Abstract
The discovery of small molecule inhibitors requires suitable binding pockets on protein surfaces. Proteins lacking this feature are considered undruggable and require innovative strategies for therapeutic targeting. The active state of mutant KRAS, the most frequently activated oncogene in cancer, is such a recalcitrant target. We designed a natural product-inspired small molecule that remodels the surface of cyclophilin A (CYPA) to create a neomorphic interface with high affinity and selectivity for the active state of KRASG12C. The resulting CYPA:drug:KRASG12C tri-complex inactivated oncogenic signaling and led to tumor regressions in multiple human cancer models. This inhibitory strategy can be used to target additional KRAS mutants and other undruggable cancer drivers. Tri-complex inhibitors selectively targeting active KRASG12C or multiple RAS mutants are in clinical trials (NCT05462717, NCT05379985).
One Sentence Summary:
Small molecules recruit cyclophilin A to the active state of mutant KRAS to disrupt oncogenic signaling and tumor growth.
KRAS is a small GTPase that cycles between an inactive (GDP-bound, OFF) state and an active (GTP-bound, ON) state (1). Active KRAS binds to and activates several effector proteins to regulate cell growth and proliferation (2). KRAS mutations act as oncogenic drivers that stimulate excessive downstream signaling and proliferation (3). KRASG12C is the most frequent KRAS mutation in non-small cell lung cancer (NSCLC) (4, 5), and 37-43% of patients with NSCLC harboring this variant respond to treatment with inhibitors targeting the inactive state of KRASG12C, such as sotorasib and adagrasib (6-9). The clinical benefits of these agents represent an important advance in precision oncology. Nevertheless, both are limited with regard to the depth and duration of response. Although the reasons for such limitations are multifactorial, cancer cells appear to bypass inactive state selective inhibition by increasing the amount of drug-insensitive/GTP-bound KRASG12C (10-15).
To date, efforts to target the active state of KRAS by traditional small molecule drug discovery strategies have been unsuccessful, suggesting that innovative approaches are needed for its inhibition. One such approach is inspired by natural products like rapamycin and FK506, which engage the immunophilin FKBP12 to inhibit mTOR or calcineurin, respectively (16-18). While the targets of these natural products are dictated by evolution (19), we now used structure-based redesign of an immunophilin ligand to direct its paired endogenous immunophilin to target the active state of mutant KRAS.
Results:
Remodeling cyclophilin A to generate a neomorphic interface that binds to active KRAS
We began by focusing on the immunophilin cyclophilin A (CYPA) because of its favorable electrostatic surface charge complementarity with residues on the effector binding interface of KRAS, which is not a feature of FKBP12 (fig. S1A). Sanglifehrin A (fig. S1B) is a natural product known to bind to CYPA with high affinity (20). A tool ligand (Compound-1) containing a minimal CYPA-binding motif of sanglifehrin A and a promiscuous cysteine-reactive warhead (Fig. 1A, B) was synthesized to facilitate covalent tethering and de novo tri-complex formation (Fig. 1A). The crystal structure of the primitive CYPA:Compound-1:KRASG12C tri-complex (Fig. 1A and table S1, 1.40 Å) suggested that macrocyclization of the ligand (fig. S1C) would decrease conformational entropy and increase protein contacts. Macrocyclization through a substituted indole linker introduced specific interactions with KRAS (see below), while the phenolic hydroxyl was removed to reduce the number of hydrogen bond donors/acceptors. The resulting cyclized core was modified with either an acetamide moiety that enables reversible binding (Compound-2, Fig. 1B) or an acrylamide warhead to covalently engage the C12 residue (Compound-3, Fig. 1B). As expected, Compound-2 induced CYPA complexes with GMPPNP-bound wild-type (WT) as well as G12C-mutant KRAS (Fig. 1C, IC50: 510 nM or 180 nM, respectively). By comparison, Compound-3 had increased potency for inducing CYPA complexes with active KRASG12C (IC50: 52 nM), while retaining weak reversible binding to KRASWT (IC50: 520 nM). Compound-3 was modified to further increase the selectivity for KRASG12C. Replacement of the acrylamide in Compound-3 with an ynamide warhead generated Compound-4, which had greater potency and selectivity for KRASG12C-CYPA tri-complex formation (Fig. 1C, IC50: 28 nM, 39-fold more selective). Due to the limit of detection in the tri-complex formation assay (see Methods), we elected to further optimize the warhead/linker using a kinetic target engagement assay, which affords higher resolution for potent compounds. The maximal rate of KRASG12C target engagement for Compound-4 was 26-fold higher than Compound-3 (Fig. 1B, D and S1D). Installation of a conformationally optimized linker improved the maximal rate a further 14-fold and resulted in RMC-4998 (Fig. 1B, D and S2A). The covalent engagement efficiency of RMC-4998 (kinact/KI: 272,000 M−1s−1) was greater than that of existing inactive state selective inhibitors (21-23), even though RMC-4998 targets the active or GTP-bound state of KRASG12C.
