Abstract
The Rat Genome Database (RGD, https://rgd.mcw.edu) has evolved from simply a resource for rat genetic markers, maps, and genes, by adding multiple genomic data types and extensive disease and phenotype annotations and developing tools to effectively mine, analyze, and visualize the available data, to empower investigators in their hypothesis-driven research. Leveraging its robust and flexible infrastructure, RGD has added data for human and eight other model organisms (mouse, 13-lined ground squirrel, chinchilla, naked mole-rat, dog, pig, African green monkey/vervet, and bonobo) besides rat to enhance its translational aspect. This article presents an overview of the database with the most recent additions to RGD’s genome, variant, and quantitative phenotype data. We also briefly introduce Virtual Comparative Map (VCMap), an updated tool that explores synteny between species as an improvement to RGD’s suite of tools, followed by a discussion regarding the refinements to the existing PhenoMiner tool that assists researchers in finding and comparing quantitative data across rat strains. Collectively, RGD focuses on providing a continuously improving, consistent, and high-quality data resource for researchers while advancing data reproducibility and fulfilling Findable, Accessible, Interoperable, and Reusable (FAIR) data principles.
Keywords: Rattus norvegicus, Rat Genome Database, genomics, quantitative phenotype, FAIR data, rat genetics, rat strain, comparative genome
Introduction
The laboratory rat (Rattus norvegicus) is an essential human disease model commonly used in translational research for understanding human physiology and afflictions (Amberger and Hamosh 2017; Justice and Sanchez 2018; Smith et al. 2019; Carter et al. 2020; Smith et al. 2020). With the ongoing efforts on the rat reference genome assembly (current—mRatBN7.2) (Howe et al. 2021; De Jong et al. 2022), strain-specific sequencing [Wistar Kyoto (WKY), spontaneously hypertensive (SHR), and spontaneously hypertensive stroke-prone (SHRSP) have been sequenced recently], and new variant discovery, the Rat Genome Database (RGD) (https://rgd.mcw.edu), which was established in 1999 (Shimoyama et al. 2015) as a centralized public source of these types of information on the laboratory rat, has undergone continuous additions and improvements. As such, RGD has become the principal source of information on rat genetic, genomics, and phenotypic data for the rat research community and other researchers (Fang et al. 2022; Ran et al. 2022; Wood et al. 2022; Ye et al. 2022). RGD offers information on genes, strains, markers, variants, quantitative trait loci (QTLs), expression data, protein structures, pathways, simple sequence length polymorphisms (SSLPs), ontologies, and sequences (Table 1). RGD is also the official nomenclature authority for rat genes, strains, and QTLs, coordinating nomenclature with the HUGO Gene Nomenclature Committee (HGNC) and the Mouse Genome Database (Bruford et al. 2020; Blake et al. 2021) with rat and mouse sharing nomenclature guidelines. As a major model organism knowledgebase, RGD is a founding member of the Alliance of Genome Resources (Alliance of Genome Resources Consortium 2022) and has recently been designated as a Global Core Biodata Resource by the Global Biodata Coalition.
Table 1.
Number of total data records for all the species in the Rat Genome Database (retrieved Jan 2023).
| Type | Total | Species | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rat | Human | Mouse | Chinchilla | Bonobo | Dog | Squirrel | Pig | African green monkey | Naked mole-rat | ||
| Genes | 680,219 | 67,810 | 158,097 | 76,851 | 33,347 | 41,717 | 53,152 | 36,372 | 32,237 | 41,662 | 40,394 |
| SSLPs/markers | 425,484 | 50,171 | 320,148 | 55,165 | |||||||
| Strains | 4,218 | 4,218 | |||||||||
| QTLs | 11,134 | 2,395 | 1,911 | 6,828 | |||||||
| Proteins | 968,668 | 55,351 | 217,011 | 88,912 | 25,257 | 43,653 | 129,485 | 25,491 | 336,793 | 19,526 | 27,189 |
| Sequences | 619,833 | 233,697 | 323,726 | 58,378 | |||||||
| Maps | 81 | 17 | 21 | 14 | 2 | 4 | 9 | 3 | 4 | 4 | 2 |
| Cell lines | 144,609 | 2,326 | 106,506 | 25,300 | 33 | 864 | 2 | 304 | 53 | 12 | |
| References | 139,526 | ||||||||||
| Exons | 33,813,533 | 2,997,016 | 12,478,387 | 7,299,221 | 1,027,506 | 1,446,827 | 4,072,276 | 964,798 | 2,260,326 | 727,477 | 539,699 |
| Promoters | 147,219 | 12,720 | 66,331 | 60,623 | 7,545 | ||||||
| 5′ UTRs | 2,010,944 | 262,085 | 470,177 | 397,267 | 102,417 | 104,892 | 331,475 | 94,710 | 98,870 | 96,028 | 53,023 |
| 3′ UTRs | 1,503,009 | 209,318 | 359,794 | 293,239 | 78,324 | 75,538 | 222,868 | 74,571 | 85,176 | 66,680 | 37,501 |
| Transcripts | 4,540,795 | 352,207 | 1,754,553 | 899,996 | 101,546 | 191,321 | 673,128 | 113,173 | 220,473 | 135,412 | 98,986 |
| Variants | 107,686,428 | 77,394,323 | 1,823,947 | 28,468,158 | |||||||
In addition to being the rat model organism database, RGD is an integrated comparative genomics resource that includes additional species (human, mouse, 13-lined ground squirrel, chinchilla, naked mole-rat, dog, pig, African green monkey/vervet, and bonobo) (Shimoyama et al. 2016; Kaldunski et al. 2022). While RGD's paradigm is to connect rat data with that of human and mouse, RGD's infrastructure makes the addition of other mammalian species relatively straightforward. Additional species have been included in RGD based on user requests with the criteria that they must be model organisms for human disease-related research which RGD actively curates (Kaldunski et al. 2022) and that they do not have a dedicated genome database.
RGD provides 15 Disease Portals (https://rgd.mcw.edu/rgdweb/portal/index.jsp) including Aging & Age-Related Disease, Cancer & Neoplastic Disease, Cardiovascular Disease, COVID-19 (Wang et al. 2022), Developmental Disease, Diabetes, Hematologic Disease, Immune & Inflammatory Disease, Infectious Disease, Liver Disease, Neurological Disease, Obesity & Metabolic Syndrome, Renal Disease, Respiratory Disease, and Sensory Organ Disease, as integrated disease research platforms.
Substantial data at RGD come from the literature through manually curated gene, strain, and QTL annotations using gene, disease, phenotype, gene–chemical interaction (ChEBI), and molecular pathway ontologies for rat; disease, ChEBI, and pathway ontologies for mouse and human; and mammalian and human phenotype ontologies (MP and HP) for rat and human, using RGD's curation tool (Hayman et al. 2016). The annotations from rat, mouse, and human at RGD are propagated to all orthologs in other RGD species via the Inferred from Sequence Orthology (ISO) evidence code where appropriate (Kaldunski et al. 2022). Moreover, RGD integrates additional data through automatic pipelines from various resources, and experimental results are also submitted directly to RGD by research community members, with the data being updated weekly. The electronic data resources at RGD, along with their sources, are listed in Table 2. Additionally, Table 3 lists the total number of genes (protein-coding and non–protein-coding) for each species at RGD, the number of genes that are unique to the NCBI and Ensembl (includes genes that have only one of them and not the other as the source of the gene record in RGD), and the number of genes with IDs shared across both NCBI and Ensembl (includes genes present in both NCBI and Ensembl). To analyze this rich resource of integrated data, RGD provides a suite of analysis and visualization tools (https://rgd.mcw.edu/wg/tool-menu/) for comparative genomics that includes OntoMate for literature search (Liu et al. 2015), Multi Ontology Enrichment Tool (MOET) for ontology enrichment (Vedi et al. 2022), PhenoMiner for finding rat quantitative data (Wang et al. 2015), Variant Visualizer for exploring variants, and JBrowse and GViewer genome browsers to view genes and other data mapped to the genome in their genomic context (Skinner et al. 2009; Laulederkind et al. 2019). Furthermore, RGD has now updated both the PhenoMiner tool (https://rgd.mcw.edu/rgdweb/phenominer/ontChoices.html), which offers more functionality and added data, and Virtual Comparative Map (VCMap) (https://rgd.mcw.edu/vcmap/, currently a beta version), a web-based tool to explore synteny between rat, mouse, and human. This article focuses on updates of RGD disease, phenotypic and genomic data, and RGD tools to analyze that information.
