Abstract
Although miR-5195-3p has been acknowledged for its tumor suppressor role in diverse cancer categories, its precise functions and mechanisms concerning melanoma have not been comprehensively elucidated. In this study, we employed quantitative reverse transcription PCR, Western blot analysis, and immunohistochemistry staining to investigate the expression patterns of miR-5195-3p and poly (rC) binding protein 2 (PCBP2) in melanoma tissues compared to adjacent tissues. Our findings revealed downregulation of miR-5195-3p and upregulation of PCBP2 in melanoma tissues. Through the implementation of a luciferase reporter assay, we successfully identified PCBP2 as a newly discovered target of miR-5195-3p in melanoma cells. Enforced expression of miR-5195-3p via mimics inhibited cell proliferation and migration in A375 and A2058 cells, as demonstrated by CCK-8 and transwell migration assays. In melanoma cells, reintroduction of PCBP2 partially reversed the inhibitory effects of miR-5195-3p overexpression. Treatment with LY294002, an inhibitor of the PI3K/AKT signaling pathway, also reversed the effects of PCBP2 in melanoma cells. Furthermore, our results suggest that miR-5195-3p inhibits the activation of the PI3K/AKT signaling pathway in melanoma by inhibiting PCBP2. In conclusion, our research has identified the miR-5195-3p targeting of the PCBP2-mediated PI3K/AKT signaling pathway as a potential therapeutic target for melanoma treatment.
Keywords: Melanoma, MIRN5195-3p, Poly(rC) binding protein 2, PI3K/AKT signaling
1. Introduction
Melanoma is a highly malignant skin cancer that originates from melanocytes located in the epidermis and considered to be one of the most aggressive forms of skin cancer worldwide [1,2]. Over the past few years, there has been a significant surge in the occurrence of melanoma, a type of skin cancer that represents around 3% of all malignant skin tumors [3]. Despite notable advancements in the treatment modalities for melanoma, including surgical excision, radiotherapy, chemotherapy, and immunotherapy, both as standalone treatments and in combination [[4], [5], [6]], the unfavorable prognosis linked to the disease underscores the importance of exploring the underlying mechanisms that drive melanoma progression and metastasis [7].
MicroRNAs (miRNAs/miRs) are an abundant group of short non-coding RNAs, typically ranging from 19 to 25 nucleotides in length, with the ability to regulate over 30% of the protein-coding genes in the human genome [8,9]. MiRNAs act as gene regulators by binding to the 3′-untranslated region (3′-UTR) of the target mRNA, leading to the inhibition of mRNA translation or mRNA degradation [10]. Dysregulated expression of miRNAs in cancers can result in their classification as either oncogenes or tumor suppressors, thereby influencing cancer cell proliferation, migration, and apoptosis [11,12]. A miRNA of particular interest in this study is miR-5195-3p, which has been found to play a role in cellular processes associated with tumor progression. For example, Yang and colleagues [13] reported that miR-5195-3p acts as a tumor suppressor by inhibiting the proliferation and cell cycle progression of glioma cells through the targeting of BIRC2. Downregulation of NEDD9 by ectopically expressed miR-5195-3p resulted in the attenuation of proliferative activity and induction of apoptosis in osteosarcoma cells [14]. Similarly, miR-5195-3p has been identified as a tumor suppressor in various cancers, including non-small cell lung cancer [15], prostate cancer [16] and ovarian cancer [17]. However, the biological functions of miR-5195-3p in melanoma remain poorly understood to date.
PCBP2 is an RNA-binding protein that specifically binds to single-stranded poly(C) with a strong affinity, which plays a crucial role in regulating various aspects of RNA, including stabilizing RNA molecules, processing mRNA, and controlling translation processes [18]. Previous studies have shown that PCBP2 plays a role in promoting cell proliferation and tumor progression, such as gastric cancer [19] and glioma [20]. A recent review by Yuan et al. [21] provided a comprehensive explanation of the molecular mechanisms of PCBP2 in cancer and concluded that PCBP2 acts as an oncogene by promoting tumorigenesis and cancer metastasis. PCBP2, working together with lnc030, exerts a notable influence on the maintenance of breast cancer stem cell stemness, which is accomplished by activating the PI3K/AKT signaling pathway downstream and boosting cholesterol synthesis [22]. The PI3K/AKT signaling pathway has been identified as playing a significant role in both the prevention and treatment of melanoma [23]. Through the utilization of TargetScan software, we successfully identified a binding site for miR-5195-3p within the 3′-UTR region of PCBP2 at position 374–380 bp. The functional understanding of the miR-5195-3p/PCBP2 axis in melanoma cells is limited, despite the known association between PCBP2 and the PI3K/AKT signaling pathway. Therefore, we proposed a hypothesis suggesting that miR-5195-3p could play a crucial role in melanoma cells by regulating the PCBP2-mediated PI3K/AKT signaling pathway.
Our study aimed to investigate the potential role of miR-5195-3p/PCBP2 axis in melanoma by analyzing their expression levels in melanoma tissues. Subsequently, we examined the relationship between miR-5195-3p and PCBP2, followed by evaluating the functional impact of the miR-5195-3p/PCBP2 axis on the proliferation and migration of human melanoma cells. Moreover, we aimed to explore the association between miR-5195-3p/PCBP2 and the PI3K/AKT molecular signaling pathway.
2. Materials and methods
2.1. Tissue specimens
Between October 2019 and May 2021, we gathered primary human melanomas and their corresponding adjacent skin specimens from 45 melanoma patients. The patients underwent pathological diagnosis by two pathologists at Guangdong Provincial People's Hospital (Guangdong, China). Prior to the tissue collection, none of the 45 melanoma patients had received any anticancer therapies, such as radio-chemotherapy, radiotherapy, immunotherapy, or other similar treatments. Before being enrolled in the study, all participants were provided with the research protocol and required to give written informed consent. The authors of the study did not have access to any personally identifiable information about the participants either during or after the data collection process. The study received approval from the Ethics Committee of the Guangdong Provincial People's Hospital (Approval number: CP-2019A7D, Guangdong, China).
