Abstract
Objective
To better understand the microbial profile of complicated parapneumonic effusions and empyema, and to evaluate whether antimicrobial selection would differ if guided by targeted metagenomic sequencing (tMGS) vs conventional cultures (CCs) alone.
Patients and Methods
We analyzed the pleural fluid of a cohort of 47 patients undergoing thoracentesis from January 1, 2017 to August 31, 2019, to characterize their microbial profile. All samples underwent 16S ribosomal ribonucleic acid gene polymerase chain reaction, followed by tMGS.
Results
Pleural space infection was deemed clinically present in 20 of the 47 (43%) participants. Of those, n=7 (35%) had positive pleural fluid cultures and n=14 (70%) had positive tMGS results. The organisms identified by tMGS were concordant with CCs; however, tMGS detected additional bacterial species over CCs alone. Streptococcus and Staphylococcus species were the most common organisms identified, with Streptococcus intermedius/constellatus identified in 5 patients. Polymicrobial infections were found in 6 of the 20 patients, with anaerobes being the most common organisms identified in these cases.
Conclusion
Streptococci and staphylococci were the most common organisms identified in infected pleural fluid. Anaerobes were common in polymicrobial infections. When compared with CCs, tMGS had higher sensitivity than CCs. Targeted metagenomic sequencing identified additional organisms, not identified by CCs, with associated potential management implications.
In community-acquired pneumonia, between 30%-50% of cases requiring hospitalization have an accompanying pleural effusion.1,2 Complicated parapneumonic effusion and empyema, the latter diagnosed when pleural fluid (PF) appears purulent and microbes spread into the normally-sterile pleural space, are associated with increased length of hospital stay, prolonged use of antibiotics, need for chest tube placement and surgical decortication, and increased morbidity and mortality.3,4 Therefore, assessing the pleural space is vital in patients with pneumonia to ensure adequate effusion management and antimicrobial treatment.
Considering the pleural space milieu, which favors the growth of organisms that may be difficult to grow in culture, particularly anaerobes, and the frequent administration of antibiotics before PF collection, the yield of conventional cultures (CCs) in pleural infections has been low.3,5 Thus, molecular methods are increasingly considered potential alternatives/adjuncts to CCs. Insa et al5 assessed the diagnostic performance of 16S ribosomal ribonucleic acid (rRNA) gene polymerase chain reaction (PCR)/Sanger sequencing of PF, reporting high sensitivity (82%) and specificity (99%) for diagnosis of infection. In a retrospective study comprising 64 empyema cases from Norway, Dyrhovden et al6 reported that 16S rRNA gene PCR/next-generation sequencing (targeted metagenomic sequencing [tMGS]) had an even higher yield than Sanger sequencing and identified a subset of empyema cases exhibiting a microbial profile similar to that of brain abscesses, with Streptococcus intermedius and/or Fusobacterium nucleatum underlying most cases. The same group has also reported S. intermedius and/or F. nucleatum as a dominant component in community-acquired pleural infection.7 Moreover, Kanellakis et al8 found that polymicrobial pleural infections were most common in their sample group from the United Kingdom, highlighting the need for broad-spectrum antibiotics. The applicability of these data in other populations is not clearly known.
Herein, a prospective study was performed to (1) better understand the microbial profile of hospitalized patients with complicated parapneumonic effusions and empyema; and (2) evaluate whether antimicrobial selection would differ if guided by tMGS plus CC vs CC alone.
Patients and Methods
The PF samples were prospectively collected from patients referred for diagnostic thoracentesis to the interventional pleural practice at our tertiary center between January 2017 and August 2019. For inclusion, PF samples had to have been obtained during thoracentesis or pleural catheter placement; samples obtained from indwelling pleural catheters were not included. This study was approved by the Mayo Clinic Institutional Review Board (IRB#16-001206). All participants provided informed consent.
If a patient was suspected of having infected PF, aerobic and anaerobic cultures were obtained by directly inoculating PF into culture bottles at the bedside at the time of PF sample collection.3 Ordering of cultures was left to the discretion of the thoracentesis-performing interventional pleural service and the primary requesting teams. Standard methods were used for culture work-up and gram stain of PF. For tMGS, PF samples were transported in a low-temperature transport box and stored at -80 °C until further processing. Patients with chronic effusions because of congestive heart failure, inflammatory processes, and malignant effusions without suspected infection served as controls.