Table 2.
A list of external data resources for the Rat Genome Database.
| Imported data type | Resource |
|---|---|
| Gene (GO) annotations | Gene Ontology Consortium (human and mouse) (The Gene Ontology Consortium 2021) and UniProt-GOA (rat) (The Gene Ontology Consortium 2019), Mouse Genome Informatics (MGI) (mouse) (BLAKE et al. 2021), and European Bioinformatics Institute (EBI) (human, 13-lined ground squirrel, chinchilla, naked mole-rat, dog, pig, African green monkey/vervet, and bonobo) (Cantelli et al. 2022) |
| Mammalian phenotype (MP) annotations | MGI (mouse) (Blake et al. 2021) and Online Mendelian Inheritance in Animals (OMIA) (dog) (Nicholas 2021) |
| Disease (DO) annotations | Online Mendelian Inheritance in Man (OMIM) (human) (Amberger and Hamosh 2017), MGI (human and mouse), OMIA (dog and pig), ClinVar (human) (Landrum et al. 2020), and Comparative Toxicogenomics Database (CTD) (human) (Davis et al. 2019) |
| Gene–chemical interaction (ChEBI) annotations | CTD (human) (Davis et al. 2019) |
| Human phenotype (HP) annotations | Human Phenotype Ontology group (Köhler et al. 2021) and ClinVar (human) (Landrum et al. 2020) |
| Pathway (PW) annotations | Small Molecule Pathway Database (SMPDB) (human) (Jewison et al. 2014) |
| RNA-Seq expression data | Gene Expression Omnibus (GEO) (rat and human) (Clough and Barrett 2016), Expression Atlas (rat, mouse, human dog, African green monkey, pig) (Moreno et al. 2022), and Genotype-Tissue Expression (GTEx) (human) (GTEx Consortium 2020) |
| Predicted protein structures | AlphaFold (all RGD species) (Jumper et al. 2021) |
| miRNA | mRNA targets—miRGate (all RGD species) (Andrés-Leó et al. 2015) |
| Variants | Genome-wide association studies (GWAS) Catalog (human) (Sollis et al. 2022), European Variation Archive (EVA) (rat, dog, African green monkey, mouse, and pig) (Cezard et al. 2022), and Dog10K genomes project (Ostrander et al. 2019) |
| Gene records, gene positions, gene model definitions (the intron/exon structures for the genes) | NCBI (Brown et al. 2015) and Ensembl (Cunningham et al. 2022) |
| Gene nomenclature | The primary sources: HGNC (human), MGI (mouse), and VGNC (other RGD species; https://vertebrate.genenames.org/) (Bruford et al. 2020); the secondary sources if primary unavailable—NCBI and Ensembl |
| Retired/archived data | From resources that are no longer freely accessible, such as the Pathway Interaction Database (Schaefer et al. 2009), Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathway annotations (Kanehisa and Goto 2000), and the Genetic Association Database (GAD) (Becker et al. 2004) |
Table 3.
The total number of genes at the Rat Genome Database by species and the number of genes from NCBI and Ensembl (retrieved Feb 2023).
| Species | Total number of genes by species | Genes unique to NCBI | Genes unique to Ensembl | a Genes shared between NCBI and Ensembl | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All gene types | Protein-coding only | Non–protein-coding | All gene types | Protein-coding only | Non–protein-coding | All gene types | Protein-coding only | Non–protein-coding | All gene types | Protein-coding only | Non–protein-coding | |
| Human | 158,609 | 21,077 | 137,532 | 97,826 | 1,175 | 96,651 | 18,330 | 438 | 17,892 | 42,453 | 19,464 | 22,989 |
| Mouse | 76,816 | 26,675 | 50,141 | 18,778 | 3,388 | 15,390 | 15,430 | 329 | 15,101 | 42,608 | 22,958 | 19,650 |
| Rat | 66,780 | 27,858 | 38,922 | 27,060 | 3,127 | 23,933 | 13,494 | 3,572 | 9,922 | 26,225 | 21,159 | 5,066 |
| Chinchilla | 33,350 | 20,869 | 12,481 | 13,261 | 4,102 | 9,159 | 3,363 | 333 | 3,030 | 16,726 | 16,434 | 292 |
| Bonobo | 41,718 | 22,519 | 19,199 | 18,619 | 5,272 | 13,347 | 3,486 | 165 | 3,321 | 19,613 | 17,082 | 2,531 |
| Dog | 53,154 | 24,198 | 28,956 | 28,273 | 4,797 | 23,476 | 2,748 | 490 | 2,258 | 22,133 | 18,911 | 3,222 |
| Squirrel | 36,379 | 20,818 | 15,561 | 17,245 | 3,694 | 13,551 | 1,961 | 69 | 1,892 | 17,173 | 17,055 | 118 |
| Pig | 32,240 | 21,107 | 11,133 | 10,817 | 1,858 | 8,959 | 1,717 | 253 | 1,464 | 19,706 | 18,996 | 710 |
| Vervet | 41,664 | 23,712 | 17,952 | 19,121 | 5,976 | 13,145 | 6,776 | 2,054 | 4,722 | 15,767 | 15,682 | 85 |
| Naked mole-rat | 40,397 | 20,295 | 20,102 | 20,518 | 3,741 | 16,777 | 2,950 | 307 | 2,643 | 16,929 | 16,247 | 682 |
Refers to the number of genes with shared IDs.
Data
Genome sequence and variants
The rat research community requires continuously updated and improved genomic resources to strengthen their studies. The first rat genome sequence, RGSC/Rnor 2.0, was that of the BN/NHsdMcwi strain, released in November 2002, followed by the first published version of the rat reference genome sequence, RGSC3.1 (completed by the Rat Genome Sequencing Project Consortium) (Gibbs et al. 2004; Lindblad-Toh 2004), and subsequently, several improved versions have been released. In 2020, the latest enhanced rat reference genome, mRatBN7.2, was sequenced and assembled by the Darwin Tree of Life Project at the Wellcome Sanger Institute as a part of the Vertebrate Genomes Project (VGP) (Howe et al. 2021) (Fig. 1a). The assembly was derived from an inbred male from the same colony of Brown Norway rats (BN/NHsdMcwi) used in the earlier rat reference genome assembly versions. The new BN rat reference genome used a variety of sequencing technologies, including Pacific Biosciences continuous long reads, 10× linked reads, BioNano maps, and Arima Hi-C super scaffoldings. This latest version of the assembly was corrected by manual curation for misjoins and removal of haplotypic duplications which allowed a mean genome coverage of ∼92×. This assembly is substantially improved compared with previous assemblies, comprising 175 scaffolds and a Benchmarking Universal Single-Copy Orthologs (BUSCO) completeness score of 96.25%, (the previous assembly Rnor_6.0 BUSCO score is 93.76%), and contiguity is much improved (Worley et al. 2008; Howe et al. 2021; De Jong et al. 2022). In comparison, the BUSCO scores for the human (assembly name: GRCh38.p14) and mouse genome assemblies (assembly name: GRCm39) are 99.2 and 99.5% (https://www.ncbi.nlm.nih.gov/assembly/), respectively (Church et al. 2011; Chin and Khalak 2019; Nurk et al. 2022).
