Summary
In most eukaryotes, meiotic crossovers (COs) are limited to 1–3 per chromosome, and are prevented from occurring close to one another by CO interference. The fission yeast Schizosaccharomyces pombe, an exception to these general rules, was reported to have the highest CO number per chromosome and no or weak interference. However, global CO frequency was indirectly estimated, calling for confirmation. Here, we used an innovative strategy to determine COs genome-wide in S. pombe. We confirmed weak CO interference, acting at physical distances compatible with the patterning of recombination precursors. We revealed a slight co-variation in CO number between chromosomes, suggesting that a limiting pro-CO factor varies between meiocytes. CO number per chromosome varies proportionally with chromosome size, with the three chromosomes having, on average, 15.9, 12.5, and 7.0 COs, respectively. This reinforces S. pombe’s status as the eukaryote with the highest CO number per chromosome described to date.
Subject areas: Cell biology
Graphical abstract

Highlights
-
•
Genome-wide analysis of meiotic crossovers in the fission yeast S. pombe
-
•
S. pombe has an exceptionally high number of crossovers per chromosome
-
•
Weak crossover interference, compatible with interfering precursors
-
•
Slight co-variation in crossover number between chromosomes
Cell biology
Introduction
During meiosis, recombination is initiated by the formation of numerous programmed DNA double-strand breaks (DSBs). The repair of DSBs results in crossovers (COs), which are instances of reciprocal recombination between homologous chromosomes, and non-crossovers (NCOs), which are local tracts of non-Mendelian segregation not associated with a CO event. Two main conserved pathways were described for CO formation. In most examined eukaryotes, including mammals, worms, flowering plants, and the yeast Saccharomyces cerevisiae, the majority of COs, called class I COs, are specifically promoted by the group of ZMM proteins (Zip1-4, Msh4/5, Mer3, and Spo16 in S. cerevisiae), while a minority of class II COs are dependent on other factors, including the structure-specific nuclease Mus81-Eme1/Mms4. Besides differences in the molecular pathways, class I COs are sensitive to CO interference, a phenomenon that prevents COs from being positioned close to each other along chromosomes, while class II COs are not, or much less, subjected to interference.1,2,3 The fission yeast Schizosaccharomyces pombe has long been used as a model to gain insights into meiotic mechanisms.4,5,6 S. pombe appears to be an outlier in terms of CO control. First, CO number per chromosome was described to be markedly elevated, with a total genetic size of 2,250 cM for three chromosomes. This corresponds to a total of 45 COs per meiosis, and 19:15:11 COs from the largest to the smallest chromosome, respectively.6,7 Nineteen COs per meiosis on a single chromosome is exceptional, when considering that 80% of the analyzed chromosomes in eukaryotes experience less than three.2,8 Second, CO interference was reported to be absent in S. pombe, with closely spaced, double COs occurring as expected if these were independent events.9,10 CO interference weak signal is detected in S. pombe as a result of interference between Spo11-DSBs, the CO precursors, and is mediated by the DNA damage-response protein kinase Tel1.11,12,13 The meiotic machinery is also unusual in S. pombe: The ZMM proteins that define the class I CO pathway are lacking, with COs being solely dependent on the Mus81 pathway. The tripartite synaptonemal complex (SC) is also absent in S. pombe meiosis, though linear elements that are reminiscent of the lateral element of the SC are formed.10,11,14,15,16,17 However, pervasive hybrid sterility and large chromosome rearrangements18,19,20,21 prevent the use of hybrids obtained by crossing natural isolates for accurate measurement of meiotic recombination in S. pombe.
Results
A method for genome-wide analysis of meiotic crossovers in S. pombe
Here, we used EMS mutagenesis to introduce a few hundred SNPs genome-wide in an isogenic strain and explored the meiotic recombination landscape in S. pombe. First, two independent isogenic haploid strains with the same background as the reference strain and with opposite mating types were subjected to moderate EMS treatment to introduce mutations genome-wide. Then, they were mated and sporulated to produce tetrads through meiosis (Figure 1A). To have a robust estimate of the recombination frequency, we generated and sequenced a total of 92 tetrads of three independent crosses named Pombe2_Tet, Pombe4_Tet, and Pombe9_Tet, with 29, 30, and 33 tetrads, respectively. These crosses bear a private set of heterozygous mutations subsequently used as SNPs in segregation analysis (Table S3). All the crosses had high levels of spore viability (>94% of the spores were viable, n = 43, 37, and 41 tetrads, respectively), suggesting that meiosis was unaffected by the heterozygous mutations (Tables S7, S8, and S9). Two tetrads gave incomplete sequencing data and were dismissed, providing a final dataset of 90 tetrads (29, 30, and 31 tetrads, for populations Pombe2_Tet, Pombe4_Tet, and Pombe9_Tet, respectively. Table S1).
Figure 1.
Experimental design for meiotic recombination analysis in Schizosaccharomyces pombe through mutagenesis of isogenic strains and tetrad sequencing
(A) SNPs (red and blue vertical bars) were introduced into the S. pombe genome (horizontal bar) by EMS mutagenesis in parallel in h+ and h-cells. After mutagenesis, de novo mutations were identified by whole genome sequencing. Cells of opposite mating type with optimal SNP distributions were mated to generate a hybrid and then tetrads after meiosis. Tetrads were dissected by micromanipulation and spores were allowed to form colonies, from which DNA was extracted and sequenced. Three independent repeats were performed.
(B) CO and GC detection in a representative tetrad (Pombe2_Tet_10). The detected COs and GCs are indicated by vertical dashed lines connecting the two exchanged chromatids. A GC is indicated by a triangle. The four chromatids are color-coded according to the parental genotypes.
