0.8583 | 0.9644 | 0.9529 | 0.9789 |
| ResNet50 | 0.8755 | 0.9612 | 0.9610 | 0.9658 |
| ResNet50V2 | 0.6957 | 0.8352 | 0.9256 | 0.9509 |
| Xception | 0.8001 | 0.8369 | 0.9311 | 0.8935 |
Table 6.
Comparison of scores of CNN models at various image resolutions for whole wheat bread.
| CNN Model | Image Resolution (pixels) |
|||
|---|---|---|---|---|
| 224 224 | 448 448 | 672 672 | 896 896 | |
| DenseNet121 | 0.9653 | 0.9856 | 0.9906 | 0.9947 |
| EfficientNetV2B0 | 0.9750 | 0.9903 | 0.9909 | 0.9954 |
| EfficientNetV2B1 | 0.9826 | 0.9879 | 0.9941 | 0.9972 |
| EfficientNetV2B2 | 0.9887 | 0.9844 | 0.9903 | 0.9910 |
| EfficientNetV2B3 | 0.9718 | 0.9846 | 0.9868 | 0.9972 |
| EfficientNetV2L | 0.9379 | 0.9873 | 0.9923 | 0.9906 |
| EfficientNetV2M | 0.9341 | 0.9748 | 0.9880 | 0.9941 |
| EfficientNetV2S | 0.9726 | 0.9887 | 0.9924 | 0.9960 |
| InceptionResNetV2 | 0.8996 | 0.9399 | 0.9619 | 0.9649 |
| InceptionV3 | 0.9091 | 0.9600 | 0.9699 | 0.9770 |
| MobileNet | 0.8976 | 0.9768 | 0.9776 | 0.9893 |
| MobileNetV2 | 0.9243 | 0.9662 | 0.9852 | 0.9861 |
| MobileNetV3Large | 0.9637 | 0.9838 | 0.9872 | 0.9972 |
| MobileNetV3Small | 0.9659 | 0.9866 | 0.9932 | 0.9972 |
| NASNetMobile | 0.8687 | 0.9126 | 0.9511 | 0.9441 |
| RegNetX016 | 0.9808 | 0.9896 | 0.9912 | 0.9853 |
| RegNetX032 | 0.9856 | 0.9895 | 0.9887 | 0.9892 |
| RegNetY016 | 0.9842 | 0.9931 | 0.9960 | 0.9947 |
| RegNetY032 | 0.9858 | 0.9941 | 0.9972 | 0.9972 |
| ResNet50 | 0.9825 | 0.9775 | 0.9944 | 0.9944 |
| ResNet50V2 | 0.9054 | 0.9540 | 0.9657 | 0.9816 |
| Xception | 0.8881 | 0.9608 | 0.9787 | 0.9870 |
Fig. 6.
Effect of image resolution on the classification performance of the top model.
The best models varied slightly across different image resolutions and types of bread, with RegNetY, EfficientNetV2, and MobileNetV3 often being the top models.