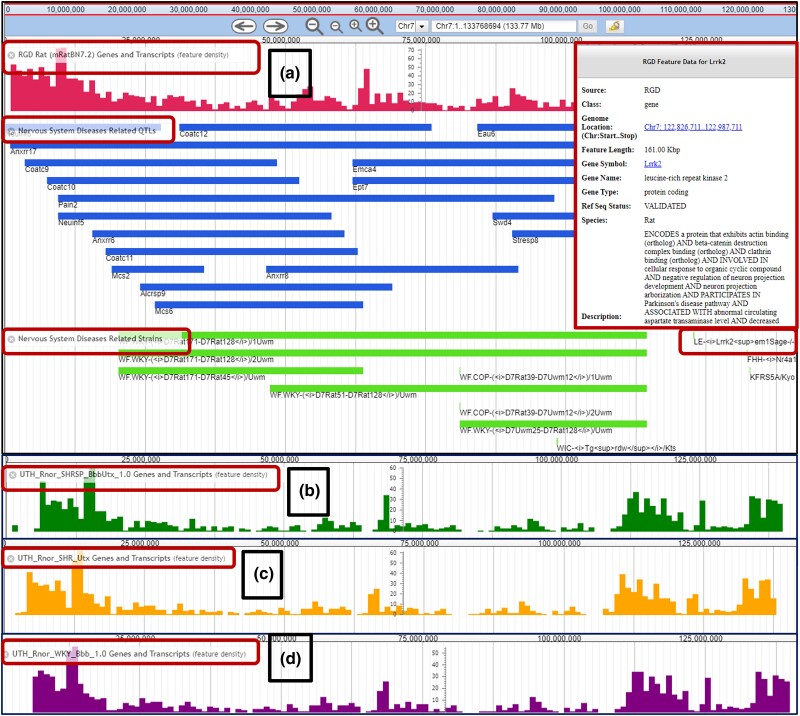
Fig. 1.
The Rat Genome Database JBrowse genome browser displaying a) feature density of mRatBN7.2 genes and transcripts, nervous system disease-related QTLs and strains, and feature data of the Lrrk2 gene; b) feature density of SHRSP/BbbUtx genes and transcripts; c) feature density of SHR/Utx; and d) feature density of WKY/Bbb in chromosome 7.
It is reported that the Brown Norway strain is separated phylogenetically from other rat strains (Saar et al. 2008). Therefore, other reference-quality rat strain genome assemblies for strains of rats that are more closely related to one another were developed (Fig. 1b–d) (Kalbfleisch et al. 2023). Among those, the three rat strain genome assemblies added to RGD since 2020 include SHRSP/BbbUtx (Fig. 1b) (NCBI Accession: PRJNA793432; Strain ID: RGD:8142383), SHR/Utx (Fig. 1c) (NCBI Accession: PRJNA825507; Strain ID: RGD:8142385), and WKY/Bbb (Fig. 1d) (NCBI Accession: PRJNA825508; Strain ID: RGD:1581635). These rat strains were selectively bred to fully explore the genetic basis of hypertension, cardiovascular disease, cerebrovascular disease, and many other polygenic or complex diseases (Doris 2017). Their assemblies were de novo compiled reference-quality genomes and were not constructed by aligning reads to the latest rat reference genome assembly, mRatBN7.2.
When compared with mRatBN7.2, the three assemblies have identical GC content (41.5%) but are larger in genome size (2.9 Gb vs 2.6 Gb). The SHR/Utx assembly, derived from the spontaneously hypertensive male rat of the stroke- and renal injury-resistant SHR-B2 lineage, has 2151 scaffolds and 3407 contigs. The genome assembly of the stroke- and renal injury-prone spontaneously hypertensive rat strain SHRSP/BbbUtx (BUSCO score 96.22%, 2734 scaffolds and 4344 contigs) is similar in contiguity and completeness to the current rat reference genome, mRatBN7.2 (Kalbfleisch et al. 2023). The WKY/Bbb genome assembly, which is closely related to SHR and SHRSP and is an accepted control strain for studying hypertension and cardiovascular disease, has the highest number of contigs (5009) and is derived from a 52-week-old male R. norvegicus sample. All these assemblies were generated by the University of Kentucky as a part of the Inbred Rat Genome Sequencing Project (UTH/UK/UofL).
Extensive genome data are available for use and download at RGD and are supported by the tools provided for genome visualization, such as JBrowse (https://rgd.mcw.edu/jbrowse/) (Skinner et al. 2009; Laulederkind et al. 2011). RGD currently maintains JBrowse instances for all versions of the rat reference assembly, the newly added strain-specific assemblies (SHR, SHRSP, WKY), and assemblies for all other species represented in RGD. For all other rat assemblies and human and mouse genomes, instances of JBrowse at RGD include disease-related tracks that show data objects annotated to diseases in particular categories, tracks showing genes that interact with classes of drugs and chemicals, genes and transcript locations and structures from both NCBI and Ensembl, gene–chemical interactions tracks, QTLs, reference sequences, and variants that are available (Shimoyama et al. 2016; Smith et al. 2020). In Fig. 1a, nervous system-related disease strains and QTLs present on chromosome 7 in the mRatBN7.2 genome assembly are shown as an example. Of note, the Lrrk2 gene was one of the genes associated with nervous system-related diseases and was found at this locus using JBrowse disease-related tracks. We will use the Lrrk2 gene as a use case throughout the manuscript.
Variation information in JBrowse includes rat variants recently acquired from the European Variant Archive (EVA) (Cezard et al. 2022) (also available for most assemblies for mouse, pig, vervet, and dog), microsatellite markers (also for human), and rat strain-specific variants. Strain-specific variants for the mRatBN7.2 assembly are derived through a collaboration with the Hybrid Rat Diversity Program (HRDP) (Tabakoff et al. 2019). Variants for human in JBrowse are from ClinVar (Landrum et al. 2020) and dbSNP (Sherry et al. 2001).
In addition to the variant information available in JBrowse, RGD's Gene/Variant report pages and Variant Visualizer tool offer the ability to explore rat strain-specific, dog breed-specific, and human clinical and genome-wide association study (GWAS) variants available in RGD data. Rat variants that appear in Variant Visualizer and on the main RGD website encompass data for unique strains and sub-strains submitted by researchers and derived from bioinformatic analyses at RGD. Notably, the data for the mRatBN7.2 assembly include variants from strains that are part of the HRDP (Tabakoff et al. 2019) and the Heterogeneous Stock (HS) Founder Strains (Hansen and Spuhler 1984; Solberg Woods et al. 2010). Variant Visualizer also includes rat variants mapped to the Rnor5.0, Rnor6.0 and mRatBN7.2 assemblies imported from EVA. RGD's human Variant Visualizer provides data from both ClinVar and, most recently, the National Human Genome Research Institute (NHGRI)–European Bioinformatics Institute (EBI) GWAS Catalog (Buniello et al. 2019) (https://www.ebi.ac.uk/gwas/). For dog, breed-specific variants were imported from the first phase of the Dog10K genomes project (http://www.dog10kgenomes.org/). Table 4 provides an overview of the volume and sources of the types of variant data available in RGD.
Table 4.
Number of variants in the Rat Genome Database (retrieved Jan 2023).