Deoxyribonucleic acid extraction was performed using 100 μL of the specimen, after proteinase K digestion and sterile bead-beating, on a bioMérieux easyMAG. Polymerase chain reaction was performed using primers designed to amplify the V1-V3 region of the bacterial 16S rRNA gene, as previously described.6 All samples underwent next-generation sequencing for this study using library preparation, normalization, and sequencing according to a modified 16S metagenomic sequencing library preparation protocol from Illumina. The DNA concentrations were measured using a QuantiFluor ONE dsDNA kit on a Quantus fluorometer (Promega).
Onboard Illumina processing included adapter trimming, index demultiplexing, and fastq generation. Files were processed using Pathogenomix RipSeq NGS software. Sequences ≥210 bases in length were used for analysis. Forward and reverse reads were queried (separately) against the Pathogenomix PRIME database with ∼54,000 clinically relevant references. Homology of ≥99.0% was required for species-level identification, whereas 98%-98.9% homology was necessary for genus-level identification. The clinical laboratory director interpreted sequencing data (R.P.), blinded to CC results and clinical findings. The sequencing data is available under the following BioProject ID: PRJNA999256.
Case ascertainment (ie, presence of pleural space infection) was determined by 2 researchers (L.G. and D.A.) who independently reviewed participants’ electronic medical records with attention to symptomatology, ancillary testing, whether pleural space infection had been suspected before PF sampling, receipt of antimicrobial therapy, and clinical course. In instances of disagreement, case ascertainment was determined on the basis of an independent review by a third researcher (E.M.C.) blinded to the tMGS results.
Statistical analysis was performed using IBM SPSS Statistics v26 software. Suitable hypothesis-contrast tests were applied according to the sample distribution. Confidence intervals were calculated with the exact Clopper-Pearson method; only asymptotic approximation was performed for the area under the curve in the receiver operating curve analysis.
Results
Pleural fluid was collected from 47 participants; 32 (68%) were men, and the mean age was 69±2 years. Conventional cultures were performed on 34 (72%) participants, with tMGS performed on all. Pleural space infection was deemed clinically present in 20 of the 47 (43%) participants.
Clinically infected patients had a lower PF pH (median, 7.3 vs 7.4; P=.008), higher PF white blood cell count (median, 3090 vs 525 ×109/L; P=.005), higher frequency of positive PF culture (35% vs 0%; P=.011) and tMGS (70% vs 22%; P=.003), and nonsignificantly higher lactate dehydrogenase (median, 545 vs 160 U/L; P=.091) than clinically uninfected patients. Clinically infected patients were more likely to have received antimicrobial therapy before sample collection (90% vs 33%; P=.001) and to have undergone prior PF evacuation procedures (95% vs 59%; P=.015). Additional demographic characteristics and other PF parameters are described in Table 1.
Table 1.
Demographic Characteristics and Pleural Fluid Parametersa
| Characteristic | Total | Infected | Noninfected | P |
|---|---|---|---|---|
| Individuals, n (%) | 47 (100) | 20 (42.6) | 27 (57.4) | – |
| Sex, man n (%) | 32 (68.1) | 16 (80.0) | 16 (59.0) | .233b |
| Age (y), mean ± SD | 69.5±1.7 | 70.2±2.4 | 68,9±2.4 | .716c |
| Exudate, n (%) | 28 (82.4) | 12 (75.0) | 16 (88.9) | .387d |
| LDHp (U/L), median (IQR) | 203.5-555.8 | 544.5-844.8 | 160.0-298.8 | .091e |
| LDHs (U/L), median (IQR) | 203.0-63.5 | 217.5-114.8 | 202.0-68.0 | .414e |
| Proteinsp (g/dL), median (IQR) | 3.9-1.4 | 4.2-2.0 | 3.5-1.4 | .636e |
| Proteinss (g/dL), median (IQR) | 5.8-1.2 | 5.7-1.3 | 6.0-1.3 | .429e |
| pH, median (IQR) | 7.4-0.1 | 7.3-0.2 | 7.4-0.1 | .008e |
| Glucose (mg/dL), median (IQR) | 106.0-66.8 | 104.0-148.0 | 107.0-36.0 | .683e |
| WBC (×109/L), median (IQR) | 856.0-4181.5 | 3090.0-19,391.0 | 525.0-1029.0 | .005e |
| Antimicrobial therapy, n (%) | 27 (57.4) | 18 (90.0) | 9 (33.3) | <.001b |
| Prior manipulation, n (%) | 20 (42.6) | 11 (55.0) | 9 (33.3) | .235b |
| Fluid evacuation, n (%) | 35 (74.5) | 19 (95.0) | 16 (59.3) | .015b |
| Positive Gram Stain, n (%) | 2 (4.3) | 2 (11.8) | 0 (0.0) | .488d |
| Positive culture, n (%) | 7 (20.6) | 7 (35.0) | 0 (0.0) | .011d |
| Positive tMGS, n (%) | 20 (42.6) | 14 (70.0) | 6 (22.2) | .003b |
IQR, interquartile range; LDHp, lactate dehydrogenase in pleural fluid; LDHs, lactate dehydrogenase in serum; tMGS, targeted metagenomic sequencing; Proteinsp, proteins in pleural fluid; Proteinss, proteins in serum; WBC, white blood cells in pleural fluid.