To define the informative SNPs for each population, we deep-sequenced the parental genomes (25-91x, Table S2), and identified 161, 121 and 172 well-supported and randomly distributed mutation markers that provided good coverage of the three chromosomes (Table S3; Figure S1). The median/mean inter-marker distance is 56/83 kb and corresponds to the resolution at which recombination sites could be detected and assigned (equivalent to 7.8/11.6 cM, see below). Based on recombination landscapes from other species, such a resolution is suitable for detecting most COs because of the associated rearrangement of flanking markers, but could detect only a small proportion of NCOs that are in the kilobase range in budding and fission yeasts.22,23,24,25
An exceptionally high number of crossovers per chromosome in S. Pombe
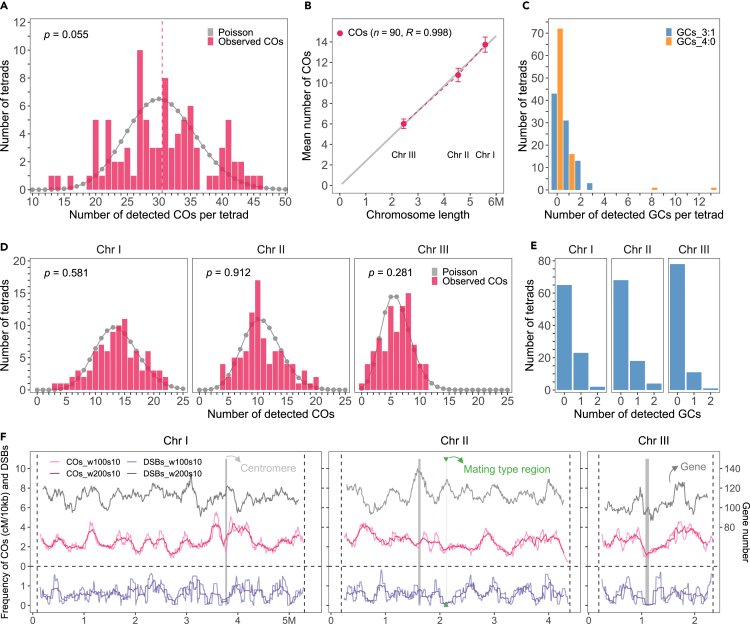
Using tetrad-based segregation profiles of the informative SNPs, we identified a total of 2,746 COs across the 90 meioses (median resolution 113.5 kb, Figures S1F and S2; Table S4). This corresponds to an average of 30.5 COs per tetrad (Figure 2A), and an average of 13.7, 10.8, and 6.0 COs for chromosome I, II and III, respectively (Figures 2B, 2D, and S3C). Similar numbers of COs were identified in the three replicates (Figure S3), suggesting that the introduced heterozygous mutations do not affect recombination (Tables S7, S8, and S9). The distribution of the total CO number per tetrad ranged from 13 to 46 and is not significantly different from the Poisson distribution (Figure 2A). For individual chromosomes, the number of COs also follows a Poisson distribution (Figure 2D), which contrasts with many species where CO interference reduces the variance in the number of COs per chromosome.26 Accordingly, we found that this variance of CO number per chromosome is larger in S. pombe than in S. cerevisiae (Figure S4). We detected at least one CO in all chromosomes in each tetrad except one where no crossover was detected on chromosome III (Figures 2D and S3C). In species in which the mean number of CO is low and achiasmate chromosomes are rare (e.g., mammals and flowering plants studied so far), a specific mechanism must be in place to ensure the “obligatory crossover”.27 In S. pombe, the elevated number of COs per chromosome provides an immediate assurance without the need for a specific additional mechanism, as the probability of an achiasmate chromosome per chance is ∼1/1000.
Figure 2.
The number and distribution of COs and GCs in S. pombe
(A) The distribution of CO numbers per tetrad. The gray points and line represent the Poisson fitted distribution. The Mann–Whitney test was used to evaluate the differences in distributions of CO numbers between the observed and Poisson distributions. The vertical dashed line indicates the mean value.
(B) Analysis of CO numbers and chromosome sizes (Mb). Crossover number and chromosome size are linearly correlated (Mb). The fitted linear regression is indicated by the gray line. The sample sizes and Pearson’s correlation coefficient are indicated in parentheses. The mean number of COs is shown with 90% confidence interval.
(C) The distribution of number of GCs with 3:1 and 4:0 allelic segregation per tetrad, separately. (D) The distribution of CO numbers per chromosome and per tetrad. The gray points and line represent the Poisson fitted distribution. The Mann–Whitney test was used to evaluate the differences between observed and Poisson distributions.
(E) The distribution of number of 3:1 GCs per chromosome and per tetrad.
(F) The chromosomal distribution of COs and DSBs28 with 100 kb (pink and light purple, w100s10) and 200 kb (red and purple, w200s10) sliding window size and 10 kb step size, and genes (black) with 200 kb sliding window size and 10 kb step size, separately. The centromeric regions are indicated by gray shading. The mating type position is indicated with green shading.
Although most detected events appear as a single crossover per interval, with two chromatids showing exchange and two unmodified chromatids, we identified 108 cases of double-crossovers (DCOs) in a single interval with the four chromatids involved (Figure S5). Because of our relatively low marker density (median/mean inter-marker distance = 56/83 kb), some DCOs may be missed in some intervals. Only DCOs involving the four chromatids of a bivalent (4C-DCO events) occurring in one interval can be detected with the flanking markers (Figure S6). However, a DCO that involves the same two chromatids (2C-DCO events) is undetectable with the flanking markers. If three chromatids are involved in a DCO event in a given interval (3C-DCO events), it is detected as a single CO (Figure S6). As chromatid interference is absent (see below), the number of two- and three-chromatid DCOs, and thus of missed DCOs, can be estimated from the number of observed four-chromatid DCOs. A total of 108 4C-DCOs were observed, leading to an estimate of 108 2C-DCOs (216 missed COs) events and 216 3C-DCOs (216 DCOs that were detected as single COs) (Table S5). We thus added 432 COs to our CO count, resulting in a total estimate of 3,178 COs in 90 meioses, corresponding to a corrected average of 35.3 COs per meiosis: 15.9 COs (793 cM), 12.5 COs (625 cM) and 7.0 COs (347 cM) for chromosome I, II and III, respectively (Figure 3; Table S5). This corresponds to 0.14 cM/kb or 7.1 kb/cM. A previous estimate of the genetic size, based on the compilation of segregation analyses of pairs of phenotypic markers, was ∼20% larger.9 In another study, crossover counts were measured by sequencing ten spores produced by a hybrid between two closely related S. pombe strains, leading to an estimate of 9.6 COs per tetrad for chromosome I, 10.2 for chromosome II, and 6.8 for chromosome III.18 This is compatible with our results, except for chromosome I where the reduced recombination frequency likely results from the presence of a large genomic inversion between these two strains. This inversion may directly suppress recombination and/or lead to inviable gametes when an odd number of COs takes place. CO numbers per chromosome are linearly correlated with the chromosome length (Pearson’s R = 0.998), with an intercept of −0.066 and a slope of 2.44 COs per megabase (Figure 2B).
Figure 3.