| Species | Assembly (name) | Source | Number of variants from each source |
|---|---|---|---|
| Human | Human Genome Assembly GRCh37 (GRCh37) | ClinVar GRCh37 | 1,452,414 |
| Human Genome Assembly GRCh38 (GRCh38) | ClinVar GRCh38 | 1,451,585 | |
| GWAS Catalog GRCh38 | 70,460 | ||
| Rat | RGSC Genome Assembly v3.4 (RGSC_v3.4) | European Variation Archive | 16,656,349 |
| RGSC Genome Assembly v5.0 (Rnor_5.0) | European Variation Archive | 55,064 | |
| RGSC Genome Assembly v6.0 (Rnor_6.0) | Hybrid Rat Diversity Panel—Rnor_6.0 | 4,706,582 | |
| mRatBN7.2 Assembly (mRatBN7.2) | European Variation Archive | 9,632,470 | |
| Hybrid Rat Diversity Panel—Rnor_6.0 | 18,627,685 | ||
| Hybrid Rat Diversity Panel—mRatBN7.2 | 14,489,795 | ||
| Dog | Dog CanFam3.1 Assembly (CanFam3.1) | Variants from the Dog10K genomes project | 28,468,158 |
Rat quantitative phenotype data
In addition to qualitative annotations using established ontologies, RGD also curates quantitative phenotype data for rat strains and developed the PhenoMiner tool to query and display this data (Wang et al. 2015). In order to organize the quantitative phenotype data, RGD has implemented five different ontologies: Rat Strain Ontology (RS), Clinical Measurement Ontology (CMO), Measurement Method Ontology (MMO), Vertebrate Trait Ontology (VT), and Experimental Condition Ontology (XCO) (Laulederkind et al. 2013; Smith et al. 2013). The definition of each of these ontology terms can be found on their respective ontology browsers that can be reached from https://rgd.mcw.edu/rgdweb/ontology/search.html. Originally, the data were derived from large-scale projects such as the PhysGen Program for Genomic Applications (PGA) (Dwinell 2010) and the National BioResource Project for the Rat in Japan (NBRP-Rat) (Serikawa et al. 2009). More recently, similar large-scale data sets have been submitted directly by researchers to RGD, loaded in bulk into the database, and made available in the PhenoMiner tool (Keele et al. 2018, 2021). In addition, on an ongoing basis, RGD curators manually extract quantitative phenotype data from the literature, including a recently completed project to comprehensively curate the literature references from an extensive review article on rat models of human diseases and related phenotypes (Szpirer 2020) and a targeted curation project to expand RGD's data for the HRDP founder strains. The data in the PhenoMiner tool are updated weekly from the new data that will be added from bulk submissions and/or manual curation. Updates in the PhenoMiner tool are discussed later in this manuscript in the “Tools updates” section.
Tool updates
New VCMap tool (beta version)
RGD's VCMap (https://rgd.mcw.edu/vcmap/) tool explores the genomic positions of genes and conserved synteny between different species. Currently, only rat, mouse, and human assemblies are available, but we will include other species and assemblies in the future for comparative genomics (Kwitek et al. 2001; Twigger et al. 2002; Laulederkind et al. 2011). Using VCMap, the relative locations of multiple genes can be seen in one window and compared across these species or assemblies. The current web-based version VCMap (0.7) is an update from the previous Java-based applet, expanding the utility of this valuable tool and making it more intuitive for users (software available from RGD GitHub). Currently, we use synteny net data (pairwise alignment data) from the University of California Santa Cruz (UCSC) to determine synteny across species (Lee et al. 2022).
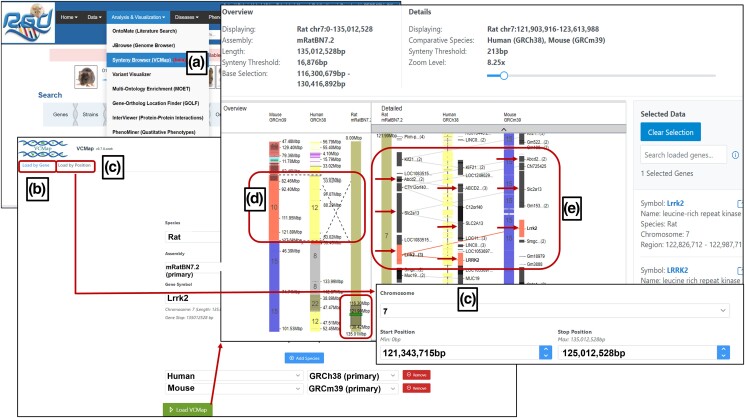
VCMap is accessible from RGD in the “Analysis and Visualization” options in the menu toolbar (Fig. 2a) and on the home page. There are two ways to begin exploring the synteny between species with VCMap, using either “Load by Gene” (Fig. 2b) or “Load by Position” (Fig. 2c) as an option from the top of the VCMap browser. Using the “Load by Gene” choice, the user selects the backbone configuration for which synteny is to be explored (includes the anchor species and assembly version) and enters the gene symbol. After that, the comparative species between rat, human, and mouse can be chosen to assess synteny. As shown in Fig. 2d and e, loading the VCMap results page displays the overview and details of the comparison information at the top, including assembly, length, comparative species, and base selection. The chromosome with the entered gene is displayed vertically, with the anchor chromosome shown (chromosome 7 in rat) rightmost in the overview and leftmost in the detailed view, arranged in increasing coordinate configuration from top to bottom. Figure 2d and e show that the human region has major syntenic blocks on chromosomes 12, 8, and 22 and the mouse has major blocks on chromosomes 10 and 15 in conserved synteny with rat chromosome 7. When mousing over the large section of the human chromosome 12 block, users can see the dashed lines linking that section to the rat chromosome are crossed, indicating that those synteny blocks are reversed relative to each other for those regions between human and rat (Fig. 2d). The circled darker area on rat chromosome 7 in the overview panel is the area in focus shown as a detailed view in Fig. 2e. Similarly, any of the regions in the overview can be viewed in detail by selecting them. However, it should be noted that the current VCMap beta version is still in active development. There will be regular updates with additional species, data types, and user interface and feature improvements.
Fig. 2.
Using Virtual Comparative Map (VCMap) to explore the Lrrk2 gene in rat, human, and mouse; a) VCMap is found in the “Analysis & Visualization” menu on the Rat Genome Database homepage; b) using the “Load by Gene” option to explore the Lrrk2 gene by gene name; c) using the “Load by Position” to explore the Lrrk2 gene by its genomic position; d) the “Overview” section displays an overview of the gene and its syntenic region; and e) the “Detailed” section provides more detailed information of the gene and syntenic region with Slc2a13 and Abcd2 genes upstream of Lrrk2 gene.
Gene/variant report pages
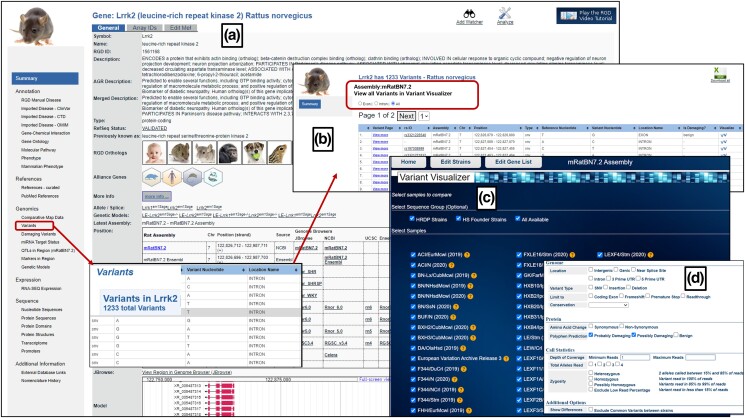
RGD users can find all gene-related information in RGD gene report pages that were recently updated and improved with more information and better navigation. RGD gene report pages show the gene name, description, RGD ID, orthologs, annotations, references, genomics, expression data, sequences, and links to additional information at other databases. In Fig. 3, the Lrrk2 gene report page (https://rgd.mcw.edu/rgdweb/report/gene/main.html?id=1561168) is presented. The left side of the page displays a scrollable summary of all the information available. The rat gene report pages provide information on orthologs for the gene with links to the corresponding species-specific gene pages in RGD. The Alliance Genes section links to pages on the Alliance of Genome Resources website for the corresponding rat gene and orthologous genes for all Alliance species (Kishore et al. 2020; Alliance of Genome Resources Consortium 2022) (Fig. 3a). Where mutations within the gene are known, as in the case of Lrrk2, the gene report links to both allele and Genetic Model (i.e. mutant strain) report pages for more information about what is known about these. A view of the gene location in the Genome Browser (JBrowse) is shown (Fig. 3a), which can be opened in full screen. The Lrrk2 gene report page also shows RGD's manual gene-disease, gene-phenotype, gene-pathway, and gene-chemical interaction annotations, along with imported annotations from Gene Ontology (GO), ClinVar, Comparative Toxicogenomics Database (CTD), Genetic Association Database (GAD), and Online Mendelian Inheritance in Man (OMIM). Users can also see the comparative map data of the different orthologs with their chromosome numbers and positions for other assemblies on the gene report pages.