Pearson’s χ2 test with Yates continuity correction.
t test.
Fisher’s exact test.
Mann-Whitney U test.
Among the 20 clinically infected patients, 7 (35%) had positive PF cultures and 14 (70%) had positive tMGS results (Table 2). In the 7 cases with positive CCs, tMGS identified the same organisms in all 7; however, tMGS detected additional bacterial species (Enterobacteriaceae, Streptococcus mitis group, and Fusobacterium nucleatum) in 3 of those cases (Table 2). Of the 14 patients with positive CCs or tMGS, 12 underwent pleural pigtail catheter placement and 8 received lytic therapy. As shown in Table 3, 6 patients in the clinically infected group had negative CCs and tMGS. Although they all received broad-spectrum antibiotics, none underwent pleural catheter placement. Other clinical characteristics are described in Table 3.
Table 2.
Bacterial Organisms Identified by Targeted Metagenomic Sequencing (tMGS) or Conventional Cultures in Clinically Infected Patients (n=14) and Clinically Uninfected Patients (n=6), With a Brief Description of the Clinical Context and Antibiotics at the Time of Thoracentesis
|
tMGS |
Cycle threshold (Ct) value | Culture | Clinical background/Intervention | Initial Antibiotic Regimen |
|---|---|---|---|---|
| Clinical evidence of pleural space infection with positive tMGS and/or conventional culture (n=14) | ||||
|
Staphylococcus aureus complex Enterobacteriaceae |
35.06 | Staphylococcus aureus | Mediastinal S. aureus surgical site infection complicated by empyema; pigtail catheter placed. No lytic therapy. | Cefepime + vancomycin + metronidazole |
| S. aureus complex | 34.61 | S. aureus | Postobstructive pneumonia and metastatic renal cell carcinoma with recurrent effusion. Indwelling pleural catheter removed days before study sample collection; no pigtail catheter placed. | Cefepime + vancomycin + metronidazole |
|
S. aureus complex Streptococcus mitis group |
35.21 | S. aureus | Hepatic hydrothorax in setting of alcoholic cirrhosis; no pigtail catheter placed. | Ceftriaxone |
| Staphylococcus epidermidis | No Ct | S. epidermidis | Chronic empyema; pigtail catheter placed. No lytic therapy. | Cefepime + vancomycin + metronidazole |
| Streptococcus intermedius/constellatus | 19.06 | Streptococcus intermedius | Community-acquired pneumonia (CAP) with empyema; pigtail catheter placed. Lytic therapy. | Piperacillin/tazobactam + vancomycin |
| S. intermedius/constellatus | 23.86 | Streptococcus anginosus group | Hospital-acquired pneumonia with empyema; pigtail catheter placed. Lytic therapy. | Piperacillin/tazobactam + vancomycin |
|
Streptococcus anginosus Fusobacterium nucleatum |
25.70 | S. anginosus group | Empyema in setting of long-term alcohol use; pigtail catheter placed. Lytic therapy. | Ceftriaxone + metronidazole |
| F. nucleatum | 37.00 | Negative | Recurrent aspiration pneumonia with empyema; Head and neck cancer resected; pigtail catheter placed. Lytic therapy. | Piperacillin/tazobactam + vancomycin |
| S. intermedius/constellatus | 30.78 | Negative | Lung abscess; pigtail catheter placed. Lytic therapy. | Piperacillin/tazobactam + vancomycin |
|
S. intermedius/constellatus Dialister invisus Prevotella denticola |
27.15 | Negative | CAP; pigtail catheter placed. Lytic therapy. | Piperacillin/tazobactam + levofloxacin |
| Haemophilus influenzae | 26.6 | Negative | CAP; pigtail catheter placed. Lytic therapy. | Levofloxacin |
| S. aureus complex | 34.62 | Negative | Methicillin resistant S. aureus–pneumonia with complicated parapneumonic effusion; pigtail catheter placed. No lytic therapy. | Vancomycin |