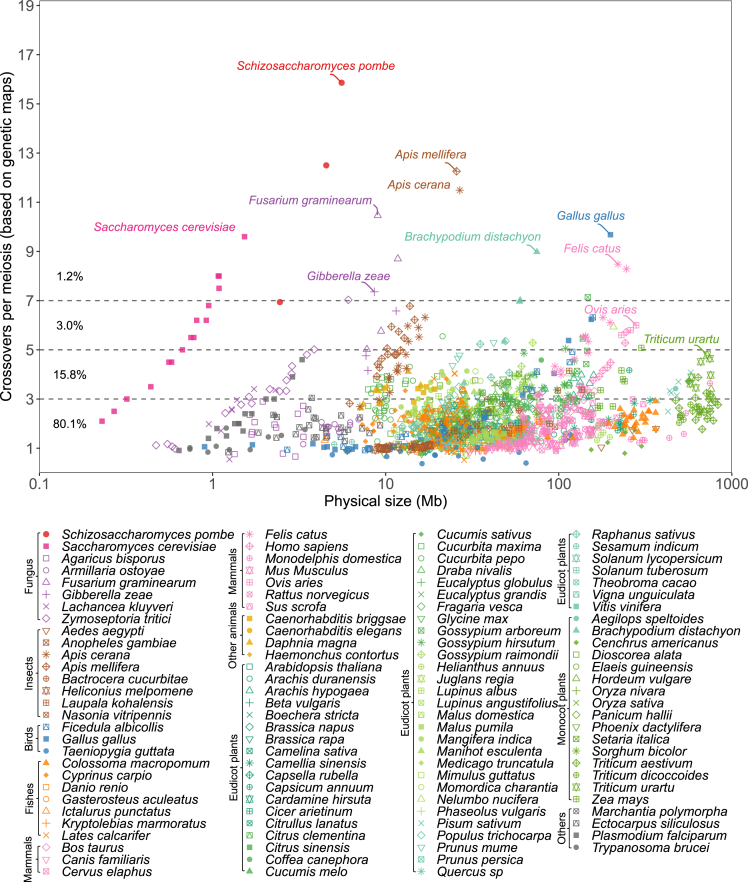
S. pombe has an exceptional high number of meiotic CO compared to other eukaryotes
The crossover number per meiosis (y axis, based on genetic maps. 50 cM = 1 CO) of 114 species (8 fungi, 8 insects, 3 birds, 7 fishes, 10 mammals, 4 other animals, 54 eudicot plants, 16 monocot plants, 1 other plant and 3 other species) is plotted against the physical size (x axis, DNA Mb, log scale) of each chromosome. The estimation of CO number of S. pombe (this study) is indicated by red dots. Plots separating the different eukaryotic clades are provided in Figures S7 and S8 and the source data in Table S6 (data from29,30,31,32,33,34,35,36,37). An earlier version of this figure, with fewer species represented, was published by Fernandes et al.8
We then compared the crossover number per chromosome in a large range of eukaryotes (Figure 3). The number of COs per meiosis appears to be constrained: while typically at least one CO per chromosome is present, likely associated with the crucial role of COs in promoting balanced chromosome segregation at meiosis, larger numbers are rarely seen. In our survey of 1,547 chromosomes from 114 species, 80.1% of chromosomes have an average of less than three COs and 95.9% less than five (Figure 3; Table S6). CO number tends to be correlated with the physical size of the chromosomes within a species (Figures S7–S9), and like in S. pombe, are linearly correlated in diverse species such as S. cerevisiae, Arabidopsis thaliana, Ovis aries, Gallus gallus, and Homo sapiens (Figures S7–S9). In sharp contrast, this is not the case when comparing chromosomes from different species: for example, the physical size of chromosome 3B of wheat (804 Mb, Triticum aestivum) is 2,977-fold longer than the chromosome 6 of budding yeast (0.27 Mb, Saccharomyces cerevisiae), although they have similar genetic sizes (2.8 vs. 2.5 COs per meiosis, Figure 3). Several species have chromosomes with unusually large genetic sizes, like chromosome 1 of European honeybee (12.3 COs, Apis mellifera, Insects), chromosome 2 of Fusarium graminearum (10.5 COs, Fungus), and chromosome 1 of chicken (9.7 COs, Gallus gallus, Birds). S. pombe appears exceptional, with chromosomes I and II experiencing the highest number of COs and chromosome I having more than two times more COs than 99% of the eukaryotic chromosomes described so far. Chromosome III, the chromosome of S. pombe that undergoes the least recombination, still receives more COs than 98% of thus far studied chromosomes (Figure 3). It is possible that other eukaryotes that have not yet been analyzed would show unusual numbers of COs per chromosome. Aspergillus fumigatus appears to be a serious challenger for the status of the champion with the highest number of CO per chromosome.38
Relatively homogeneous distribution of crossovers along chromosomes in S. Pombe
We next analyzed the CO distribution along S. pombe chromosomes. We observed that COs are relatively homogeneously distributed along chromosomes, largely distributing within the 2-fold standard deviation of the random distribution, without very hot or cold regions (Figures 2F and S10). Notably, we do not detect a strong effect of the centromeres on CO rates. However, it should be noted that the limited resolution of our study may hide strong local variations in recombination rates (Figure S1F). Such variations include the recombination suppression next to the centromeres as previously reported in S. pombe and observed in many other eukaryotes.22,39,40,41,42,43,44,45,46 At that resolution, the DSB distribution28 and CO distribution (Figure 2F) are only weakly correlated (Pearson’s correlation: 0.21 and 0.17, in 100 kb and in 200 kb window size, respectively) despite the existence of DNA double-strand break hotspots at higher resolution.28,47
Gene conversions in S. Pombe
In addition to COs, we identified a total of 66 gene conversions (GCs) characterized by a 3:1 segregation profile of a single SNP, which corresponds to an average of 0.7 per tetrad (Figures 2C, 2E, and S3). Among them, 37 GCs were flanked by a CO event (Table S4) and could be either associated to the CO, or be an independent NCO recombination event in the same interval (median size = 134 kb). We thus estimated that between 29 and 66 NCOs were detected in our dataset. In total, we analyzed the segregation of 13,975 SNPs in 90 tetrads, which resulted in an estimation of an average NCO conversion rate of 0.21–0.47% per base pair per meiosis. With a genome size of 12.57 Mb, this predicts that on average 26–59 kb are converted by NCOs per meiosis. In comparison, in S. cerevisiae 66.1 NCOs per meiosis with a median of 1.8 kb were observed,22 corresponding to 119 kb undergoing NCO-associated gene conversion per meiosis. Although individual gene conversion tract lengths are highly heterogeneous in S. pombe, the estimate of the minimal gene conversion tract length from three different constructs is about 1 kb.25 This size estimate would predict an average of 26–59 NCOs per meiosis, which is similar to our estimation of CO number (35 per meiosis), suggesting that DSB are repaired as either COs or NCOs with similar frequencies in S. pombe. Previous studies reported that 65–70% of GCs have an associated CO.16,25
We also observed 37 gene conversions (GCs) with a 4:0 allelic segregation. Remarkably, 13 and 8 of these GCs occurred in only two independent tetrads of the Pombe2_Tet population, and a common GC occurred in 16 tetrads of the Pombe4_Tet population (Figures 2C, S3, and S11). Although these events can result from the GC-mediated repair of two DSBs at the same locus on two sister chromatids during meiotic prophase, such a scenario is unlikely. Instead, the fact that two tetrads out of 90 contain 21 of the 37 4:0 segregating NCOs while all the other tetrads have never more than three such NCOs could reflect the existence of a post-zygotic diploid stage in a small fraction of cells from the population with an elevated genetic instability, at least in the Pombe2_Tet population.22,23,24 The recurrent 4:0 segregating NCOs in the 16 tetrads of the Pombe4_Tet population could reflect either an elevated meiotic or mitotic instability of a given locus.