3.4. Experiment 3: comparing data augmentation methods
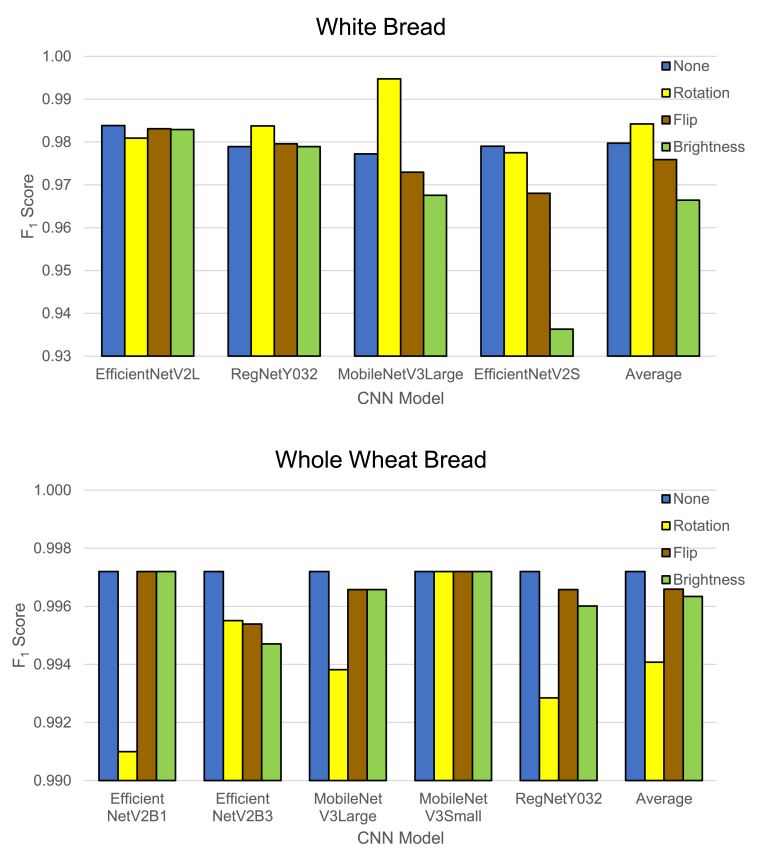
Data augmentation can aid in artificially generating more training data, which in turn can help models generalize better. Our datasets were relatively small; thus, data augmentation could improve classification accuracy. We chose the following three data augmentation methods for our task: brightness adjustment, flipping, and rotation, as specified in Section 2.3. For this experiment, we used the best image representation for each type of bread, included only the top four to five models from the previous experiment, and fixed the image resolution at 896 × 896 pixels.
The results are depicted in Fig. 7. Data augmentation did not improve classification performance in most cases for white bread. However, the results of MobileNetV3Large significantly improved when paired with random image rotation augmentation, with an impressive score of 0.9948 (precision=0.9896, recall=1). In all other cases, the classification accuracy either barely improved or decreased instead. In the case of whole wheat bread, all top models already achieved a near-perfect score of 0.9972 (precision=1, recall=0.9944), and all image augmentation methods did not improve classification accuracy.
Fig. 7.
Effects of data augmentation on the classification performance of the top models.
3.5. Experiment 4: effect of transfer learning
In this experiment, we evaluated the degree to which transfer learning affected classification accuracy. We used the best configurations from previous experiments, as follows:
White bread: MobileNetV3Large CNN model, grayscale with normalized mean representation, image resolution of 896 × 896 pixels, and image rotation augmentation.
Whole wheat bread: MobileNetV3Large CNN model, RGB representation without brightness normalization, image resolution of 896 × 896 pixels, and no data augmentation.
Without transfer learning, the CNN models suffered from severe overfitting, where they could correctly classify training images but failed to generalize to unseen images. We alleviated the problem and improved the results by using much lower learning rates and setting the value of the weight decay parameter at 0.005 to act as a regularizer. The results are shown in Table 7. The scores for both types of bread were poor without transfer learning, with whole wheat bread yielding better results than white bread as before. However, in both cases, the classification accuracy was too low for these models to be useful for consumers. Therefore, we conclude that transfer learning significantly improves classification accuracy and is necessary when the dataset is small, such as in our case.
Table 7.
Effects of transfer learning on scores.
| Type of Bread | With Transfer Learning | Without Transfer Learning |
|---|---|---|
| White | 0.9948 | 0.6212 |
| Whole Wheat | 0.9972 | 0.7594 |
3.6. Experiment 5: cross-dataset prediction
In this experiment, we aimed to evaluate how well the top model performed when the test images were of a different type of bread than the types in the training images. Considering such scenarios is essential because obtaining model training images to cover all varieties of bread that consumers may encounter is a practical challenge. To be feasible, the model must be able to correctly classify images of bread that may be slightly different from what it has been trained on. For this experiment, the classification model was MobileNetV3Large, the top model for both types of bread. The image resolution was fixed at 896 × 896 pixels.
The results are presented in Table 8. We found that scores for cross-dataset prediction were noticeably lower than for same-dataset prediction. In addition, when the models were trained with whole wheat bread and tested with white bread, the score was lower than when the models were trained with white bread and tested with whole wheat bread. This is consistent with earlier results showing that mold is more challenging to detect on white bread than on whole wheat bread. Training with the more challenging white bread images exposed the model to more complex patterns, resulting in a better classifier. The high score of 0.9402 for training with white bread and testing with whole wheat bread suggests that if the training dataset were sufficiently challenging, the model could accurately classify different types of bread. This is a fortunate scenario because, in practice, one can reasonably expect the user’s bread images to be similar to those of at least one of the common types of bread.