|
S. intermedius/constellatus Fusobacterium nucleatum |
18.63 | Negative | Bronchopleural fistula complicated by empyema; pigtail catheter placed. Lytic therapy. | Ceftriaxone |
| S. mitis group | 35.18 | Negative | Parapneumonic effusion; pigtail catheter placed. No lytic therapy. | Ceftriaxone + levofloxacin |
| No clinical evidence of pleural space infection with positive tMGS (n=6). | ||||
| Enterobacteriaceae | 34.77 | Not done | Pleural effusion following mitral/tricuspid valve operation. No evidence of subsequent infection. | None |
| Capnocytophaga granulosa | 35.01 | Not done | Malignant effusion due to marginal zone lymphoma. Pleural space instrumentation before study sample collection. No evidence of subsequent infection. | None |
| S. mitis group | 36.23 | Not done | Malignant effusion due to metastatic colon cancer. Pleural space instrumentation before study sample collection. No evidence of subsequent infection. | None |
|
Enterobacteriaceae Corynebacterium freneyi |
35.88 | Not done | Malignant effusion due to breast cancer. Pleural space instrumentation before study sample collection. Subsequent admission with concern for infection; received antibiotics. | None |
| S. mitis group | 34.40 | Not done | Malignant effusion due to lung adenocarcinoma. No evidence of subsequent infection. | None |
| Enterobacteriaceae | 34.08 | Negative | Malignant effusion due to anaplastic thyroid carcinoma. Initial concern for neutropenic fever. Suspicion for infected pleural effusion was low. No evidence of subsequent infection. | Cefepime + vancomycin |
Table 3.
Samples from Patients with Low Clinical Suspicion for Infection and Negative Targeted Metagenomic Sequencing (tMGS)
| tMGS | Culture | Antibiotics | Clinical Background |
|---|---|---|---|
| Negative | Negative | Piperacillin/tazobactam + vancomycin | Suspected malignant effusion vs infection, cytology was positive for adenocarcinoma. Previous week had pneumonia treated with broad-spectrum antibiotics. Low suspicion of infected pleural space. |
| Negative | Not obtained | Vancomycin | Thoracoabdominal aortic aneurysm repair and pigtail catheter 7 d before symptomatic effusion. Culture then grew Staphylococcus epidermidis. Pigtail catheter was placed again for symptom control. Low suspicion of infected pleural space. |
| Negative | Negative | Ceftriaxone + azithromycin | Community-acquired pneumonia suspected; patient dismissed on levofloxacin. Likely parapneumonic effusion. Low suspicion of infected pleural space. |
| Negative | Negative | Cefepime + metronidazole | Suspected aspiration, bronchial washes negative for bacterial infection but devoid of secretions. Low suspicion of infected pleural space. |
| Negative | Negative | Cefepime + Metronidazole + vancomycin | Suspected malignant effusion vs infection, in the setting of metastatic ileocecal adenocarcinoma. |
| Negative | Negative | Cefepime + metronidazole + vancomycin | Cytology was positive for adenocarcinoma. Antibiotics stopped after 24 hours. Low suspicion of infected pleural space. |
In the clinically uninfected group, CCs were performed on 14 of the 27 patients, none of which were positive. However, tMGS returned positive in 6 of the 27 (22%), among whom 1 had received antibiotics before PF collection for neutropenic fever (antibiotics were discontinued 2 days after admission) and 5 were considered to have a (para)malignant effusion, 3 of whom had undergone pleural space instrumentation within 3 weeks preceding PF collection (Table 2). Pleural space infection was not suspected in any of those patients.