Weak crossover interference in S. Pombe
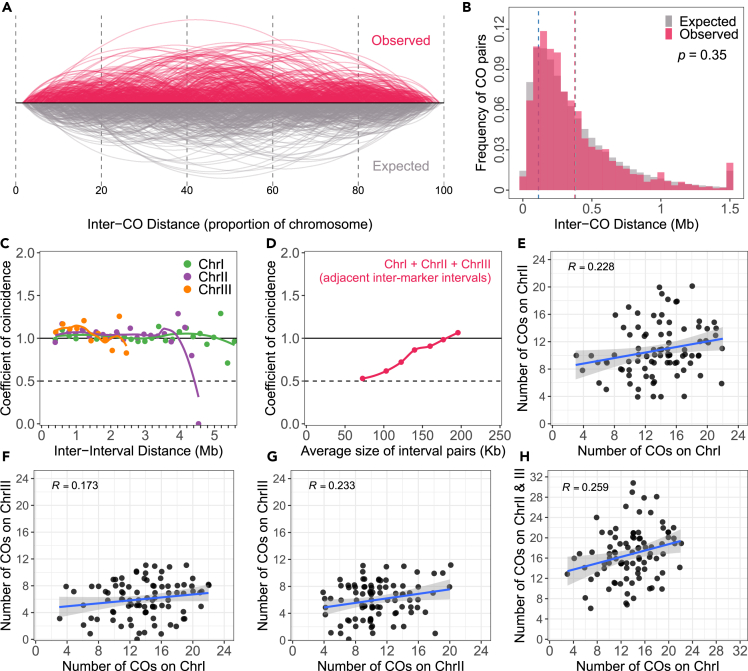
Along chromosomes, COs are not independently distributed in most eukaryotes, as CO interference prevents close positioning.48,49 Based on analysis of pairs or trios of markers, S. pombe was reported to lack CO interference.9 However, short-range (∼20 cM/150kb) CO interference was subsequently reported in S. pombe.11 To further explore CO interference in S. pombe, we first examined the distribution of the distance between adjacent COs on the same chromosome, comparing with the expected distribution in the absence of CO interference (Figures 4A, 4B, S12, and S13A–S13F). We simulated the expected distribution by using the same marker list, and kept the same sample size and CO number for each chromosome, tetrad and population, but randomly distributed the COs between tetrads. The observed distribution was found to be remarkably similar to that expected for independent COs, with numerous pairs of COs placed less than 200 kb (∼28 cM) apart. (Figures 4A and 4B).
Figure 4.
CO interference at short physical distances and co-variation of CO frequencies in S. pombe
(A) Comparison of the observed (red) and expected (under the hypothesis of independence, gray) distribution of inter-CO distances along the proportional scale of chromosomes.
(B) Distribution of observed (red) and expected in absence of interference (gray) distances between COs. The expected CO list was obtained with 1000 simulations by sampling the observed CO number per chromosome and tetrad in the corresponding scale of genomic regions covered by EMS-induced SNPs. The blue vertical dotted line indicates the median CO resolution of the whole population. The p value from a Mann-Whitney test between the expected and observed distributions is shown.
(C) The CoC curves of the observed COs. Chromosomes were divided into ∼200 kb intervals, and the coefficient of coincidence was calculated for each possible pair of intervals (observed frequency of concomitant CO in both intervals/product of frequency of CO in each interval. A coefficient of coincidence of 1 indicates absence of interference). Note that the last point of the curves tends to be noisy as a single pair of intervals represent the largest distance. The coefficient of coincidence was averaged for pairs of intervals separated by the same distance, and plotted versus the physical distance between external borders of the two intervals.
(D) CoC analysis using adjacent inter-marker intervals. CoC was calculated for pairs of adjacent intervals of selected physical sizes and plotted versus the mean total size of the pair of intervals (Table 1; Tables S10, S11, S12, S13, S14, S15, S16, and S17).
(E–H). Correlation analysis of the numbers of COs on different chromosomes in the same tetrad. The blue line indicates the linear regression, and the gray zone represents the 95% confidence interval. The data points were vertical and horizontal jitter-shifted for improving visualization.
To further describe CO interference, we performed coefficient of coincidence (CoC) analysis, which is the ratio of observed frequency of double-COs to the frequency of expected double-COs in a pair of defined intervals.12,50 In brief, the expected frequency of two COs happening concomitantly in two intervals, assuming they occur independently, is the product of the frequency of single COs in each interval. The CoC is calculated by dividing the observed frequency of double-COs by this expectation. A CoC <1 indicates that the two COs do not occur independently, but that the presence of one CO in one of the considered intervals prevents - interferes with - the presence of a CO in the other interval. We divided each chromosome in intervals of fixed size (∼200 kb in Figures 4C and S13G–S13I), and calculated the CoC for all pairs of intervals. A CoC curve is obtained by plotting the mean CoC for pairs of intervals separated by a given distance versus this distance (Figures 4C and S12). In presence of interference, the CoC curve is expected to deviate from 1, approaching zero for short inter-interval distances. We found that the CoC curves are flat (close to 1) for each chromosome, when considering intervals size of 200 kb or more, supporting an absence of CO interference in S. pombe at this scale which represents 4–8% of the chromosome length (Figures 4C, S12, and S13G–S13I). It should be noted that because of limited resolution of CO positions (median resolution ∼113 kb, Figure S2), these global CoC curves, using the entire dataset, cannot exclude CO interference acting at shorter physical distances.11 In this analysis, the shortest considered distance corresponds to two adjacent 200 kb intervals (total size of the pair of intervals = 400 kb, first point in Figure 4C), which corresponds to a genetic size of ∼28 cM for each interval (and ∼56 cM for the pair of intervals). It seems reasonable to suppose that a CO interferes only with a potential neighboring CO, and probably not several COs away. Given the high frequency of COs per chromosome in S. pombe, it might have been expected that interference is not detected beyond 400 kb/50 cM.
To explore interference at shorter distances, we selected exclusively the pairs of adjacent intervals (three consecutive markers, excluding the intervals in which zero CO were observed) and sorted them by physical size before calculating the CoC (Table 1; Figure 4D; Tables S10, S11, S12, S13, S14, S15, S16, and S17). The selection was based on the total size of the pair of adjacent intervals. When considering all adjacent interval pairs, or pairs of adjacent intervals of 200, 180, 160 or 140 ± 20 kb in total size, we detected as many events of double-CO (one in each interval in the same tetrad) as expected under the hypothesis of independence (CoC >0.85, Table 1; Figure 4D; Tables S10, S11, S12, S13, and S14). When selecting for shorter sizes (total interval size ∼120 kb or less), we detected fewer double-COs than expected, going down to a CoC ∼0.5 for the smallest analyzed interval pairs. (Table 1; Figure 4D; Tables S15, S16, and S17). Thus, CO interference is detected when considering adjacent pairs of intervals of a total size of up to ∼150 kb (∼4% of chromosome physical length) (Table 1; Figure 4D). It should be noted that this corresponds to ∼20 cM, roughly half of the genetic distance at which interference acts in other species like Drosophila.51 CO interference ranging over such a short physical distance could be entirely imposed by interference of DSBs.11,12,13,52,53
Table 1.
Analysis of CO interference between adjacent pairs of intervals
| Range of interval size (total size of the pair, kb) | Average size of interval pairs (kb) | Expected number of tetrads with COs in both intervals | Observed number of tetrads with COs in both intervals | CoC | p-value |
|---|---|---|---|---|---|
| 31–850 | 205.1 | 606.5 | 647 | 1.07 | 0.088 |
| 200 ± 20 | 195.8 | 84.5 | 90 | 1.07 | 0.534 |
| 180 ± 20 | 176.8 | 95.6 | 94 | 0.98 | 0.867 |
| 160 ± 20 | 159.8 | 82.6 | 75 | 0.91 | 0.387 |
| 140 ± 20 | 139.7 | 70.7 | 61 | 0.86 | 0.238 |
| 120 ± 20 | 122.4 | 63.8 | 46 | 0.72 | 0.023 |
| 100 ± 20 | 103.1 | 38.9 | 24 | 0.62 | 0.016 |
| 60 ± 40 | 72.6 | 20.7 | 11 | 0.53 | 0.031 |
Pairs of adjacent intervals were selected with decreasing total size. CoC were calculated by dividing the number of observed tetrads with COs in both intervals by their expected number under the hypothesis of independence (Tables S10, S11, S12, S13, S14, S15, S16, and S17). p-values were calculated by Chi-squared Test.