2.2. Immunohistochemistry (IHC) staining
Some of the collected tissue samples were used to create paraffin-embedded sections, which were then cut into sections that were 4 μm in thickness. The sections were deparaffinized using xylene and rehydrated using graded ethanol. To retrieve antigens, the sections were treated with 10 mmol/L sodium citrate buffer (pH = 6.0) and heated for 15 min in a microwave. To block endogenous peroxidase activity, the sections were immersed in a 3% hydrogen peroxide solution for 10 min. Subsequently, the sections were labeled using a rabbit anti-human PCBP2 antibody (obtained from Abcam, Cambridge, MA, USA) and incubated overnight at 4 °C. They were then washed with PBST and treated with a secondary antibody conjugated with streptavidin-horseradish peroxidase for 1 h. Following that, the segments were briefly counterstained with hematoxylin for 30 s, covered with a slip and viewed under a light microscope at a magnification of 250 × .
2.3. Cell transfection and treatments
The human melanoma cell lines A375 and A2058 were procured from the American Type Culture Collection (Manassas, VA, USA) and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) at 37 °C under 5% CO2. RiboBio Company (Guangzhou, China) provided the synthetic miR-5195-3p mimics, negative control mimics (miR-NC), PCBP2 overexpression plasmid pcDNA3.1-PCBP2, and empty pcDNA3.1 plasmid. The PI3K specific inhibitor, LY294002, was obtained from Sigma-Aldrich. For the analysis of PI3K/AKT signaling, A375 and A2058 cells were transfected with 2.5 μg of pcDNA3.1-PCBP2 or empty pcDNA3.1 for 48 h, and subsequently treated with 50 μmol/L LY294002 at 37 °C for another 48 h. The transfection of oligonucleotides and plasmids was carried out using the Lipofectamine 3000 transfection reagent (Invitrogen, USA) following the protocols provided by the manufacturer.
2.4. TargetScan prediction
We first visited the TargetScan website at https://www.targetscan.org/vert_71/to access the online tool for miRNA target prediction. By choosing the organism “human” and entering the miRNA “miR-5195-3p”, we started the search and selected PCBP2 as a potential target of miR-5195-3p.
2.5. Dual-luciferase reporter assay
According to the predicted complementary binding of miR-5195-3p located at 374–380 bp of PCBP2 3′-UTR using TargetScan software (https://www.targetscan.org/vert_71/), GenePharma (Shanghai, China) chemically synthesized pGL3-PCBP2-3′-UTR wild type (WT) and pGL3-PCBP2-3′-UTR mutant (MUT) plasmids by inserting the sequences containing the WT binding site and the sequences with MUT binding site into the pGL3 Dual-Luciferase reporter plasmid. Ninety-six-well plates were used to plate A375 and A2058 cells at a density of 1 × 104 cells per well and these cells were then allowed to grow until they reached 70% confluence. Subsequently, Lipofectamine 3000 reagent was utilized to co-transfect the cells with miR-5195-3p mimics or miR-NC (100 nmol/L) and pGL3 vectors (100 ng) containing PCBP2 sequences (WT/MUT) for 48 h at 37 °C. After being harvested, the cells were analyzed for their relative luciferase activities using a kit for dual-luciferase reporter assay system (Promega, Madison, WI, USA).
2.6. Quantitative reverse transcription PCR
TRIzol reagent (Thermo Fisher Scientific, Inc.) was employed for the extraction of total RNA from both tissue samples and cells, followed by reverse transcription using SuperScript IV reverse transcriptase (Thermo Fisher Scientific, Inc.). For detecting miR-5195-3p expression and PCBP2 mRNA expression, TaqMan MicroRNA PCR Kit (from Applied Biosystems) and SYBR Premix Ex Taq™ Kit (from Takara Bio) were utilized, respectively, for carrying out quantitative reverse transcription PCR. The reaction conditions were established as follows: 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. The relative expression fold of miR-5195-3p and PCBP2 was calculated using the 2−ΔΔCt method and was normalized with respect to U6 and GAPDH, respectively. The primers used for the PCR reaction included miR-5195-3p forward: 5′-AACCCCTAAGGCAACTGGAT-3′ and reverse: 5′-GAACATGTCTGCGTATCTC-3′; U6 forward: 5′-CTCGCTCGGCAGAACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′; PCBP2 forward: 5′-CTCCGGTCATCTTTGCAGGT-3′ and reverse: 5′-CTGAGCAGATGCATCCAAACC-3′; GAPDH forward: 5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse: 5′-GCATCGCCCCACTTGATTTT-3′.
2.7. Cell counting Kit-8 (CCK-8) assay
Cellular proliferation was assessed by carrying out CCK-8 assay, following the instructions provided by the manufacturer. In brief, A375 and A2058 cells were seeded at a density of 3000 cells per well in 96-well plates. After 24, 48, and 72 h, 10 μl of CCK-8 solution (Beyotime, China) was added to cells in each well of the plates. The plates were then incubated at 37 °C for 2 h, after which the optical density (OD) of the cells was measured at 450 nm using a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA).
2.8. Transwell migration assay
To assess migration ability, transwell assay was conducted using transwell chambers (Corning, NY, USA) with 8-μm pore size. In short, we seeded 1 × 105 cells in 200 μl of serum-free medium into the upper chamber of the transwell inserts. The lower chambers were filled with 600 μl of medium supplemented with 20% FBS, which served as a chemoattractant. After incubation for 24 h at 37 °C, the cells that migrated to the bottom chambers were fixed with 100% methanol, stained with 0.1% crystal violet for 20 min at room temperature, and photographed. We counted the migrated cells in five randomly selected visual fields using an inverted microscope (Olympus, Tokyo, Japan).
2.9. Western blot analysis
The protein sample was obtained using RIPA lysis buffer (Beyotime, China), and the concentration of proteins was determined using the BCA assay (Beyotime, China). Following dilution in loading buffer, an equal amount of protein (30 μg) was separated via 8–12% SDS gel electrophoresis and subsequently transferred onto a PVDF membrane (Millipore, USA). The PVDF membranes were incubated with 5% non-fat milk for 2 h to block non-specific binding, and then probed with primary antibodies against PCBP2, PCNA, E-cadherin, p-PI3K, PI3K, p-AKT, AKT, and GAPDH (Abcam, Cambridge, UK) at 4 °C overnight. Before visualized with an enhanced chemiluminescence detection kit (Santa Cruz, TX, USA), protein bands were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature.
2.10. Statistical analysis
Data were analyzed by GraphPad Prism version 8.0 (GraphPad Software Inc.) and presented as mean ± standard deviation (SD). Statistical analysis involved the use of Student's t-test to compare two groups, while one-way analysis of variance (ANOVA) followed by Tukey's post hoc test was applied for comparisons among multiple groups. Spearman's correlation analysis was employed to determine the correlation between miR-5195-3p and PCBP2 mRNA expression. The p-values less than 0.05 wereconsidered to be statistically significant.
3. Results
3.1. The expression levels of miR-5195-3p and PCBP2 in melanoma tissues
Tumor tissues and corresponding adjacent tissues were obtained from melanoma patients in a paired manner. These tissues were then subjected to quantitative reverse transcription PCR to assess the expression level of miR-5195-3p. Fig. 1A displayed a significant reduction in miR-5195-3p expression in human melanoma tissues compared to adjacent tissues. Subsequently, the protein expression of PCBP2 was evaluated in both tumor and adjacent tissues. Immunohistochemical staining for PCBP2 demonstrated significantly higher positive staining in melanoma tissues compared to adjacent normal tissue samples (Fig. 1B). Consistently, representative images obtained from Western blot analysis confirmed the upregulation of PCBP2 in tumor tissues compared to normal tissues (Fig. 1C).
Fig. 1.
The expression levels of miR-5195-3p and PCBP2 in melanoma tissues. (A) The expression level of miR-5195-3p was determined in 45 paired melanoma tissues and adjacent tissues using quantitative reverse transcription PCR. ***p < 0.001, by Student's t-test; (B) Representative immunohistochemical staining of PCBP2 in melanoma and normal tissues (Scale bar: 100 μm; 250 × magnification). (C) Representative images of Western blot analysis on four paired tumor and normal tissues derived from melanoma patients were shown. **p < 0.01, by Student's t-test.
3.2. PCBP2 expression was negatively regulated by miR-5195-3p through targeted binding
Next, we examined the association between miR-5195-3p and PCBP2. By employing TargetScan software, we generated predictions for the binding sequences of PCBP2 to miR-5195-3p (Fig. 2A). To validate this interaction, we conducted dual-luciferase reporter assays. As shown in Fig. 2B, the relative luciferase activity of A375 and A2058 cells, transfected with miR-5195-3p mimics, was significantly reduced in the PCBP-WT transfection group compared to the control-mimics group. However, no significant change in luciferase activity was observed in the PCBP2-MUT transfection group, indicating the specificity of the interaction. Furthermore, we investigated the regulatory influence of miR-5195-3p on PCBP2 expression in melanoma cells. Our findings demonstrated that the transfection of miR-5195-3p mimics significantly suppressed both the mRNA levels (Fig. 2C) and protein expression (Fig. 2D) of PCBP2 in A375 and A2058 cells. Furthermore, we observed a notable increase in PCBP2 mRNA expression in tumor tissues compared to adjacent tissues (Fig. 2E). Additionally, a significant negative correlation was detected between miR-5195-3p and PCBP2 mRNA expression in melanoma tumor tissues (Fig. 2F, p = 0.0072).
Fig. 2.
Targeted binding of miR-5195-3p to PCBP2. (A) Sequence analysis of binding sites between miR-5195-3p and PCBP2. (B) Dual-luciferase reporter assay was performed in A375 and A2058 cells after co-transfection with miR-5195-3p mimics or miR-NC and PCBP2-WT or PCBP2-MUT plasmids. (C–D) The expression levels of PCBP2 mRNA and protein were determined in A375 and A2058 cells transfected with miR-5195-3p mimics or miR-NC. (E) The expression level of PCBP mRNA was determined in 45 paired melanoma tissues and adjacent tissues using quantitative reverse transcription PCR. (F) Spearman's correlation analysis was performed to assess the association between miR-5195-3p and PCBP2 mRNA expression in melanoma tissues. The experiments were replicated three times and presented as mean ± SD. **p < 0.01, ***p < 0.001, by Student's t-test.