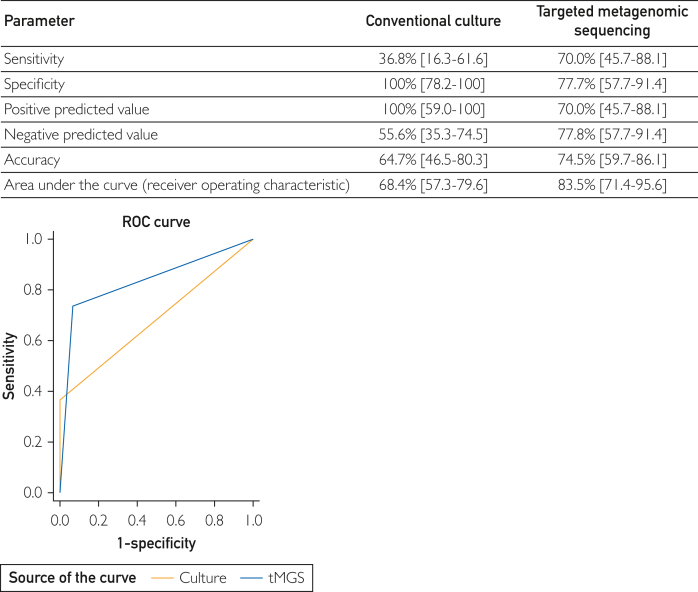
Overall, tMGS had a higher sensitivity (70% vs 37%) and accuracy (75% vs 65%) with a wider area under the curve (0.84 vs 0.68; P=.022), albeit with lower specificity than CC (78% vs 100%) (Figure).
Figure.
Receiver operating characteristic curve for prediction of pleural fluid infection on the basis of conventional cultures (CCs) and targeted metagenomic sequencing (tMGS).
Discussion
Pleural space infections are commonly because of Streptococcus pneumoniae, Staphylococcus aureus, and other aerobic bacteria, which can cause both monomicrobial and polymicrobial empyema.6,9 In the present study, pleural space infection was monomicrobial in 9 of the 14 cases, with Streptococcus and Staphylococcus species, including S. aureus (found in 4 cases), being among the most common organisms identified by CCs and tMGS. Interestingly, however, S. intermedius/constellatus was detected by tMGS in 5 cases (2 of which were culture-positive), including 2 community-acquired cases of pneumonia (Dialister invisus and Prevotella denticola were codetected in 1 case each), 1 hospital-acquired pneumonia, 1 bronchopleural fistula, and 1 lung abscess. The addition of tMGS identified 3 extra cases over CCs alone, suggesting that tMGS may indeed inform clinical decisions. The results of this study further support the growing evidence implicating S. intermedius/constellatus as a causative agent of pneumonia.6,10
Previous studies have reported that the rate of polymicrobial infections was ∼23%; most of these studies relied mainly on CCs, with the newer use of next-generation sequencing–metagenomics studies reporting that polymicrobial infections are possibly more common and even predominant.8,11 Herein, nearly half of the cases were polymicrobial, supporting the concept that polymicrobial infections are likely unrecognized if only CCs are used and reinforcing the need for better evaluation of PF using advanced molecular diagnostics.
Expectedly, there was good correlation between positive CCs and tMGS, with tMGS detecting the same organisms identified in all positive CCs. Nevertheless, tMGS detected additional bacterial species in 43% of those cases. Importantly, tMGS identified an organism in 7 patients with high suspicion of infection but negative CCs. Conceivably, with a more complete microbial profile generated by tMGS, antimicrobial therapy could be more appropriately tailored, but further studies are needed to clarify this concept further.
