Abstract
Background
An accurate prediction of mortality in end-of-life care is crucial but presents challenges. Existing prognostic tools demonstrate moderate performance in predicting survival across various time frames, primarily in in-hospital settings and single-time evaluations. However, these tools may fail to capture the individualized and diverse trajectories of patients. Limited evidence exists regarding the use of artificial intelligence (AI) and wearable devices, specifically among patients with cancer at the end of life.
Objective
This study aimed to investigate the potential of using wearable devices and AI to predict death events among patients with cancer at the end of life. Our hypothesis was that continuous monitoring through smartwatches can offer valuable insights into the progression of patients at the end of life and enable the prediction of changes in their condition, which could ultimately enhance personalized care, particularly in outpatient or home care settings.
Methods
This prospective study was conducted at the National Taiwan University Hospital. Patients diagnosed with cancer and receiving end-of-life care were invited to enroll in wards, outpatient clinics, and home-based care settings. Each participant was given a smartwatch to collect physiological data, including steps taken, heart rate, sleep time, and blood oxygen saturation. Clinical assessments were conducted weekly. The participants were followed until the end of life or up to 52 weeks. With these input features, we evaluated the prediction performance of several machine learning–based classifiers and a deep neural network in 7-day death events. We used area under the receiver operating characteristic curve (AUROC), F1-score, accuracy, and specificity as evaluation metrics. A Shapley additive explanations value analysis was performed to further explore the models with good performance.
Results
From September 2021 to August 2022, overall, 1657 data points were collected from 40 patients with a median survival time of 34 days, with the detection of 28 death events. Among the proposed models, extreme gradient boost (XGBoost) yielded the best result, with an AUROC of 96%, F1-score of 78.5%, accuracy of 93%, and specificity of 97% on the testing set. The Shapley additive explanations value analysis identified the average heart rate as the most important feature. Other important features included steps taken, appetite, urination status, and clinical care phase.
Conclusions
We demonstrated the successful prediction of patient deaths within the next 7 days using a combination of wearable devices and AI. Our findings highlight the potential of integrating AI and wearable technology into clinical end-of-life care, offering valuable insights and supporting clinical decision-making for personalized patient care. It is important to acknowledge that our study was conducted in a relatively small cohort; thus, further research is needed to validate our approach and assess its impact on clinical care.
Trial Registration
ClinicalTrials.gov NCT05054907; https://classic.clinicaltrials.gov/ct2/show/NCT05054907
Keywords: artificial intelligence, end-of-life care, machine learning, palliative care, survival prediction, terminal cancer, wearable device
Introduction
Survival Prediction Tools in End-of-Life Care
Survival prediction is a critical aspect of end-of-life care. Knowing the likely clinical course allows the medical team to create care plans and avoid the overuse of aggressive care. It also helps patients and their families in other ways, such as by allowing the families to fulfill the patients’ final wishes. The most widely recognized and commonly used tools for survival prediction in end-of-life care include the Palliative Performance Scale [1-4], Palliative Prognostic Index [5-8], and Palliative Prognostic Score [9-12], which had been developed and validated in the past decades.
These tools typically rely on clinical symptoms, signs, and functional levels to estimate prognosis. Some tools incorporate blood tests and clinician predictions to enhance the evaluation process (see Multimedia Appendix 1 [1-16] for further detail). Although these tools have shown fair performance in predicting short-term survival lengths ranging from 7 to 60 days, studies validating these tools have generally been conducted in inpatient settings and have usually considered only a single evaluation upon the patient’s admission [17,18].
The Need for Anticipating and Identifying the Dying Process in Outpatient Care
End-of-life care encompasses various scenarios and preferences. Many patients express a desire for home care, seeking a sense of safety and comfort during their final weeks of life [19-23]. However, this phase can involve a range of potential events, varying from mild discomfort to urgent medical needs to, ultimately, the death event.
The existing tools may perform well in predicting survival from a statistical and population perspective but fail to capture the diverse trajectories of individual patients. For example, patients with high prognostic scores may pass away more rapidly than predicted, whereas those with low scores may survive longer than expected. Given the emphasis on outpatient and home care to reduce hospitalization and alleviate the burden on patients and their families, it is crucial to anticipate and identify impending changes in advance, including the dying process. This allows for adequate preparation and support to be provided promptly.
The Application of Wearable Devices and Artificial Intelligence in End-of-Life Care
Digital health technology has been widely adopted in end-of-life care for various purposes, such as education, telemedicine, and prognosis prediction using electronic health records [24-31]. Although wearable devices have also been demonstrated to help monitor and predict physical conditions [32-40], there have been only a few studies on the use of wearable devices in end-of-life care. One study focused on the feasibility and acceptability of the devices [41,42], whereas another reported that a deep learning model could predict future outcomes using noncommercial actigraphy data in an in-hospital setting [43]. We believe that the prediction of death or emergent events using wearable devices and artificial intelligence (AI) could enable the medical team to provide timely and high-quality care. However, there is currently no research examining the use of commercial wearable devices in a general setting.
This pilot study aimed to combine wearable devices and machine learning to develop a prediction model for the death event of patients with cancer at the end of life. We hypothesized that using a smartwatch for continuous monitoring may provide greater insight into the progress of patients with terminal cancer and, with the aid of machine learning techniques, may be able to predict changes in their conditions.
Methods
Study Design and Participants
This is a prospective observational pilot study conducted at the National Taiwan University Hospital from September 2021 to August 2022. Patients who were receiving or going to receive outpatient or home-based end-of-life care were referred by their medical staff. The eligibility criteria were (1) age >20 years and (2) terminal cancer diagnosis (incurable cancer with limited life expectancy, judged by the physician of the primary team). The exclusion criterion was the inability to use smartphones because patients or their caregivers needed to use a smartphone to upload the wearable device data.
Ethics Approval
The study protocol was approved by the institutional review board of the National Taiwan University Hospital (RIND202105097) and registered on ClinicalTrials.gov (NCT05054907).
Data Collection
The following data were collected during the study: basic demographic data, clinical assessment data, and wearable device data. Basic demographic data were evaluated upon enrollment, including age, sex, cancer diagnosis, presence of metastasis, systemic disease, and material use.
Clinical assessments were conducted on a weekly basis, evaluating consciousness, appetite, symptoms, functional level (using the Australia-modified Karnofsky Performance Status [AKPS]) [44], and clinical care phase [45] (Multimedia Appendix 2 [44,45]). The evaluation was usually performed by the research assistant face to face or via a phone call, depending on whether the patient had a clinic visit that week. However, the patient’s condition was assessed by the inpatient care staff if they were admitted to the hospital or by the home care team if they received a home visit that week.
All participants were provided with a smartwatch, Garmin VivoSmart 4 (Garmin), and asked to wear it all day as long as they could tolerate it. The wearable device data were collected continuously. Participants or their caregivers were taught to operate a smartphone app to synchronize the wearable data on a regular basis (at least once every 7 days). Physiological data, including steps walked, heart rate (HR), sleep status, and blood oxygen saturation (measured during sleep time), were collected. The data were presented as a total sum or an average value of 1 day.
Most patients who received end-of-life care in Taiwan had a life expectancy of just a few weeks or months [46]. To ensure that we could follow most of these patients until the end of their lives, we set a follow-up duration of up to 52 weeks. We hypothesized that physiological changes, which can be detected through wearable device measurements, may occur within a shorter time frame, ideally less than 2 weeks before the death event [47]. Consequently, we chose a 7-day prediction interval, as it is an intuitively manageable time frame that enables clinicians and caregivers to communicate essential information to families in advance.
Data Processing
The data set was a combination of basic demographic data, clinical assessment data, and wearable device data. As shown in Figure 1, one day corresponded to one data point, which served as an individual observation for model training. The label was a recent death event, defined based on whether the patient died within the next 7 days. As the clinical assessment was performed once a week, we used forward filling until the next assessment. For any data point of any case, if some of the wearable data were missing, we used interpolation to fill in the missing value. However, days without any wearable device data uploaded were directly excluded from the analysis.
Figure 1.