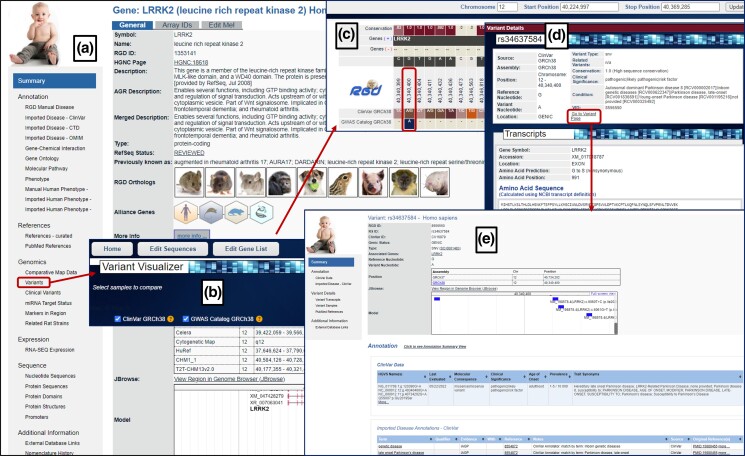
Fig. 3.
Exploring the Lrrk2 gene and its variants in rat; a) the Rat Genome Database rat gene report page for the Lrrk2 gene linking to its variants; b) variant overview page that shows all the rat Lrrk2 gene variants; c) Variant Visualizer tool to select rat strain-specific variants; d) option to choose polyphen prediction type.
Below the comparative map data section, a link to species-specific variants at RGD for the gene is presented (https://rgd.mcw.edu/rgdweb/report/gene/main.html?id=1561168#cnVariants; Fig. 3a). RGD has improved the variant data already present with recently included additional data as mentioned above (Table 4). The “Variants in Lrrk2” icon (Fig. 3a) shows an overview of the total number of variants present for the gene. Clicking on this icon takes the user to the variant overview page where all the variants associated with the gene are listed (https://rgd.mcw.edu/rgdweb/report/rsId/main.html?geneId=1561168; Fig. 3b). The list has multiple columns that include information related to the assembly, chromosome number, position, type, and reference and variant nucleotides, with each variant linked to its variant page. Where a Polyphen prediction for whether the variant is damaging to the protein function is available, this prediction is included in the table. In the future, we are considering expanding the prediction algorithms for finding the damaging variants. Links are provided to view each variant individually in Variant Visualizer. Alternatively, selecting the option to view all the variants in the Variant Visualizer tool provided at the top of this variant overview page brings the user to the Variant Visualizer landing page. Figure 3c shows the Variant Visualizer (https://rgd.mcw.edu/rgdweb/front/config.html) landing page for rat, where the option is available to select strains either individually or by sequence groups (HRDP, HS Founder strains). Selecting all available strains for the Lrrk2 gene on this page and choosing probably/possibly damaging as the option in Polyphen predictions on the following page (Fig. 3d) shows that there are two variants in the gene that are predicted to be damaging and are present in sequenced rat strains with variant data available at RGD (Fig. 4a). Clicking on the specific variant, for example, for rat strain LE/Stm, the Lrrk2 gene variant shows a Variant Details popup that includes information such as the variant type, conservation, sequencing depth, variant nucleotide, and reference nucleotide (Fig. 4b). The Variant Details pane also shows transcript location for each transcript of the gene, i.e. whether the variant is in an exon, intron, UTR, etc. For variants in the protein-coding sequence, the predicted amino acid position, change, and functional consequence, in this case, a nonsynonymous change from phenylalanine (F) to cysteine (C) and the predicted protein sequence with the variant amino acid embedded are provided. Where available, Polyphen predictions and the detailed output of the tool are also shown. For each variant in the Variant Visualizer, the details popup links to the corresponding variant report page, which gives further information (Fig. 4c).
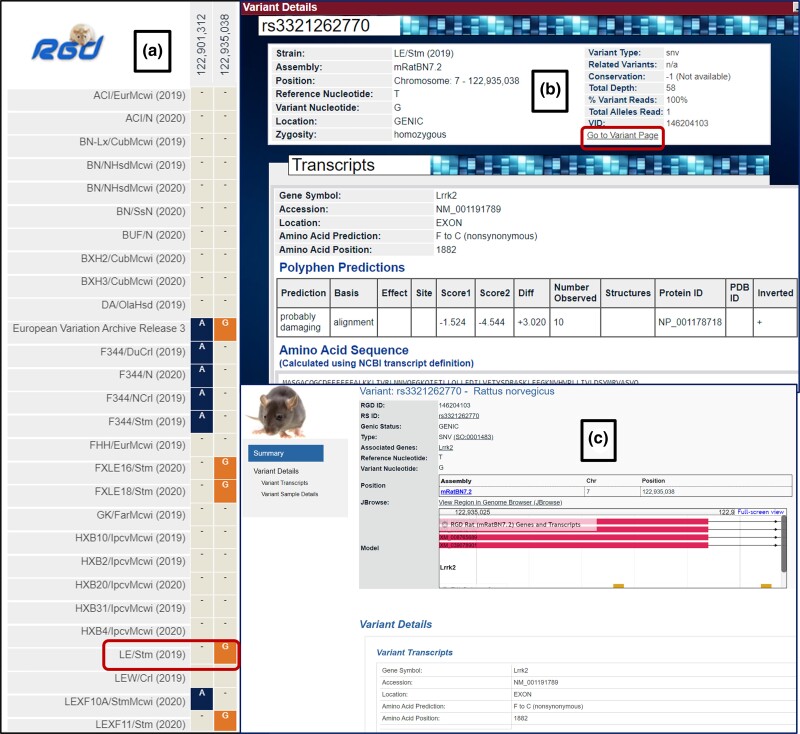
Fig. 4.
Exploring the Lrrk2 gene variants in rat using the Variant Visualizer tool; a) results page from Variant Visualizer tool displays the rat strain-specific damaging variants for the Lrrk2 gene; b) clicking on the LE/Stm rat strain opens the Variant Details display that has the “Go to Variant Page” link that; c) opens the variant report page for that variant.
The rat variant report pages (https://rgd.mcw.edu/rgdweb/report/variants/main.html?id=146204103) display the variant with its unique RGD ID (146204103 in Fig. 4c) and reference SNP (rs) ID (that links to EVA for that variant). This page provides the choice to view the variant position in the Genome Browser (JBrowse) and includes information such as transcripts and variant sample details. The sample detail section shows the strains that the variant has been found in, along with the information on the variant allele depth and zygosity. The strain/sample names link to Variant Visualizer for that variant in that strain.
Similarly, using the human gene report page (https://rgd.mcw.edu/rgdweb/report/gene/main.html?id=1353141), human variants in the LRRK2 gene can be explored (Fig. 5a and b). Figure 5c and d show that variant rs34637584 (https://rgd.mcw.edu/rgdweb/report/variants/main.html?id=8556550), which can be found in both the ClinVar and GWAS Catalog data sets, is designated as pathogenic in ClinVar. The variant is predicted to be linked with several nervous system diseases. The variant report page can be accessed using the “Go to variant report page” link. The page also previews the variant's location in RGD's Genome Browser (JBrowse). The annotation details imported from ClinVar present their clinical significance and disease associations. These disease associations have been translated at RGD into the Disease Ontology (DO) annotations shown in Fig. 5e. Similar information about the variant's associations with disease traits imported from the GWAS Catalog is also provided (not shown). In addition, the Variant Details section listed in the summary on the left side of the page provides the same information seen in the Variant Visualizer's Variant Details popup mentioned previously. Finally, for researchers interested in more information, links to references and external databases such as OMIM and dbSNP are available (Fig. 5e).
Fig. 5.
Exploring the LRRK2 gene and its variants in human with a) its gene report page; b) Variant Visualizer; c) results from the Variant Visualizer showing variants from ClinVar and GWAS Catalog; d) Variant Details display and “Go to Variant Page” that; e) opens up the variant report page containing additional information.