Table 8.
scores for cross-dataset prediction (same-dataset results given as reference).
| Training Dataset | Test Dataset |
|
|---|---|---|
| White | Whole Wheat | |
| White | 0.9948 | 0.9402 |
| Whole Wheat | 0.8591 | 0.9972 |
3.7. Discussions
3.7.1. Computational requirements in mobile devices
The model with the highest classification accuracy, MobileNetV3Large, is a relatively small CNN model with low computational requirements that can be used in real-time on smartphones. It only takes 0.08 and 1.2 s to classify a 224 × 224 and an 896 × 896 pixel image on a 2016 Google Pixel 1 smartphone (Howard et al., 2019). Therefore, the CNN model can be embedded in a smartphone as an application. Once the user captures a microscopic image of the bread, the classification result will be presented after a short processing time. Thus, the low computational requirements make checking the bread convenient for consumers.
3.7.2. Detection of mold inside bread
A smartphone’s camera with a clip-on lens can only capture the surface of the bread. If mold exists deep below the bread surface, it may either be seen as blurry dark spots or be invisible, depending on how prominent the colony is. This is a limitation of the proposed method. However, it is much more common for mold to grow on the surface of bread because the primary source of fungal contamination is spores in the ambient air that land on the bread and form colonies if the conditions are suitable (Bernardi et al., 2019). Most fungal spores in flour and other raw materials are eliminated during bread baking; thus, the chance of mold growing only internally remains low (Garcia et al., 2019).
4. Conclusions
In this paper, we proposed a novel method for consumers to detect mold in bread via microscopic images taken using clip-on lenses, with deep convolutional neural networks as the image classifier. We evaluated the method using white and whole wheat sandwich bread with natural mold growth. We found that RGB image representation worked best for whole wheat bread, while grayscale with normalized mean representation worked best for white bread. Image resolution significantly affected the classification accuracy for white bread, where a resolution of 896 × 896 pixels was necessary to obtain satisfactory performance. By contrast, mold could be accurately detected in whole wheat bread even at a low resolution of 224 × 224 pixels. Data augmentation could further improve the results in some cases. Moreover, transfer learning was necessary as the classification accuracy was poor without it. Overall, the best models were highly accurate, achieving an score of 0.99 or higher. The models could classify images of a different type of bread than the types they were trained on, with challenging training datasets resulting in higher accuracy. Research questions for the next step include how to use the clip-on lenses to take microscopic images of opaque food without cutting it and whether such images can be used to detect disease-causing microorganisms in food. Answering such questions would enable application of the proposed method to a broader variety of foods.
Funding
This work was supported by the College of Computing, Prince of Songkla University (Grant No. COC6604008S and COC6604032S).
CRediT authorship contribution statement
Panisa Treepong: Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Visualization, Writing – original draft. Nawanol Theera-Ampornpunt: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling Editor: Dr. Maria Corradini
Footnotes
The datasets are available at https://github.com/NawanolT/Bread-Mold-Datasets.
Data availability
The data used in this study is available at the link indicated in the manuscript.
References
- Alzubaidi L., Fadhel M.A., Al-Shamma O., Zhang J., Duan Y. Deep learning models for classification of red blood cells in microscopy images to aid in sickle cell anemia diagnosis. Electronics. 2020;9(3):427. [Google Scholar]
- Amodio M.L., Ceglie F., Chaudhry M.M.A., Piazzolla F., Colelli G. Potential of NIR spectroscopy for predicting internal quality and discriminating among strawberry fruits from different production systems. Postharvest Biol. Technol. 2017;125:112–121. [Google Scholar]
- Apexel . 2023. Microscope.https://www.apexeloptic.com/product-category/microscope-archives/ [Google Scholar]
- Bernardi A.O., Garcia M.V., Copetti M.V. Food industry spoilage fungi control through facility sanitization. Curr. Opin. Food Sci. 2019;29:28–34. [Google Scholar]
- Bonah E., Huang X., Aheto J.H., Yi R., Yu S., Tu H. Comparison of variable selection algorithms on vis-NIR hyperspectral imaging spectra for quantitative monitoring and visualization of bacterial foodborne pathogens in fresh pork muscles. Infrared Phys. Technol. 2020;107 [Google Scholar]
- Chollet F. Xception: deep learning with depthwise separable convolutions. Proceed. IEEE Conf.Comput.Vis. Pattern Recog. 2017:1251–1258. doi: 10.1109/cvpr.2017.195. [DOI] [Google Scholar]
- Chu X., Wang W., Ni X., Li C., Li Y. Classifying maize kernels naturally infected by fungi using near-infrared hyperspectral imaging. Infrared Phys. Technol. 2020;105 [Google Scholar]
- Degirmencioglu N., Göcmen D., Inkaya A.N., Aydin E., Guldas M., Gonenc S. Influence of modified atmosphere packaging and potassium sorbate on microbiological characteristics of sliced bread. J. Food Sci. Technol. 2011;48:236–241. doi: 10.1007/s13197-010-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.V., da Pia A.K.R., Freire L., Copetti M.V., Sant’Ana A.S. Effect of temperature on inactivation kinetics of three strains of Penicillium paneum and P. roqueforti during bread baking. Food Control. 2019;96:456–462. [Google Scholar]
- Godbole V., Dahl G.E., Gilmer J., Shallue C.J., Nado Z. 2023. Deep Learning Tuning Playbook.http://github.com/google-research/tuning_playbook [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proceed. IEEE Conf.Comput.Vis. Pattern Recog. 2016:770–778. doi: 10.1109/cvpr.2016.90. [DOI] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Identity mappings in deep residual networks. Eur.Conf. Comput. Vis. 2016:630–645. doi: 10.1007/978-3-319-46493-0_38. [DOI] [Google Scholar]
- Howard A.G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H. 2017. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. [DOI] [Google Scholar]
- Howard A., Sandler M., Chu G., Chen L.C., Chen B., Tan M., Wang W., Zhu Y., Pang R., Vasudevan V., Le Q.V., Adam H. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2019. Searching for MobileNetV3. [DOI] [Google Scholar]
- Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Densely connected convolutional networks. Proceed. IEEE Conf.Comput.Vis. Pattern Recog. 2017:4700–4708. doi: 10.1109/cvpr.2017.243. [DOI] [Google Scholar]
- Iqbal M.S., El-Ashram S., Hussain S., Khan T., Huang S., Mehmood R., Luo B. Efficient cell classification of mitochondrial images by using deep learning. J. Opt. 2019;48:113–122. [Google Scholar]
- Jubayer F., Soeb J.A., Mojumder A.N., Paul M.K., Barua P., Kayshar S., Akter S.S., Rahman M., Islam A. Detection of mold on the food surface using YOLOv5. Curr. Res. Food Sci. 2021;4:724–728. doi: 10.1016/j.crfs.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D.P., Ba J. 2014. Adam: A Method for Stochastic Optimization. [DOI] [Google Scholar]
- knowthystore . 2023. TINYSCOPE™ Smartphone Microscope Lens.https://knowthystore.com/products/microscope-camera-lens [Google Scholar]
- Liu R., Dai W., Wu T., Wang M., Wan S., Liu J. AIMIC: deep learning for microscopic image classification. Comput. Methods Progr. Biomed. 2022;226:1–12. doi: 10.1016/j.cmpb.2022.107162. [DOI] [PubMed] [Google Scholar]
- Mahbod A., Schaefer G., Ecker R., Ellinger I. Pollen grain microscopic image classification using an ensemble of fine-tuned deep convolutional neural networks. Proceed. Int.Confe.Pattern Recog. Int.Workshops Challeng.: Virtual Event, Part I. 2021:344–356. [Google Scholar]