The combination of wearable device data and clinical assessments. The figure illustrates the process of data combination. Each column represents 1 data point. Each row represents a different kind of data. Days without any wearable device data uploaded were directly excluded from the analysis.
For cases that received a follow-up period longer than 50 days, we kept all the data labeled as positive and randomly sampled the remaining negative data points to bring the total number of data points to 50 in each case. For cases with a follow-up period shorter than 50 days, we included all of the data points. The data set was then divided into testing (25%) and training-validation (75%) sets, stratified by label and sex. To improve the model training, we upsampled the positive data points in the training-validation set. The entire flowchart of the study is shown in Figure 2.
Figure 2.

Flowchart of the study. The figure illustrates the flowchart of the entire study from data collection and data processing to the training and evaluation of machine learning models. CV: cross-validation; ML: machine learning; NTU: National Taiwan University.
Feature Engineering
The original features collected are listed in Textbox 1. To account for changes in an individual’s condition, we calculated the differences between the original features and their past averages in wearable data and clinical data and used these differences as new features.
All the original features collected in the study.
Physiological factor
Steps
Minimum heart rate (HR)
Maximum HR
Average HR
Resting HR
Average stress level
Maximum stress level
Sleep duration
Deep sleep duration
Light sleep duration
Rapid eye movement (REM) sleep duration
Awake duration
SpO2 (expressed in average)
Clinical assessment
Consciousness
Appetite
Urination
Edema
Pain score
Sleep
Drowsiness
Nausea
Constipation
Diarrhea
Dyspnea
Fatigue
Fever
Functional level (using Australia-modified Karnofsky Performance Status)
Care phase
Pain control change
Basic demographic
Sex
Age
Cancer diagnosis
Diagnosis time
Confirmed metastasis
Past history
Alcohol use
Betel nut use
Cigarette use
For the feature selection, 2 demographic features (sex and age) were directly included to account for the physical differences between the individuals. Most wearable device features were directly included in the model, except for 3. “Sleep duration” parameters were transformed into the ratio of wakefulness to sleep. “Stress” parameters were excluded owing to there being no clear definition of their value. “Resting heart rate” data were excluded considering the similarity of “resting heart rate” to “minimal heart rate” and a lack of a clear definition of “resting.” As for clinical assessment features, we used the SelectKBest method built into the Scikit-learn package and selected the top 10 potential candidate features using ANOVA on the training set. A correlation analysis between the selected features was performed on the training-validation set.
Classification Model
We chose the following machine learning–based classifiers for supervised learning: logistic regressions, support vector machine, decision trees, random forests, k-nearest neighbor (KNN), adaptive boosting (AdaBoost), and extreme gradient boosting (XGBoost). We proposed a multiperceptron deep neural network to compare the performance of deep learning models with that of machine learning models on death prediction. The models were implemented using the Python library Scikit-learn.
Model Assessment
Three-fold cross-validation was used in the training-validation set for hyperparameter tuning. The grid search parameters are listed in Table 1. Once the optimal hyperparameters were selected, we trained each model on the full training-validation set and evaluated its performance on the testing set. We used the area under the receiver operating characteristic curve (AUROC), precision, recall (sensitivity), F1-score, specificity, and accuracy as evaluation metrics. To reduce the impact of randomness on the algorithms, we repeated the training process 100 times for each model and reported the average values of the evaluation metrics. To investigate the impact of clinical assessment on prediction, we also trained and evaluated the models using only wearable device parameters, sex, and age as input features.
Table 1.
Grid search parameters in hyperparameter tuning.
| Classifiera | Parameters_grid | Selected value |
| LogisticRegression |
|
|
| SVMb (kernel = “rbf,” degree = 3) |
|
|
| DecisionTree |
|
|
| RandomForest |
|
|
| KNeighborsClassifier |
|
|
| AdaBoostClassifier |
|
|
| XGBClassifier (eval_metric = “aucpr,” n_estimators = 100, booster = “gbtree,” colsample_bytree = 1, learning_rate = 0.3) |
|
|
| MLPClassifier (hidden_layer_sizes = (64, 64, 64), activation = “relu”) |
|
|
aThe algorithms were performed using the Python package Scikit-learn 0.24.2, and all other parameters not shown in this table were set to their default values.
bSVM: support vector machine.
Model Explanation
To explore the machine learning model more deeply, a Shapley additive explanations (SHAP) algorithm was applied [48,49]. The impact of the feature values on each prediction of a recent death event in the testing set was obtained.
User Feedback on Wearable Devices
To evaluate the feasibility of wearable devices in the population with terminal cancer, we surveyed the participants during weekly follow-ups to assess any difficulties or discomfort they experienced while using the wearable device. At the end of the study, we also solicited feedback from the family or main caregivers of the participants who had passed away about their experience with the wearable device and their willingness to use similar devices in the future.
Results
Patient Demographics
Between September 2021 and August 2022, a total of 45 patients were enrolled. Data from 11% (5/45) of patients were unavailable; therefore, these patients were excluded from the data analysis: 4% (2/45) left the study early owing to personal reasons, 4% (2/45) died soon after the enrollment, and 2% (1/45) did not use the wearable device owing to a personal reason. In all, 89% (40/45) of patients were included in the analysis.
Table 2 illustrates the demographics of the study participants, with approximately one-third (12/45, 27%) being diagnosed with lung cancer. The median age of the patients was 70.5 years. At the time of enrollment, most participants had limited function, as determined by the AKPS assessment. The median care duration was 34 days, ranging from a minimum of 6 days to a maximum of 271 days. Except for 5% (2/45) of patients who withdrew from the study for personal reasons, most patients were followed until their death, at which point their care time was equal to their survival period.
Table 2.
Basic demographics of the participants (n=40).
| Characteristic | Participant, n (%) | |
| Age group (years) | ||
|
|
≤40 | 1 (2) |
|
|
41-65 | 12 (30) |
|
|
65-80 | 15 (38) |
|
|
≥80 | 12 (30) |
| Sex | ||
|
|
Male | 17 (42) |
|
|
Female | 23 (58) |
| Cancer diagnosis | ||
|
|
Lung | 12 (30) |
|
|
Colorectal | 6 (15) |
|
|
Head and neck | 5 (12) |
|
|
Pancreas | 4 (10) |
|
|
Liver | 3 (8) |
|
|
Breast | 2 (5) |
|
|
Prostate | 2 (5) |
|
|
Other | 6 (15) |
| AKPSa score on initial assessment | ||
|
|
≤20 | 12 (30) |
|
|
30-50 | 20 (50) |
|
|
60-70 | 7 (18) |
|
|
≥80 | 1 (2) |
| Follow-up time or survival lengthb | ||
|
|
<7 days | 2 (5) |
|
|
8-30 days | 14 (35) |
|
|
1-2 months | 15 (38) |
|
|
2-3 months | 5 (12) |
|
|
>3 months | 4 (10) |
| Wearable device use (%) | ||
|
|
<25 | 3 (8) |
|
|
25-50 | 3 (8) |
|
|
50-75 | 6 (15) |
|
|
75-90 | 9 (22) |
|
|
>90 | 19 (48) |
| Mortality during follow-up timeb | 38 (95) | |
| Participants whose wearable device data were collected until at least 7 days before their death events | 28 (70) | |
aAKPS: Australia-modified Karnofsky Performance Status.
bOverall, 2 (5%) patients left the study for personal reasons. Most patients were followed until their death, for whom the follow-up time represented their survival period.
Completeness of Data
The wearable devices were worn, on average, for 77.42% (n=1657) of the total 2140 study days. Of the 40 participants, 34 (85%) wore the devices for more than half of their study days. Moreover, 28 (70%) out of 40 participants wore the devices until at least 7 days before their deaths. Sleeping-related parameters were missing for approximately 14.67% (243/1657) of the collected data points, which indicates that the devices were not worn at night on those days.
Figure 2 shows the study flowchart and data processing. A total of 1657 data points were collected, of which 177 (10.68%) were labeled as positive. After a downsampling of the participants with follow-up time longer than 50 days, a total of 1135 (68.5%) out of 1657 data points were included, with 177 (15.59%) positive labels. One-fourth of the data (284/1135, 25.02% data points) were randomly assigned to the testing set, and the remaining were assigned to the training set. Random upsampling of the positive data points resulted in a total of 1436 data points in the training-validation set, with an equal number of positive and negative data points.