PhenoMiner
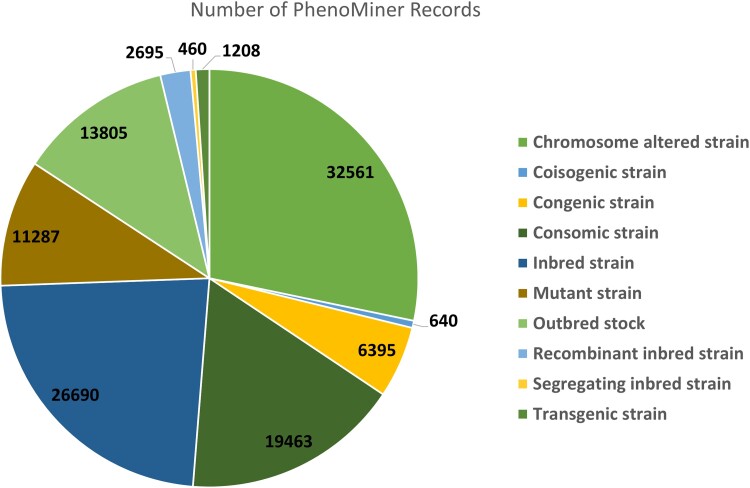
The PhenoMiner tool (https://rgd.mcw.edu/rgdweb/phenominer/ontChoices.html) is a unique quantitative phenotypic data repository and search engine developed at RGD to complement the qualitative phenotype data that RGD has accumulated and presented since its establishment (Laulederkind et al. 2013; Wang et al. 2015; Zhao et al. 2019). It contains quantitative phenotypic data from about 1400 rat strains categorized into chromosome-altered, coisogenic, congenic, consomic, inbred, mutant, recombinant inbred, segregating inbred and transgenic strains, and outbred stocks, quantitated by different measurement methods in various experimental conditions (Fig. 6). The PhenoMiner tool includes results from high-throughput phenotyping projects (PGA, NBRP, HRDP), bulk data loads submitted by research groups (Keele et al. 2018, 2021), and manual curation efforts using the in-house PhenoMiner curation tool (Wang et al. 2015).
Fig. 6.
Number of PhenoMiner records for different rat strain categories.
PhenoMiner facilitates data exploration within and across different studies and strains by querying using standardized Rat Strain (RS), Clinical Measurement (CMO), Measurement Method (MMO), Experimental Condition (XCO), and Vertebrate Trait (VT) ontology terms. RS provides a structured representation of the relationships between strains to facilitate selections, CMO contains terms that represent what was measured, MMO terms represent how the measurement was done, XCO terms represent the conditions under which the measurements were made, and VT terms are used to categorize measurable or observable characteristics of a vertebrate organism (Laulederkind et al. 2013; Laulederkind et al. 2019). Between 2020 and 2022, RGD curators created nearly 200 new terms for PhenoMiner.
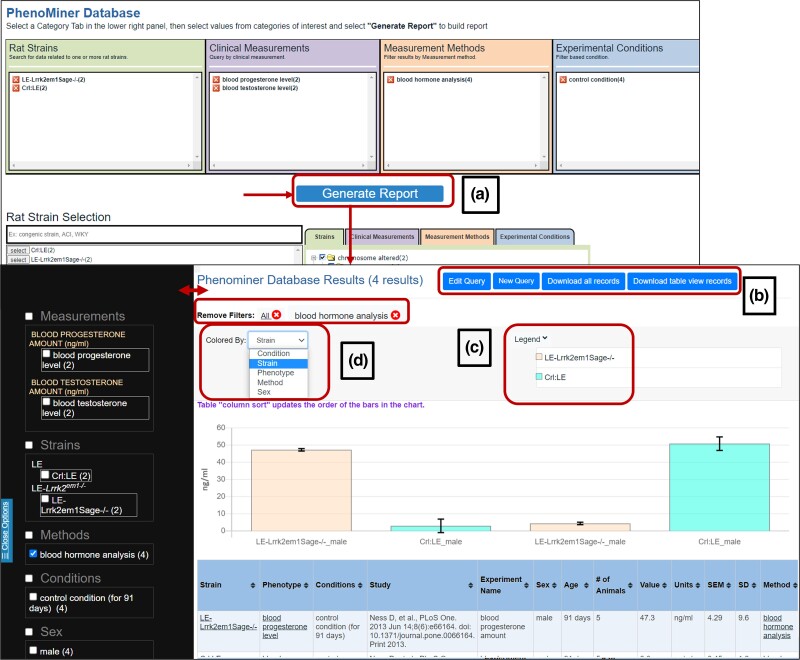
Users can reach the PhenoMiner user interface using the “Analysis & Visualization” tab in the menu at the top of any RGD page (Fig. 2a). This newer version of PhenoMiner has all the ontology sections on its main page. In the bottom section of the page, options in the Rat Strain Selections load from the Rat Strain Ontology automatically, although users can start with any of the other ontologies by selecting the appropriate tabs below the “Generate Report” button on the same page. The selections that are made in the bottom panels appear in the boxes at the top in their respective ontology group. The “Generate Report” button shows the results from the selected options (Fig. 7a).
Fig. 7.
Exploring different rat strain measurements using PhenoMiner: a) PhenoMiner interface that shows options to select strains, clinical measurements, measurement methods, and experimental methods to “Generate Report”; b) option to edit query and download results; c) legend reflects the color represented in the graph; and d) the graph can be colored by strain, condition, phenotype, method, and sex.
Using gene/variant report pages and Variant Visualizer, we established that the Lrrk2 gene contains a damaging variant in LE rats. Also, in human, the LRRK2 gene variant is predicted to be linked with nervous system-related diseases (Figs. 3–5). To explore this further, we searched in PhenoMiner for brain function-related hormones progesterone and testosterone levels in the blood in Crl:LE and LE-Lrrk2em1Sage−/− rat strains (Fig. 7a). The results page shows the table and graph of the results (Fig. 7). Options for further filtering the results are provided in the left sidebar (Fig. 7). When the results use more than one unit, a graph is not displayed on the page until appropriate selections to limit the results to those with the same units are made. The filters on the left side of the page are linked to the graph and table, and selections that are made appear concurrently in both the graph and table. There are options to download using “Download all records” to retrieve the unfiltered result set and “Download table view records” to save the results of a filtered query (Fig. 7b). The graph provides the option to color the bars based on several choices with the legend displaying what each color corresponds to. The bars in Fig. 7c are colored by the strain as seen from the legend, but options to color by condition, method, phenotype, and sex (Fig. 7d) are also available. The table can be sorted by the values in any of the columns, as indicated by the up and down arrows in the column headers.
Disease Portal updates
RGD has focused almost since its inception on providing its users with disease-related data and resources for their research. Therefore, Disease Portals (https://rgd.mcw.edu/rgdweb/portal/index.jsp) were created that consolidate disease-associated data, including genes, strains, and QTLs for a selected disease category integrated at RGD (Hayman et al. 2016), were created. The customized ontology browser in each of the 15 Disease Portals shows relevant disease terms with less specific parent and more specific child terms. The annotations associated with the selected disease term can also be viewed with links to the ontology report page, allowing users to find and compare annotations across different RGD data types and species. RGD Disease Portals also have RGD tools that provide disease-associated gene analysis and visualization, such as MOET (Vedi et al. 2022) and GViewer.
With the continual and rapid progress in the discovery of gene–disease associations and ontology development, the disease gene data at RGD also need to be constantly updated. RGD curators regularly update the information in the Disease Portals that was created in the past. Following a recently completed project to expand the data in the Cancer & Neoplastic Disease Portal, an update of the Cardiovascular Disease Portal is currently in progress.
Being FAIR at RGD
The Findable, Accessible, Interoperable, and Reusable (FAIR) data principles are essential for data repositories to improve their discovery and usability (Wilkinson et al. 2016). The findability aspect requires data to have unique identifiers used explicitly and recorded systematically in searchable resources for easy retrieval. Accessibility is ensured by having the data open, free, and available to everyone. The use of standardized vocabularies and data maintains interoperability. Reusability requires data to have a clear and accessible license for its usage and to maintain provenance (Jauer and Deserno 2020).