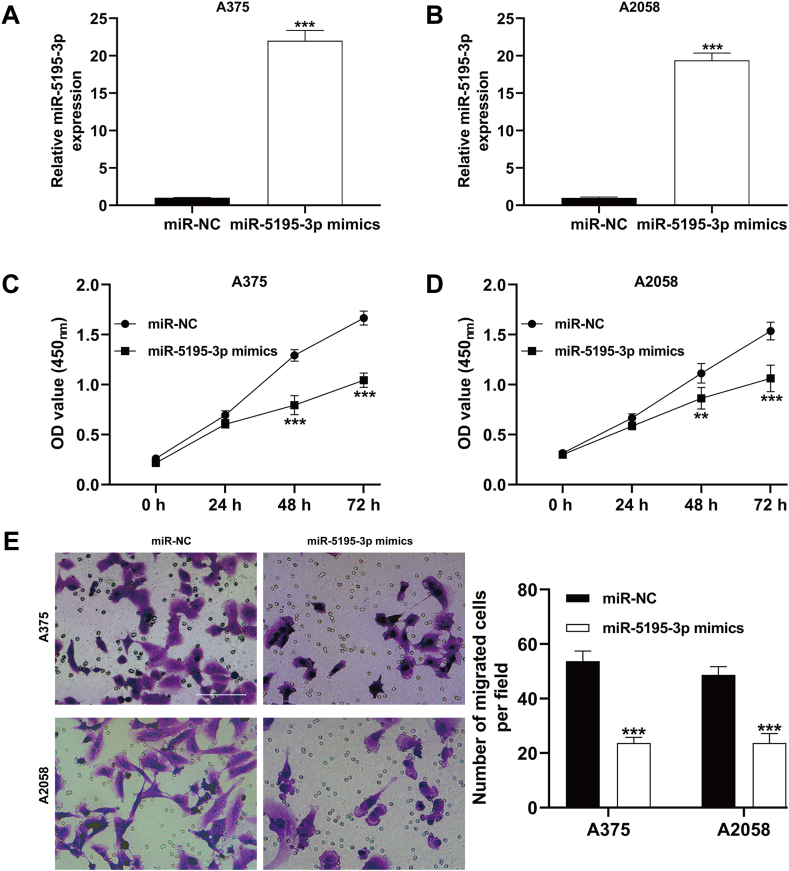
3.3. Overexpression of miR-5195-3p led to a reduction in cell proliferation and migration in melanoma cells
To examine the regulatory function of miR-5195-3p in melanoma progression, we conducted proliferation and migration assays on melanoma cells following miR-5195-3p overexpression. We transfected A375 and A2058 cells with miR-5195-3p mimics, and verified the transfection efficiency using quantitative reverse transcription PCR (Fig. 3A and B). The CCK-8 assay demonstrated that miR-5195-3p overexpression significantly suppressed the proliferation of A375 and A2058 cells compared to control cells (Fig. 3C and D). In addition, the transwell migration assay revealed that the number of migrated cells per field was significantly decreased in the miR-5195-3p overexpression group compared to the control group in both A375 (miR-5195-3p mimics vs. miR-NC: 23.7 ± 2.1 vs. 53.7 ± 3.8, p < 0.001) and A2058 (miR-5195-3p mimics vs. miR-NC: 23.3 ± 3.5 vs. 48.7 ± 3.1, p < 0.001) cells (Fig. 3E).
Fig. 3.
Overexpression of miR-5195-3p suppressed cell proliferation and migration in melanoma cells. A375 and A2058 cells were transfected with miR-5195-3p mimics or miR-NC for 48 h. (A–B) The expression level of miR-5195-3p was determined in transfected A375 and A2058 cells using quantitative reverse transcription PCR. (C–D) CCK-8 assay was performed to analyze the effects of miR-5195-3p overexpression on A375 and A2058 cell proliferation. (E) Cell migration was assessed in A375 and A2058 cells after miR-5195-3p overexpression by transwell assay. Scale bars = 50 μm. The experiments were replicated three times and presented as mean ± SD. **p < 0.01, ***p < 0.001, by Student's t-test.
3.4. The inhibitory effect of miR-5195-3p on melanoma cells was partially reversed by overexpression of PCBP2
Next, we conducted rescue experiments to assess the involvement of PCBP2 as a downstream regulator in the miR-5195-3p-mediated regulation of melanoma cell proliferation and migration. Firstly, we confirmed the overexpression of PCBP2 in A375 and A2058 cells after transfection with the pcDNA3.1-PCBP2 vector using Western blot analysis (Fig. 4A). Next, A375 and A2058 cells were co-transfected with pcDNA3.1-PCBP2 together with or without miR-5195-3p mimics. Following transfection, the expression of PCBP2 was significantly reduced in miR-5195-3p overexpression cells, while the decreased PCBP2 expression was restored following pcDNA3.1-PCBP2 co-transfection in both A375 and A2058 cells (Fig. 4B). We observed that the restoration of PCBP2 partially rescued the decreased proliferation rate (Fig. 4C and D) and impaired migration ability (Fig. 4E) induced by miR-5195-3p overexpression in A375 and A2058 cells. Taken together, the data from the rescue experiments indicate that PCBP2 overexpression can partially reverse the inhibitory effect of miR-5195-3p, providing further evidence of the targeting relationship between miR-5195-3p and PCBP2 in melanoma cells.
Fig. 4.
Overexpression of PCBP2 partially reversed the inhibitory effect of miR-5195-3p in melanoma cells. (A) Protein level of PCBP2 was measured in A375 and A2058 cells after transfection with pcDNA3.1-PCBP2 or pcDNA3.1. (B) Protein level of PCBP2 was detected in A375 and A2058 cells after co-transfected with pcDNA3.1-PCBP2 in combination with or without miR-5195-3p mimics. (C–D) CCK-8 assay was performed to analyze cell proliferation in A375 and A2058 cells after co-transfected with pcDNA3.1-PCBP2 in combination with or without miR-5195-3p mimics. (E) Cell migration was assessed in A375 and A2058 cells after co-transfected with pcDNA3.1-PCBP2 in combination with or without miR-5195-3p mimics. Scale bars = 50 μm. The experiments were replicated three times and presented as mean ± SD. **p < 0.01, ***p < 0.001, compared with miR-NC + pcDNA3.1; ##p < 0.01, ###p < 0.001, compared with miR-5195-3p mimics + pcDNA3.1, by ANOVA with Tukey's post hoc test.