7-Day Death Event Prediction Model
A total of 24 features were included in the model (Table 3). The correlation matrix of all the selected features in the training-validation set is shown in Figure 3.
Table 3.
Selected features in the models.
|
|
Featurea |
| Basic demographic |
|
| Clinical assessments |
|
| Wearable device parameters |
|
aFeatures with name ending with “_to_mean” represent the calculated difference between the original feature and past-4-week average in clinical assessment and between the original feature and past-7-day average in wearable device data.
b“Spo2_average” represents the average blood oxygen saturation measured during the nighttime.
Figure 3.
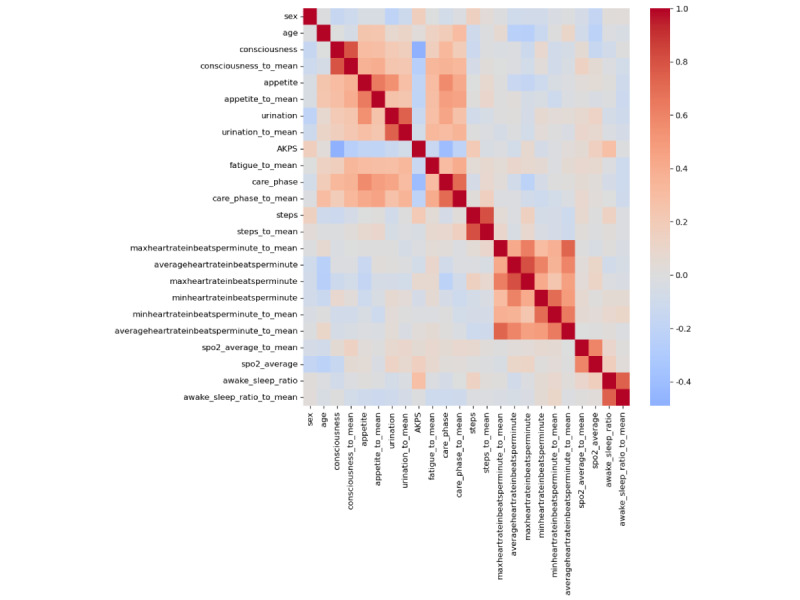
The correlation matrix of the selected features on the training-validation set. Features with name ending with “_to_mean” represent the calculated difference between the original feature and past 4 week average in clinical assessment and between the original feature and past 7 day average in wearable device data. AKPS: Australia-modified Karnofsky Performance Status.
Figure 4 and Table 4 show the performance of the machine learning models in terms of the AUROC and other evaluation metrics. Among all the models, XGBoost yielded the best result, with an AUROC of 96%, an F1-score of 78.5%, an accuracy of 93%, and a specificity of 97% on the testing set, whereas deep neural network achieved an AUROC of 93.3%, an F1-score of 76.8%, an accuracy of 92.7%, and a specificity of 96.2%.Random forests and KNN also yielded fair performances. The result of the models without any clinical assessment features is shown in Table 5.
Figure 4.

Area under the receiver operating characteristic curve (AUROC) of each model on the testing set. AdaBoost: adaptive boosting; DNN: deep neural network; DT: decision tree; KNN: k-nearest neighbor; LR: logistic regression; RF: random forest; SVM: support vector machine; XGBoost: extreme gradient boost.
Table 4.
Model performances (based on evaluation metrics) on the testing set.
| Model name | Accuracy | Precision | Recall | F1-score | Specificity | AUROCa |
| LRb | 0.750 | 0.360 | 0.720 | 0.540 | 0.750 | 0.832 |
| SVMc | 0.860 | 1.000 | 0.130 | 0.565 | 1.000 | 0.920 |
| KNNd | 0.910 | 0.660 | 0.910 | 0.785 | 0.910 | 0.938 |
| Decision tree | 0.877 | 0.624 | 0.605 | 0.615 | 0.930 | 0.767 |
| Random forest | 0.923 | 0.777 | 0.733 | 0.755 | 0.959 | 0.962 |
| XGBooste | 0.930 | 0.850 | 0.720 | 0.785 | 0.970 | 0.960 |
| AdaBoostf | 0.900 | 0.700 | 0.650 | 0.675 | 0.950 | 0.900 |
| DNNg | 0.927 | 0.790 | 0.746 | 0.768 | 0.962 | 0.933 |
aAUROC: area under the receiver operating characteristic curve.
bLR: logistic regression.
cSVM: support vector machine.
dKNN: k-nearest neighbor.
eXGBoost: extreme gradient boost.
fAdaBoost: adaptive boosting.
gDNN: deep neural network.
Table 5.
Model performance without the clinical assessment features.
| Model name | Accuracy | Precision | Recall | F1-score | Specificity | AUROCa |
| LRb | 0.720 | 0.350 | 0.870 | 0.610 | 0.690 | 0.814 |
| SVMc | 0.840 | 0.600 | 0.070 | 0.335 | 0.990 | 0.824 |
| KNNd | 0.800 | 0.430 | 0.700 | 0.565 | 0.820 | 0.821 |
| Decision tree | 0.835 | 0.494 | 0.530 | 0.512 | 0.894 | 0.712 |
| Random forest | 0.868 | 0.610 | 0.501 | 0.556 | 0.938 | 0.893 |
| XGBooste | 0.890 | 0.720 | 0.500 | 0.610 | 0.960 | 0.868 |
| AdaBoostf | 0.830 | 0.470 | 0.570 | 0.520 | 0.880 | 0.815 |
| DNNg | 0.893 | 0.687 | 0.632 | 0.660 | 0.944 | 0.899 |
aAUROC: area under the receiver operating characteristic curve.
bLR: logistic regression.
cSVM: support vector machine.
dKNN: k-nearest neighbor.
eXGBoost: extreme gradient boost.
fAdaBoost: adaptive boosting.
gDNN: deep neural network.
Explainable AI
Figure 5 and Table 6 show the results of the SHAP value analysis for the top-performing model, XGBoost, on the testing set. We also analyzed the SHAP values for the random forest, KNN, and deep learning models, which demonstrated similar performance levels (the results are shown in Multimedia Appendix 3). Among these models, the feature “average heart rate” consistently had the highest impact on prediction. Other features that were ranked in the top 5 by at least 2 of the models included “care_phase,” “urination,” “appetite,” “sex,” and “steps.”
Figure 5.

The Shapley additive explanations (SHAP) value of the extreme gradient boosting model on the testing set. The summary plot illustrates the feature values and the SHAP values of individual points. Features with name ending in “_to_mean” represent the calculated difference between the original feature and past 4 week average in clinical assessment and between the original feature and past 7 day average in wearable device data. AKPS: Australia-modified Karnofsky Performance Status.
Table 6.
The mean absolute Shapley additive explanations (SHAP) value of the extreme gradient boosting model on the testing set.
| Feature | Mean absolute SHAP value (SD) |
| averageheartrateinbeatsperminute | 0.1454 (0.088) |
| care_phase | 0.0600 (0.040) |
| steps | 0.0533 (0.054) |
| urination | 0.0356 (0.032) |
| appetite | 0.0328 (0.025) |
| consciousness_to_mean | 0.0307 (0.023) |
| steps_to_mean | 0.0273 (0.025) |
| minheartrateinbeatsperminute | 0.0269 (0.028) |
| age | 0.0268 (0.034) |
| averageheartrateinbeatsperminute_to_mean | 0.0265 (0.017) |
| awake_sleep_ratio_to_mean | 0.0248 (0.026) |
| care_phase_to_mean | 0.0231 (0.023) |
| AKPSa | 0.0218 (0.016) |
| sex | 0.0217 (0.023) |
| fatigue_to_mean | 0.0193 (0.017) |
| appetite_to_mean | 0.0175 (0.033) |
| spo2_average | 0.0163 (0.018) |
| awake_sleep_ratio | 0.0157 (0.016) |
| spo2_average_to_mean | 0.0143 (0.017) |
| minheartrateinbeatsperminute_to_mean | 0.0140 (0.013) |
| urination_to_mean | 0.0134 (0.021) |
| consciousness | 0.0119 (0.012) |
| maxheartrateinbeatsperminute | 0.0077 (0.010) |
| maxheartrateinbeatsperminute_to_mean | 0.0077 (0.008) |
aAKPS: Australia-modified Karnofsky Performance Status.
For the best performing model, XGBoost, the distribution of the testing set data points in terms of “time before death events” and the prediction results is shown in Figure 6. A few samples of the SHAP analysis in the wrong prediction cases are shown in Figure 7.
Figure 6.

Testing data distribution based on time before death (extreme gradient boosting). This figure illustrates a bar plot depicting the distribution of data based on the time before death. The x-axis represents the time before death events in days, whereas the y-axis represents the count of specific prediction results. Within the 7-day range, the data points were labeled as positive, resulting in either true-positive or false-negative outcomes. Conversely, data points occurring >8 days before death were labeled as either true-negative or false-positive outcomes. Therefore, the pink and red bars in the figure represent the distributions of false-negative and false-positive cases, respectively, as predicted by extreme gradient boosting.
Figure 7.

The Shapley additive explanations (SHAP) value analysis of 4 wrong prediction cases. This figure shows the SHAP value analysis of 4 wrong prediction cases. The upper 2 cases demonstrate the substantial impact of the average heart rate on the prediction, resulting in false-negative or false-positive outcomes. By contrast, the lower 2 cases exhibit the minimal or negligible effects of the average heart rate, with other factors dominating the prediction results. Note: features with name ending in “_to_m” represent the calculated difference between the original feature and past 4 week average in clinical assessment and between the original feature and past 7 day average in wearable device data. HR: heart rate.
User Feedback on Wearable Devices
User feedback was collected when the patient left the study, mostly owing to death. All the participants relied on their caregivers to update the data from the wearable device and charge the device. The demographics of the caregivers and feedback details are presented in Table 7 and Textbox 2. Overall, 30% (12/40) of the caregivers were aged ≥65 years, and only 32% (13/40) had experience in using wearable devices. Most of the caregivers could operate the device well after being instructed, and 70% (28/40) of the participants expressed willingness to participate in a similar study in the future. The most frequently reported problem was forgetting to charge the wearable device, which needed to be done once every 5 days in our study. Redness and itchiness of the skin were the main side effects reported, but only in a few patients (2/40, 5%).
Table 7.
Demographics of the caregivers (n=40) and feedback on wearable device use.
| Characteristic | Participants, n (%) | |
| Age group (years) | ||
|
|
<35 | 3 (8) |
|
|
35-50 | 8 (20) |
|
|
50-65 | 17 (42) |
|
|
65-80 | 10 (25) |
|
|
>80 | 2 (5) |
| Educational level | ||
|
|
Elementary school | 6 (15) |
|
|
Junior high school | 4 (10) |
|
|
Senior high school | 9 (22) |
|
|
College or university | 20 (50) |
|
|
Unclear | 1 (2) |
| Previous experience in using wearable devices | ||
|
|
Yes | 13 (32) |
|
|
No | 27 (68) |
| Cooperation with wearable device and smartphone use | ||
|
|
No problem with manipulating the device or apps | 30 (75) |
|
|
Need some help but generally fine | 8 (20) |
|
|
Need a lot of help or completely unable to cooperate | 2 (5) |
| Willing to participate in a similar study in the future (answer from the caregiver or wearable device user) | ||
|
|
Yes | 28 (70) |
|
|
No | 12 (30) |
| Problem when using the device | ||
|
|
Forget to charge the device | 6 (15) |
|
|
Discomfort (itch or redness) on the skin | 2 (5) |
|
|
Forget to wear | 2 (5) |
Opinions and feedback regarding the wearable device and the study.
“Want a larger monitor on the watch.”
“Want to directly manipulate and see data on the watch.”
“Hope more detailed description on the parameter, and thus to help the user understand its meaning.”
“Patient felt bothered by the vibration of watch.”
“Not wearing the device since the patient is in delirium and kept scratching the wristband unconsciously.”
“Not wearing due to admission and wrist was inserted with IV catheter.”
“Not wearing due to admission and there is only foreign caregiver at bedside. Families could not help charge the watch or update the data.”
Discussion
Principal Findings
Although we are not the first to explore the use of wearable devices in end-of-life care, we are unique in our approach. Pavic et al [41,42] demonstrated the feasibility of wearable devices and compared the data of patients with readmission with those of patients without readmission. Yang et al [43] used actigraphy along with deep learning to predict the prognosis of patients who were hospitalized. However, actigraphy recorded only wrist movements, and their study was confined to a hospital setting. Our study extends this area of research by developing prediction models using data from commercially available wearable devices in a broader, general end-of-life care setting. In addition, we demonstrated the potential of AI in predicting death events among patients with cancer at the end of life.
Wearable Device Parameters in Death Event Prediction
Our study has proved that the physiological data measured by a wearable device can be used in clinical prediction models. The SHAP values further provide explanations for the predictions of the models. Among all the parameters in our study, “average heart rate” was identified as the most important feature in predicting a recent death event. This is consistent with previous studies, as an increased HR was noticed before an emergency visit to a hospital or in the last days of patients with a terminal illness [41,47].
Besides HR, Pavic et al [41] also found that “heart rate variation” and “steps speed,” but not “steps count,” were significantly different between patients with and patients without emergency visits to hospitals. Contrary to their result, “steps count” was identified as an indicating parameter in some of our models. We suspect that the difference came from the relatively poor functional level of our population. Ideally speaking, individuals who are ambulatory would be the most suitable participants for wearable device studies, as certain parameters such as “steps count,” “steps speed,” and “climbing stairs” could yield more diverse and enriching data in these populations. However, to faithfully reflect the demographics that we typically care for, we did not establish any exclusion criteria based on the functional level. As a result, most participants in our study were patients who were not ambulatory, as evidenced by their AKPS scores upon enrollment—over 80% (32/40) of the patients scored <50.
Although our findings may have limitations when applied to individuals with better functional status, we believe that the results are pertinent and applicable to the patients we typically care for. Interestingly, even though our patients had limited functional abilities, we discovered that the ambulatory parameter “steps” still held predictive value for imminent mortality. This could indirectly reflect the patient’s status in very basic activities, such as changing posture, going to the bathroom, and sitting up. Patients tend to stay in bed more and decrease these activities when they enter the terminal phase.
The wearable device used in our study can measure blood oxygen saturation (SpO2), which is an increasingly common function for new wearable devices in the market. As it has been reported that saturation decreases in the last days of patients [47], it is a little surprising that the parameter does not seem to have a role in any of our models. A reasonable explanation for this is varied data quality. Until now, there have been no commercial smartwatches or wristbands that are well validated for measuring SpO2 [50]. Aside from improving the quality of hardware measurement, a possible solution is to consider the change and variation in oxygen saturation on a second or minute scale, rather than the daily average value. Deep learning models for high-dimensional time-series data, such as convolutional neural network and long short-term memory, provide good performances for this type of data [43,51,52] and will be considered in our future work.
Clinical Assessments in Death Event Prediction
As our explainable AI models have shown, clinical assessment features play a role in predicting death events. Among the models with a good performance, we found that a sign of reduced urine output, poor appetite, and deteriorating clinical care phase suspected by the clinical caregiver were ranked as high-impact features.
To further investigate the impacts, we reran the models using only wearable device parameters, sex, and age as input features. This resulted in lower precision and recall in predicting events while maintaining fair specificity, as shown in Table 5. When building the data set, we used forward filling in clinical assessment data to ensure that it reflects the most recent evaluation of the patient’s condition at that time point. In our opinion, the clinical assessment features might help the models determine whether the physiological changes detected by the wearable device are meaningful indicators of a potential event. These results suggest that clinical assessment is just as important as wearable device monitoring.
Wrong Prediction Case Analysis
We further examined the worst case of the XGBoost model prediction. First, we examined the distribution of “time before death events” in our testing set and identified that all the wrong predictions fell within 2 ranges of time: 1 to 7 days (false negatives) and 7 to 12 days (false positives) before death, as shown in Figure 6. These results reinforce the high specificity of our model, as patients who were far from the death event (eg, 20 days or longer before death) were rarely misclassified as “going to die.” Among the 284 data points in the testing set, only 6 (2.1%) false-positive predictions occurred, and in all cases, the patients died within 12 days.
During the review of the SHAP value analysis for incorrect predictions, 2 main types of explanations emerged, with the key feature being the “average heart rate.” Among the 6 false-positive predictions, 3 (50%) were primarily influenced by an elevated average HR, whereas the other 3 (50%) were driven by factors such as a decreased appetite or deteriorating care phase. A similar pattern was observed for the false-negative predictions within 1 to 7 days before death. In approximately half of the cases, a relatively low average HR played a significant role in predicting a negative outcome, accompanied by other parameters, including “care phase,” “appetite,” “consciousness,” and “steps,” aligning in the same direction. In the remaining half of the cases, the average HR had a small positive impact, whereas the other parameters contributed to a negative prediction. Some case results are depicted in Figure 7, and a comprehensive analysis of all the incorrect cases can be found in Multimedia Appendix 4.
Our analysis suggests that the average HR is an important indicator but not an absolute one in the 7-day death prediction model. In a vital sign study conducted in a palliative care unit, an increasing trend in HR was observed up to 2 weeks before death [47]. An increased HR may indicate a natural dying process, but it could also signify complications such as occult infection or sepsis. However, this trend was not observed in all patients. Conversely, certain causes of death, such as cardiac death, may be very acute events and not exhibit signs days before death. Different causes of death may contribute to the model’s false predictions.
Unknown Interactions Between Parameters and Models
It is interesting to investigate whether age plays a role in the prediction. We observed that the average HR had more positive impacts on the prediction results in the younger patient group (aged <65 years). When reviewing the wrong predictions made for the data points belonging to younger patients, especially for the false-negative cases, we found that the average HR increased in all cases, leading to a small positive impact on the prediction of death events. As for the false-negative cases from patients aged >80 years, only 1 (25%) out of the 4 cases showed a small positive impact from the average HR, whereas the other 3 (75%) cases had a strong negative impact from it. This finding aligns with the well-known concept that the maximal HR and HR stress response decrease with age [53,54]. However, the model still ranked the average HR as the most important feature in both the aged 65-80 years and aged >80 years patient groups. It remains unclear whether there are more relationships between age and other parameters. Our analysis regarding different age groups is shown in Multimedia Appendix 5.
Although explainable models can provide high-impact features and, therefore, seem to be more reliable for clinical use, there have been debates over the use of explainable AI in health care [55]. One issue with these models is that they may not always provide accurate or clinically reasonable interpretations of data. For instance, in our deep learning model, the feature “sex” was ranked as having a high impact, and male data points had a higher SHAP value. However, this does not necessarily mean that male patients are more likely to die within 7 days, as our data were stratified by sex. There may be interactions between the “sex” feature and other features that contribute to the model’s prediction, but we lack the means to adequately explore this. Current explanation methods are approximations of a model’s decision-making process and may not accurately reflect the true underlying logic. Therefore, it is important to interpret the SHAP values and other explanations from explainable AI models with caution.
Feasibility of Wearable Devices and AI in End-of-Life Care
Based on the feedback from our users, caregivers play a crucial role in the operation of these devices, as many of our patients were too weak to operate the devices as their illness progressed. However, to our surprise, many of them had no difficulty using the devices and expressed willingness to participate in similar studies in the future. Despite this, the results showed that, in the 38 patients who passed away during their follow-up time, only 28 (74%) out of wore their devices until 7 days before their death. This suggests that there are still many situations in which wearable devices become a burden for patients at the end of their lives. For example, patients may continuously try to remove the devices when they are in a delirium state. Another scenario is that when patients need to be admitted to the hospital, their family caregivers may not be able to stay with them and help charge the device or update the data. In 1 instance, although uncommon in end-of-life care, a caregiver reported having to remove the device because the patient had intravenous catheters on both hands while in the hospital. These issues highlight the limitations of the device hardware for these patients, but we expect these limitations to decrease as wearable technology continues to advance in size, form, and function.
Potentials of Environmental Factors in Predicting the End-of-Life Status
In addition to the clinical conditions and personal physiological changes detectable through wearable device data, environmental factors have been shown to benefit the models in predicting the disease status [32]. It is intriguing to explore whether external environmental variations, such as temperature or air quality, can impact the condition of patients at the end of life, particularly in the face of the current challenges posed by climate change. We obtained temperature data for Northern Taiwan from a public data set as a new input feature and examined their effect on model performance [56]. However, owing to limitations in interpretation and the fact that this feature was not originally included in our study design, we have included the analysis results in Multimedia Appendix 6. Nevertheless, we believe that the inclusion of environmental factors in prediction models holds promise, provided that it is done within the framework of a well-designed study and a larger cohort in the future.
Limitations
Our study is subject to several limitations that should be acknowledged. First, there was a large variation in survival time among the patients, resulting in data imbalances within our data set. We attempted to mitigate this issue by performing downsampling on individuals with longer survival times. Second, the nature of our study resulted in a class imbalance between the positive and negative labels. We intentionally did not perform any processing on the testing set to maintain the real-world distribution of the data. We believe that the performance of the models can be adequately evaluated using precision, recall, and F1-score. Third, it is important to note that our study was conducted on a relatively small cohort. Therefore, further validation is necessary to ensure the robustness of the prediction models.
Future Scope of Wearable Devices and AI in End-of-Life Care
Many patients with terminal cancer expressed a preference for living and dying at home, as it provides comfort, autonomy, security, and social interaction [19-22,57]. High-quality home-based and outpatient end-of-life care relies on timely medical and emotional support from the care team and good preparedness for death [23,58]. This study presents a prototype that demonstrates the potential of combining wearable devices and machine learning models to perform noninvasive, real-time monitoring of physical conditions and provide an early warning system for impending death events. To further advance this field, we propose three key directions for future research:
External validation: validate the robustness of prediction models in different health care facilities and among diverse populations, including patients at the end of life without a cancer diagnosis, to ensure the generalizability of the models.
Prediction of other important events: explore the prediction of crucial events other than death in end-of-life care, such as emergent medical needs that require timely intervention. These predictions can significantly impact the quality of care provided to home-based patients.
Real-world application: assess the benefits of integrating wearable devices and AI into real-world end-of-life care settings. For example, determine whether AI-powered predictions can help reduce emergency department visits or whether wearable devices enhance patients’ sense of safety during home care. Further research is needed to examine the practical application of wearable devices and AI in end-of-life care, with a focus on improving the overall quality of the care provided.
Conclusions
The findings of this study suggest that it is possible to predict death events among patients with terminal cancer using wearable devices and machine learning techniques. Although our prototype demonstrates the potential of these approaches in end-of-life care, further research is needed to confirm the robustness of the models and their effectiveness in real-world settings.
Acknowledgments
The authors would like to thank Zhi-Lei Yu for his help with case management and follow-up assessments. The authors would like to thank Hue-Chun Yeh and Wei-Fang Hsu for their help with referring and assessing home care patients. The authors thank the patients, caregivers, families, health professionals, and all others who participated in the study.
This study was supported by the Taiwan Ministry of Science and Technology under the grant MOST 110-2634-F-002-032.
Abbreviations
- AdaBoost
adaptive boosting
- AI
artificial intelligence
- AKPS
Australia-modified Karnofsky Performance Status
- AUROC
area under the receiver operating characteristic curve
- HR
heart rate
- KNN
k-nearest neighbor
- SHAP
Shapley additive explanations
- SpO2
blood oxygen saturation
- XGBoost
extreme gradient boosting
Survival prediction tools in end-of-life care.
Clinical assessment items (translated to English).
Shapley additive explanations value analysis of different models (extreme gradient boosting, deep neural network, random forest, and k-nearest neighbor).
Shapley additive explanations value analysis of the wrong prediction cases (extreme gradient boosting).
The extreme gradient boosting model’s predictions in different age groups.
Using environmental factors in predicting the end-of-life status.
Data Availability
Deidentified data and a data dictionary supporting the findings of this study will be made available upon request to researchers who submit a methodologically sound proposal and have obtained approval from the study authors. The data are not publicly available because of the limited consent obtained from the participants, who agreed to use the data only for research in palliative medicine or end-of-life care. The data will be made available immediately following publication, and we guarantee its availability until at least December 31, 2027, with no anticipated end date. Researchers interested in accessing the data should submit their proposals to JST on his email ID.
Footnotes
Authors' Contributions: J-HL conceived the main idea and developed the study protocol in discussion with FL, S-YC, J-ST, C-YS, H-LH, and J-KP. FL acquired funding from the Taiwan Ministry of Science and Technology and provided the wearable devices and data-collecting platform. J-ST, C-YS, H-LH, and J-KP referred qualified participants to the study. FL, S-YC, and J-ST supervised the study. J-HL performed the data analysis and wrote the manuscript. All other authors provided feedback and helped modify the manuscript.
Conflicts of Interest: None declared.
References
- 1.Anderson F, Downing GM, Hill J, Casorso L, Lerch N. Palliative performance scale (PPS): a new tool. J Palliat Care. 1996;12(1):5–11. [PubMed] [Google Scholar]
- 2.Harrold J, Rickerson E, Carroll JT, McGrath J, Morales K, Kapo J, Casarett D. Is the palliative performance scale a useful predictor of mortality in a heterogeneous hospice population? J Palliat Med. 2005 Jun;8(3):503–9. doi: 10.1089/jpm.2005.8.503. [DOI] [PubMed] [Google Scholar]
- 3.Lau F, Downing GM, Lesperance M, Shaw J, Kuziemsky C. Use of Palliative performance scale in end-of-life prognostication. J Palliat Med. 2006 Oct;9(5):1066–75. doi: 10.1089/jpm.2006.9.1066. [DOI] [PubMed] [Google Scholar]
- 4.Baik D, Russell D, Jordan L, Dooley F, Bowles KH, Masterson Creber RM. Using the palliative performance scale to estimate survival for patients at the end of life: a systematic review of the literature. J Palliat Med. 2018 Nov;21(11):1651–61. doi: 10.1089/jpm.2018.0141. https://europepmc.org/abstract/MED/30129809 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morita T, Tsunoda J, Inoue S, Chihara S. The Palliative Prognostic Index: a scoring system for survival prediction of terminally ill cancer patients. Support Care Cancer. 1999 May;7(3):128–33. doi: 10.1007/s005200050242. [DOI] [PubMed] [Google Scholar]
- 6.Morita T, Tsunoda J, Inoue S, Chihara S. Improved accuracy of physicians' survival prediction for terminally ill cancer patients using the Palliative Prognostic Index. Palliat Med. 2001 Sep;15(5):419–24. doi: 10.1191/026921601680419474. [DOI] [PubMed] [Google Scholar]
- 7.Stone CA, Tiernan E, Dooley BA. Prospective validation of the Palliative Prognostic Index in patients with cancer. J Pain Symptom Manage. 2008 Jun;35(6):617–22. doi: 10.1016/j.jpainsymman.2007.07.006. https://linkinghub.elsevier.com/retrieve/pii/S0885-3924(07)00798-1 .S0885-3924(07)00798-1 [DOI] [PubMed] [Google Scholar]
- 8.Kao CY, Hung YS, Wang HM, Chen JS, Chin TL, Lu CY, Chi CC, Yeh YC, Yang JM, Yen JH, Chou WC. Combination of initial palliative prognostic index and score change provides a better prognostic value for terminally ill cancer patients: a six-year observational cohort study. J Pain Symptom Manage. 2014 Nov;48(5):804–14. doi: 10.1016/j.jpainsymman.2013.12.246. https://linkinghub.elsevier.com/retrieve/pii/S0885-3924(14)00158-4 .S0885-3924(14)00158-4 [DOI] [PubMed] [Google Scholar]
- 9.Pirovano M, Maltoni M, Nanni O, Marinari M, Indelli M, Zaninetta G, Petrella V, Barni S, Zecca E, Scarpi E, Labianca R, Amadori D, Luporini G. A new palliative prognostic score: a first step for the staging of terminally ill cancer patients. Italian multicenter and study group on palliative care. J Pain Symptom Manage. 1999 Apr;17(4):231–9. doi: 10.1016/s0885-3924(98)00145-6. https://linkinghub.elsevier.com/retrieve/pii/S0885392498001456 .S0885392498001456 [DOI] [PubMed] [Google Scholar]
- 10.Maltoni M, Nanni O, Pirovano M, Scarpi E, Indelli M, Martini C, Monti M, Arnoldi E, Piva L, Ravaioli A, Cruciani G, Labianca R, Amadori D. Successful validation of the palliative prognostic score in terminally ill cancer patients. Italian multicenter study group on palliative care. J Pain Symptom Manage. 1999 Apr;17(4):240–7. doi: 10.1016/s0885-3924(98)00146-8. https://linkinghub.elsevier.com/retrieve/pii/S0885392498001468 .S0885392498001468 [DOI] [PubMed] [Google Scholar]
- 11.Glare P, Virik K. Independent prospective validation of the PaP score in terminally ill patients referred to a hospital-based palliative medicine consultation service. J Pain Symptom Manage. 2001 Nov;22(5):891–8. doi: 10.1016/s0885-3924(01)00341-4. https://linkinghub.elsevier.com/retrieve/pii/S0885-3924(01)00341-4 .S0885-3924(01)00341-4 [DOI] [PubMed] [Google Scholar]
- 12.Glare P, Eychmueller S, Virik K. The use of the palliative prognostic score in patients with diagnoses other than cancer. J Pain Symptom Manage. 2003 Oct;26(4):883–5. doi: 10.1016/s0885-3924(03)00335-x. https://linkinghub.elsevier.com/retrieve/pii/S088539240300335X .S088539240300335X [DOI] [PubMed] [Google Scholar]
- 13.Forrest LM, McMillan DC, McArdle CS, Angerson WJ, Dunlop DJ. Evaluation of cumulative prognostic scores based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. Br J Cancer. 2003 Sep 15;89(6):1028–30. doi: 10.1038/sj.bjc.6601242. https://europepmc.org/abstract/MED/12966420 .6601242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roxburgh CS, McMillan DC. Role of systemic inflammatory response in predicting survival in patients with primary operable cancer. Future Oncol. 2010 Jan;6(1):149–63. doi: 10.2217/fon.09.136. [DOI] [PubMed] [Google Scholar]
- 15.Hiratsuka Y, Yamaguchi T, Maeda I, Morita T, Mori M, Yokomichi N, Hiramoto S, Matsuda Y, Kohara H, Suzuki K, Tagami K, Yamaguchi T, Inoue A. The Functional Palliative Prognostic Index: a scoring system for functional prognostication of patients with advanced cancer. Support Care Cancer. 2020 Dec;28(12):6067–74. doi: 10.1007/s00520-020-05408-x.10.1007/s00520-020-05408-x [DOI] [PubMed] [Google Scholar]
- 16.Chen PY, Huang CH, Peng JK, Yeh SY, Hung SH. Prediction accuracy between terminally ill patients' survival length and the estimations made from different medical staff, a prospective cohort study. Am J Hosp Palliat Care. 2023 Apr;40(4):440–6. doi: 10.1177/10499091221108507. [DOI] [PubMed] [Google Scholar]
- 17.Hui D, Ross J, Park M, Dev R, Vidal M, Liu D, Paiva CE, Bruera E. Predicting survival in patients with advanced cancer in the last weeks of life: how accurate are prognostic models compared to clinicians' estimates? Palliat Med. 2020 Jan;34(1):126–33. doi: 10.1177/0269216319873261. [DOI] [PubMed] [Google Scholar]
- 18.Hiratsuka Y, Suh SY, Hui D, Morita T, Mori M, Oyamada S, Amano K, Imai K, Baba M, Kohara H, Hisanaga T, Maeda I, Hamano J, Inoue A. Are prognostic scores better than clinician judgment? A prospective study using three models. J Pain Symptom Manage. 2022 Oct;64(4):391–9. doi: 10.1016/j.jpainsymman.2022.06.008.S0885-3924(22)00784-9 [DOI] [PubMed] [Google Scholar]
- 19.Oosterveld-Vlug MG, Custers B, Hofstede J, Donker GA, Rijken PM, Korevaar JC, Francke AL. What are essential elements of high-quality palliative care at home? An interview study among patients and relatives faced with advanced cancer. BMC Palliat Care. 2019 Nov 06;18(1):96. doi: 10.1186/s12904-019-0485-7. https://bmcpalliatcare.biomedcentral.com/articles/10.1186/s12904-019-0485-7 .10.1186/s12904-019-0485-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nysæter TM, Olsson C, Sandsdalen T, Wilde-Larsson B, Hov R, Larsson M. Preferences for home care to enable home death among adult patients with cancer in late palliative phase - a grounded theory study. BMC Palliat Care. 2022 Apr 11;21(1):49. doi: 10.1186/s12904-022-00939-y. https://bmcpalliatcare.biomedcentral.com/articles/10.1186/s12904-022-00939-y .10.1186/s12904-022-00939-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng A, Maitland E, Wang S, Nicholas S, Lan K, Wang J. Preferences for End-of-Life care among patients with terminal cancer in China. JAMA Netw Open. 2022 Apr 01;5(4):e228788. doi: 10.1001/jamanetworkopen.2022.8788. https://europepmc.org/abstract/MED/35467732 .2791282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes B, Calanzani N, Curiale V, McCrone P, Higginson IJ. Effectiveness and cost-effectiveness of home palliative care services for adults with advanced illness and their caregivers. Cochrane Database Syst Rev. 2013 Jun 06;2013(6):CD007760. doi: 10.1002/14651858.CD007760.pub2. https://europepmc.org/abstract/MED/23744578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kastbom L, Milberg A, Karlsson M. A good death from the perspective of palliative cancer patients. Support Care Cancer. 2017 Mar;25(3):933–9. doi: 10.1007/s00520-016-3483-9.10.1007/s00520-016-3483-9 [DOI] [PubMed] [Google Scholar]
- 24.Finucane AM, O'Donnell H, Lugton J, Gibson-Watt T, Swenson C, Pagliari C. Digital health interventions in palliative care: a systematic meta-review. NPJ Digit Med. 2021 Apr 06;4(1):64. doi: 10.1038/s41746-021-00430-7. doi: 10.1038/s41746-021-00430-7.10.1038/s41746-021-00430-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avati A, Jung K, Harman S, Downing L, Ng A, Shah NH. Improving palliative care with deep learning. BMC Med Inform Decis Mak. 2018 Dec 12;18(Suppl 4):122. doi: 10.1186/s12911-018-0677-8. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-018-0677-8 .10.1186/s12911-018-0677-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou TJ, Wu YR, Tsai JS, Cheng SY, Yao CA, Peng JK, Chiu TY, Huang HL. Telehealth-based family conferences with implementation of shared decision making concepts and humanistic communication approach: a mixed-methods prospective cohort study. Int J Environ Res Public Health. 2021 Oct 14;18(20):10801. doi: 10.3390/ijerph182010801. https://www.mdpi.com/resolver?pii=ijerph182010801 .ijerph182010801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuit AS, Holtmaat K, Lissenberg-Witte BI, Eerenstein SE, Zijlstra JM, Eeltink C, Becker-Commissaris A, van Zuylen L, van Linde ME, Menke-van der Houven van Oordt CW, Sommeijer DW, Verbeek N, Bosscha K, Tewarie RN, Sedee RJ, de Bree R, de Graeff A, de Vos F, Cuijpers P, Verdonck-de Leeuw IM. Efficacy of the eHealth application Oncokompas, facilitating incurably ill cancer patients to self-manage their palliative care needs: a randomized controlled trial. Lancet Reg Health Eur. 2022 Apr 21;18:100390. doi: 10.1016/j.lanepe.2022.100390. https://linkinghub.elsevier.com/retrieve/pii/S2666-7762(22)00083-7 .S2666-7762(22)00083-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirozawa T, Yamada T, Ohwada H. New survival prediction system for terminal patients based on machine learning. Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine; BIBM '18; December 3-6, 2018; Madrid, Spain. 2018. pp. 2756–8. https://ieeexplore.ieee.org/document/8621357 . [DOI] [Google Scholar]
- 29.Leger S, Zwanenburg A, Pilz K, Lohaus F, Linge A, Zöphel K, Kotzerke J, Schreiber A, Tinhofer I, Budach V, Sak A, Stuschke M, Balermpas P, Rödel C, Ganswindt U, Belka C, Pigorsch S, Combs SE, Mönnich D, Zips D, Krause M, Baumann M, Troost EG, Löck S, Richter C. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017 Oct 16;7(1):13206. doi: 10.1038/s41598-017-13448-3. doi: 10.1038/s41598-017-13448-3.10.1038/s41598-017-13448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Jing X, Cao W, Zeng Y, Wu C, Zeng W, Chen W, Hu X, Zhou Y, Cai X. A non-lab nomogram of survival prediction in home hospice care patients with gastrointestinal cancer. BMC Palliat Care. 2020 Dec 07;19(1):185. doi: 10.1186/s12904-020-00690-2. https://bmcpalliatcare.biomedcentral.com/articles/10.1186/s12904-020-00690-2 .10.1186/s12904-020-00690-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills J, Fox J, Damarell R, Tieman J, Yates P. Palliative care providers' use of digital health and perspectives on technological innovation: a national study. BMC Palliat Care. 2021 Aug 07;20(1):124. doi: 10.1186/s12904-021-00822-2. https://bmcpalliatcare.biomedcentral.com/articles/10.1186/s12904-021-00822-2 .10.1186/s12904-021-00822-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu CT, Li GH, Huang CT, Cheng YC, Chen CH, Chien JY, Kuo PH, Kuo LC, Lai F. Acute exacerbation of a chronic obstructive pulmonary disease prediction system using wearable device data, machine learning, and deep learning: development and cohort study. JMIR Mhealth Uhealth. 2021 May 06;9(5):e22591. doi: 10.2196/22591. https://mhealth.jmir.org/2021/5/e22591/ v9i5e22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Qiu H, Xiong S. A novel hybrid deep neural network to predict pre-impact fall for older people based on wearable inertial sensors. Front Bioeng Biotechnol. 2020 Feb 12;8:63. doi: 10.3389/fbioe.2020.00063. https://europepmc.org/abstract/MED/32117941 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nait Aicha A, Englebienne G, van Schooten KS, Pijnappels M, Kröse B. Deep learning to predict falls in older adults based on daily-life trunk accelerometry. Sensors (Basel) 2018 May 22;18(5):1654. doi: 10.3390/s18051654. https://www.mdpi.com/resolver?pii=s18051654 .s18051654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Low CA. Harnessing consumer smartphone and wearable sensors for clinical cancer research. NPJ Digit Med. 2020 Oct 27;3:140. doi: 10.1038/s41746-020-00351-x. doi: 10.1038/s41746-020-00351-x.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pardoel S, Kofman J, Nantel J, Lemaire ED. Wearable-sensor-based detection and prediction of freezing of gait in Parkinson’s disease: a review. Sensors (Basel) 2019 Nov 24;19(23):5141. doi: 10.3390/s19235141. https://www.mdpi.com/resolver?pii=s19235141 .s19235141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai CH, Chen PC, Liu DS, Kuo YY, Hsieh TT, Chiang DL, Lai F, Wu CT. Panic attack prediction using wearable devices and machine learning: development and cohort study. JMIR Med Inform. 2022 Feb 15;10(2):e33063. doi: 10.2196/33063. https://medinform.jmir.org/2022/2/e33063/ v10i2e33063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabry F, Eltaras T, Labda W, Alzoubi K, Malluhi Q. Machine learning for healthcare wearable devices: the big picture. J Healthc Eng. 2022 Apr 18;2022:4653923. doi: 10.1155/2022/4653923. doi: 10.1155/2022/4653923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitratza M, Goodale BM, Shagadatova A, Kovacevic V, van de Wijgert J, Brakenhoff TB, Dobson R, Franks B, Veen D, Folarin AA, Stolk P, Grobbee DE, Cronin M, Downward GS. The performance of wearable sensors in the detection of SARS-CoV-2 infection: a systematic review. Lancet Digit Health. 2022 May;4(5):e370–83. doi: 10.1016/S2589-7500(22)00019-X. https://linkinghub.elsevier.com/retrieve/pii/S2589-7500(22)00019-X .S2589-7500(22)00019-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radin JM, Wineinger NE, Topol EJ, Steinhubl SR. Harnessing wearable device data to improve state-level real-time surveillance of influenza-like illness in the USA: a population-based study. Lancet Digit Health. 2020 Feb;2(2):e85–93. doi: 10.1016/S2589-7500(19)30222-5. https://linkinghub.elsevier.com/retrieve/pii/S2589-7500(19)30222-5 .S2589-7500(19)30222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavic M, Klaas V, Theile G, Kraft J, Tröster G, Blum D, Guckenberger M. Mobile health technologies for continuous monitoring of cancer patients in palliative care aiming to predict health status deterioration: a feasibility study. J Palliat Med. 2020 May;23(5):678–85. doi: 10.1089/jpm.2019.0342. [DOI] [PubMed] [Google Scholar]