RGD offers a centralized platform of resources and tools that can be found and interrogated to unravel innovative prospects and develop scalability. The goal of RGD has been to provide open access standardized data, and it has been working all along to be a “FAIR resource” following the FAIR data principles (https://fairsharing.org/1951) to enhance its usability. RGD data are findable and accessible through multiple channels, including the RGD web application (https://rgd.mcw.edu), representational state transfer application programming interfaces (REST APIs) (https://rest.rgd.mcw.edu/rgdws/swagger-ui.html), file download site (https://download.rgd.mcw.edu/data_release), and through multiple third parties that import RGD data (including NCBI, Monarch, Ensembl, GO) (Wilkinson et al. 2016). In addition, RGD assigns globally unique persistent identifiers for genes, strains, and other genomic objects. The identifier format at RGD is registered at https://bioregistry.io/ (Hoyt et al. 2022) and https://identifiers.org/ (Wimalaratne et al. 2018) to avoid any potential conflicts. All RGD data are open and also available on the download site (https://download.rgd.mcw.edu/data_release), including genes for all the RGD species; miRNA targets for human, rat, and mouse; QTLs for rat, mouse, and human; genome annotation (in GFF format) and rat strains; SSLPs for rat, mouse, and human; and variants as VCF files for rat. Orthologs for all the RGD species are in their respective folders. All ontology annotations across ontologies, species, and data types are provided in both a format that closely follows the Gene Ontology's GAF2.2 specification and in expanded “with_terms” files that include additional information. For ontologies developed by RGD, the ontology files themselves in both the OBO (Jackson et al. 2021) and OWL (https://www.w3.org/2001/sw/wiki/OWL) formats are available on the download site and in GitHub (https://github.com/rat-genome-database). Software is licensed under the GPL v3 and is available for download via GitHub, which is updated weekly. The RGD GitHub link is present at the top right of the RGD homepage near the search bar. Data interoperability at RGD is realized using standardized ontologies for curation in addition to the use of standard biological file formats whenever possible. Reusability is ensured using the Creative Commons (CC) BY 4.0 explicit license (https://creativecommons.org/licenses/by/4.0/). RGD's publicly available REST APIs provide programmatic access to stored RGD information and annotations. Also, requests for data are repeatable and, wherever possible, include provenance for traceability. As a result of adhering to these standards and practices, RGD data can integrate with modernized informatics infrastructure for use by the research community. Thus, RGD has enhanced the exposure of its data and tools to be utilized globally.
To further data FAIRification efforts, RGD participates with other databases, such as the Gene Ontology Consortium, DO, and Human Phenotype Ontology (HPO), in developing additional and existing ontologies/vocabularies by requesting new terms, edits to existing terms, and updates to the structure of the ontologies (i.e. new term–term relationships) in addition to the expansion of five RGD-developed ontologies. At present, RGD group members have been working on mapping RGD's instance of Disease Ontology (RDO) terms to terms of the Experimental Factor Ontology (EFO) in order to integrate EFO associations from the GWAS Catalog into RGD's larger disease-related infrastructure. Additionally, RGD not only acquires and integrates data from other groups mentioned at the beginning of this article (Table 2), but also makes its data available to other groups, including the NCBI, GO, Monarch Initiative, Ensembl, and others for use by the broader biomedical research and clinical communities. RGD has facilitated bulk download and automated access to curated and other data. RGD also offers support to investigators to provide them with information on specific data sets or building data pipelines using the “Contact” link found in the menu bar on the top right of most RGD web pages or the “Send Message” option that appears on most RGD pages. RGD is a founding member of the Alliance of Genome Resources (https://www.alliancegenome.org), a consortium of the major model organism databases focused on harmonizing and presenting cross-species information. Being a member of the Alliance, rat and human data from RGD are submitted for integration into the Alliance database with data for six other essential model organisms from the other member groups, including FlyBase, Mouse Genome Informatics (MGI), Saccharomyces Genome Database (SGD), WormBase, XenBase, ZFIN, and GO (Alliance of Genome Resources Consortium 2022). RGD also participates in the Research Resource Identification Initiative (RRID: SCR_006444), an initiative of The FAIR Data Informatics Lab at UCSD (Wilkinson et al. 2016), which facilitates the identification of RGD rat strains with unique Research Resource Identifiers (RRIDs) based on the strain RGD IDs. RGD displays the unique RRIDs associated with the strains as citation IDs on the strain report pages. Using such unique identifiers allows specific resources employed within a study to be unambiguously identified, enabling the reuse of specific resources and improving the reproducibility of results. Due to the importance, and extensive use, of RGD data and tools globally, RGD has been recently identified as a Global Core Biodata Resource (https://globalbiodata.org/) by the Global Biodata Coalition, adding RGD to the “collection of 37 resources whose long-term funding and sustainability is deemed critical to life science and biomedical research worldwide.”
Future directions and conclusion
RGD is focused on integrating genetic, multi-omics, and biological data across multiple rat strains and populations and extending data integration across species to human, mouse, and other model organisms of human disease. RGD aims to create an integrated data environment for functional annotations to genes, genetic variation, genomic features, strains, and genetic loci for rats using our well-established automated and manual processes employing multiple ontologies. As part of the Genome Reference Consortium (GRC), RGD will maintain and curate the rat reference genome (currently mRatBN7.2), add sequence variant data from strains undergoing whole genome sequencing, and incorporate new strain-specific de novo assemblies and annotations. Robust transcriptome data from sequencing of total RNA, full-length transcripts, and single cell/nuclear RNA and epigenomic data from assay for transposase-accessible chromatin with sequencing (ATAC-seq), reduced representation bisulfite sequencing (RRBS), and others will be brought into RGD from many of the sequenced strains. These data, along with the robust mining, visualization, and analysis tools at RGD, will be leveraged to integrate multiple phenotypes (via phenotype and strain annotations) with genetic information (gene function, expression, regulation) within the Disease Portals. Precision Model Portals for HRDP and HS rats are being developed with integrated data across multiple strains from whole animal phenotypes to transcriptomes and epigenomes to genetic variants and individual rat genotypes.
RGD plans to create a comparative platform to integrate these data across species through their respective genomes and predict precision preclinical models through in silico comparative studies. Where not available from other sources, pairwise genome synteny maps will be generated using established methods between rat, human, mouse, and other species in RGD and the Alliance of Genome Resources to complement the existing ortholog information. Data having associated genomic locations in each species will be entered into RGD's cross-species integration tools, including genes, variants, gene regulatory regions, epigenome marks, QTLs, expression quantitative trait loci (eQTLs), GWAS, spontaneous and induced mutants. Users will be able to query across species for various data types or upload data sets for comparative analysis and visualization. RGD is also planning to integrate its data with the newer version of JBrowse (JBrowse 2.0) that offers many new features including interactive editing of configuration, ability to search by gene name/ID, and displaying multiple chromosomes in a single view. The addition of more options in gene/variant report pages at RGD is planned, along with the continuous improvement of ontologies and Disease Portals. Together, these integrated ecosystems will lead to new hypotheses about complex disease genetics and disease mechanisms. Furthermore, we have begun to assess how to make the RGD website more mobile-friendly, and we are currently working to improve the user experience on mobile devices. Overall, RGD will continue to integrate genetic and biological data across species in model organisms of human disease and into RGD tools, enhancing RGD's value as a resource for computational, translational, and clinical research.
Contributor Information
Mahima Vedi, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Jennifer R Smith, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
G Thomas Hayman, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Monika Tutaj, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Kent C Brodie, Clinical and Translational Science Institute, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Jeffrey L De Pons, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Wendy M Demos, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Adam C Gibson, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Mary L Kaldunski, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Logan Lamers, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Stanley J F Laulederkind, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Jyothi Thota, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Ketaki Thorat, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Marek A Tutaj, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Shur-Jen Wang, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Stacy Zacher, Finance and Administration, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Melinda R Dwinell, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Anne E Kwitek, The Rat Genome Database, Department of Physiology, Medical College of Wisconsin, Milwaukee, WI 53226, USA; Department of Biomedical Engineering, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The data (https://rgd.mcw.edu/wg/data-menu/) and tools (https://rgd.mcw.edu/wg/tool-menu/) described in the article are available from the RGD website (https://rgd.mcw.edu), REST APIs (https://rest.rgd.mcw.edu/rgdws/swagger-ui.html), and file download site (https://download.rgd.mcw.edu/data_release), and the software is available on the GitHub (https://github.com/rat-genome-database) link at the top right of the RGD homepage. For more information on the RGD webpages and tools, there are help pages (https://rgd.mcw.edu/wg/help3/) and video tutorials (https://rgd.mcw.edu/wg/home/rgd_rat_community_videos/rgd-tutorials/). RGD also offers virtual office hours that are available by appointment using the “Contact” link on the homepage (https://rgd.mcw.edu/rgdweb/contact/contactus.html).