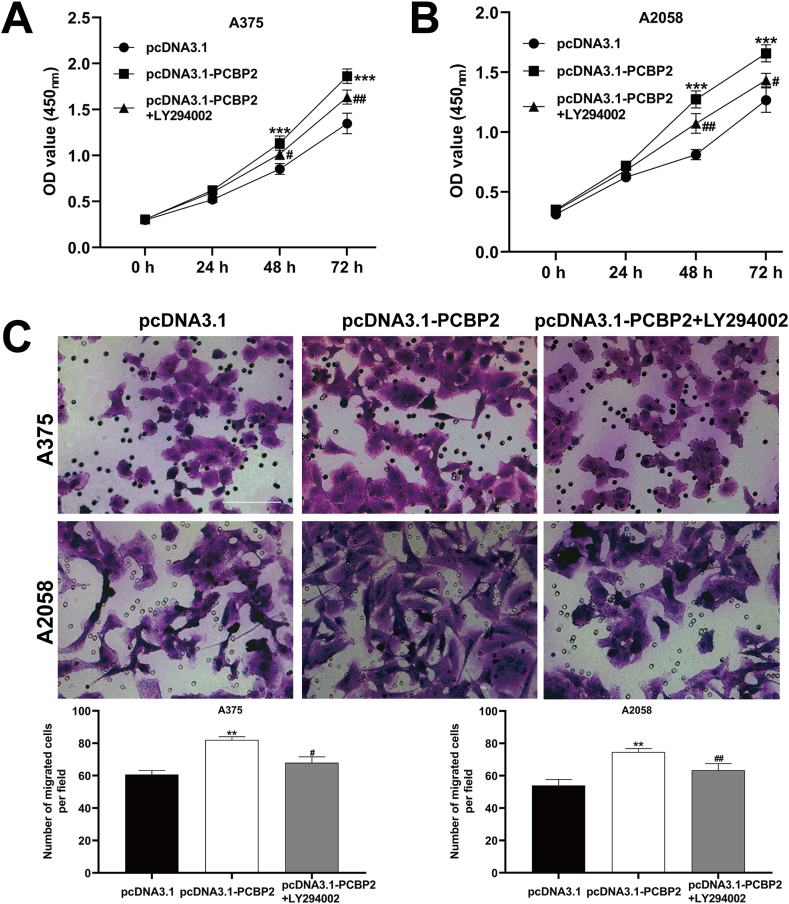
3.5. Blocking PI3K/AKT pathway attenuated the effects of PCBP2 on melanoma cell proliferation and migration
To determine the role of the PI3K/AKT pathway in PCBP2-mediated melanoma cell proliferation and migration, we treated A375 and A2058 cells transfected with pcDNA3.1-PCBP2 with the PI3K/AKT pathway inhibitor LY294002. The CCK-8 assay showed that the increased cell proliferation rate induced by pcDNA3.1-PCBP2 was rescued by LY294002 treatment in both A375 (Fig. 5A) and A2058 (Fig. 5B) cells. Additionally, the transwell assay revealed that PCBP2 overexpression significantly increased the number of migrated cells, which was attenuated by LY294002 treatment in both A375 and A2058 cells (Fig. 5C). These findings suggest that the PI3K/AKT pathway is involved in the PCBP2-mediated effects on melanoma cell proliferation and migration.
Fig. 5.
The effects of inhibited PI3K/AKT pathway on PCBP2-mediated cell proliferation and migration in melanoma cells. A375 and A2058 cells were transfected with 2.5 μg pcDNA3.1-PCBP2, followed by treatment with LY294002 (50 μmoL/l) for 48 h at 37 °C. (A–B) Cell proliferation rate was determined using CCK-8 assay in A375 and A2058 cells. (C) Transwell assay was applied to analyze cell migration ability in A375 and A2058 cells. Scale bars = 50 μm. The experiments were replicated three times and presented as mean ± SD. **p < 0.01, ***p < 0.001, compared with pcDNA3.1; #p < 0.05, ##p < 0.01, compared with pcDNA3.1-PCBP2, by ANOVA with Tukey's post hoc test.
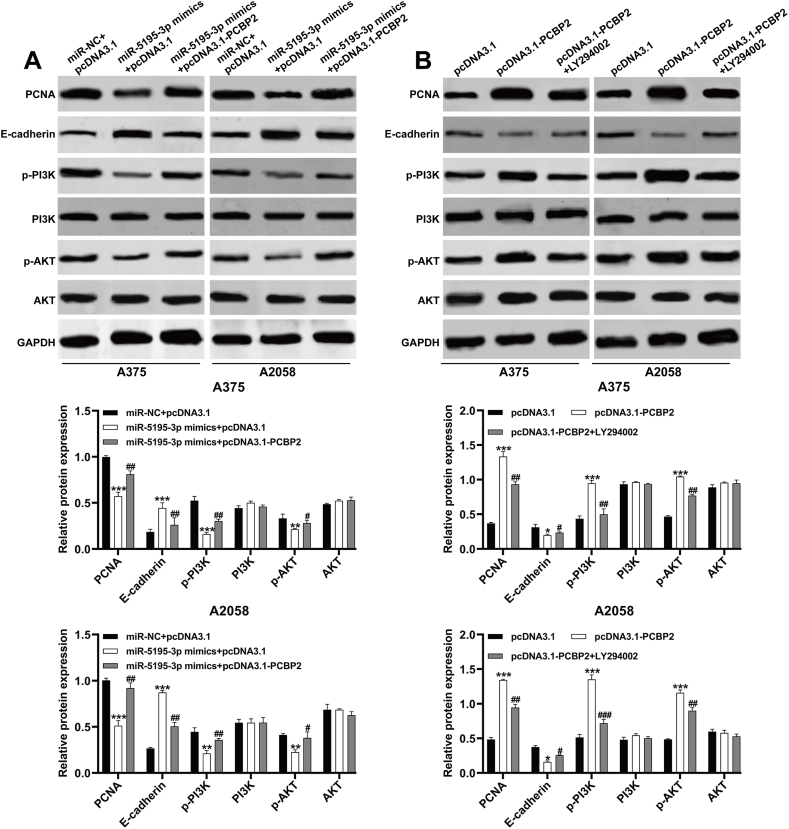
3.6. miR-5195-3p suppressed the PI3K/AKT signaling pathway in melanoma cells via PCBP2
To elucidate the regulatory role of the miR-5195-3p/PCBP2 axis on the PI3K/AKT signaling pathway in melanoma cells, we first examined the expression levels of PI3K/AKT pathway-related proteins in A375 and A2058 cells co-transfected with miR-5195-3p mimics and pcDNA3.1 or pcDNA3.1-PCBP2. As shown in Fig. 6A, the decreased expression levels of p-PI3K and p-AKT due to miR-5195-3p were restored in A375 and A2058 cells upon co-transfection with pcDNA3.1-PCBP2. However, the total expression of PI3K and AKT remained unchanged in all groups. Additionally, the downregulation of PCBA and upregulation of E-cadherin induced by miR-5195-3p mimics were reversed in A375 and A2058 cells after co-transfection with pcDNA3.1-PCBP2. Furthermore, we observed that overexpression of PCBP2 resulted in increased expression levels of PCNA, p-PI3K, and p-AKT, as well as decreased expression of E-cadherin, which were all attenuated in A375 and A2058 cells after treatment with LY294002 (Fig. 6B). These findings suggest that miR-5195-3p inhibits the activation of the PI3K/AKT signaling pathway by downregulating PCBP2 in melanoma cells.
Fig. 6.
miR-5195-3p suppressed the PI3K/AKT signaling pathway in melanoma cells via PCBP2. (A) The protein expression levels of PCNA, E-cadherin, p-PI3K, PI3K, p-AKT and AKT were measured in A375 and A2058 cells after co-transfection with miR-5195-3p mimics and pcDNA3.1-PCBP2 or pcDNA3.1. **p < 0.01, ***p < 0.001, compared with miR-NC + pcDNA3.1; #p < 0.05, ##p < 0.01, compared with miR-5195-3p mimics + pcDNA3.1, by ANOVA with Tukey's post hoc test. (B) The protein expression levels of PCNA, E-cadherin, p-PI3K, PI3K, p-AKT and AKT were measured in A375 and A2058 cells transfected with pcDNA3.1-PCBP2, followed by treatment with LY294002. *p < 0.05, ***p < 0.001, compared with pcDNA3.1; #p < 0.05, ##p < 0.01, ###p < 0.001, compared with pcDNA3.1-PCBP2, by ANOVA with Tukey's post hoc test. The experiments were replicated three times and presented as mean ± SD.
4. Discussion
Growing evidence suggests that dysregulation of miRNAs expression and function plays a critical role in the malignant progression of melanoma [24]. Hence, the identification of novel miRNAs and their functional targets involved in melanoma progression presents an opportunity to develop promising therapeutic targets for this disease [25]. This study has uncovered the mechanism underlying the miR-5195-3p/PCBP2 axis in melanoma, which can serve as a valuable theoretical foundation for early diagnosis and potential therapeutic interventions for this disease. Here, we demonstrated that the expression of miR-5195-3p was decreased in melanoma tissues compared to adjacent normal tissues. This finding is consistent with previous studies that have also observed reduced expression of miR-5195-3p in ovarian cancer samples, and its downregulation has been associated with metastasis [26]. MiR-5195-3p expression was also reported to be downregulated in acute myeloid leukemia [27] and non-small-cell lung cancer [28]. Nevertheless, there exist reports that indicated that miR-5195-3p expression was increased in hepatocellular carcinoma [29] and glioma [30], which are contrary to our current results. The inconsistent findings regarding miR-5195-3p expression in various tumors may be attributed to its context-dependent functions in different cancer types. When considering our current findings along with the results of previous studies, it seems that changes in miR-5195-3p expression may play a critical role in the development and malignant progression of melanoma.
We further investigated the functional role of miR-5195-3p in the malignant progression of melanoma. Our results demonstrated that overexpression of miR-5195-3p using mimics led to a significant reduction in proliferation and migration of A375 and A2058 cells. Recent studies have demonstrated that therapeutic miRNAs, including miRNA mimics and miRNA inhibitors, have the potential to treat melanoma by restoring dysregulated miRNAs to their normal levels [31]. Our findings are consistent with previous studies showing that the transfection of miR-5195-3p mimic significantly increased the expression level of miR-5195-3p and effectively reduced the proliferation and invasion of bladder cancer cells [32]. The similar anti-tumor effects of miR-5195-3p have been also observed in several other tumors, including prostate cancer [16], ovarian cancer [17], colon cancer [33] and pituitary adenoma [34]. Conversely, the study by Zhang et al. [30] showed that increased miR-5195-3p expression induced by p72 overexpression was involved in the promotive effects of p72 in malignant migration and invasion of glioma cells. It is possible that the varying results from previous studies and our current findings may be attributed to differences in tumor origins and the diverse mechanisms involved in tumorigenesis. Based on our results, it appears that miR-5195-3p functions as a tumor suppressor in melanoma by negatively regulating cell proliferation and migration.
To further investigate the underlying molecular mechanisms of miR-5195-3p in regulating melanoma cells, we utilized bioinformatics tools to predict potential target genes of miR-5195-3p and confirmed that PCBP2 was a novel direct target of miR-5195-3p. After confirming the binding between miR-5195-3p and PCBP2, we proceeded to investigate the functional role of the miR-5195-3p/PCBP2 axis in melanoma progression. Consistent with our prediction, we observed that overexpression of PCBP2 counteracted the suppressive effects on melanoma cell proliferation and migration mediated by miR-5195-3p. To the best of our knowledge, PCBP2 has been shown to promote the malignant properties of tumor cells and has been identified as a target of various miRNAs, such as miR-490-3p in bladder cancer [35] and miR-34a in gastric cancer [36]. In line with our data, Huang et al. [37] also demonstrated that miR-5195-3p mimics reduced cell proliferation, migration, and invasion by directly targeting PCBP2 in colorectal cancer. In terms of molecular mechanisms, our study demonstrated that miR-5195-3p targets PCBP2 in melanoma cells, resulting in downregulation of PCNA, p-PI3K, and p-AKT expression, as well as upregulation of E-cadherin expression. Increasing evidence suggests that the activation of the PI3K/AKT signaling pathway is significantly involved in the initiation of melanoma and in the development of therapeutic resistance [38]. Hence, therapeutic miRNAs that block the PI3K/AKT pathway have been extensively investigated as a strategy to partially reverse the promotion of melanoma malignancy. Importantly, the activation of the PI3K/AKT pathway by PCBP2 is a crucial step in the lnc030-SQLE-cholesterol synthesis pathway, which has potential as a therapeutic target for breast cancer stem cells (BCSCs) [22]. Our rescue experiments consistently demonstrated that PI3K/AKT pathway inhibitor LY294002 reversed the effects of PCBP2 overexpression on melanoma cell proliferation and migration. Herein, we discovered that miR-5195-3p exerting suppressive effects on melanoma cell proliferation and migration through targeting PCBP2-mediated PI3K/AKT pathway. Certainly, our present study had several limitations. These limitations include the absence of relevant in vivo data, the lack of further investigation into the long-term effects of targeting the miR-5195-3p/PCBP2 axis on melanoma cell behavior or survival, and the unavailability of northern blot analysis for detecting miR-5195-3p expression in tissue specimens.
In conclusion, our study suggests that miR-5195-3p functions as a therapeutic suppressive miRNA in melanoma cells. It achieves this by inhibiting cellular behaviors through the inactivation of the PI3K/AKT signaling pathway, primarily by targeting PCBP2. These findings underscore the promising prospects of targeted therapies involving miR-5195-3p and PCBP2 for the effective treatment of melanoma.
Ethics approval and consent to participate
All the experiments were performed in accordance with the Declaration of Helsinki. This study was approved by the Ethics Committee of the Guangdong Provincial People's Hospital (Approval number: CP-2019A7D, Guangdong, China).
Author contribution statement
Jing Liu: Conceived and designed the experiments.
Xushan Cha: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Botao Yang; Yucai Wu: Performed the experiments.
Yang Chen: Analyzed and interpreted the data.
Yongshuang Li; Jinhua Wang: Analyzed and interpreted the data; Wrote the paper.
Data availability statement
Data will be made available on request.
Consent for publication
Not applicable.
Funding
This work is supported by the Scientific Research Subject of Traditional Chinese Medicine Bureau of Guangdong Province (NO. 20201345).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e19227.
Contributor Information
Xushan Cha, Email: cha_xushan1215cxs@yeah.net.
Jing Liu, Email: liujing1350@gzucm.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- 1.Brancaccio G., Russo T., Lallas A., Moscarella E., Agozzino M., Argenziano G. Melanoma: clinical and dermoscopic diagnosis. G. Ital. Dermatol. venereol. Organo Uff. Soc. Ital. Dermatol. Sifilogr. 2017;152:213–223. doi: 10.23736/S0392-0488.17.05571-7. [DOI] [PubMed] [Google Scholar]
- 2.Yde S.S., Sjoegren P., Heje M., Stolle L.B. Mucosal melanoma: a literature review. Curr. Oncol. Rep. 2018;20:28. doi: 10.1007/s11912-018-0675-0. [DOI] [PubMed] [Google Scholar]
- 3.Li L., London N.R., Chen X. Malignant mucosal melanoma of the eustachian tube with extension into the ipsilateral external ear canal: a case report and review of the literature. Ear Nose Throat J. 2021;(100):730S–733S. doi: 10.1177/0145561320904813. [DOI] [PubMed] [Google Scholar]
- 4.Domingues B., Lopes J.M., Soares P., Populo H. Melanoma treatment in review. ImmunoTargets Ther. 2018;(7):35–49. doi: 10.2147/ITT.S134842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolchok J.D., Chiarion-Sileni V., Gonzalez R., Rutkowski P., Grob J.J., Cowey C.L., Lao C.D., Wagstaff J., Schadendorf D., Ferrucci P.F., et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber J., Mandala M., Del Vecchio M., Gogas H.J., Arance A.M., Cowey C.L., Dalle S., Schenker M., Chiarion-Sileni V., Marquez-Rodas I., et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med. 2017;(377):1824–1835. doi: 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- 7.Tonella L., Pala V., Ponti R., Rubatto M., Gallo G., Mastorino L., Avallone G., Merli M., Agostini A., Fava P., et al. Prognostic and predictive biomarkers in stage III melanoma: current insights and clinical implications. Int. J. Mol. Sci. 2021;(22) doi: 10.3390/ijms22094561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu T.X., Rothenberg M.E. MicroRNA. J. Allergy Clin. Immunol. 2018;(141):1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Fabian M.R., Sundermeier T.R., Sonenberg N. Understanding how miRNAs post-transcriptionally regulate gene expression. Prog. Mol. Subcell. Biol. 2010;(50):1–20. doi: 10.1007/978-3-642-03103-8_1. [DOI] [PubMed] [Google Scholar]
- 11.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Kim J., Yao F., Xiao Z., Sun Y., Ma L. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev. 2018;(37):5–15. doi: 10.1007/s10555-017-9712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Yan D.M., Xhu L.X., Si D.M., Liang Q.H. MiR-5195-3p inhibits the proliferation of glioma cells by targeting BIRC2. Eur. Rev. Med. Pharmacol. Sci. 2020;(24):267–273. doi: 10.26355/eurrev_202001_19921. [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Shi G., Zhu D., Jin Y., Yang X. miR-5195-3p suppresses cell proliferation and induces apoptosis by directly targeting NEDD9 in osteosarcoma. Cancer Biother. Radiopharm. 2019;(34):405–412. doi: 10.1089/cbr.2018.2761. [DOI] [PubMed] [Google Scholar]
- 15.Yang Q. MicroRNA-5195-3p plays a suppressive role in cell proliferation, migration and invasion by targeting MYO6 in human non-small cell lung cancer. Biosci. Biotechnol. Biochem. 2019;(83):212–220. doi: 10.1080/09168451.2018.1540288. [DOI] [PubMed] [Google Scholar]
- 16.Zeng X., Hu Z., Shen Y., Wei X., Gan J., Liu Z. MiR-5195-3p functions as a tumor suppressor in prostate cancer via targeting CCNL1. Cell. Mol. Biol. Lett. 2022;(27):25. doi: 10.1186/s11658-022-00326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Zhang X., Liu Y., Shi X., Li L., Jia Y., Wu F., Cui H., Li L. MiR-5195-3p functions as a tumor suppressor by targeting RHBDD1 in ovarian cancer. Histol. Histopathol. 2023 doi: 10.14670/HH-18-595. [DOI] [PubMed] [Google Scholar]