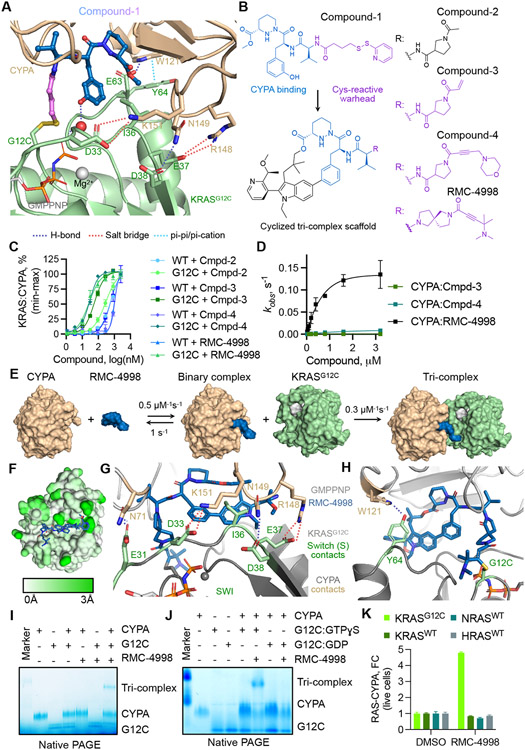
Fig. 1. Development of a tri-complex inhibitor targeting the active state of KRASG12C.
(A) Tertiary structure of the primitive CYPA:Compound-1:KRASG12C-GMPPNP complex at 1.40 Å resolution. (B) The chemical structure of Compound-1 and the macrocyclized scaffold for subsequent compounds, with the sanglifehrin-derived CYPA-binding moiety shown in blue. R denotes divergent chemical moieties comprising non-covalent (black) and covalent (pink) derivatives. (C) The interaction between the indicated KRAS proteins (50 nM) and CYPA (20 μM) in the presence of increasing compound concentrations was determined by TR-FRET (mean ± SEM, n=3, N=2). (D) The rate (kobs) of covalent modification of KRASG12C in the presence of CYPA (25 μM) and the indicated compounds (mean ± SEM, N=3). (E) Schematic of tri-complex formation by RMC-4998 along with rate constants for each reaction. (F) Mapped pairwise atomic distances between structures of apo-CYPA (PDB: 3K0N) and RMC-4998-bound CYPA showing the structural rearrangements (white to green gradient) that gave rise to the high-affinity neomorphic binding interface for KRASG12C. Interactions between RMC-4998, CYPA, and the Switch I (G) or Switch II (H) regions of KRASG12C in the crystal structure of the mature tri-complex (1.53 Å). (I) The interaction between KRASG12C (2 μM), RMC-4998 (10 μM), and CYPA (2 μM) was determined by native PAGE. (J) As in J, but KRASG12C was loaded with non-hydrolysable GTPɣS (active state) or GDP (inactive state). A representative of at least two independent experiments is shown in (I) and (J). (K) HEK293 cells co-expressing small bit luciferase-tagged RAS variants and large-bit luciferase tagged CYPA were treated with RMC-4998 (100 nM) for 2h followed by determination of luciferase activity (mean ± SEM, N=4).
We next sought to determine the structural basis for tri-complex formation and KRAS inhibition. RMC-4998 bound to CYPA reversibly to form a low affinity binary complex (Kd = 1.09 μM, koff = 1.0 s−1, Fig. 1E and fig. S1E). The crystal structure of the mature CYPA:RMC-4998:KRASG12C-GMPPNP tri-complex (1.53 Å, table S1) revealed that the aforementioned chemical optimizations increased the number of contacts between RMC-4998 and CYPA, compared to those observed in the primitive complex (fig. S2A and S2B). RMC-4998 retained the piperazic acid and peptidic linker of Compound-1 to maintain interactions with Q63 and the N102 backbone, but the substituted indole in the macrocycle of RMC-4998 introduced an additional cation-pi interaction with R55 as well as numerous favorable hydrophobic contacts (fig. S2A-G). Through these interactions, RMC-4998 remodeled the molecular surface of CYPA to create a neomorphic interface with affinity for KRASG12C (Fig. 1F, Movie S1).
The covalent engagement of KRASG12C by CYPA:RMC-4998 led to the formation of a stable tri-complex (Fig. 1E) with a half-life that was greater than 8 hours after compound washout in biophysical assays (fig. S3A) and cellular experiments (fig. S3B). The crystal structure of the tri-complex revealed several key interactions enabling stable binding (Fig. 1G and 1H): (1) non-covalent contacts between RMC-4998 and the Switch I and II motifs of KRASG12C. These were mediated by the indole introduced during macrocyclization and included lipophilic interactions with I36 on the Switch I (Fig. 1G) and a pi-pi interaction with Y64 on the Switch II motif of KRAS (Fig. 1H). (2) Neomorphic contacts between CYPA and KRASG12C, including hydrogen bonds between N71/K151/R148 in CYPA and E31/D33/E37 in the Switch I motif of KRAS (Fig. 1G), as well as W121 in CYPA and Y64 in KRAS (Fig. 1H). (3) Covalent modification of C12 by the ynamide warhead in RMC-4998 (Fig. 1H and S2D).
The remodeled molecular surface of inhibitor-bound CYPA enabled selective and active state-specific binding to KRASG12C. All three components were required for tri-complex formation (Fig. 1I and S4A); neither RMC-4998 nor CYPA were able to bind to KRASG12C alone. No complex was detected with GDP-loaded KRASG12C, suggesting a selectivity for the active (GTP-bound) state of the mutant (Fig. 1J). RMC-4998 induced a tri-complex between KRASG12C and CYPA in live cells without affecting wild-type KRAS, NRAS, or HRAS (Fig. 1K). RMC-4998 had comparable activity on G12C-mutant HRAS, NRAS and KRAS, with little, if any, activity on KRASG13C (fig. S4B and S4C).
Tri-complex formation disrupts effector binding to active KRAS
Structural superpositions suggested that, when bound in the tri-complex, CYPA occludes the effector binding interface of KRASG12C, including that occupied by the RAS-binding and cysteine-rich domains (RBD-CRD) of CRAF (Fig. 2A), the catalytic subunit (p110) of PI3K (fig. S5A), the allosteric site of the catalytic domain of SOS1 (fig. S5B, SOScat), and the RAS-interacting domain (RID) of RALGDS (fig. S5C). Indeed, the formation of the CYPA:RMC-4998:KRASG12C tri-complex led to a concentration-dependent dissociation of the BRAF RBD and RALGDS RID from mutant KRAS in biochemical assays (Fig. 2B and fig. S5D). In a live-cell kinetic assay, RMC-4998 treatment led to rapid association of KRASG12C with CYPA, an effect that was paralleled by the dissociation of full-length CRAF from KRASG12C (Fig. 2C and D). CYPA was indispensable for inhibition by RMC-4998, as evidenced in biochemical assays (Fig. 2B) as well as in CYPA-null cells and those with rescued expression (Fig. 2E and fig. S6A, B). By comparison, CYPA loss had no effect on the cellular activity of the inactive state selective KRASG12C inhibitor adagrasib (fig. S6C and D). It is possible that a dependency on CYPA expression could limit the therapeutic utility of tri-complex inhibitors like RMC-4998. This is unlikely to be true, however, when considering that CYPA is highly abundant across various cancer types (fig. S6E), has low inter-patient variation in expression (fig. S6F), and has higher expression in tumors as compared to normal tissue (fig. S6G).