Funding
RGD is grateful for funding support from the National Heart, Lung, and Blood Institute (R01HL064541) on behalf of the National Institutes of Health (NIH) and from the National Human Genome Research Institute (NHGRI) as a founding member of the Alliance of Genome Resources (U24HG010859).
Literature cited
- Alliance of Genome Resources Consortium . Harmonizing model organism data in the Alliance of Genome Resources. Genetics. 2022;220(4):iyac022. doi: 10.1093/genetics/iyac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinformatics. 2017;58(1):1.2.1–1.2.12. doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-León E, González Peña D, Gómez-López G, Pisano DG. miRGate: a curated database of human, mouse and rat miRNA–mRNA targets. Database (Oxford). 2015;2015:bav035. doi: 10.1093/database/bav035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36(5):431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Blake JA, Baldarelli R, Kadin JA, Richardson JE, Smith CL, Bult CJ; Mouse Genome Database Group . Mouse Genome Database (MGD): knowledgebase for mouse–human comparative biology. Nucleic Acids Res. 2021;49(D1):D981–D987. doi: 10.1093/nar/gkaa1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GR, Hem V, Katz KS, Ovetsky M, Wallin C, Ermolaeva O, Tolstoy I, Tatusova T, Pruitt KD, Maglott DR, et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43(D1):D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford EA, Braschi B, Denny P, Jones TEM, Seal RL, Tweedie S. Guidelines for human gene nomenclature. Nat Genet. 2020;52(8):754–758. doi: 10.1038/s41588-020-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantelli G, Bateman A, Brooksbank C, Petrov AI, Malik-Sheriff RS, Ide-Smith M, Hermjakob H, Flicek P, Apweiler R, Birney E, et al. The European Bioinformatics Institute (EMBL-EBI) in 2021. Nucleic Acids Res. 2022;50(D1):D11–D19. doi: 10.1093/nar/gkab1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CS, Richardson A, Huffman DM, Austad S. Bring back the rat!. J Gerontol A Biol Sci Med Sci. 2020;75(3):405–415. doi: 10.1093/gerona/glz298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezard T, Cunningham F, Hunt SE, Koylass B, Kumar N, Saunders G, Shen A, Silva AF, Tsukanov K, Venkataraman S, et al. The European Variation Archive: a FAIR resource of genomic variation for all species. Nucleic Acids Res. 2022;50(D1):D1216–D1220. doi: 10.1093/nar/gkab960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Khalak A. Human genome assembly in 100 minutes. bioRxiv 705616. 10.1101/705616, 17 July 2019, preprint: not peer reviewed. [DOI]
- Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9(7):e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E, Barrett T. The Gene Expression Omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, et al. Ensembl 2022. Nucleic Acids Res. 2022;50(D1):D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res. 2019;47(D1):D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong TV, Chen H, Brashear WA, Kochan KJ, Hillhouse AE, Zhu Y, Dhande IS, Hudson EA, Sumlut MH, Smith ML, et al. mRatBN7.2: familiar and unfamiliar features of a new rat genome reference assembly. Physiol Genomics. 2022;54(7):251–260. doi: 10.1152/physiolgenomics.00017.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doris PA. Genetics of hypertension: an assessment of progress in the spontaneously hypertensive rat. Physiol Genomics. 2017;49(11):601–617. doi: 10.1152/physiolgenomics.00065.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwinell MR. Online tools for understanding rat physiology. Brief Bioinformatics. 2010;11(4):431–439. doi: 10.1093/bib/bbp069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Wu J, Reho JJ, Lu KT, Brozoski DT, Kumar G, Werthman AM, Silva SD Jr, Muskus Veitia PC, Wackman KK, et al. RhoBTB1 reverses established arterial stiffness in angiotensin II-induced hypertension by promoting actin depolymerization. JCI Insight. 2022;7(9):e158043. doi: 10.1172/jci.insight.158043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium . The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium . The Gene Ontology Resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428(6982):493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium . The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res. 1984;8(5):477–479. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- Hayman GT, et al. The Disease Portals, disease-gene annotation and the RGD disease ontology at the Rat Genome Database. Database (Oxford). 2016;2016:baw034. doi: 10.1093/database/baw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Dwinell M, Shimoyama M, Corton C, Betteridge E, Dove A, Quail MA, Smith M, Saba L, Williams RW. The genome sequence of the Norway rat, Rattus norvegicus Berkenhout 1769. Wellcome Open Res. 2021;6:118. doi: 10.12688/wellcomeopenres.16854.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt CT, Balk M, Callahan TJ, Domingo-Fernández D, Haendel MA, Hegde HB, Himmelstein DS, Karis K, Kunze J, Lubiana T, et al. Unifying the identification of biomedical entities with the Bioregistry. Sci Data. 2022;9(1):714. doi: 10.1038/s41597-022-01807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R, Matentzoglu N, Overton JA, Vita R, Balhoff JP, Buttigieg PL, Carbon S, Courtot M, Diehl AD, Dooley DM. OBO Foundry in 2021: operationalizing open data principles to evaluate ontologies. Database (Oxford). 2021;2021:baab069. doi: 10.1093/database/baab069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauer ML, Deserno TM. Data provenance standards and recommendations for FAIR data. Stud Health Technol Inform. 2020;270:1237–1238. doi: 10.3233/SHTI200380. [DOI] [PubMed] [Google Scholar]
- Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, et al. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42(D1):D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice JA, Sanchez RM. A rat model of perinatal seizures provoked by global hypoxia. Methods Mol Biol. 2018;1717:155–159. doi: 10.1007/978-1-4939-7526-6_13. [DOI] [PubMed] [Google Scholar]
- Kalbfleisch TS, Hussien AbouEl Ela NA, Li K, Brashear WA, Kochan KJ, Hillhouse AE, Zhu Y, Dhande IS, Kline EJ, Hudson EA. et al. et al. The assembled genome of the stroke-prone spontaneously hypertensive rat. Hypertension. 2023; 80(1):138–146. doi: 10.1161/HYPERTENSIONAHA.122.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldunski ML, Smith JR, Hayman GT, Brodie K, De Pons JL, Demos WM, Gibson AC, Hill ML, Hoffman MJ, Lamers L, et al. The Rat Genome Database (RGD) facilitates genomic and phenotypic data integration across multiple species for biomedical research. Mamm Genome. 2022;33(1):66–80. doi: 10.1007/s00335-021-09932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele GR, Prokop JW, He H, Holl K, Littrell J, Deal A, Francic S, Cui L, Gatti DM, Broman KW, et al. Genetic fine-mapping and identification of candidate genes and variants for adiposity traits in outbred rats. Obesity (Silver Spring). 2018;26(1):213–222. doi: 10.1002/oby.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele GR, Prokop JW, He H, Holl K, Littrell J, Deal AW, Kim Y, Kyle PB, Attipoe E, Johnson AC, et al. Sept8/SEPTIN8 involvement in cellular structure and kidney damage is identified by genetic mapping and a novel human tubule hypoxic model. Sci Rep. 2021;11(1):2071. doi: 10.1038/s41598-021-81550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore R, Arnaboldi V, Van Slyke CE, Chan J, Nash RS, Urbano JM, Dolan ME, Engel SR, Shimoyama M, Sternberg PW. Automated generation of gene summaries at the Alliance of Genome Resources. Database (Oxford). 2020;2020:baaa037. doi: 10.1093/database/baaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Gargano M, Matentzoglu N, Carmody LC, Lewis-Smith D, Vasilevsky NA, Danis D, