Summary
Hypoplastic left heart syndrome (HLHS) is a severe congenital heart defect (CHD) characterized by hypoplasia of the left ventricle and aorta along with stenosis or atresia of the aortic and mitral valves. HLHS represents only ∼4%–8% of all CHDs but accounts for ∼25% of deaths. HLHS is an isolated defect (i.e., iHLHS) in 70% of families, the vast majority of which are simplex. Despite intense investigation, the genetic basis of iHLHS remains largely unknown. We performed exome sequencing on 331 families with iHLHS aggregated from four independent cohorts. A Mendelian-model-based analysis demonstrated that iHLHS was not due to single, large-effect alleles in genes previously reported to underlie iHLHS or CHD in >90% of families in this cohort. Gene-based association testing identified increased risk for iHLHS associated with variation in CAPN2 (p = 1.8 × 10−5), encoding a protein involved in functional adhesion. Functional validation studies in a vertebrate animal model (Xenopus laevis) confirmed CAPN2 is essential for cardiac ventricle morphogenesis and that in vivo loss of calpain function causes hypoplastic ventricle phenotypes and suggest that human CAPN2707C>T and CAPN21112C>T variants, each found in multiple individuals with iHLHS, are hypomorphic alleles. Collectively, our findings show that iHLHS is typically not a Mendelian condition, demonstrate that CAPN2 variants increase risk of iHLHS, and identify a novel pathway involved in HLHS pathogenesis.
Keywords: congenital heart defect, exome sequencing, association, oligogenic, development, frog, Xenopus
Blue et al. characterize the genetic basis of isolated hypoplastic left heart syndrome (iHLHS), implicating hypomorphic CAPN2 variants through association testing and functional validation in a Xenopus laevis model. Their results suggest that iHLHS is not typically a Mendelian condition and identify a novel pathway involved in HLHS pathogenesis.
Introduction
Over the past decade, the use of next-generation sequencing (NGS) accompanied by ever-improving computational strategies to identify candidate genes and predict pathogenicity of variants has led to discovery of the genetic basis of thousands of Mendelian conditions (MCs) and transformed the practice of genetic medicine.1 Many of these MCs are characterized by multiple malformations, so-called multiple-malformation syndromes (MMSs), and the discovery of genes underlying MMSs has in turn led to much deeper knowledge of development and disease pathogenesis. Yet, the vast majority of malformations occur in persons without a known MMS, are isolated (i.e., a person has only one malformation), and are not inherited in Mendelian patterns. Moreover, assuming that most major birth defects are malformations, about 1 of every 33 children born in the United States has a malformation.2 Collectively, birth defects represent the most common cause of death in the first year of life3 and are responsible for more than 3 million pediatric deaths annually. Yet, in contrast to MMSs, progress toward understanding the genetic basis of isolated malformations has been slow.4
Among malformations, the most frequent causes of morbidity and mortality are congenital heart defects (CHDs), and among CHDs, hypoplastic left heart syndrome (HLHS) is the most common cause of early death.3,5,6 HLHS is characterized by marked abnormalities of the left-sided heart structures, with a critically underdeveloped left ventricle and aorta along with a combination of aortic and mitral valve stenosis or atresia. HLHS occurs in ∼1/4,000 live births in the United States, the majority of which are isolated (i.e., the HLHS is not a feature of an MMS), simplex cases.7 The genetic basis of risk of HLHS is well established. Family studies have revealed a high risk of recurrence of CHDs among siblings, a high relative risk (∼37-fold) for first-degree relatives, and a heritability of 0.71–0.90.8,9,10 Multiplex HLHS families have been reported, although most of these featured reduced penetrance and/or variable expressivity with other left-sided heart defects, such as bicuspid aortic valve.11,12
Genetic studies of candidate genes have identified several “known genes” underlying risk for HLHS, including NOTCH1 (MIM: 190198), GJA1 (MIM: 121014), MYH6 (MIM: 160710), HAND1 (MIM: 602406), RBFOX2 (MIM: 612149), and NKX2-5 (MIM: 600584). However, many of the putative pathogenic variants in these genes are observed in reportedly unaffected relatives, suggesting that penetrance is reduced and/or effect size is modest.13,14,15,16 Enrichment for putatively damaging de novo variants has been observed in other genes involved in heart development, brain development, and chromatin modification.17,18 But many of these de novo events occurred in individuals with features suggestive of an MMS (e.g., Kabuki [MIM: 147920], Adams-Oliver [MIM: 100300], Noonan [MIM: 163950], and Jacobsen [MIM: 147791]) that includes HLHS, or so-called syndromic HLHS. Collectively, variants in these genes account for only a small fraction (∼2%–10%) of families with syndromic HLHS17,19 and an even smaller fraction of isolated HLHS (iHLHS).20,21,22,23
Based in part on these observations, it has been hypothesized that the majority of iHLHS might be explained by an oligogenic inheritance model.8,19 Oligogenic models occupy the genetic risk space between MCs and polygenic or complex traits and may include an interaction between, or combinatorial effects among, several moderate-effect alleles and/or environmental factors. Oligogenic inheritance of iHLHS is supported by evidence from a mouse model of iHLHS and the identification of genes underlying murine iHLHS.24 However, empirical evidence for oligogenic inheritance of iHLHS in humans has proven challenging to generate, limited in large part by the relatively modest sample size of cohorts with iHLHS.24
To identify moderate-risk alleles for iHLHS, we organized a multinational network of investigators and assembled a cohort representing 331 families with iHLHS who underwent exome sequencing (ES). Analysis of exome data using Mendelian models identified four ClinVar likely pathogenic (LP) or pathogenic (P) variants in three “known genes” in 4 families, LP or P variants meeting American College of Medical Genetics (ACMG) criteria in 23 families, and no novel genes for iHLHS. Gene-based association testing of rare variants identified a single novel gene, CAPN2 (MIM: 114230), with a genome-wide suggestive association with iHLHS. Functional studies in a vertebrate animal model (Xenopus laevis) confirmed that capn2 is essential for cardiac ventricle morphogenesis and suggest that the CAPN2 variants we identified act as hypomorphic alleles in vivo.
Material and methods
Samples
This study was approved by the institutional review board (IRB) of Seattle Children’s Research Institute; the IRB of each of three independent studies in the United States, Canada, and Europe; and the Centers for Disease Control and Prevention IRB, along with the IRBs for each participating site of the National Birth Defects Prevention Study (NBDPS). Written informed consent was obtained for each child, their parents, and, in some cases, their relatives. All affected individuals were clinically diagnosed with iHLHS following best practices, summarized below. A total of 336 families with simplex cases of iHLHS were ascertained from four separate studies.
Three-hundred and seventy-five participants (129 families) were ascertained and sampled from the NBDPS between 1997 and 2011. The NBDPS is a population-based case-control study that included women with pregnancies affected by a major structural birth defect with estimated dates of delivery between October 1997 and December 2011.25,26 Affected individuals were ascertained from birth defect surveillance systems in 10 US states, and a clinical geneticist and/or pediatric cardiologist reviewed data abstracted from medical records to determine eligibility.27 Persons with known chromosomal or single-gene disorders and known syndromes for which the genetic basis is unknown were excluded. All CHDs were centrally reviewed and classified by a team of pediatric cardiologists. HLHS was clinically defined by extreme smallness of the left-sided heart structures (mitral valve and left ventricle) and aorta (including the aortic valve, ascending aorta, arch, and sometimes descending aorta [coarctation]). A Z-score threshold was not considered when assessing cardiac morphology and assigning the diagnosis of HLHS. The ventricular septum could be intact, or a ventricular septal defect could be present. NBDPS cardiac defects were confirmed by echocardiography, catheterization, surgery, or autopsy; affected individuals diagnosed by prenatal ultrasound/echocardiography were included only if the procedure was completed by a pediatric cardiologist or in a prenatal diagnosis center with expertise in this area.28
One hundred and sixty-seven participants (71 families) were ascertained and sampled under an IRB-approved protocol at Nationwide Children’s Hospital between 2004 and 2016. Children and adults with CHD were approached at their outpatient visit or were referred by other institutions. Additional family members were recruited via the index subject. Medical records were reviewed, and cardiac defects were confirmed by echocardiography, operative note, or procedure report, and the records were searched for any additional health problems, including the presence of other birth defects, specific syndrome diagnosis, or neurodevelopmental disorders. HLHS was defined as mitral valve atresia or stenosis along with aortic valve atresia or stenosis with hypoplasia of the left ventricle and aortic arch (Z-score < −2). Individuals were included if HLHS co-occurred with bicuspid aortic valve but excluded if they had a complex cardiac defect (e.g., presence of left-sided defect and second major cardiovascular malformation) or additional malformations or a known chromosomal abnormality or were diagnosed with an MMS.