- Meng N., Lam E.Y., Tsia K.K., So H.K.H. Large-scale multi-class image-based cell classification with deep learning. IEEE J.Biomed. Health Inform. 2018;23(5):2091–2098. doi: 10.1109/JBHI.2018.2878878. [DOI] [PubMed] [Google Scholar]
- Meng R., Lv Z., Yan J., Chen G., Zhao F., Zeng L., Xu B. Development of spectral disease indices for southern corn rust detection and severity classification. Rem. Sens. 2020;12(19):3233. [Google Scholar]
- Mishra B.K., Rath A.K., Tripathy P.K. Detection of fungal contagion in food items using enhanced image segmentation. Int. J. Eng. Adv. Technol. 2019;8(6):1748–1757. [Google Scholar]
- Nguyen L.D., Lin D., Lin Z., Cao J. Deep CNNs for microscopic image classification by exploiting transfer learning and feature concatenation. IEEE Int.Sympos.Cir.Syst. 2018:1–5. [Google Scholar]
- Pissard A., Marques E.J.N., Dardenne P., Lateur M., Pasquini C., Pimentel M.F., Pierna J.A.F., Baeten V. Evaluation of a handheld ultra-compact NIR spectrometer for rapid and non-destructive determination of apple fruit quality. Postharvest Biol. Technol. 2021;172:1–10. [Google Scholar]
- Qin J., Chao K., Kim M.S., Lu R., Burks T.F. Hyperspectral and multispectral imaging for evaluating food safety and quality. J. Food Eng. 2013;118(2):157–171. [Google Scholar]
- Qin F., Gao N., Peng Y., Wu Z., Shen S., Grudtsin A. Fine-grained leukocyte classification with deep residual learning for microscopic images. Comput. Methods Progr. Biomed. 2018;162:243–252. doi: 10.1016/j.cmpb.2018.05.024. [DOI] [PubMed] [Google Scholar]
- Radosavovic I., Kosaraju R.P., Girshick R., He K., Dollár P. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2020. Designing network design spaces; pp. 10428–10436. [DOI] [Google Scholar]
- Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. [Google Scholar]
- Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. MobileNetV2: inverted residuals and linear bottlenecks. Proceed. IEEE Conf.Comput.Vis. Pattern Recog. 2018:4510–4520. doi: 10.1109/cvpr.2018.00474. [DOI] [Google Scholar]
- SmartLens C.U. 2023. All Products.http://chula-smartlens.lnwshop.com/category [Google Scholar]
- SmartMicroOptics . 2023. Homepage - SmartMicroOptics.https://smartmicrooptics.com/ [Google Scholar]
- Sun K., Wang Z., Tu K., Wang S., Pan L. Recognition of mould colony on unhulled paddy based on computer vision using conventional machine-learning and deep learning techniques. Sci. Rep. 2016;6(1):1–14. doi: 10.1038/srep37994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the inception architecture for computer vision. Proceed. IEEE Conf.Comput.Vis. Pattern Recog. 2016:2818–2826. doi: 10.1109/cvpr.2016.308. [DOI] [Google Scholar]
- Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. Proceeding of the Thirty-First AAAI Conference on Artificial Intelligence. 2017. Inception-v4, Inception-ResNet and the impact of residual connections on learning. [DOI] [Google Scholar]
- Tan M., Le Q. EfficientNetV2: smaller models and faster training. Int.Conf. Mach. Learn. 2021:10096–10106. doi: 10.48550/arXiv.2104.00298. [DOI] [Google Scholar]
- Teerachaichayut S., Ho H.T. Non-destructive prediction of total soluble solids, titratable acidity and maturity index of limes by near infrared hyperspectral imaging. Postharvest Biol. Technol. 2017;133:20–25. [Google Scholar]
- Wahid M.F., Hasan M.J., Alom M.S. Deep convolutional neural network for microscopic bacteria image classification. 5th Int.Conf. Adv. Electric. Eng. 2019:866–869. [Google Scholar]
- Xie C., Yang C., He Y. Hyperspectral imaging for classification of healthy and gray mold diseased tomato leaves with different infection severities. Comput. Electron. Agric. 2017;135:154–162. [Google Scholar]
- Zoph B., Vasudevan V., Shlens J., Le Q.V. Learning transferable architectures for scalable image recognition. Proceed. IEEE Conf.Comput.Vis. Pattern Recog. 2018:8697–8710. doi: 10.1109/cvpr.2018.00907. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in this study is available at the link indicated in the manuscript.