A subject of controversy is whether anaerobic coverage should be routinely administered to those with infected PF not known to have an anaerobic infection. The American Thoracic Society 2017 consensus guidelines state, “Even when there is a positive, monomicrobial, aerobic culture to direct therapy, it often is reasonable to continue anaerobic coverage.”3 The 2010 British Thoracic Society guidelines make an even stronger recommendation, stating “…antibiotics to cover anaerobic infection should be used in all patients except those with culture-proven pneumococcal infection”. Continuing with anaerobic coverage despite negative cultures is also recommended in the most recently updated guidelines from European Respiratory Society/European Society of Thoracic Surgeons.2,9 In this regard, anaerobes were the most common organisms in polymicrobial PF infections, although in 1 case, they were the only organisms identified. F. nucleatum was detected in 3 cases: 1 bronchopleural fistula (with S. intermedius/constellatus), 1 aspiration pneumonia, and 1 empyema in the setting of long-term alcohol use (with Streptococcus anginosus). All cases were identified by tMGS only. All but 1 patient were on antibiotics with anaerobic coverage. In contrast, 5 patients with no anaerobes detected received anaerobic coverage. Furthermore, 1 patient had S. aureus identified by CCs and tMGS; considering that no anaerobic bacteria were identified in this case, adding anaerobic coverage (per guidelines’ recommendations) may have been unnecessary. Although this study was not aimed at investigating treatment efficacy, our results suggest that tMGS may help formulate antimicrobial regimens in a more targeted way than those guided by CCs alone.3,9
The data presented shows that tMGS has double the sensitivity of CCs. Nevertheless, tMGS, which was positive in 6 cases in which infection was not clinically suspected, had a lower specificity than CCs. However, it should be noted that 3 of those patients with false positive tMGS had pleural space instrumentation, rendering it plausible that external contamination could be responsible for seemingly false results. Remarkably, another subject with false positive tMGS was subsequently admitted for concern about pleural space infection, raising the possibility that tMGS could be used as an early infection detection tool. On the contrary, 6 patients were considered clinically infected but had negative CCs and tMG; none of those patients had subsequent clinical worsening, and, eventually, the risk of infection was considered low in all.
Limitations and Future Directions
There are technical limitations to this study. All samples underwent next-generation sequencing, regardless of the associated cycle threshold (Ct) value; all samples with positive results but no clinical evidence of infection had Ct values above 34 cycles, whereas half of those with positive results and clinical evidence of infection had Ct values below 34 cycles. Interpretation of results in the context of the organism(s) detected and abundance thereof, alongside specific Ct values, should be evaluated in future studies. A better understanding of the panoply of pathogens involved in pleural space infections, and, conversely, which organisms may not be clinically relevant, may also inform results reporting and interpretation. Finally, given the microbiology defined here and elsewhere, the construction of a multiplex PCR panel for testing PF, possibly with certain antimicrobial resistance genes (eg, mecA [for expansion of gene symbols, use search tool at www.genenames.org]) included, may bear consideration to provide rapid results.
This study is also limited by its single-center design and small sample size. The study design did not enable assessment as to whether using tMGS for clinical decision-making would impact patient outcomes. However, the data presented contribute to accumulating evidence that molecular techniques like tMGS may add to CCs and aid antibiotic management in patients with parapneumonic effusions and empyemas. Specifically, the use of tMGS can further inform the development of multiplex PCR-based pathogen detection panels, which generally have a quicker turnaround time (1-2 hours compared with 3-5 days for CC and tMGS) and are less costly than tMGS. In addition, such assays could include assessment of select resistance genes (eg, mecA). In addition, although further prospective studies are needed to address optimization of antimicrobial stewardship in pleural infections, it is plausible that tMGS or PCR-based pathogen-guided treatment will improve antimicrobial stewardship, with decreased antimicrobial consumption and health care costs. New prospective studies using advanced molecular diagnostics are, therefore, needed to evaluate antimicrobial stewardship, clinical decision-making, and patient outcomes when compared with current CC-guided practice.
Conclusion
In pleural infections, tMGS had higher accuracy and sensitivity but possibly lower specificity than CCs. Targeted metagenomic sequencing identified additional organisms not identified by CCs with associated potential management implications. Whether implementing tMGS testing of PF in clinical practice translates into superior patient outcomes deserves further study.