Crossover number covaries between chromosomes in S. Pombe
We then tested the existence of covariation of CO numbers between chromosomes within nuclei, as observed in humans and other eukaryotes.54 We observed that CO number tends to covary across chromosomes within individual tetrads, (Figures 4E–4H and S13J–S13M). The numbers of COs per chromosome are positively correlated between chromosomes (Figures 4E–4G) and also between two groups of chromosomes with comparable physical lengths (ChrI v.s. ChrII and III, Figure 4H). This indicates that if one cell has a large number of COs on one chromosome, it weakly tends to have also a large number of COs on the other chromosomes. This suggests that a limiting factor for CO formation stochastically varies between cells, causing some cells to have more COs than others, either conjointly on different chromosomes or at different positions on the same chromosome. As the SC is absent and the interference is weak, we speculate that variation in DSB frequencies may be the source of cell-to-cell variations in COs, potentially through variation in the condensation of the loop-axis structure.54
Absence of chromatid interference in S. Pombe
Finally, we explored chromatid interference. As described above, when considering two adjacent COs, three types of outcomes would be expected depending on how many chromatids are affected (2C-DCOs, 3C-DCOs and 4C-DCOs, Figure S6). In the absence of chromatid interference, DCOs are randomly distributed among the four chromatids of the bivalent, and the three types of DCOs will give a ratio of 1:2:1 (2C:3C:4C DCOs).9 If the chromatid implicated in a CO affects somehow the choice of the chromatid of the adjacent CO, we would expect a deviation from this ratio. In our tetrad analysis, the ratio of 2C:3C:4C DCOs for each individual chromosome (1,012, 768 and 399 pairs of COs were analyzed separately) and the whole genome (2,179 CO pairs), did not significantly deviate from 1:2:1 (Chi-Square test, Table 2). To explore the potential centromere effect on chromatid interference, we further quantified the deviation from the expected 1:2:1 ratio of 2C:3C:4C DCOs for two groups of DCOs on the same chromosome arms or that span the centromere, separately. In all cases, no significant deviation was detected (Table 2). These results showed an absence of chromatid interference in fission yeast, confirming the previous report which had less statistical power.9
Table 2.
Chromatid interference is absent in S. pombe
| Whole chromosome |
Same arm |
Different arm |
||||
|---|---|---|---|---|---|---|
| Total DCOs | Observed 2C:3C:4C ratio | Total DCOs | Observed 2C:3C:4C ratio | Total DCOs | Observed 2C:3C:4C ratio | |
| I | 1012 | 247:502:263 (p = 0.821) | 930 | 222:467:241 (p = 0.821) | 82 | 25:35:22 (p = 0.668) |
| II | 768 | 184:384:200 (p = 0.821) | 688 | 164:346:178 (p = 0.821) | 80 | 20:38:22 (p = 0.861) |
| III | 399 | 95:187:117 (p = 0.668) | 338 | 87:156:95 (p = 0.668) | 61 | 8:31:22 (p = 0.479) |
| Total | 2179 | 526:1073:580 (p = 0.668) | 1956 | 473:969:514 (p = 0.668) | 223 | 53:104:66 (p = 0.668) |
2C DCOs mean two COs use the same non-sister chromatids; 3C DCOs mean two COs share one chromatid; 4C DCOs mean two COs use the different pair of non-sister chromatids.
Chi-Square test of goodness-of-fit was performed for each ratio, and no significant deviation from the 1:2:1 ratio was detected. The significant p value was corrected by the fdr method.
Discussion
In summary, regulation of meiotic recombination appears to be relaxed in S. pombe, resulting in an exceptionally high number of COs per chromosome and weak crossover interference. The high frequency of COs, even if randomly distributed among chromosomes, makes it highly unlikely that a given chromosome will lack a CO. In many species, but not all, balanced chromosome distribution relies on the physical link between homologs provided by a CO.26 It is intriguing that the simple strategy of having many COs, and thus at least one connection per chromosome that support balanced segregation is observed in very few species. Most eukaryotes have a low number of COs and evolved sophisticated mechanisms to ensure their non-random distribution among and along chromosomes. This suggests that a pervasive evolutionary constraint imposes a low CO number in eukaryotes and that S. pombe has escaped this constraint and may have gained advantage from achieving lots of COs.
Limitation of the study
The number of markers introduced by mutagenesis is relatively low, limiting the resolution at which COs can be detected. For the same reason, multiple crossovers and crossovers close to chromosome ends could be missed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| EMS - ethyl methanesulfonate | sigma | M0880 |
| Deposited data | ||
| Whole-genome resequencing data | This study | EMBL-EBI ArrayExpress: E-MTAB- 12514 and E-MTAB-12516 |
| Related code | This study | Zenodo/GitHub https://doi.org/10.5281/zenodo.8204099 |
| Experimental models: Organisms/strains | ||
| Schizosaccharomyces pombe (h- leu1-32 ura4-D18) | Paul Russell | PR109 |
| Schizosaccharomyces pombe (h+ leu1-32 ura4-D18) | Paul Russell | PR110 |
| PR109 EMS mutagenized | This study | BLP49 |
| PR109 EMS mutagenized | This study | BLP59 |
| PR109 EMS mutagenized | This study | BLP64 |
| PR110 EMS mutagenized | This study | BLP18 |
| PR110 EMS mutagenized | This study | BLP23 |
| PR110 EMS mutagenized | This study | BLP33 |
| Software and algorithms | ||
| BWA v0.7.15-r1140 | Li and Durbin55 | https://github.com/lh3/bwa |
| Sambamba v0.6.8 | Tarasov et al.56 | https://github.com/biod/sambamba |
| inGAP-family | Lian et al.57 | https://sourceforge.net/projects/ingap-family/ |
| Tandem Repeats Finder v4.09 | Benson G.58 | https://tandem.bu.edu/trf |
| mosdepth v0.3.3 | Pedersen and Quinlan59 | https://github.com/brentp/mosdepth |
| MADpattern v.1.1 | Zhang et al. and White et al.12,50 | https://github.com/mwhite4/MADpatterns |
| R v3.6.2 | https://cran.r-project.org/ | |
| ggplot2 v3.4.0 | https://ggplot2.tidyverse.org/ | |
| Pombe_Recombination-v1.0 | This study | https://doi.org/10.5281/zenodo.8204099 |
| Other | ||
| Schizosaccharomyces pombe reference genome | PomBase database | https://www.pombase.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contacts Bertrand Llorente (bertrand.llorente@inserm.fr) and Raphael Mercier (mercier@mpipz.mpg.de).
Materials availability
S. pombe stains generated in this study will be distributed upon request.
Experimental model and study participant details
Strain construction, mating, growth conditions and tetrad dissection
Media and methods for studying and mutagenizing S. pombe were as described in.60 S. pombe strains PR109 (h-leu1-32 ura4-D18) and PR110 (h+ leu1-32 ura4-D18) were used for three successive rounds of mutagenesis with ethylmethane sulfonate mutagenesis (EMS from Sigma) based on.61 Briefly, S. pombe cells were inoculated in liquid YES medium overnight at 32°C, diluted to 2-5 × 10exp6 cells/ml the next morning and grown for 6 h. Next, cells were resuspended in liquid EMM at 1 × 10exp8 cells/ml and 1 mL was treated or not with 20 μL of EMS (i.e., 2% final) for 3 h in 10 mL tubes at 32°C. Next, cells were washed three times with 5 mL of EMM, resuspended in 1 mL of EMM, plated on YES plates and incubated at 32°C until individual colonies formed. After such EMS treatment, cell viability was estimated by plating untreated cells and was around 20%. Five independent clones of the first round of mutagenesis were randomly chosen to be at the root of two subsequent similar rounds of mutagenesis. Each clone used showed normal colony size and was checked for its ability to mate and sporulate without further phenotyping. To test sporulation, h+ and h-cells were mixed in water, plated on malt extract (3% weight/volume, Formedium ME medium) supplemented with adenine (0.05 g/L), uracil (0.05 g/L), L-histidine (0.05 g/L) and L-leucine (0.05 g/L), incubated at 25°C and asci were observed after 3 days. Eventually, five mutagenized clones from each of the PR109 and PR110 backgrounds were sequenced to identify de novo mutations and determine the optimal combinations of mutation patterns for recombination analyses. Because not all the pairwise crosses yielded good sporulation efficiencies (e.g., BLP59 x BLP23 gave relatively poor sporulation efficiency), we selected the following h-x h+ crosses that showed good sporulation efficiencies and dissected and sequenced the corresponding four-viable spores tetrads: BLP49 (A) x BLP23 (I) (Pombe2_Tet), 29 tetrads; BLP59 (C) x BLP18 (H) (Pombe4_Tet), 30 tetrads and BLP64 (D) x BLP33 (K) (Pombe9_Tet), 33 tetrads. The number of viable spores per tetrad was 94% for Pombe2_Tet out of 43 tetrads (34 four-viable spores tetrads, seven three-viable spores tetrads and two two-viable spore tetrad), 95% for Pombe4_Tet out of 37 tetrads (30 four-viable spores tetrads, six three-viable spores tetrads and one two-viable spore tetrad) and 95% for Pombe9_Tet out of 41 tetrads (33 four-viable spores tetrads, seven three-viable spores tetrads and one two-viable spore tetrad).
Genomic DNA extraction and sequencing
S. pombe spores were grown to saturation in 1.6 mL of YES medium at 32°C in 96-well plates (2 mL deep well plates). Cells were harvested by centrifugation, washed once with TE and lyzed during 2 h at 37°C using 30 units of zymolyase in Y1 Qiagen buffer. Samples were centrifuged at 2,700 g for 5 min and supernatants were treated with 0.5 mg/mL of RNaseA for 10 min at room temperature. DNA was next purified from these lysates using the E-Z 96 Tissue DNA Kit from Omega Bio-tek following the manufacturer’s instructions except for DNA binding to the columns that was done by gravity flow instead of centrifugation. The sequencing library was then prepared for 2 × 150 bp HiSeq 3000 Illumina sequencing.62 The Mendelian segregation of the mating type locus in each tetrad was verified by PCR as in.63
Method details
High quality EMS-induced mutation marker calling
The reference genome and genomic annotations of the fission yeast Schizosaccharomyces pombe were downloaded from the PomBase database (https://www.pombase.org).64,65 The whole genome resequencing short reads of parental samples of the three independent populations (Pombe2_Tet, Pombe4_Tet and Pombe9_Tet, Table S2) were aligned to the reference genome using BWA v0.7.15-r114055 with the default parameters. A command set of Sambamba v0.6.856 was used to format the read alignment file. The variant calling and filtering processes were largely performed following a previously described pipeline.45,57,66,67,68 First, variations were called for each individual parental sample by inGAP-family.57 Then, non-allelic, low-quality and artificial variants were selected and filtered by checking the effects of tandem repeats, small indels and structural variations predicted by Tandem Repeats Finder v4.0958 and inGAP-family, separately. Finally, mutations (G to A and C to T) specific to each parental genome were kept as high-quality markers (Table S3) for subsequent analysis.
Identification of meiotic recombination events
The sequencing reads of the 92 tetrads (Table S1) were aligned to the reference genome by BWA and then formatted by Sambamba as described before. The sequencing depths were measured using mosdepth v0.3.359 with “-n --fast-mode --by 100”. For each spore, high-quality mutation markers were genotyped by inGAP-family. The sequencing depths, reads mapping ratio, the covered marker number and the allele frequency of mutation markers were estimated and examined to remove potentially contaminated and problematic samples (Table S1). Two tetrads (Pombe9_Tet_10 and Pombe9_Tet_14) were discarded, because more than one problematic spore was detected (Table S1). During the genotyping process, markers that were not covered or poorly covered in only one of the four spores was imputed and re-genotyped based on the segregation pattern. Meiotic CO events were defined as two (or more) nearby markers showing different genotype patterns in the four spores and each flanking marker having a segregation ratio of 2:2. GC events were identified as markers with 3:1 or 4:0 segregation pattern among the four spores. For each tetrad, identified CO and GC events were manually checked by visualization of the marker genotype and chromosomal genotypes. For those unsolved double-CO events (Figure S5), which are two COs occurred within the interval of two adjacent markers and involve all four chromatids, and the pair of homolog chromosomes that CO occurred cannot be differentiated, these unsolved DCOs were removed in the subsequent CO and chromatid interference analysis.
Sparse coverage and unreliable genotyping may cause biased calling of COs. Therefore, we examined the effect of sequencing depths on CO detection. We found that the resolution and numbers of detected COs were independent of sequencing depth (mean = 19.1×, Pearson’s correlation coefficient, R = 0.03 and −0.03), suggesting sufficient sequencing depths and the absence of bias in CO identification among tetrads (Figures S3B and S14).
CO and chromatid interference analysis
To simulate the CO landscape in the absence of interference, COs were randomly sampled in the genomic range of first and last markers in the population along chromosomes. To match the contribution of COs per chromosome and tetrad, we kept the exact same sample size in the simulation. The distances between simulated COs were calculated as the observed dataset. The CoC analysis was performed using MADpattern v.1.1 (https://github.com/mwhite4/MADpatterns).12,50 For the chromatid interference analysis, the Chi-Square test of goodness-of-fit was performed to evaluate the deviation from the random 1:2:1 ratio for each observed 2C:3C:4C ratio, using the chisq.test function (stats v3.6.2 package) in R environment. Adjacent double COs were not included in the CO and chromatid interference analysis.
Quantification and statistical analysis
The statistical details can be found in the figure legends. In Figures 2A–2D, 4B, S1B–S1E, and S13D–S13F, the Mann–Whitney test was used. In Figures 4E–4H, the Pearson’s correlation was performed. In Figures 2B, 4E–4H, S4, S7, S8, and S13J–S13M, the linear model was fitted. In Figures 4C, S12, and S13G–S13I, the loess smoothing method was used. In Tables S1, S2, S10, S11, S12, S13, S14, S15, S16, and S17, the Chi-Square test and goodness-of-fit was performed. The significant p value was corrected by the fdr method. R (v3.6.2) was used for data processing and visualization.