Ninety-five DNA participants (84 families) that were ascertained for iHLHS and sampled under an IRB-approved protocol at the Hospital for Sick Children were recruited to the Heart Centre Biobank Registry between 2007 and 2016.29 Probands with clinical diagnosis of HLHS, along with their relatives, were ascertained and consented to participate in the biobank. Those with clinical or genetic diagnosis of syndromic HLHS were excluded to increase power to identify genetic variants influencing iHLHS.
Ninety-five participants (52 families) were included from “The genetics of LVOTO defects” study at Helsinki University Children’s Hospital, Finland, which included pediatric patients with left-ventricular outflow tract obstruction (LVOTO) defects and their family members. Only probands with HLHS were selected, and HLHS was defined as hypoplastic left ventricle not forming the apex of the heart, with aorta and/or mitral hypoplasia or atresia. Patients with malformations of other organ systems or a known MMS were not included. All participants were of self-reported Finnish ancestry. Blood or saliva samples were collected between 2014 and 2015, and the DNA was extracted at the Finnish Institute of Molecular Medicine. The study protocol was approved by the Ethics Board of Helsinki and Uusimaa Hospital District, and the guidelines of the Declaration of Helsinki were followed.
Control samples (n = 2,192) were selected from independent projects in the University of Washington Center for Mendelian Genomics. The controls represent unrelated individuals ascertained as unaffected parents of children with rare MCs for which clinical characteristics did not include CHD. They had no known history of CHD. These samples were sequenced, and data were processed via the same pipeline and at a similar time as samples in the iHLHS cohort.
Sequencing and data quality control
Exome capture was completed with the Nimblegen SeqCap EZ Human Exome Library v.2.0 kit (Roche) according to the manufacturer’s protocol with minor custom additions as previously described.30 Minimum sample quality thresholds included mean depth of 50× with >90% of targeted sites exhibiting >8× coverage. Reads were aligned to the reference human genome (hg19) with the Burrows-Wheeler Aligner.31 Variant calling was performed using the Genome Analysis Toolkit (GATK) HaplotypeCaller (v.4.0.5.0) in accordance with GATK best practices.32 The joint-called multisample variant call format file was normalized and decomposed by Vt v.0.5733 to ensure a unified representation of variants and then annotated with Variant Effect Predictor v.95.3.34 Copy-number variants (CNVs) were called from ES data using CoNIFER.35 Exome analyses were performed between 2019 and 2022.
Following ES, Peddy36 was used to identify unexpected and incorrect familial relationships, to confirm self-reported ancestry, and to identify samples with sequencing quality control (QC) metrics indicative of poor-quality DNA and/or contamination (i.e., high/low heterozygosity, low overall depth, and skewed allele balance). Five families (5/336) were subsequently excluded from further analysis: three families due to duplicate submission of the same subjects and two due to unresolvable sample misidentification. Parental sequence data were available for half of all families (i.e., trios).
The final cohort consisted of 331 families: 125 singletons, 39 parent-offspring pairs (i.e., duos), 159 trios, and 8 families with additional family members with non-HLHS CHDs.
GEMINI37 v.0.30.1 was used to filter out low-quality variant calls, requiring each variant to have a minimum read depth of 6, a maximum read depth of 500, missingness less than 10%, and a minimum genotype quality of 20 in at least 95% of individuals, while excluding variants with non-PASS GATK filter flags and greater than one alternate allele. To remove potential pipeline-specific calling artifacts, single-nucleotide variants with minor allele frequencies >5% in our internal database of >4,200 exomes were excluded. Similarly, to remove poorly called indels located in highly polymorphic, multiply mapping, or complex regions, we excluded insertions or deletions that overlapped a 9 bp window that contained indels in >5% of individuals in our internal database. CNVs were called along with ∼1,000 contemporaneously sequenced samples using the same pipeline and were filtered to include only high-confidence events with svd_rpkm values >1 or <−1, to reduce systematic noise in read depths due to differences in exon size and targeting efficiency, and coverage of >4 probes/exons.
Identification of variants in known HLHS or left-sided heart defect genes
We reviewed the HLHS/CHD literature to create a list of 58 “known candidate genes” for putatively monogenic forms of HLHS/CHD. This list included 7 genes previously implicated in iHLHS, 13 genes implicated in syndromic HLHS, 17 genes implicated in isolated CHDs, and 21 genes implicated in syndromic left-sided CHDs (Table S1). We identified variants in these 58 candidate genes that are listed in ClinVar with at least one LP or P interpretation or met ACMG criteria for LP or P. To identify additional candidate variants in these genes that have not yet been reported, we categorized variants as predicted pathogenic (PP) if a variant met the following criteria: had a Phred-scaled Combined Annotation Dependent Depletion (CADD)-Splice v.1.6 score38 ≥15; was predicted to have protein-coding impact (i.e., moderate or high impact according to the GEMINI schema, which excludes synonymous and non-coding variants); did not have an alternate allele frequency (maxAAF) ≥0.001 in any single population within the EVS/ESP6500, 1000 Genomes (phase 3 release), or gnomAD Browser v.2.0.1; and, for multiplex families, co-segregated with the phenotype.
Mendelian-model-based analysis
To search exome-wide for novel iHLHS candidate genes, we imposed more stringent filters on PP variants. We removed poorly called insertion/deletions (indels) that overlapped with any 9 bp window that contained indels in >0.01% of individuals in our internal database; we applied a more stringent frequency cutoff than used in QC because the aim was to identify large-effect alleles with high confidence in the variant calls, and such alleles are expected to be very rare. Proband genotypes were filtered against parental or other available familial genotypes assuming complete penetrance (i.e., unaffected parents could not be carriers for a putatively dominant variant) under de novo, autosomal-dominant (for three families with an affected child and affected parent), autosomal-recessive (homozygous and compound heterozygous), and X-linked inheritance patterns. Approximately half (167/331) of the iHLHS families were “complete” trios, allowing for the identification of de novo events. For the 164 incomplete trios lacking sequence data for one parent (i.e., preventing variant filtration under a de novo model), variants were filtered against any unaffected family members for whom we had sequence data. For example, homozygous and compound heterozygous variants in incomplete trios were identified by assuming that the second allele was transmitted from the parent who was unavailable for study or by identifying genes in which two variants had the potential to be present in trans. Potential de novo variants in the remaining “incomplete” trios were not considered in this analysis due to the large number of rare heterozygous PP variants expected exome wide. Finally, to prioritize among the putative novel candidate genes that might underlie monogenic forms of iHLHS, we considered genes with PP variants segregating in a pattern consistent with the same mode of inheritance in three or more families. Variants in these genes were then manually reviewed in Integrated Genome Viewer39 for quality.
Sample selection and QC for association testing
Subjects were defined as the proband from each of the 331 families used in the Mendelian-model-based analysis. Samples underwent extended QC to align sample sequencing quality and depth between cases and controls. We used PLINK v.1.940 to identify case or control samples with high genotype missingness and Peddy to identify samples with outlier values for heterozygosity, sequencing depth, or skewed allele balance.36 Pairs of samples with estimated kinship values >0.1 were flagged, and one sample per pair was removed to reduce cryptic relatedness.
To reduce confounding due to population stratification, Peddy was used to predict sample ancestry. Due to lack of ancestry-matched controls, samples were restricted to those confidently assigned to European ancestry (probability of EUR ancestry >0.75). Principal components (PCs) for the remaining samples were estimated using PC-Air as implemented in GENESIS26 (Figure S1). The first four PCs were selected as covariates based on their position in a scree plot, none of which showed significant correlation with outcome. A genetic relatedness matrix of kinship coefficients estimated using KING-Robust was adjusted for those first four PCs using the PC-Relate approach in GENESIS.