- 42.Pavic M, Klaas V, Theile G, Kraft J, Tröster G, Guckenberger M. Feasibility and usability aspects of continuous remote monitoring of health status in palliative cancer patients using wearables. Oncology. 2020;98(6):386–95. doi: 10.1159/000501433. doi: 10.1159/000501433.000501433 [DOI] [PubMed] [Google Scholar]
- 43.Yang TY, Kuo PY, Huang Y, Lin HW, Malwade S, Lu LS, Tsai LW, Syed-Abdul S, Sun CW, Chiou JF. Deep-learning approach to predict survival outcomes using wearable actigraphy device among end-stage cancer patients. Front Public Health. 2021 Dec 09;9:730150. doi: 10.3389/fpubh.2021.730150. https://europepmc.org/abstract/MED/34957004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abernethy AP, Shelby-James T, Fazekas BS, Woods D, Currow DC. The Australia-modified Karnofsky Performance Status (AKPS) scale: a revised scale for contemporary palliative care clinical practice [ISRCTN81117481] BMC Palliat Care. 2005 Nov 12;4:7. doi: 10.1186/1472-684X-4-7. https://bmcpalliatcare.biomedcentral.com/articles/10.1186/1472-684X-4-7 .1472-684X-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masso M, Allingham SF, Banfield M, Johnson CE, Pidgeon T, Yates P, Eagar K. Palliative care phase: inter-rater reliability and acceptability in a national study. Palliat Med. 2015 Jan;29(1):22–30. doi: 10.1177/0269216314551814.0269216314551814 [DOI] [PubMed] [Google Scholar]
- 46.Lin CP, Tsay MS, Chang YH, Chen HC, Wang CY, Chuang YS, Wu CY. A comparison of the survival, place of death, and medical utilization of terminal patients receiving hospital-based and community-based palliative home care: a retrospective and propensity score matching cohort study. Int J Environ Res Public Health. 2021 Jul 07;18(14):7272. doi: 10.3390/ijerph18147272. https://www.mdpi.com/resolver?pii=ijerph18147272 .ijerph18147272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruera S, Chisholm G, Dos Santos R, Crovador C, Bruera E, Hui D. Variations in vital signs in the last days of life in patients with advanced cancer. J Pain Symptom Manage. 2014 Oct;48(4):510–7. doi: 10.1016/j.jpainsymman.2013.10.019. https://linkinghub.elsevier.com/retrieve/pii/S0885-3924(14)00046-3 .S0885-3924(14)00046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundberg SM, Lee S. A unified approach to interpreting model predictions. Proceedings of the 31st International Conference on Neural Information Processing Systems; NIPS'17; December 4-9, 2017; Long Beach, CA. 2019. pp. 4768–77. https://dl.acm.org/doi/10.5555/3295222.3295230 . [Google Scholar]
- 49.Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, Katz R, Himmelfarb J, Bansal N, Lee SI. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020 Jan;2(1):56–67. doi: 10.1038/s42256-019-0138-9. https://europepmc.org/abstract/MED/32607472 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Khatami R. Can we trust the oxygen saturation measured by consumer smartwatches? Lancet Respir Med. 2022 May;10(5):e47–8. doi: 10.1016/S2213-2600(22)00103-5.S2213-2600(22)00103-5 [DOI] [PubMed] [Google Scholar]
- 51.Fedorin I, Slyusarenko K. Consumer smartwatches as a portable PSG: LSTM based neural networks for a sleep-related physiological parameters estimation. Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society; EMBC '21; November 1-5, 2021; Mexico, FD. 2021. pp. 449–52. https://ieeexplore.ieee.org/document/9629597 . [DOI] [PubMed] [Google Scholar]
- 52.Naemi A, Schmidt T, Mansourvar M, Wiil UK. Personalized predictive models for identifying clinical deterioration using LSTM in emergency departments. Stud Health Technol Inform. 2020 Nov 23;275:152–6. doi: 10.3233/SHTI200713.SHTI200713 [DOI] [PubMed] [Google Scholar]
- 53.Tanaka H, Monahan KD, Seals DR. Age-predicted maximal heart rate revisited. J Am Coll Cardiol. 2001 Jan;37(1):153–6. doi: 10.1016/s0735-1097(00)01054-8. https://linkinghub.elsevier.com/retrieve/pii/S0735-1097(00)01054-8 .S0735-1097(00)01054-8 [DOI] [PubMed] [Google Scholar]
- 54.Kudielka BM, Buske-Kirschbaum A, Hellhammer DH, Kirschbaum C. Differential heart rate reactivity and recovery after psychosocial stress (TSST) in healthy children, younger adults, and elderly adults: the impact of age and gender. Int J Behav Med. 2004;11(2):116–21. doi: 10.1207/s15327558ijbm1102_8. [DOI] [PubMed] [Google Scholar]
- 55.Ghassemi M, Oakden-Rayner L, Beam AL. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit Health. 2021 Nov;3(11):e745–50. doi: 10.1016/S2589-7500(21)00208-9. https://linkinghub.elsevier.com/retrieve/pii/S2589-7500(21)00208-9 .S2589-7500(21)00208-9 [DOI] [PubMed] [Google Scholar]
- 56.CWB observation data inquire system. Central Weather Bureau, Taiwan. [2023-06-18]. https://e-service.cwb.gov.tw/HistoryDataQue ry/
- 57.Nilsson J, Blomberg C, Holgersson G, Carlsson T, Bergqvist M, Bergström S. End-of-life care: where do cancer patients want to die? A systematic review. Asia Pac J Clin Oncol. 2017 Dec;13(6):356–64. doi: 10.1111/ajco.12678. [DOI] [PubMed] [Google Scholar]
- 58.Danielsen BV, Sand AM, Rosland JH, Førland O. Experiences and challenges of home care nurses and general practitioners in home-based palliative care - a qualitative study. BMC Palliat Care. 2018 Jul 18;17(1):95. doi: 10.1186/s12904-018-0350-0. https://bmcpalliatcare.biomedcentral.com/articles/10.1186/s12904-018-0350-0 .10.1186/s12904-018-0350-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival prediction tools in end-of-life care.
Clinical assessment items (translated to English).
Shapley additive explanations value analysis of different models (extreme gradient boosting, deep neural network, random forest, and k-nearest neighbor).
Shapley additive explanations value analysis of the wrong prediction cases (extreme gradient boosting).
The extreme gradient boosting model’s predictions in different age groups.
Using environmental factors in predicting the end-of-life status.
Data Availability Statement
Deidentified data and a data dictionary supporting the findings of this study will be made available upon request to researchers who submit a methodologically sound proposal and have obtained approval from the study authors. The data are not publicly available because of the limited consent obtained from the participants, who agreed to use the data only for research in palliative medicine or end-of-life care. The data will be made available immediately following publication, and we guarantee its availability until at least December 31, 2027, with no anticipated end date. Researchers interested in accessing the data should submit their proposals to JST on his email ID.