Potential Competing Interests
Dr Patel reports receiving grants from ContraFect, TenNor Therapeutics Limited, and BioFire. Dr Patel is a consultant for PhAST, Torus Biosystems, Day Zero Diagnostics, Mammoth Biosciences, and HealthTrackRx; monies are paid to Mayo Clinic. Mayo Clinic and Dr Patel have a relationship with Pathogenomix. Dr Patel has research supported by Adaptive Phage Therapeutics. Mayo Clinic has a royalty-bearing know-how agreement and equity in Adaptive Phage Therapeutics. Dr Patel is also a consultant for Netflix, Abbott Laboratories, Oxford Nanopore Technologies, and CARB-X. In addition, Dr Patel has a patent on Bordetella pertussis/parapertussis PCR issued, a patent on a device/method for sonication, with royalties paid by Samsung to Mayo Clinic, and a patent on an anti-biofilm substance issued. Dr Patel receives honoraria from the NBME, Up-to-Date and the Infectious Diseases Board Review Course. Dr Samhouri reports providing consultation (unpaid) to AI Therapeutics that is unrelated to the submitted work. Dr Carmona reports providing consultation to Boehringer Ingelheim for sarcoidosis that is unrelated to the submitted work.
Acknowledgments
The authors acknowledge the excellent support from Ms Koscianski for her assistance in submitting the sequencing data, the interventional pulmonary practice for collecting the samples, and Dr Ali for processing the samples for the study. Luis Gimenez-Miranda and Bilal F. Samhouri contributed equally to this study.
References
- 1.Jain S., Self W.H., Wunderink R.G., et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedawi E.O., Ricciardi S., Hassan M., et al. ERS/ESTS statement on the management of pleural infection in adults. Eur Respir J. 2023;61(2) doi: 10.1183/13993003.01062-2022. [DOI] [PubMed] [Google Scholar]
- 3.Shen K.R., Bribriesco A., Crabtree T., et al. The American Association for Thoracic Surgery consensus guidelines for the management of empyema. J Thorac Cardiovasc Surg. 2017;153(6):e129–e146. doi: 10.1016/j.jtcvs.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Sogaard M., Nielsen R.B., Nørgaard M., Kornum J.B., Schonheyder H.C., Thomsen R.W. Incidence, length of stay, and prognosis of hospitalized patients with pleural empyema: a 15-year Danish nationwide cohort study. Chest. 2014;145(1):189–192. doi: 10.1378/chest.13-1912. [DOI] [PubMed] [Google Scholar]
- 5.Insa R., Marín M., Martín A., et al. Systematic use of universal 16S rRNA gene polymerase chain reaction (PCR) and sequencing for processing pleural effusions improves conventional culture techniques. Medi (Baltim) 2012;91(2):103–110. doi: 10.1097/MD.0b013e31824dfdb0. [DOI] [PubMed] [Google Scholar]
- 6.Dyrhovden R., Nygaard R.M., Patel R., Ulvestad E., Kommedal O. The bacterial aetiology of pleural empyema. A descriptive and comparative metagenomic study. Clin Microbiol Infect. 2019;25(8):981–986. doi: 10.1016/j.cmi.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 7.Dyrhovden R., Eagan T.M., Fløtten O., et al. Pleural empyema caused by Streptococcus intermedius and Fusobacterium nucleatum–a distinct entity of pleural infections. Clin Infect Dis. 2023 doi: 10.1093/cid/ciad378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanellakis N.I., Wrightson J.M., Gerry S., et al. The bacteriology of pleural infection (TORPIDS): an exploratory metagenomics analysis through next generation sequencing. Lancet Microbe. 2022;3(4):e294–e302. doi: 10.1016/S2666-5247(21)00327-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong H.L., Flurin L., Thoendel M.J., et al. Targeted versus shotgun metagenomic sequencing-based detection of microorganisms in sonicate fluid for periprosthetic joint infection diagnosis. Clin Infect Dis. 2023;76(3):e1456–e1462. doi: 10.1093/cid/ciac646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiraishi Y., Kryukov K., Tomomatsu K., et al. Diagnosis of pleural empyema/parapneumonic effusion by next-generation sequencing. Infect Dis (Lond) 2021;53(6):450–459. doi: 10.1080/23744235.2021.1892178. [DOI] [PubMed] [Google Scholar]
- 11.Hassan M., Cargill T., Harriss E., et al. The microbiology of pleural infection in adults: a systematic review. Eur Respir J. 2019;54(3) doi: 10.1183/13993003.00542-2019. [DOI] [PubMed] [Google Scholar]