Acknowledgments
We thank the anonymous reviewers for their comments and suggestions to improve the manuscript. We would like to thank the Max Planck Genome center for library preparation and sequencing. We thank Neysan Donnelly for proofreading the manuscript. This work was supported by core funding from the Max Planck Society and an Alexander von Humboldt Fellowship to Q.L. B.L. lab was supported by the ANR grant ANR-18-CE12-0013-01.
Author contributions
Q.L., B.L., and R.M. designed the research. B.L., L.M., and M.G. generated the biological material. Q.L., B.L., and R.M. analyzed the data. Q.L. and R.M. wrote the article with input from B.L.
Declaration of interests
The authors declare no competing interests.
Published: August 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107614.
Contributor Information
Bertrand Llorente, Email: bertrand.llorente@inserm.fr.
Raphael Mercier, Email: mercier@mpipz.mpg.de.
Supplemental information
Data and code availability
-
•
The raw sequencing data can be accessed in EMBL-EBI ArrayExpress under the accession numbers E-MTAB-12514 https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12514 and E-MTAB-12516 https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12516.
-
•
The list of identified COs and GCs can be found in Table S4. Accession numbers are listed in the key resources table.
-
•
The related code is available at Zenodo/GitHub https://doi.org/10.5281/zenodo.8204099
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Hunter N. Meiotic Recombination: The Essence of Heredity. Csh Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Copenhaver G.P. Meiotic Recombination: Mixing It Up in Plants. Annu. Rev. Plant Biol. 2018;69:577–609. doi: 10.1146/annurev-arplant-042817-040431. [DOI] [PubMed] [Google Scholar]
- 4.Martín-Castellanos C., Blanco M., Rozalén A.E., Pérez-Hidalgo L., García A.I., Conde F., Mata J., Ellermeier C., Davis L., San-Segundo P., et al. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 2005;15:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cromie G., Smith G.R. Meiotic Recombination in Schizosaccharomyces pombe: A Paradigm for Genetic and Molecular Analysis. Genome Dyn. Stab. 2008;3:195. doi: 10.1007/7050_2007_025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munz P., Wolf K., Kohli J., Leupold U. In: Molecular Biology of the Fission Yeast. Nasim A., Young P., Johnson B.F., editors. Academic Press; 1989. Genetics Overview; pp. 1–30. [Google Scholar]
- 7.Egel R. In: Molecular Biology of the Fission Yeast. Nasim A., Young P., Johnson B.F., editors. Academic Press; 1989. Mating-Type Genes, Meiosis, and Sporulation; pp. 31–73. [Google Scholar]
- 8.Fernandes J.B., Séguéla-Arnaud M., Larchevêque C., Lloyd A.H., Mercier R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. USA. 2018;115:2431–2436. doi: 10.1073/pnas.1713078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohli J., Bähler J. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia. 1994;50:295–306. doi: 10.1007/BF01924013. [DOI] [PubMed] [Google Scholar]
- 11.Fowler K.R., Hyppa R.W., Cromie G.A., Smith G.R. Physical basis for long-distance communication along meiotic chromosomes. Proc. Natl. Acad. Sci. USA. 2018;115:E9333–E9342. doi: 10.1073/pnas.1801920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Liang Z., Hutchinson J., Kleckner N. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berchowitz L.E., Copenhaver G.P. Genetic interference: don't stand so close to me. Curr. Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollingsworth N.M., Brill S.J. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith G.R., Boddy M.N., Shanahan P., Russell P. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osman F., Dixon J., Doe C.L., Whitby M.C. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 17.Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- 18.Clément-Ziza M., Marsellach F.X., Codlin S., Papadakis M.A., Reinhardt S., Rodríguez-López M., Martin S., Marguerat S., Schmidt A., Lee E., et al. Natural genetic variation impacts expression levels of coding, non-coding, and antisense transcripts in fission yeast. Mol. Syst. Biol. 2014;10:764. doi: 10.15252/msb.20145123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanders S.E., Eickbush M.T., Yu J.S., Kang J.W., Fowler K.R., Smith G.R., Malik H.S. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. Elife. 2014;3 doi: 10.7554/eLife.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naumov G.I., Kondratieva V.I., Naumova E.S. [Hybrid Sterility of the Yeast Schizosaccharomyces pombe: Genetic Genus and Many Species in statu nascendi?] Mikrobiologiia. 2015;84:159–169. [PubMed] [Google Scholar]
- 21.Brown W.R.A., Liti G., Rosa C., James S., Roberts I., Robert V., Jolly N., Tang W., Baumann P., Green C., et al. A Geographically Diverse Collection of Schizosaccharomyces pombe Isolates Shows Limited Phenotypic Variation but Extensive. G3 (Bethesda) 2011;1:615–626. doi: 10.1534/g3.111.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L.M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brion C., Legrand S., Peter J., Caradec C., Pflieger D., Hou J., Friedrich A., Llorente B., Schacherer J. Variation of the meiotic recombination landscape and properties over a broad evolutionary distance in yeasts. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutreux F., Dutta A., Peltier E., Bibi-Triki S., Friedrich A., Llorente B., Schacherer J. Lessons from the meiotic recombination landscape of the ZMM deficient budding yeast Lachancea waltii. PLoS Genet. 2023;19 doi: 10.1371/journal.pgen.1010592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm C., Bähler J., Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;136:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto S.P., Payseur B.A. Crossover Interference: Shedding Light on the Evolution of Recombination. Annu. Rev. Genet. 2019;53:19–44. doi: 10.1146/annurev-genet-040119-093957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zickler D., Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fowler K.R., Sasaki M., Milman N., Keeney S., Smith G.R. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24:1650–1664. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazier T., Glémin S. Diversity and determinants of recombination landscapes in flowering plants. PLoS Genet. 2022;18 doi: 10.1371/journal.pgen.1010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avia K., Coelho S.M., Montecinos G.J., Cormier A., Lerck F., Mauger S., Faugeron S., Valero M., Cock J.M., Boudry P. High-density genetic map and identification of QTLs for responses to temperature and salinity stresses in the model brown alga Ectocarpus. Sci. Rep. 2017;7 doi: 10.1038/srep43241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinzelmann R., Rigling D., Sipos G., Münsterkötter M., Croll D. Chromosomal assembly and analyses of genome-wide recombination rates in the forest pathogenic fungus Armillaria ostoyae. Heredity. 2020;124:699–713. doi: 10.1038/s41437-020-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diop S.I., Subotic O., Giraldo-Fonseca A., Waller M., Kirbis A., Neubauer A., Potente G., Murray-Watson R., Boskovic F., Bont Z., et al. A pseudomolecule-scale genome assembly of the liverwort Marchantia polymorpha. Plant J. 2020;101:1378–1396. doi: 10.1111/tpj.14602. [DOI] [PubMed] [Google Scholar]
- 33.Kreplak J., Madoui M.A., Cápal P., Novák P., Labadie K., Aubert G., Bayer P.E., Gali K.K., Syme R.A., Main D., et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019;51:1411–1422. doi: 10.1038/s41588-019-0480-1. [DOI] [PubMed] [Google Scholar]
- 34.Hazzouri K.M., Gros-Balthazard M., Flowers J.M., Copetti D., Lemansour A., Lebrun M., Masmoudi K., Ferrand S., Dhar M.I., Fresquez Z.A., et al. Genome-wide association mapping of date palm fruit traits. Nat. Commun. 2019;10:4680. doi: 10.1038/s41467-019-12604-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haenel Q., Laurentino T.G., Roesti M., Berner D. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Mol. Ecol. 2018;27:2477–2497. doi: 10.1111/mec.14699. [DOI] [PubMed] [Google Scholar]
- 36.Abeyratne C.R., Macaya-Sanz D., Zhou R., Barry K.W., Daum C., Haiby K., Lipzen A., Stanton B., Yoshinaga Y., Zane M., et al. High-resolution mapping reveals hotspots and sex-biased recombination in Populus trichocarpa. G3 (Bethesda) 2023;13:jkac269. doi: 10.1093/g3journal/jkac269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi Y.Y., Sun L.X., Huang Z.Y., Wu X.B., Zhu Y.Q., Zheng H.J., Zeng Z.J. A SNP based high-density linkage map of Apis cerana reveals a high recombination rate similar to Apis mellifera. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auxier B., Becker F., Nijland R., Debets A.J.M., van den Heuvel J., Snelders E. Meiosis in the human pathogen Aspergillus fumigatus has the highest known number of crossovers. bioRxiv. 2022 doi: 10.1101/2022.01.14.476329. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung V.G., Burdick J.T., Hirschmann D., Morley M. Polymorphic variation in human meiotic recombination. Am. J. Hum. Genet. 2007;80:526–530. doi: 10.1086/512131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chowdhury R., Bois P.R.J., Feingold E., Sherman S.L., Cheung V.G. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritz K.R., Noor M.A.F., Singh N.D. Variation in Recombination Rate: Adaptive or Not? Trends Genet. 2017;33:364–374. doi: 10.1016/j.tig.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Stapley J., Feulner P.G.D., Johnston S.E., Santure A.W., Smadja C.M. Variation in Recombination Frequency and Distribution across Eukaryotes: Patterns and Processes. Philos. Trans. R. Soc. B. 2017;372:20160455. doi: 10.1098/rstb.2016.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Maclean C.J., Zhang J. Evolution of the Yeast Recombination Landscape. Mol. Biol. Evol. 2019;36:412–422. doi: 10.1093/molbev/msy233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowan B.A., Heavens D., Feuerborn T.R., Tock A.J., Henderson I.R., Weigel D. An Ultra High-Density Arabidopsis thaliana Crossover Map That Refines the Influences of Structural Variation and Epigenetic Features. Genetics. 2019;213:771–787. doi: 10.1534/genetics.119.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lian Q., Solier V., Walkemeier B., Durand S., Huettel B., Schneeberger K., Mercier R. The megabase-scale crossover landscape is largely independent of sequence divergence. Nat. Commun. 2022;13:3828. doi: 10.1038/s41467-022-31509-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellermeier C., Higuchi E.C., Phadnis N., Holm L., Geelhood J.L., Thon G., Smith G.R. RNAi and heterochromatin repress centromeric meiotic recombination. Proc. Natl. Acad. Sci. USA. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyppa R.W., Smith G.R. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Diezmann L., Rog O. Let's get physical - mechanisms of crossover interference. J. Cell Sci. 2021;134 doi: 10.1242/jcs.255745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pazhayam N.M., Turcotte C.A., Sekelsky J. Meiotic Crossover Patterning. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.681123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White M.A., Wang S., Zhang L., Kleckner N. Quantitative Modeling and Automated Analysis of Meiotic Recombination. Methods Mol. Biol. 2017;1471:305–323. doi: 10.1007/978-1-4939-6340-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foss E., Lande R., Stahl F.W., Steinberg C.M. Chiasma interference as a function of genetic distance. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson C.M., Oke A., Yam P., Zhuge T., Fung J.C. Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia V., Gray S., Allison R.M., Cooper T.J., Neale M.J. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature. 2015;520:114–118. doi: 10.1038/nature13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S., Veller C., Sun F., Ruiz-Herrera A., Shang Y., Liu H., Zickler D., Chen Z., Kleckner N., Zhang L. Per-Nucleus Crossover Covariation and Implications for Evolution. Cell. 2019;177:326–338.e16. doi: 10.1016/j.cell.2019.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lian Q., Chen Y., Chang F., Fu Y., Qi J. inGAP-family: Accurate Detection of Meiotic Recombination Loci and Causal Mutations by Filtering Out Artificial Variants due to Genome Complexities. Dev. Reprod. Biol. 2022;20:524–535. doi: 10.1016/j.gpb.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedersen B.S., Quinlan A.R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreno S., Klar A., Nurse P. Volume 194. Academic Press; 1991. [56] Molecular genetic analysis of fission yeast Schizosaccharomyces pombe; pp. 795–823. (Guide to Yeast Genetics and Molecular Biology). [DOI] [PubMed] [Google Scholar]
- 61.Abt T.D., Souffriau B., Foulquié-Moreno M.R., Duitama J., Thevelein J.M. Genomic saturation mutagenesis and polygenic analysis identify novel yeast genes affecting ethyl acetate production, a non-selectable polygenic trait. Microb. Cell. 2016;3:159–175. doi: 10.15698/mic2016.04.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rowan B.A., Patel V., Weigel D., Schneeberger K. Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. G3 (Bethesda) 2015;5:385–398. doi: 10.1534/g3.114.016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ekwall K., Thon G. Mating-Type Determination in Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2017;2017 doi: 10.1101/pdb.prot091728. pdb.top079772. [DOI] [PubMed] [Google Scholar]
- 64.Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 65.Harris M.A., Rutherford K.M., Hayles J., Lock A., Bähler J., Oliver S.G., Mata J., Wood V. Fission stories: using PomBase to understand Schizosaccharomyces pombe biology. Genetics. 2022;220 doi: 10.1093/genetics/iyab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H., Xu W., Sun Y., Lian Q., Wang C., Yu C., He C., Wang J., Ma H., Copenhaver G.P., Wang Y. The cohesin loader SCC2 contains a PHD finger that is required for meiosis in land plants. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capilla-Pérez L., Durand S., Hurel A., Lian Q., Chambon A., Taochy C., Solier V., Grelon M., Mercier R. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023613118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Durand S., Lian Q., Jing J., Ernst M., Grelon M., Zwicker D., Mercier R. Joint control of meiotic crossover patterning by the synaptonemal complex and HEI10 dosage. Nat. Commun. 2022;13:5999. doi: 10.1038/s41467-022-33472-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequencing data can be accessed in EMBL-EBI ArrayExpress under the accession numbers E-MTAB-12514 https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12514 and E-MTAB-12516 https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-12516.
-
•
The list of identified COs and GCs can be found in Table S4. Accession numbers are listed in the key resources table.
-
•
The related code is available at Zenodo/GitHub https://doi.org/10.5281/zenodo.8204099
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.