- 18.Choi H.S., Hwang C.K., Song K.Y., Law P.Y., Wei L.N., Loh H.H. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun. 2009;(380):431–436. doi: 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Lei J., Zheng Q., Tan S., Ding K., Yu C. Poly(rC) binding protein 2 (PCBP2) promotes the viability of human gastric cancer cells by regulating CDK2. FEBS Open Bio. 2018;(8):764–773. doi: 10.1002/2211-5463.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han W., Xin Z., Zhao Z., Bao W., Lin X., Yin B., Zhao J., Yuan J., Qiang B., Peng X. RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3. J. Clin. Invest. 2013;(123):2103–2118. doi: 10.1172/JCI61820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan C., Chen M., Cai X. Advances in poly(rC)-binding protein 2: structure, molecular function, and roles in cancer. Biomed. Pharmacother. = Biomedecine & pharmacotherapie. 2021;(139) doi: 10.1016/j.biopha.2021.111719. [DOI] [PubMed] [Google Scholar]
- 22.Qin Y., Hou Y., Liu S., Zhu P., Wan X., Zhao M., Peng M., Zeng H., Li Q., Jin T., et al. A novel long non-coding RNA lnc030 maintains breast cancer stem cell stemness by stabilizing SQLE mRNA and increasing cholesterol synthesis. Adv. Sci. 2021;(8) doi: 10.1002/advs.202002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farhan M., Silva M., Xingan X., Zhou Z., Zheng W. Artemisinin inhibits the migration and invasion in uveal melanoma via inhibition of the PI3K/AKT/mTOR signaling pathway. Oxid. Med. Cell. Longev. 2021;(2021) doi: 10.1155/2021/9911537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J.Y., Zheng L.L., Wang T.T., Hu M. Functional annotation of metastasis-associated MicroRNAs of melanoma: a meta-analysis of expression profiles. Chin. Med. J. 2016;129:2484–2490. doi: 10.4103/0366-6999.191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korfiati A., Grafanaki K., Kyriakopoulos G.C., Skeparnias I., Georgiou S., Sakellaropoulos G., Stathopoulos C. Revisiting miRNA association with melanoma recurrence and metastasis from a machine learning point of view. Int. J. Mol. Sci. 2022;(23) doi: 10.3390/ijms23031299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebrahimi S.O., Reiisi S. Downregulation of miR-4443 and miR-5195-3p in ovarian cancer tissue contributes to metastasis and tumorigenesis. Arch. Gynecol. Obstet. 2019;(299):1453–1458. doi: 10.1007/s00404-019-05107-x. [DOI] [PubMed] [Google Scholar]
- 27.Wang D., Ming X., Xu J., Xiao Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol. Oncol. 2021;(39):390–400. doi: 10.1002/hon.2874. [DOI] [PubMed] [Google Scholar]
- 28.Li G., Li X., Yuan C., Zhou C., Li X., Li J., Guo B. Long non-coding RNA JPX contributes to tumorigenesis by regulating miR-5195-3p/VEGFA in non-small cell lung cancer. Cancer Manag. Res. 2021;(13):1477–1489. doi: 10.2147/CMAR.S255317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Li M., Liao H., Wu J., Chen B., Pang R., Huang J., Zhu Y. Long noncoding RNA matrilineal expression gene 3 inhibits hepatocellular carcinoma progression by targeting microRNA-5195-3p and regulating the expression of forkhead box O1. Bioengineered. 2021;(12):12880–12890. doi: 10.1080/21655979.2021.2005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Tian H., Miao Y., Feng X., Li Y., Wang H., Song X. Upregulation of p72 enhances malignant migration and invasion of glioma cells by repressing Beclin1 expression. Biochemistry. Biokhimiia. 2016;81:574–582. doi: 10.1134/S0006297916060031. [DOI] [PubMed] [Google Scholar]
- 31.Yang C., Wang R., Hardy P. Potential of miRNA-based nanotherapeutics for uveal melanoma. Cancers. 2021;(13) doi: 10.3390/cancers13205192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Z., Zhang Y., Cao R., Li L., Zhong K., Chen Q., Xiao J. miR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol. Res. 2017;(25):1081–1087. doi: 10.3727/096504016X14831120463349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jahangiri Moez M., Bjeije H., Soltani B.M. Hsa-miR-5195-3P induces downregulation of TGFbetaR1, TGFbetaR2, SMAD3 and SMAD4 supporting its tumor suppressive activity in HCT116 cells. Int. J. Biochem. Cell Biol. 2019;109:1–7. doi: 10.1016/j.biocel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Zhang C., Zong X., Wang Z., Ren R., Wang L., Sun P., Zhu C., Guo M., Guo G., et al. ST8SIA6-AS1 promotes the epithelial-to-mesenchymal transition and angiogenesis of pituitary adenoma. J. Oncol. 2022;(2022) doi: 10.1155/2022/7960261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C.M., Song L.D., Wang J.W., Ye H.B., Chen S. Role of miR-490-3p in blocking bladder cancer growth through targeting the RNA-binding protein PCBP2. Kaohsiung J. Med. Sci. 2022;(38):30–37. doi: 10.1002/kjm2.12457. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Hu C.E., Liu Y.C., Zhang H.D., Huang G.J. The RNA-binding protein PCBP2 facilitates gastric carcinoma growth by targeting miR-34a. Biochem. Biophys. Res. Commun. 2014;(448):437–442. doi: 10.1016/j.bbrc.2014.04.124. [DOI] [PubMed] [Google Scholar]
- 37.Huang C.M., Cao G.Y., Yang C.X., Chen Y., Liu G.D., Xu B.W., Zhang X. LncRNA ST8SIA6-AS1 promotes colorectal cancer cell proliferation, migration and invasion by regulating the miR-5195/PCBP2 axis. Eur. Rev. Med. Pharmacol. Sci. 2020;(24):4203–4211. doi: 10.26355/eurrev_202004_21000. [DOI] [PubMed] [Google Scholar]
- 38.Davies M.A. The role of the PI3K-AKT pathway in melanoma. Cancer J. 2012;18:142–147. doi: 10.1097/PPO.0b013e31824d448c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.