Extensive variant QC for the case-control association analyses excluded variants with (1) gross deviation from Hardy-Weinberg equilibrium (HWE); (2) inadequate read depth; (3) abnormally high read depth, because this typically is indicative of spurious read alignment; (4) poor genotyping quality; (5) low call rate; and (6) call rate differences by phenotypic status. Biallelic sites without a GATK filter flag were then filtered to exclude those with extreme deviation from HWE in the final dataset of 1,313 samples (p < 1.51 × 10−7). Fourteen subjects with previously identified likely causal variants were excluded from the sample set, reducing the case count from 213 to 199. Sites with high missingness (>0.1) as estimated in PLINK were excluded. Last, we retained only sites in which samples had genotype quality >20 and depth >6. After applying all filters, 94,762 variant sites and 1,299 (199 subjects; 1,100 control individuals) samples remained.
Gene-based association testing
Gene-based association testing was performed using the unified sequencing kernel association test with optimal kernel weighting (SKAT-O)41 via the GENESIS42 package in the R statistical language. We included low-frequency autosomal variants with a maximum population allele frequency in ExAC, or maxAAF, ≤0.01 that were predicted to have protein-coding impact (i.e., moderate or high impact according to the GEMINI schema, which excludes synonymous and non-coding variants). To limit our analysis to only genes with a sufficient number of observed alleles to provide adequate statistical power, we excluded genes with a total minor allele count <10 summed across all variants within the gene among subjects and control individuals. In total, 58,369 variant sites in 5,308 genes passed our inclusion criteria. Association testing was performed, adjusting for sex and PCs 1–4 as fixed effects, under a dichotomous trait model with variant weights following the beta distribution suggested by Madsen-Browning (0.5, 0.5) to emphasize the impact of rare variants in the aggregate analysis.43
In vivo functional studies of Capn2 in Xenopus
Xenopus laevis oocytes were fertilized and de-jellied following standard procedures.44 In preparation for microinjection, embryos were arrayed in a Petri dish lined with Nitex nylon mesh (1,000 μm) in a solution of 3% Ficoll 400 (Fisher Bioreagents BP525) in 0.75× MMR as previously described.45 After injection, embryos were gradually equilibrated with 0.1× MMR and raised to tadpole stage 43–45. For CRISPR-Cas9-mediated genome editing, 2 nL of a 300 mM KCl solution containing 300 pg/nL single guide (sg) RNA targeting Xenopus capn2, 62.5 pg/nL Alt-R S.p. HiFi Cas9 nuclease (Integrated DNA Technologies [IDT]), and 80 pg/nL GFP mRNA was injected into the left dorsolateral blastomere of eight-cell-stage embryos fated to give rise to the left side of the heart.46 For capn2 mRNA specificity rescue experiments, this pooled sgRNA was co-injected with approximately 200 pg of synthetic mRNA encoding wild-type Xenopus capn2, a Xenopus ortholog of the human CAPN2707C>T mutation, or a Xenopus ortholog of the human CAPN21112C>T mutation. For morpholino-mediated knockdown of Xenopus Capn2 protein, embryos were injected as above with 35 pg/nL GFP mRNA plus either 20 ng of morpholino oligonucleotide targeting the Xenopus capn2 translation start site (TCGGCGACCCCGCTCATGTCG; GeneTools) or 20 ng of standard control morpholino (GeneTools). For specificity rescue experiments, 1.55 ng of morpholino-resistant wild-type capn2 mRNA was co-injected with capn2 morpholino. Tadpoles with abnormal ventricles were identified by visual inspection by two independent observers. In all cases, analysis of variance was conducted to evaluate differences in the mean percentage (weighted average) of heart phenotypes, with at least three independent experiments (performed with 8–57 embryos per condition). Error bars in all graphs represent standard deviations.
Injected embryos were fixed in Dent’s fixative (80% methanol/20% dimethyl sulfoxide) for 2–3 h at room temperature. Samples were then processed for whole-mount fluorescence immunocytochemistry,47 using a mouse monoclonal antibody directed against cardiac Troponin (DSHB CT3; 1:100) and a rabbit polyclonal against GFP (Invitrogen A6455; 1:500). The secondary antibodies Alexa 555-conjugated goat anti-mouse IgG (Invitrogen A32727; 1:500) and Alexa 488-conjugated goat anti-rabbit IgG (Invitrogen A32731; 1:500) were used to visualize Troponin T and GFP, respectively. For clearing, embryos were washed five times in methanol and then transferred to Murray’s Clear (2 parts benzyl benzoate and 1 part benzyl alcohol) for imaging on Zeiss Lumar.V12 and Leica TCS SPE microscopes.
Results
Mendelian-model-based analysis
We identified 27 families (8.2%) with either a ClinVar LP or a P variant (4 families [1.2%]) or with a variant meeting ACMG criteria for LP or P (23 families [7.0%]) in genes reported to underlie iHLHS (2 families [0.6%]), an MMS of which a CHD is a clinical finding (6 families [1.8%]), or isolated CHD (15 families [4.5%]).
ClinVar known LP or P variants were found in 4 of 331 (1.2%) families (Table 1). No ClinVar known LP or P variants were found in any of the 331 families in the seven genes previously reported to underlie iHLHS (Table S1). In 3 of 331 families, ClinVar known LP or P variants were found in two genes underlying dominantly inherited MC-MMS (Table 1) in which HLHS is a clinical feature (i.e., syndromic HLHS). Of these, two individuals had variants (one confirmed de novo) in KMT2D48 (MIM: 602113) underlying Kabuki syndrome (MIM: 147920), in which CHDs are common but HLHS is rare,22 and one individual had a variant in NOTCH149 (MIM: 190198), one of several genes that underlie Adams-Oliver syndrome (MIM: 616028). Analysis of 37 genes reported to underlie isolated CHD or MCs that frequently include left-sided CHDs but not HLHS revealed a ClinVar known LP variant in ACTC1 (MIM: 102540) in one family (Table S1). P variants in ACTC1 can result in left-ventricular non-compaction, cardiomyopathy, and/or atrial septal defects.
Table 1.
Number of variants in families affected by isolated hypoplastic left heart syndrome (iHLHS) identified by Mendelian-model-based analysis
| Phenotype category | Probands | ClinVar LP/P | ACMG LP/P | PP |
|---|---|---|---|---|
| Isolated HLHS | 6 | 0 | 2 | 4 |
| MC-MMS/CHD | 12 | 3 | 5 | 4 |
| Isolated CHD | 23 | 1 | 14 | 8 |
| Multiple candidate variants | 5 | 0 | 2 | 8 |
| Total probands explained | 46 | 4 | 23 | 19a |
Two variants were identified in five families, including eight PP variants. Hence, the total number of probands potentially explained by PP variants is 19 instead of 24. Abbreviations: ACMG, American College of Medical Genetics and Genomics; CHD, congenital heart defect; LP, likely pathogenic; MC-MMS, Mendelian conditions-multiple malformation syndromes; P, pathogenic; PP, predicted pathogenic.
ACMG LP or P variants were found in a total of 23/331 (7.0%) families (Table 1). These included 7/331 (2.1%) families with variants in MYH6 (MIM: 160710, one confirmed de novo), a gene that has been associated with iHLHS and atrial septal defects50 as well as both dilated and hypertrophic cardiomyopathy.51 In genes underlying MMS, two variants were found in both KMT2D (one confirmed de novo) and NOTCH1 (one confirmed de novo) and one variant in MYRF52 (MIM: 608329). ACMG LP or P variants were found in 14 families (4.2%) in genes underlying dominantly inherited isolated CHDs or MCs with left-sided CHDs, including one variant each in MYH7 (MIM: 160760) and CREBBP (MIM: 600140).
PP variants were found in a total of 21 families, including 8/331 (2.4%) families with variants in genes that underlie either iHLHS or dominantly inherited MC-MMS of which HLHS is a clinical feature (Table 1). Specifically, four individuals had PP variants in MYH6, one person had a PP variant in KMT2D, one individual had a PP variant in NOTCH1, and two males were hemizygous for PP variants in KDM6A53 (MIM: 300128). PP variants were detected in eight (2.4%) families in genes underlying either isolated or dominantly inherited MCs with left-sided CHDs. These included individual families with variants in DTNA (MIM: 601239), NR2F2 (MIM: 107773), ZFPM2 (MIM: 603693), CHD7 (MIM: 608892), JAG1 (MIM: 601920), NOTCH2 (MIM: 618026), and CREBBP and a female proband heterozygous for a variant in ZIC3 (MIM: 300265), which underlies an X-linked disorder of left-right asymmetry including left-sided CHD.