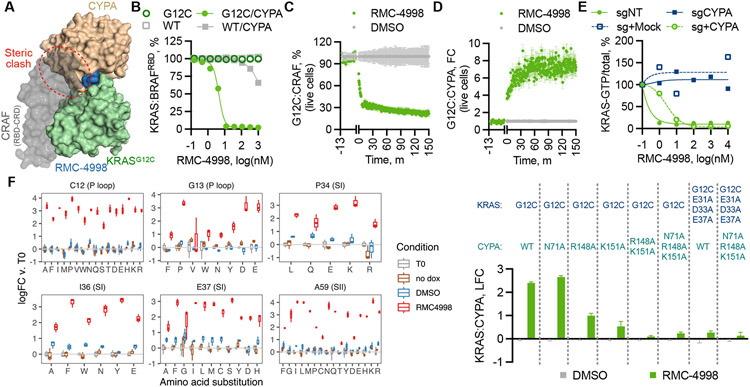
Fig. 2. Structural constraints for tri-complex formation and active state selective KRAS inhibition.
(A) Superimposition of the structures of the CYPA:RMC-4998: KRASG12C tri-complex and the KRAS:CRAFRBD/CRD complex (PDB: 6XI7) (31). (B) The effect of RMC-4998 on the interaction between the indicated KRAS proteins (12.5 nM) and BRAFRBD (50 nM) in either the presence or the absence of CYPA was determined by TR-FRET (mean ± SD, N=4). HEK293 cells co-expressing small bit luciferase-tagged KRASG12C and large bit luciferase-tagged CYPA (C) or full-length CRAF (D) were treated with RMC-4998 (100 nM) followed by determination of reconstituted luciferase activity in live cells (mean ± SEM, N=3). (E) The indicated parental, CYPA-null or CYPA rescued H358 cells were treated as shown and their extracts were analyzed by RBD-pulldown and immunoblotting to determine the effect on KRAS activation. A representative of two independent experiments is shown. (F) KRAS mutant cells were infected with a dox-inducible saturation mutagenesis library based on a KRASG12C backbone and treated with either DMSO or RMC-4885 (100 nM) for two weeks. Shown is the log(fold-change) (logFC) in abundance relative to T0 (mean ± 95%CI, N=3) for variants meeting the threshold for statistical significance (see Methods). (G) The effect of the indicated CYPA variants on tricomplex formation in live cells was determined as in C (mean ± SEM, N=3).
Functional interrogation of the neomorphic binding interface
The distinct inhibitory mechanism of RMC-4998 prompted an unbiased evaluation of the functional role of the amino acid residues in the neomorphic binding interface between CYPA and KRASG12C. To this end, KRASG12C mutant cells were infected with a doxycycline (dox)-inducible cDNA library, encoding all possible amino acid substitutions in KRASG12C, followed by treatment with either DMSO or RMC-4998 for two weeks in the presence or absence of dox (fig. S7A). The analysis confirmed the importance of the KRAS C12, I36, and E37 residues (Fig. 2F), as predicted by the structural studies above (Fig. 1G and 1H). The screen, however, revealed that mutations in additional residues, including those at G13, P34 and A59, attenuated the effect of RMC-4998 (Fig. 2F and fig. S7B). The latter likely disrupt the conformation of the switch regions to prevent CYPA:RMC-4998 from engaging active KRASG12C. Cells expressing KRASG12C containing a P34R, I36Q or A59G secondary mutation confirmed the ability of these variants to activate ERK signaling in the presence of RMC-4998 (fig. S7C). Secondary in cis mutations on KRASG12C (for example, R68, H95, and Y96) confer resistance to adagrasib and/or sotorasib (11-13). RMC-4998 occupies a distinct binding site and does not contact any of these residues, suggesting it should retain inhibitory activity against these mutants (Fig. 1G). Indeed, mutations in these residues were not identified in the saturation mutagenesis screen (fig. S7A) and had no effect on displacement of the CRAF RBD by RMC-4998 in a live-cell assay (fig. S7D).
We next used the live cell biosensor to interrogate residues in CYPA that contribute to the neomorphic interface. Alanine mutations at the N71, K151, and R148 residues of CYPA that form direct contacts with KRASG12C resulted in a loss of tri-complex formation (Fig. 2G). Mutations in their interacting partners on switch I of KRASG12C (E31A/D33A/E37A) also abolished the formation of the tri-complex, highlighting the importance of these residues in binding (Fig. 2G). These mutations had minimal effect on the enzymatic proline isomerase activity of CYPA (fig. S8).
Selective inhibition of oncogenic signaling and proliferation
Disruption of effector binding by RMC-4998 inhibited downstream ERK signaling in KRASG12C mutant cancer cell models with an IC50 ranging between 1 and 10 nM (Fig. 3A). RMC-4998 treatment also attenuated AKT/MTOR and RAL signaling (fig. S9A and S9B). The latter pathways, however, had residual activity despite the presence of the drug, suggesting that they are only partially activated by mutant KRAS in cancer cells. Targeting the GTP-bound state of KRASG12C with the tri-complex inhibitor RMC-4998 (0.1 μM) led to a faster disruption of the interaction between KRASG12C and CRAF as compared to inactive state selective inhibitors adagrasib (1 μM) and sotorasib (10 μM) (Fig. 3B). A similar disruption was observed for the interaction between KRASG12C and SOScat (fig. S9C). Complete covalent modification of KRASG12C in RMC-4998-treated MIA PaCa-2 cells occurred within 5 minutes, again faster than observed with adagrasib (fig. S9D). The kinetics of inactive state selective inhibition are limited by the rate of GTP hydrolysis by KRASG12C (24), which RMC-4998 overcomes by directly targeting the active state.
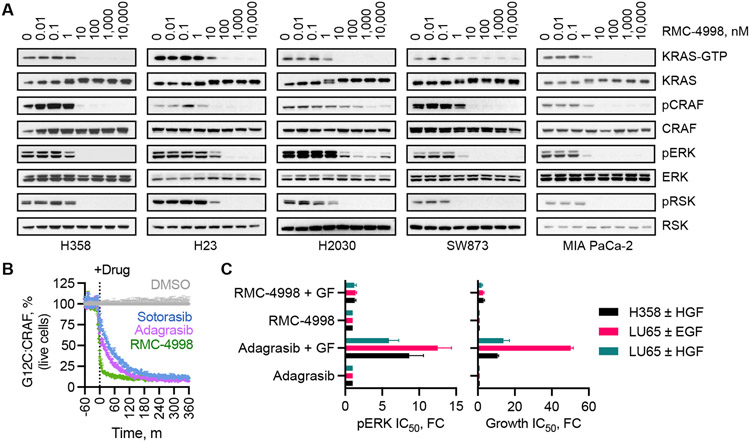
Fig. 3. Cellular effects of active state selective KRASG12C inhibition.
(A) Extracts from the indicated KRASG12C mutant cell lines were treated with RMC-4998 for 2 h before lysis and immunoblotting. The levels of active KRAS (KRAS-GTP) were determined by pull-down with the RAS-binding domain (RBD) of CRAF. A representative of at least two independent experiments for each cell line is shown. (B) The kinetics of target inhibition in live cells treated with an active state (RMC-4998, 100 nM) or an inactive state (sotorasib, 10 μM and adagrasib, 1 μM) selective inhibitor were determined as in Fig. 2C (mean ± SEM, N=3) (C) The indicated cell lines were treated as shown in the presence or absence of growth factor (GF) stimulation to determine the effect on ERK phosphorylation (pERK/total, 4 h) or cell proliferation (120 h) (mean ± SEM, N=3-4). EGF: epidermal growth factor, HGF: hepatocyte growth factor.
Another limitation of inactive state selective inhibitors is their susceptibility to cellular stimuli (10) that drive nucleotide exchange to induce GTP-loading of KRASG12C. This is particularly evident following receptor tyrosine kinase (RTK) activation (25, 26). Indeed, exposure to EGFR or MET ligands led to attenuated target engagement (fig. S9E), ERK inhibition, and antiproliferative effect by the inactive state selective inhibitor adagrasib (Fig. 3C). In contrast, RTK stimulation had a minimal effect on RMC-4998 activity. In the absence of growth factor stimulation, treatment of KRASG12C-mutant cell lines with either RMC-4998 or adagrasib produced initial pathway suppression followed by a rebound over the course of the next 72 hours. The re-addition of RMC-4998, but not adagrasib, suppressed the reactivated pathway signaling (fig. S9F). Therefore, targeting of the active state KRASG12C through tri-complex formation has distinct biological properties from existing inactive state selective clinical-stage inhibitors.
Potent anti-tumor activity in cell line and patient-derived xenografts
Further optimization of RMC-4998 led to RMC-6291, an active-state selective KRASG12C inhibitor currently undergoing clinical testing. RMC-4998 and RMC-6291 have similar chemical structures and nearly identical kinetic constants for engaging active KRASG12C (fig. S10A). Both RMC-4998 and RMC-6291 inhibited ERK signaling and induced apoptosis in KRASG12C-mutant H358 cells (fig. S10B). In a cancer cell line panel (Fig. 4A, Table S2), RMC-6291 inhibited the proliferation of KRASG12C mutant cells with a median IC50 of 0.11 nM, or 13,500 times more potently than that of non-G12C mutant models, indicating its selectivity index. RMC-4998 had median IC50 of 0.28 nM and selectivity index of 1450. Of note, both the potency and the selectivity index of RMC-4998 and RMC-6291 were greater than those of adagrasib. Selective cellular engagement of KRASG12C was further evidenced by a proteome-wide cysteine-reactivity assay, wherein RMC-6291 reacted with KRASG12C with high selectivity over other cellular proteins (fig. S11A and data S1).
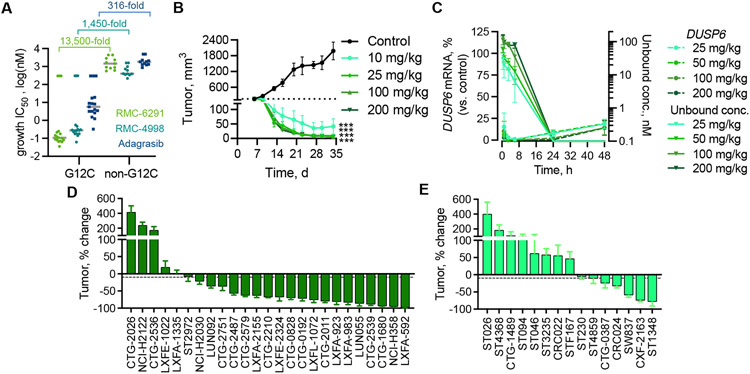
Fig. 4. Potent and selective suppression of KRASG12C-driven tumor growth by tri-complex inhibitors.
(A) The indicated cells were treated with increasing concentrations of RMC-6291, RMC-4998, or adagrasib for 120h and the effect on cell viability was determined using the 3D CellTiter-Glo assay. Each point represents an individual cell line (n = 17 for G12C, n=11 for non-G12C) and the gray line indicates the median IC50 for each group of cell lines. (B) Mice bearing H358 CDX tumors were treated with RMC-6291 at the indicated dose, administered orally once daily, and the tumor volume was assessed for 28 days. ***adj.p<0.001 for RMC-6291 (all dose groups) vs control, using repeated measures 2-way ANOVA (n=6/group for control, n=8/group for RMC-6291); adjusted based on multiple comparison via Dunnett’s test on the final tumor measurement. (C) The unbound plasma concentration of RMC-6921 and the expression of DUSP6 mRNA in H358 tumors following administration of a single oral dose of RMC-6291 at the indicated doses (mean ± SEM, n=3). (D, E) The indicated NSCLC (D) or CRC (E) xenograft models were treated with RMC-6291(200 mg/kg administered orally once daily) to determine the effect on mean tumor growth or regression after 28±2 days (% change from baseline, mean ± SEM, n as indicated in table S3). The dashed line indicates 10% reduction in tumor volume from baseline.
Daily oral administration of RMC-6291 in mice bearing NCI-H358 xenografts was well tolerated (fig. S11B) and led to near complete tumor regression (Fig. 4B). RMC-6291 achieved dose-dependent plasma concentrations and inhibition of downstream signaling output, as assessed by the effect on tumor DUSP6 mRNA expression (Fig. 4C). A single dose of RMC-6291 led to sustained engagement of KRASG12C and inhibition of ERK phosphorylation in tumors for ~24 hours, and also induced apoptosis, as evidenced by TUNEL staining (fig. S12A-D). These data indicate that the tri-complex inhibitor RMC-6291 is a potent and selective inhibitor of the active conformation of KRASG12C in vivo. Similar in vivo activity was observed with RMC-4998 (fig. S13A-C).
Next, we evaluated the anti-tumor activity of RMC-6291 across a panel of patient-derived xenograft (PDX) models of KRASG12C mutant NSCLC and CRC. Targeted exome DNA sequencing revealed that the PDX panels were representative of the genomic landscape of patients with KRASG12C mutant lung or colorectal cancers (fig. S14A and S14B). RMC-6291 treatment resulted in mean tumor regression in 76% (19/25) of NSCLC models and in 40% (6/15) of the CRC models tested after a standard 28-day treatment period (Fig. 4D and 4E, Table S3). These results indicate that RMC-6291 can drive deep anti-tumor responses following daily oral administration in preclinical studies.
Discussion:
The discovery of RMC-4998 and RMC-6291 as tri-complex inhibitors of GTP-bound KRASG12C represents the successful re-engineering of an immunophilin-binding natural product to engage a target previously thought to be undruggable. Neither CYPA nor its natural product ligand have been previously reported to interact with KRAS. Their complex formation was the product of purposeful, structure-guided chemical modifications, to mold a high-affinity binding interface between CYPA and active KRASG12C. The target-directed approach described here differs from other pharmacologic or proteomic screens describing identification of rapamycin or sangliferin analogues that engage additional targets besides MTOR or CYPA, respectively (27, 28). Selective disruption of effector binding to the active state of KRASG12C by the recruitment of CYPA is a distinct inhibitory mechanism that promises to overcome some of the limitations of inactive state selective KRASG12C inhibitors, while maintaining selective target engagement and a potentially wide therapeutic index. A phase I/II clinical trial testing RMC-6291 (NCT05462717) is currently ongoing.
The tri-complex inhibitory strategy described here has broad implications for cancer therapy. It can be utilized to develop inhibitors of additional oncogenic RAS mutants, such as through the introduction of distinct covalent warheads or functional groups that can selectively target other RAS mutants (G13C, G12D etc.) in an allele-specific manner. Alternatively, further optimization of the reversible drug binding capacity demonstrated by Compound-2 would enable the concurrent (non-discriminatory) inhibition of multiple RAS oncoproteins. A reversible tri-complex inhibitor RMC-6236 (29) (RASMULTI) developed through this approach is also in a phase I/II clinical trial (NCT05379985). Although its broader applicability requires further investigation, re-engineering of natural products to create neomorphic binding interfaces on their paired immunophilins may prove effective at targeting additional “undruggable” cancer drivers even beyond RAS.
Supplementary Material
Acknowledgements:
The authors thank Gregory Verdine, Alexis Borisy and others at Warp Drive Bio for their contributions to the early sanglifehrin analogs, and Kevan Shokat for helpful discussions. P.L. thanks Megan Mroczkowski for discussing this work and the manuscript.
Funding:
This study was funded by Revolution Medicines, Inc. P.L. is supported in part by the NIH/NCI (1R01CA23074501, 1R01CA23026701A1 and 1R01CA279264-01), The Pew Charitable Trusts, the Damon Runyon Cancer Research Foundation, and the Pershing Square Sohn Cancer Research Alliance. P.L. is also supported by the Josie Robertson Investigator Program and the Support Grant-Core Grant program (P30 CA008748) at Memorial Sloan Kettering Cancer Center.
Footnotes
Competing interests:
P.L. listed as an inventor on patents filed by MSKCC on the treatment of KRAS or BRAF mutant cancers. P.L. reports grants to his institution from Amgen, Mirati, Revolution Medicines, Boehringer Ingelheim and Virtec Pharmaceuticals. P.L. reports consulting fees from Black Diamond Therapeutics, AmMax, OrbiMed, PAQ-Tx, Repare Therapeutics and Revolution Medicines, as well as membership on the Scientific Advisory Board of Frontier Medicines and Biotheryx (consulting fees and equity in each). C.S., K.S., Y.C.Y, J.C., A.T., T.C., Z.W., A.M., M.Z., V.V., C.W., A.G., D.W., A.S., A.M., M.S-C., N.D., A.C., J.E., J.E.K., E.S.K., M.S,, R.N., D.W., A.L.G., and J.S. are current or former employees of Revolution Medicines, Inc.
Data and materials availability:
PDB files have been deposited into RCSB with the accession numbers 8G9Q and 8G9P. All other data are available in the main text or the supplementary materials.
References and notes:
- 1.Simanshu DK, Nissley DV, McCormick F, RAS Proteins and Their Regulators in Human Disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malumbres M, Barbacid M, RAS oncogenes: the first 30 years. Nat Rev Cancer 3, 459–465 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D, RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 11, 761–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riely GJ et al. , Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 14, 5731–5734 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, Liang SQ, Schmid RA, Peng RW, New Horizons in KRAS-Mutant Lung Cancer: Dawn After Darkness. Front Oncol 9, 953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lito P, Solomon M, Li LS, Hansen R, Rosen N, Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skoulidis F et al. , Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 384, 2371–2381 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janne PA et al. , Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRAS(G12C) Mutation. N Engl J Med 387, 120–131 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Xue JY et al. , Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 577, 421–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Awad MM et al. , Acquired Resistance to KRAS(G12C) Inhibition in Cancer. N Engl J Med 384, 2382–2393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y et al. , Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 599, 679–683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka N et al. , Clinical Acquired Resistance to KRAS(G12C) Inhibition through a Novel KRAS Switch-II Pocket Mutation and Polyclonal Alterations Converging on RAS-MAPK Reactivation. Cancer Discov 11, 1913–1922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho CSL et al. , HER2 mediates clinical resistance to the KRAS(G12C) inhibitor sotorasib, which is overcome by co-targeting SHP2. Eur J Cancer 159, 16–23 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Ryan MB et al. , Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res 26, 1633–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Shokat KM, Bifunctional Small-Molecule Ligands of K-Ras Induce Its Association with Immunophilin Proteins. Angew Chem Int Ed Engl 58, 16314–16319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banaszynski LA, Liu CW, Wandless TJ, Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc 127, 4715–4721 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Liu J et al. , Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Shigdel UK et al. , Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface. Proc Natl Acad Sci U S A 117, 17195–17203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanglier JJ et al. , Sanglifehrins A, B, C and D, novel cyclophilin-binding compounds isolated from Streptomyces sp. A92-308110. I. Taxonomy, fermentation, isolation and biological activity. J Antibiot (Tokyo) 52, 466–473 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Hallin J et al. , The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canon J et al. , The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Weiss A et al. , Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C. Cancer Discov 12, 1500–1517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C et al. , The G protein signaling regulator RGS3 enhances the GTPase activity of KRAS. Science 374, 197–201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santana-Codina N et al. , Defining and Targeting Adaptations to Oncogenic KRAS(G12C) Inhibition Using Quantitative Temporal Proteomics. Cell Rep 30, 4584–4599 e4584 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Lou K et al. , KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Sci Signal 12, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pua KH, Stiles DT, Sowa ME, Verdine GL, IMPDH2 Is an Intracellular Target of the Cyclophilin A and Sanglifehrin A Complex. Cell Rep 18, 432–442 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Guo Z et al. , Rapamycin-inspired macrocycles with new target specificity. Nat Chem 11, 254–263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koltun ES et al. , Direct targeting of KRASG12X mutant cancers with RMC-6236, a first-in-class, RAS-selective, orally bioavailable, tri-complex RASMULTI (ON) inhibitor. Cancer Research 82, 3597–3597 (2022). [Google Scholar]
- 30.Fraser JS et al. , Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran TH et al. , KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat Commun 12, 1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W, Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ et al. , Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long F et al. , AceDRG: a stereochemical description generator for ligands. Acta Crystallogr D Struct Biol 73, 112–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov GN et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liebschner D et al. , Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dornan J et al. , Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans. J Biol Chem 274, 34877–34883 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Szep S, Park S, Boder ET, Van Duyne GD, Saven JG, Structural coupling between FKBP12 and buried water. Proteins 74, 603–611 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pacold ME et al. , Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Sondermann H et al. , Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119, 393–405 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Huang L, Hofer F, Martin GS, Kim SH, Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol 5, 422–426 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Tang Z et al. , GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zydowsky LD et al. , Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci 1, 1092–1099 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. P. G. Consortium, AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakravarty D et al. , OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.