Several PP variants were de-prioritized due to conflicting evidence for pathogenicity. Two individuals with variants in KRAS (MIM: 190070) and one individual with a variant in MYH7 were found to have variants that met criteria for a PP variant but were annotated as benign or likely benign in ClinVar. Last, each of five individuals had either ACMG LP or P or PP variants identified in two different genes. One person had both an ACMG LP or P heterozygous variant in MYH7 and a homozygous PP variant in NOTCH2, one person was heterozygous for an ACMG LP or P variant in HRAS (MIM: 190020) and a PP variant in NOTCH1, one person was heterozygous for PP variants in MYH6 and NR2F2, one person was heterozygous for PP variants in GATA5 (MIM: 611496) and ZFPM2, and one person was heterozygous for PP variants in MED13L (MIM: 608771) and MYH6.
No putatively novel genes, defined as three or more families with compelling candidate variants in the same gene, were discovered. Collectively, these results suggest that iHLHS was, in the vast majority of families in this cohort, not due to large-effect alleles in known or novel genes.
Gene-based association testing
To discover candidate genes harboring risk variants for iHLHS with more modest effect sizes and maximize our power given the limited sample size, we performed gene-based association testing using SKAT-O,41 seeking to identify genes with an excess of rare variants (maxAAF ≤1%) predicted to have protein-altering consequences. After trios with cryptic relatedness, those in whom ClinVar LP or P variants or ACMG LP or P variants were identified, and those for whom adequate ancestry-matched controls were unavailable were removed, the sample size was reduced to 199. When the 199 cases were combined with 1,100 ancestry-matched controls, 5,357 genes had a minor allele count >10 and were included in the SKAT-O analysis. Of these, CAPN2 had suggestive evidence of association with HLHS (p = 1.8 × 10−5, Bonferroni cutoff 9.4 × 10−6; Figure 1). A sensitivity analysis repeating the analysis using a burden test rather than SKAT-O again found the strongest statistical support for CAPN2. We observed an excess of rare coding variants in cases (5.5%, 11/199) vs. controls (0.73%, 8/1100). Among the eight different variants in the case group, seven were predicted to lead to either damaging substitutions (each with Phred-scaled CADD-Splice scores >21) or protein truncation (n = 1) (Figure 2 and Table S2). Two variants were found in probands who inherited the variant from unaffected parents, four variants were found in probands from incomplete trios, one variant (c.707C>T) was found twice in two probands from incomplete trios, and one variant (c.1112C>T) was found once in a proband who inherited it from an unaffected parent and twice in probands from incomplete trios. These results suggest that rare variants in CAPN2 are associated with increased risk of iHLHS but are not large-effect alleles. No individual in either the case group or the control group had more than one rare protein-altering variant in CAPN2.
Figure 1.
Discovery association tests
(A–D) Comparison of SKAT-O (A and B) and burden (C and D) approaches to gene-based rare variant aggregate association analysis. Manhattan (B and D) and QQ (A and C) plots demonstrate that CAPN2 is the most significantly associated gene tested, regardless of approach. A low median λ value for the SKAT-O QQ plot (A) suggests genomic deflation in this approach.
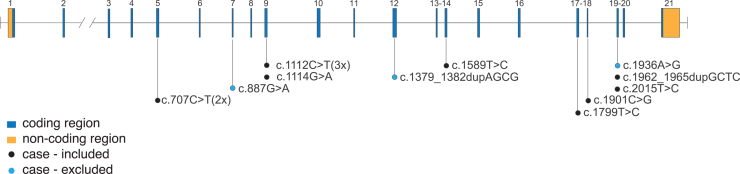
Figure 2.
CAPN2 variants identified in individuals with isolated hypoplastic left heart syndrome (iHLHS)
Gene diagram of CAPN2 (GenBank: NM_001748.4) with locations of variants identified in individuals with iHLHS. Black dots represent variants present in the association subset of the cohort, blue dots represent variants identified in samples excluded from the association analysis due to lack of ancestry-matched controls.
Two additional individuals with iHLHS who were excluded from the association analysis had both a variant in CAPN2 and a candidate variant identified under the Mendelian-model-based analysis of known genes, one in an individual with an ACMG LP/P de novo stop-gain in KMT2D and one in an individual with a heterozygous PP missense in NOTCH2. One additional individual with iHLHS was found both to have a variant in CAPN2 and to be heterozygous for a known pathogenic splice acceptor variant in DHCR7 underlying the recessively inherited Smith-Lemli-Opitz syndrome (MIM: 270400).
Replication of the observed association with CAPN2 was impaired by the limited availability of additional iHLHS cases. Instead, we attempted replication using exome data from HLHS trios and control individuals (drawn from the parents of children with other types of CHD) enrolled in the Pediatric Genomics Cardiac Consortium or Pediatric Heart Network. Only a small subset (356/1,213 = 29%) of these cases had isolated CHD. After extensive curation of data from 1,213 trios with CHD for QC, relatedness, and matching for genetic similarity, the final dataset consisted of 138 HLHS-affected individuals and 370 control individuals, and 43/138 (31%) HLHS-affected individuals had an abnormal extracardiac finding, suggesting that, at best, only approximately two-thirds of these subjects had iHLHS. CAPN2 failed to meet the minimum allele count threshold of 10, and no significant genome-wide associations were identified for any common variant or gene tested. Accordingly, we turned to testing in a model organism to validate the functional consequences of the CAPN2 variants significantly associated with iHLHS.
Functional characterization of CAPN2 and HLHS-associated variants in heart development
To assess the in vivo requirement for CAPN2 in vertebrate heart development, we employed embryos of the frog Xenopus laevis. Although this amphibian develops a three-chambered heart, many molecular, cellular, and morphogenetic aspects of Xenopus cardiogenesis are identical to those of higher vertebrates.54 Moreover, the single ventricle in the frog heart has been found to be similar to the mammalian left ventricle,55,56 making Xenopus an excellent model for testing the potential role of CAPN2 variants in left-ventricular morphogenesis and the pathogenesis of HLHS.57
To perform CRISPR-mediated editing of the Xenopus capn2 locus, sgRNAs targeting capn2 were injected, along with Cas9 protein, into a blastomere of the early embryo that is fated to contribute to the heart.46 These reagents were confirmed to produce indels within the capn2 locus in injected embryos (Figure S2). In contrast to control embryos that were injected with capn2 sgRNA alone, which rarely exhibit heart defects (4.9%, n = 125; Figures 3A and 3F), 57.3% (n = 185) of embryos injected with capn2 sgRNA plus Cas9 protein had hypoplastic ventricles with reduced ventricular cavities (Figures 3B and 3F), analogous to the underdeveloped left-ventricle morphology and small ventricular chamber found in human HLHS patients.58 Importantly, co-injection of CRISPR reagents with wild-type (WT) capn2 mRNA led to a significant (>50%) reduction in the number of embryos with abnormal hearts (27.1%, n = 151; p < 0.05; Figure 3F). The restoration of normal ventricle morphology by WT capn2 mRNA (Figure 3C) confirms that the hypoplastic ventricle phenotype is specific to the loss of capn2 function. As a further confirmation of the specificity of the capn2 CRISPR-Cas9-induced heart defect, we utilized a second loss-of-function reagent: a morpholino oligonucleotide that binds to the Capn2 translation start site to knock down levels of Capn2 protein. This independent strategy also resulted in hearts with hypoplastic ventricles rescuable by exogenous WT capn2 mRNA (Figure S3).
Figure 3.
CRISPR-Cas9-mediated functional studies of CAPN2 variants in vivo
Xenopus laevis embryos were injected with capn2 single guide RNA (sgRNA) alone (control; A), capn2 sgRNA plus Cas9 protein (to elicit capn2 indels and loss of Capn2 function; B), or capn2 sgRNA/Cas9 co-injected with WT capn2 mRNA (C), mutant capn2 mRNA engineered to contain the orthologous human CAPN2707C>T variant (707 mRNA; D), or mutant capn2 mRNA engineered to contain the orthologous human CAPN21112C>T variant (1112 mRNA; E). Injected F0 embryos were allowed to develop through cardiogenesis and immunostained for Troponin T (red) to highlight the outflow tract (oft) and ventricle (v) of the heart (top row); optical sectioning was used to visualize the internal ventricular chamber (arrows, bottom row). In contrast to controls (A), embryos with CRISPR-Cas9-mediated loss of function of Capn2 (B) develop a hypoplastic ventricle and reduced ventricular chamber. Normal phenotype (C) is restored by co-injecting exogenous WT capn2 mRNA, confirming specificity of the phenotype. Co-injection of 707 mRNA can also restore normal ventricle phenotypes (D), although the frequency of rescue was not statistically significant (F), indicating that the CAPN2707C>T variant may be a mildly hypomorphic allele. Co-injection of 1112 mRNA is unable to rescue the HLHS-like phenotype at all (E and F), indicating the CAPN21112C>T variant may represent a null allele with respect to function necessary for normal ventricular development. (F) Results were quantified from three independent trials (n = 14–57 embryos per condition, per experiment); error bars indicate standard deviation. Significance of differences between the percentage of HLHS-like phenotypes in each condition is noted. Scale bars, 100 μm.
Having confirmed by two different experimental strategies that loss of function of Capn2 is responsible for HLHS-like ventricle phenotypes in vivo, we then used the Xenopus system to explore the potential pathogenicity of individual-specific human CAPN2 variants. Two different CAPN2 variants observed in our cohort, c.707C>T/pS236F and c.1112C>T/p.A371V, were selected for further studies because they were both present in multiple affected individuals, were conserved in Xenopus, and were predicted to be damaging to the CAPN2 protein. The human CAPN2707C>T variant results in a serine (S) to phenylalanine (F) mutation within the protein coding region at position 236. This S residue and flanking residues are conserved in the Xenopus capn2 ortholog. Likewise, the human CAPN21112C>T variant results in an alanine (A) to valine (V) mutation at position 371, in a region also well conserved in the Xenopus ortholog. This evolutionary conservation of the variant domains suggests that these regions of CAPN2 may be important for protein function.
To test this prediction, we determined whether Xenopus capn2 mRNAs containing the C-to-T transitions corresponding to the human CAPN2707C>T and CAPN21112C>T variants were, like WT capn2 mRNA, capable of rescuing CRISPR-Cas9-induced HLHS-like ventricle phenotypes. We did not observe decreased viability of mutant embryos prior to termination for heart morphology studies. While the Xenopus capn2 mRNA corresponding to the CAPN2707C>T variant (“707 mRNA”) was able to restore a WT ventricle phenotype (32.8%, n = 108; Figure 3D), the frequency of rescue was slightly less than that elicited by WT capn2 mRNA and did not quite achieve statistical significance (Figure 3F), suggesting that the CAPN2707C>T variant may represent a mildly hypomorphic allele. In contrast, injection of capn2 mRNA corresponding to the human CAPN21112C>T variant (“1112 mRNA”) was unable to reduce the frequency of hypoplastic ventricle phenotypes at all (64%, n = 59, p < 0.01 vs. +WT mRNA; Figures 3E and 3F), suggesting this variant may represent a more damaging hypomorphic or null allele. Taken together, these animal models cause hypoplastic ventricle phenotypes and suggest that human CAPN2707C>T and CAPN21112C>T variants are hypomorphic alleles, supporting the hypothesis that variants in CAPN2 increase risk of iHLHS.
Discussion
Gene-based association testing of rare variants identified CAPN2 as a candidate gene for iHLHS. Functional studies of Capn2 in Xenopus laevis demonstrated that it is important for ventricle morphogenesis, as loss of capn2 function results in hypoplastic ventricles and reduced ventricular chambers, similar to the heart malformations found in human HLHS. Consistent with a role in ventricular morphogenesis and HLHS, peak expression of capn2 in Xenopus coincides with the formation of the linear heart tube and precedes looping and ventricular wall thickening and trabeculation.59 The addition of exogenous WT capn2 mRNA was sufficient to restore a normal ventricle phenotype, confirming that the morphological changes observed are specific to the loss of normal capn2 function. In contrast, attempts to rescue the phenotype with mRNAs encoding Xenopus orthologs of the CAPN2707C>T and CAPN21112C>T variants were less successful than the WT mRNA, suggesting these variants are hypomorphic alleles and insufficient to cause HLHS independently. This finding is consistent with the observation that, in all the trios where parents were available for analysis, one unaffected parent transmitted the variant to their affected child. Thus, c.1112C>T and c.707C>T appear to be risk alleles with moderate effect size. Alternatively, the unaffected parents might have mild CHDs that have, to date, gone undetected.
CAPN2 encodes a large catalytic subunit of the calpain system of calcium-activated cysteine proteases.60 CAPN2 is not known to underlie any MCs, although homozygous loss of Capn2 is embryonic lethal in mice, resulting in cardiovascular defects.61 CAPN2 protein is most strongly and consistently expressed in myoblasts engaged in muscle fiber formation in the developing heart,62 where it localizes to cellular focal adhesion sites.63 Focal adhesions are structures that physically anchor the cell to the extracellular matrix and are also used for regulatory signaling. CAPN2 plays a vital role in regulating integrin-dependent cell adhesion dynamics,64,65,66,67 which are essential for coordinated cell migration during myoblast migration and differentiation.68,69,70 CAPN2 cleaves and releases focal-adhesion-specific components, such as PTK2, TLN1, spectrins, and PXN,65 from focal adhesion complexes resulting in modulated RAC1/RHOA activity64,71,72 and increased turnover of focal contacts. Capn2 is expressed in the heart at embryonic stage 30, during linear heart tube formation (stages 29–32), and prior to looping and ventricular looping (evident at stage 35).73 This suggests the underlying defect causing iHLHS could occur early in heart development; future in vivo studies could investigate potential mitral stenosis defects or heart inflow/outflow function to establish the specific timing of Capn2-related HLHS.
Our findings suggest that one pathogenic mechanism for iHLHS may be cardiac-specific disruption of focal adhesion dynamics. Calpain activity can be activated in response to oxidative stress,74 hyperglycemia,75 physical stress (via β-integrins),76 and fasting,77 collectively linking CAPN2 with environmental stress response. Indeed, CAPN2 has been specifically implicated in the pathogenesis of adult cardiac hypertrophy following cardiac stresses such as hypertension, hypoxia, or hyperglycemia.78 Given that many birth defects, including HLHS, occur much more frequently among diabetic mothers,79,80 our results may help to explain how in utero glucose-induced Ca2+ overload or oxidative stress81 could play a role in the pathogenesis of HLHS by triggering cardiac cell apoptosis via the calpain pathway.
Analysis under Mendelian models of inheritance found an explanatory known P variant in only 4 families, explanatory ACMG LP or P variants in 23 families, and PP variants in previously reported candidate genes for HLHS/CHDs in only 19 (totaling 46/331, 13.9%) families, although this may be an underestimate due to incomplete trios in the cohort. PP variants were limited to MYH6, MYRF, NOTCH1, KDM6A, and KMT2D, each of which underlies an MC-MMS. No PP variants were found in HAND1, RBFOX2, NKX2.5, or GJA1, genes for which data supporting their roles as risk factors for HLHS are limited.14,82,83 PP variants were also not found in either SAP130 (MIM: 609697) or PCDHA13 (MIM: 606319), of which hypomorphic alleles increase the risk of HLHS in mice and have been hypothesized to play a role in HLHS in humans.24 The low frequency of known P or PP variants in known/candidate HLHS/CHD genes contrasts with the higher frequency of suspected P variants found in many studies completed to date that lump HLHS with other CHDs into broader phenotypic classes (e.g., “left-sided lesions”).17,19,84 Moreover, many if not most of these variants were found in individuals with additional clinical findings such as intellectual disability or malformations of other organ systems.20 Accordingly, the relatively low frequency of large-effect alleles found in the cohort we studied may more accurately reflect the contribution of such variants to iHLHS.
Despite the relatively modest sample size and therefore limited statistical power to survey the exome, we were able to identify a candidate gene encoding a protein involved in the focal adhesion and the calpain proteolytic pathways, implicating these pathways as potential contributors to the pathogenesis of iHLHS. Nevertheless, variants in CAPN2 and in known/candidate HLHS/CHD genes were found in only a minority of our cohort, suggesting that multiple other yet undiscovered genes contribute to risk for iHLHS. It is possible that P variants in CAPN2 may underlie an anatomic subtype of iHLHS; deep clinical phenotype data for CAPN2 variant carriers would be necessary to investigate this hypothesis but are unavailable to us after participant de-identification in this genetics study. Our modest sample size required sharing of samples and data across multiple cohorts. This highlights a strength of both careful phenotypic assessment and curation as well as the advantages of international collaboration to discover genetic risk factors for rare, isolated birth defects.
Data and code availability
For NBDPS participants, the sequence data are not publicly available due to IRB restrictions. The study questionnaires and process for accessing the data used in this study are described at https://www.cdc.gov/ncbddd/birthdefects/nbdps-public-access-procedures.html. The code book and analytic code may be made available upon request. Sequence data for the Nationwide Children’s Hospital and Hospital for Sick Children Research (Heart Centre Biobank Registry) cohorts are available in the AnVIL through dbGaP: phs000693. Sequence data for the Helsinki University Children’s Hospital (“The genetics of LVOTO defects” study) cohort and the NBDPS cohort are not permitted to be shared due to lack of consent to genomic data sharing.
Acknowledgments
We thank the families for their participation and support. We extend our gratitude and respect to our friend and colleague, Dr. Deborah A. Nickerson, who passed away in December 2021. She was a leader in human genomics research and a passionate advocate for trainees and women in science and contributed to hundreds of discoveries that have set the stage for precision medicine. We thank NBDPS scientists, staff, and the Genetics Collaborative Working Group; the California Department of Public Health, Maternal Child and Adolescent Health Division for providing surveillance data; the Labatt Family Heart Centre Biobank at the Hospital for Sick Children for access to DNA samples; the Exome Aggregation Consortium and contributors for reference exome data; and all contributors to Geno2MP for use of data included in Geno2MP. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention, the National Institutes of Health, or the California Department of Public Health. Sequencing was provided by the University of Washington Center for Mendelian Genomics and was supported by National Human Genome Research Institute and National Heart, Lung, and Blood Institute grants UM1 HG006493 and U24 HG008956. This work was also supported by Centers for Disease Control and Prevention cooperative agreements under PA #96043, PA #02081, FOA #DD09-001, FOA #DD13-003, and NOFO #DD18-001 to the Centers for Birth Defects Research and Prevention participating in NBDPS and/or the Birth Defects Study to Evaluate Pregnancy exposures, Finnish Medical Foundation, Finnish Foundation for Pediatric Research, Finnish Foundation for Cardiovascular Research, University of Helsinki, and Academy of Finland (grant 331405); National Institute of Child Health and Human Development grant HD095937; Nationwide Children’s Hospital Foundation; and NHLBI grant HL109759-A1. S.M. was funded by the Canadian Institutes of Health Research under the frame of ERA PerMed, the Ted Rogers Centre for Heart Research, and the Heart and Stroke Foundation of Canada/Robert M Freedom Chair in Cardiovascular Science.
Declaration of interests
M.J.B. and J.X.C. are the editor-in-chief and deputy editor, respectively, of Human Genetics and Genomics Advances and were recused from the editorial handling of this article. J.J.W. is an employee and shareholder of Invitae. M.J.B. is chair of the Scientific Advisory Board of GeneDx. S.M. is on the Hypertrophic Cardiomyopathy Advisory Board of Bristol-Myers Squibb.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100232.
Web resources
ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/.
Geno2MP: https://geno2mp.gs.washington.edu/Geno2MP/gnomAD: http://gnomad.broadinstitute.org.
Human Genome Variation: http://www.hgvs.org/mutnomen/.
Online Mendelian Inheritance in Man (OMIM): http://www.omim.org/.
Supplemental information
References
- 1.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., et al. The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Update on Overall Prevalence of Major Birth Defects --- Atlanta, Georgia, 1978--2005. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5701a2.htm [PubMed]
- 3.Yoon P.W., Olney R.S., Khoury M.J., Sappenfield W.M., Chavez G.F., Taylor D. Contribution of birth defects and genetic diseases to pediatric hospitalizations: a population-based study. Arch. Pediatr. Adolesc. Med. 1997;151:1096–1103. doi: 10.1001/archpedi.1997.02170480026004. [DOI] [PubMed] [Google Scholar]
- 4.Feldkamp M.L., Carey J.C., Byrne J.L.B., Krikov S., Botto L.D. Etiology and clinical presentation of birth defects: population based study. BMJ. 2017;357:j2249. doi: 10.1136/bmj.j2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyert D.L., Mathews T.J., Menacker F., Strobino D.M., Guyer B. Annual summary of vital statistics: 2004. Pediatrics. 2006;117:168–183. doi: 10.1542/peds.2005-2587. [DOI] [PubMed] [Google Scholar]
- 6.Rosano A., Botto L.D., Botting B., Mastroiacovo P. Infant mortality and congenital anomalies from 1950 to 1994: an international perspective. J. Epidemiol. Community Health. 2000;54:660–666. doi: 10.1136/jech.54.9.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker S.E., Mai C.T., Canfield M.A., Rickard R., Wang Y., Meyer R.E., Anderson P., Mason C.A., Collins J.S., Kirby R.S., et al. Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- 8.McBride K.L., Pignatelli R., Lewin M., Ho T., Fernbach S., Menesses A., Lam W., Leal S.M., Kaplan N., Schliekelman P., et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: Segregation, multiplex relative risk, and heritability. Am. J. Med. Genet. 2005;134A:180–186. doi: 10.1002/ajmg.a.30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellesøe S.G., Workman C.T., Bouvagnet P., Loffredo C.A., McBride K.L., Hinton R.B., van Engelen K., Gertsen E.C., Mulder B.J.M., Postma A.V., et al. Familial co-occurrence of congenital heart defects follows distinct patterns. Eur. Heart J. 2017;39:1015–1022. doi: 10.1093/eurheartj/ehx314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinton R.B., Martin L.J., Tabangin M.E., Mazwi M.L., Cripe L.H., Benson D.W. Hypoplastic left heart syndrome is heritable. J. Am. Coll. Cardiol. 2007;50:1590–1595. doi: 10.1016/j.jacc.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Yagi H., Liu X., Gabriel G.C., Wu Y., Peterson K., Murray S.A., Aronow B.J., Martin L.J., Benson D.W., Lo C.W. The genetic landscape of hypoplastic left heart syndrome. Pediatr. Cardiol. 2018;39:1069–1081. doi: 10.1007/s00246-018-1861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner J.I., Berg K.A., Schneider D.S., Clark E.B., Boughman J.A. Cardiac malformations in relatives of infants with hypoplastic left-heart syndrome. Am. J. Dis. Child. 1989;143:1492–1494. doi: 10.1001/archpedi.1989.02150240114030. [DOI] [PubMed] [Google Scholar]
- 13.Theis J.L., Hrstka S.C.L., Evans J.M., O’Byrne M.M., de Andrade M., O’Leary P.W., Nelson T.J., Olson T.M. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum. Genet. 2015;134:1003–1011. doi: 10.1007/s00439-015-1582-1. [DOI] [PubMed] [Google Scholar]
- 14.Dasgupta C., Martinez A.-M., Zuppan C.W., Shah M.M., Bailey L.L., Fletcher W.H. Identification of connexin43 (α1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE) Mutat. Res. 2001;479:173–186. doi: 10.1016/s0027-5107(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 15.Theis J.L., Zimmermann M.T., Evans J.M., Eckloff B.W., Wieben E.D., Qureshi M.Y., O’Leary P.W., Olson T.M. Recessive MYH6 Mutations in Hypoplastic Left Heart With Reduced Ejection Fraction. Circ. Cardiovasc. Genet. 2015;8:564–571. doi: 10.1161/circgenetics.115.001070. [DOI] [PubMed] [Google Scholar]
- 16.Elliott D.A., Kirk E.P., Yeoh T., Chandar S., McKenzie F., Taylor P., Grossfeld P., Fatkin D., Jones O., Hayes P., et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease Associations with atrial septal defect and hypoplastic left heart syndrome. J. Am. Coll. Cardiol. 2003;41:2072–2076. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- 17.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J., et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glessner J.T., Bick A.G., Ito K., Homsy J., Rodriguez-Murillo L., Fromer M., Mazaika E., Vardarajan B., Italia M., Leipzig J., et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ. Res. 2014;115:884–896. doi: 10.1161/circresaha.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A.H., Hanchard N.A., Furthner D., Fernbach S., Azamian M., Nicosia A., Rosenfeld J., Muzny D., D’Alessandro L.C.A., Morris S., et al. Whole exome sequencing in 342 congenital cardiac left sided lesion cases reveals extensive genetic heterogeneity and complex inheritance patterns. Genome Med. 2017;9:95. doi: 10.1186/s13073-017-0482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sifrim A., Hitz M.-P., Wilsdon A., Breckpot J., Turki S.H.A., Thienpont B., McRae J., Fitzgerald T.W., Singh T., Swaminathan G.J., et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digilio M.C., Marino B., Baban A., Dallapiccola B. Cardiovascular malformations in Adams–Oliver syndrome. Am. J. Med. Genet. 2015;167A:1175–1177. doi: 10.1002/ajmg.a.36764. [DOI] [PubMed] [Google Scholar]
- 22.Digilio M.C., Marino B., Toscano A., Giannotti A., Dallapiccola B. Congenital heart defects in Kabuki syndrome. Am. J. Med. Genet. 2001;100:269–274. doi: 10.1002/ajmg.1265. [DOI] [PubMed] [Google Scholar]
- 23.Schulz S., Fröber R., Kraus C., Schneider U. Prenatal diagnosis of hypoplastic left heart syndrome associated with Noonan Syndrome and de novo RAF1 mutation. Prenat. Diagn. 2012;32:1016–1018. doi: 10.1002/pd.3938. [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Yagi H., Saeed S., Bais A.S., Gabriel G.C., Chen Z., Peterson K.A., Li Y., Schwartz M.C., Reynolds W.T., et al. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017;49:1152–1159. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reefhuis J., Gilboa S.M., Anderka M., Browne M.L., Feldkamp M.L., Hobbs C.A., Jenkins M.M., Langlois P.H., Newsome K.B., Olshan A.F., et al. The national birth defects prevention study: A review of the methods. Birth Defects Res. A Clin. Mol. Teratol. 2015;103:656–669. doi: 10.1002/bdra.23384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins M.M., Almli L.M., Pangilinan F., Chong J.X., Blue E.E., Shapira S.K., White J., McGoldrick D., Smith J.D., Mullikin J.C., et al. Exome sequencing of family trios from the National Birth Defects Prevention Study: Tapping into a rich resource of genetic and environmental data. Birth Defects Res. 2019;111:1618–1632. doi: 10.1002/bdr2.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botto L.D., Lin A.E., Riehle-Colarusso T., Malik S., Correa A., National Birth Defects Prevention Study Seeking causes: Classifying and evaluating congenital heart defects in etiologic studies. Birth Defects Res. A Clin. Mol. Teratol. 2007;79:714–727. doi: 10.1002/bdra.20403. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen S.A., Olney R.S., Holmes L.B., Lin A.E., Keppler-Noreuil K.M., Moore C.A., National Birth Defects Prevention Study Guidelines for case classification for the national birth defects prevention study. Birth Defects Res. A Clin. Mol. Teratol. 2003;67:193–201. doi: 10.1002/bdra.10012. [DOI] [PubMed] [Google Scholar]
- 29.Papaz T., Safi M., Manickaraj A.-K., Ogaki C., Breaton Kyryliuk J., Burrill L., Dodge C., Chant-Gambacort C., Walter L.-L., Rosenberg H., et al. Factors influencing participation in a population-based biorepository for childhood heart disease. Pediatrics. 2012;130:e1198–e1205. doi: 10.1542/peds.2012-0687. [DOI] [PubMed] [Google Scholar]
- 30.Chong J.X., McMillin M.J., Shively K.M., Beck A.E., Marvin C.T., Armenteros J.R., Buckingham K.J., Nkinsi N.T., Boyle E.A., Berry M.N., et al. De Novo Mutations in NALCN Cause a Syndrome Characterized by Congenital Contractures of the Limbs and Face, Hypotonia, and Developmental Delay. Am. J. Hum. Genet. 2015;96:462–473. doi: 10.1016/j.ajhg.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan A., Abecasis G.R., Kang H.M. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., NHLBI Exome Sequencing Project. Quinlan A.R., Nickerson D.A., Eichler E.E. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen B.S., Quinlan A.R. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am. J. Hum. Genet. 2017;100:406–413. doi: 10.1016/j.ajhg.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paila U., Chapman B.A., Kirchner R., Quinlan A.R. GEMINI: Integrative Exploration of Genetic Variation and Genome Annotations. PLoS Comput. Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., Mesirov J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017;77:e31–e34. doi: 10.1158/0008-5472.can-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7–16. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., NHLBI GO Exome Sequencing Project—ESP Lung Project Team. Christiani D.C., Wurfel M.M., Lin X. Optimal Unified Approach for Rare-Variant Association Testing with Application to Small-Sample Case-Control Whole-Exome Sequencing Studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madsen B.E., Browning S.R. A Groupwise Association Test for Rare Mutations Using a Weighted Sum Statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Early Development of Xenopus laevis: A Laboratory Manual. https://www.cshlpress.com/default.tpl?&--eqSKUdatarq=855
- 45.Pickett M.A., Dush M.K., Nascone-Yoder N.M. Acetylcholinesterase plays a non-neuronal, non-esterase role in organogenesis. Development. 2017;144:dev.. doi: 10.1242/dev.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moody S.A. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev. Biol. 1987;122:300–319. doi: 10.1016/0012-1606(87)90296-x. [DOI] [PubMed] [Google Scholar]
- 47.Lee C., Kieserman E., Gray R.S., Park T.J., Wallingford J. Whole-Mount Fluorescence Immunocytochemistry on Xenopus Embryos. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot4957. pdb.prot4957. [DOI] [PubMed] [Google Scholar]
- 48.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C., et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Southgate L., Sukalo M., Karountzos A.S.V., Taylor E.J., Collinson C.S., Ruddy D., Snape K.M., Dallapiccola B., Tolmie J.L., Joss S., et al. Haploinsufficiency of the NOTCH1 Receptor as a Cause of Adams–Oliver Syndrome With Variable Cardiac Anomalies. Circ. Cardiovasc. Genet. 2015;8:572–581. doi: 10.1161/circgenetics.115.001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ching Y.-H., Ghosh T.K., Cross S.J., Packham E.A., Honeyman L., Loughna S., Robinson T.E., Dearlove A.M., Ribas G., Bonser A.J., et al. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat. Genet. 2005;37:423–428. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 51.Carniel E., Taylor M.R.G., Sinagra G., Di Lenarda A., Ku L., Fain P.R., Boucek M.M., Cavanaugh J., Miocic S., Slavov D., et al. α-Myosin Heavy Chain. Circulation. 2005;112:54–59. doi: 10.1161/circulationaha.104.507699. [DOI] [PubMed] [Google Scholar]
- 52.Qi H., Yu L., Zhou X., Wynn J., Zhao H., Guo Y., Zhu N., Kitaygorodsky A., Hernan R., Aspelund G., et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14:e1007822. doi: 10.1371/journal.pgen.1007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lederer D., Grisart B., Digilio M.C., Benoit V., Crespin M., Ghariani S.C., Maystadt I., Dallapiccola B., Verellen-Dumoulin C. Deletion of KDM6A, a Histone Demethylase Interacting with MLL2, in Three Patients with Kabuki Syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hempel A., Kühl M. A Matter of the Heart: The African Clawed Frog Xenopus as a Model for Studying Vertebrate Cardiogenesis and Congenital Heart Defects. J. Cardiovasc. Dev. Dis. 2016;3:21. doi: 10.3390/jcdd3020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pitcairn E., Harris H., Epiney J., Pai V.P., Lemire J.M., Ye B., Shi N.-Q., Levin M., McLaughlin K.A. Coordinating heart morphogenesis: A novel role for hyperpolarization-activated cyclic nucleotide-gated (HCN) channels during cardiogenesis in Xenopus laevis. Commun. Integr. Biol. 2017;10:e1309488. doi: 10.1080/19420889.2017.1309488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gessert S., Kühl M. Comparative gene expression analysis and fate mapping studies suggest an early segregation of cardiogenic lineages in Xenopus laevis. Dev. Biol. 2009;334:395–408. doi: 10.1016/j.ydbio.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 57.Grossfeld P., Nie S., Lin L., Wang L., Anderson R.H. Hypoplastic Left Heart Syndrome: A New Paradigm for an Old Disease? J. Cardiovasc. Dev. Dis. 2019;6:10. doi: 10.3390/jcdd6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crucean A., Alqahtani A., Barron D.J., Brawn W.J., Richardson R.V., O’Sullivan J., Anderson R.H., Henderson D.J., Chaudhry B. Re-evaluation of hypoplastic left heart syndrome from a developmental and morphological perspective. Orphanet J. Rare Dis. 2017;12:138. doi: 10.1186/s13023-017-0683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Imajoh S., Aoki K., Ohno S., Emori Y., Kawasaki H., Sugihara H., Suzuki K. Molecular cloning of the cDNA for the large subunit of the high-calcium-requiring form of human calcium-activated neutral protease. Biochemistry-us. 1988;27:8122–8128. doi: 10.1021/bi00421a022. [DOI] [PubMed] [Google Scholar]
- 61.Takano J., Mihira N., Fujioka R., Hosoki E., Chishti A.H., Saido T.C. Vital Role of the Calpain-Calpastatin System for Placental-Integrity-Dependent Embryonic Survival. Mol. Cell Biol. 2011;31:4097–4106. doi: 10.1128/mcb.05189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raynaud F., Marcilhac A., Chebli K., Benyamin Y., Rossel M. Calpain 2 expression pattern and sub-cellular localization during mouse embryogenesis. Int. J. Dev. Biol. 2008;52:383–388. doi: 10.1387/ijdb.072448fr. [DOI] [PubMed] [Google Scholar]
- 63.Beckerle M.C., Burridge K., DeMartino G.N., Croall D.E. Colocalization of calcium-dependent protease II and one of its substrates at sites of cell adhesion. Cell. 1987;51:569–577. doi: 10.1016/0092-8674(87)90126-7. [DOI] [PubMed] [Google Scholar]
- 64.Flevaris P., Stojanovic A., Gong H., Chishti A., Welch E., Du X. A molecular switch that controls cell spreading and retraction. J. Cell Biol. 2007;179:553–565. doi: 10.1083/jcb.200703185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franco S., Perrin B., Huttenlocher A. Isoform specific function of calpain 2 in regulating membrane protrusion. Exp. Cell Res. 2004;299:179–187. doi: 10.1016/j.yexcr.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 66.Chan K.T., Bennin D.A., Huttenlocher A. Regulation of Adhesion Dynamics by Calpain-mediated Proteolysis of Focal Adhesion Kinase (FAK) J. Biol. Chem. 2010;285:11418–11426. doi: 10.1074/jbc.m109.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dedieu S., Mazères G., Poussard S., Brustis J.-J., Cottin P. Myoblast migration is prevented by a calpain-dependent accumulation of MARCKS. Biol. Cell. 2003;95:615–623. doi: 10.1016/j.biolcel.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 68.Buffolo M., Batista Possidonio A.C., Mermelstein C., Araujo H. A conserved role for calpains during myoblast fusion. Genesis. 2015;53:417–430. doi: 10.1002/dvg.22870. [DOI] [PubMed] [Google Scholar]
- 69.Barnoy S., Glaser T., Kosower N.S. Calpain and calpastatin in myoblast differentiation and fusion: Effects of inhibitors. Biochim. Biophys. Acta. 1997;1358:181–188. doi: 10.1016/s0167-4889(97)00068-2. [DOI] [PubMed] [Google Scholar]
- 70.Dedieu S., Poussard S., Mazères G., Grise F., Dargelos E., Cottin P., Brustis J.-J. Myoblast migration is regulated by calpain through its involvement in cell attachment and cytoskeletal organization. Exp. Cell Res. 2004;292:187–200. doi: 10.1016/j.yexcr.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 71.Bialkowska K., Kulkarni S., Du X., Goll D.E., Saido T.C., Fox J.E. Evidence That β3 Integrin-Induced Rac Activation Involves the Calpain-Dependent Formation of Integrin Clusters That Are Distinct from the Focal Complexes and Focal Adhesions That Form as Rac and Rhoa Become Active. J. Cell Biol. 2000;151:685–696. doi: 10.1083/jcb.151.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kulkarni S., Goll D.E., Fox J.E.B. Calpain Cleaves RhoA Generating a Dominant-negative Form That Inhibits Integrin-induced Actin Filament Assembly and Cell Spreading. J. Biol. Chem. 2002;277:24435–24441. doi: 10.1074/jbc.m203457200. [DOI] [PubMed] [Google Scholar]
- 73.Zanardelli S., Christodoulou N., Skourides P.A. Calpain2 protease: A new member of the Wnt/Ca2+ pathway modulating convergent extension movements in Xenopus. Dev. Biol. 2013;384:83–100. doi: 10.1016/j.ydbio.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 74.Ray S.K., Fidan M., Nowak M.W., Wilford G.G., Hogan E.L., Banik N.L. Oxidative stress and Ca2+ influx upregulate calpain and induce apoptosis in PC12 cells. Brain Res. 2000;852:326–334. doi: 10.1016/s0006-8993(99)02148-4. [DOI] [PubMed] [Google Scholar]
- 75.Harwood S.-M., Allen D.-A., Raftery M.-J., Yaqoob M.M. High glucose initiates calpain-induced necrosis before apoptosis in LLC-PK1 cells. Kidney Int. 2007;71:655–663. doi: 10.1038/sj.ki.5002106. [DOI] [PubMed] [Google Scholar]
- 76.Suryakumar G., Kasiganesan H., Balasubramanian S., Kuppuswamy D. Lack of β3 Integrin Signaling Contributes to Calpain-Mediated Myocardial Cell Loss in Pressure-Overloaded Myocardium. J. Cardiovasc. Pharmacol. 2010;55:567–573. doi: 10.1097/fjc.0b013e3181d9f5d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ilian M.A., Forsberg N.E. Gene expression of calpains and their specific endogenous inhibitor, calpastatin, in skeletal muscle of fed and fasted rabbits. Biochem. J. 1992;287:163–171. doi: 10.1042/bj2870163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patterson C., Portbury A.L., Schisler J.C., Willis M.S. Tear Me Down. Circ. Res. 2011;109:453–462. doi: 10.1161/circresaha.110.239749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Correa A., Gilboa S.M., Besser L.M., Botto L.D., Moore C.A., Hobbs C.A., Cleves M.A., Riehle-Colarusso T.J., Waller D.K., Reece E.A., et al. Diabetes mellitus and birth defects. Am. J. Obstet. Gynecol. 2008;199:237.e1-e9. doi: 10.1016/j.ajog.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becerra J.E., Khoury M.J., Cordero J.F., Erickson J.D. Diabetes mellitus during pregnancy and the risks for specific birth defects: a population-based case-control study. Pediatrics. 1990;85:1–9. [PubMed] [Google Scholar]
- 81.Kumar S., Kain V., Sitasawad S.L. High glucose-induced Ca2+ overload and oxidative stress contribute to apoptosis of cardiac cells through mitochondrial dependent and independent pathways. Biochim. Biophys. Acta. 2012;1820:907–920. doi: 10.1016/j.bbagen.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 82.Firulli B.A., Toolan K.P., Harkin J., Millar H., Pineda S., Firulli A.B. The HAND1 frameshift A126FS mutation does not cause hypoplastic left heart syndrome in mice. Cardiovasc. Res. 2017;113:1732–1742. doi: 10.1093/cvr/cvx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomita-Mitchell A., Stamm K.D., Mahnke D.K., Kim M.-S., Hidestrand P.M., Liang H.L., Goetsch M.A., Hidestrand M., Simpson P., Pelech A.N., et al. Impact of MYH6 variants in hypoplastic left heart syndrome. Physiol. Genom. 2016;48:912–921. doi: 10.1152/physiolgenomics.00091.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jin S.C., Homsy J., Zaidi S., Lu Q., Morton S., DePalma S.R., Zeng X., Qi H., Chang W., Sierant M.C., et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For NBDPS participants, the sequence data are not publicly available due to IRB restrictions. The study questionnaires and process for accessing the data used in this study are described at https://www.cdc.gov/ncbddd/birthdefects/nbdps-public-access-procedures.html. The code book and analytic code may be made available upon request. Sequence data for the Nationwide Children’s Hospital and Hospital for Sick Children Research (Heart Centre Biobank Registry) cohorts are available in the AnVIL through dbGaP: phs000693. Sequence data for the Helsinki University Children’s Hospital (“The genetics of LVOTO defects” study) cohort and the NBDPS cohort are not permitted to be shared due to lack of consent to genomic data sharing.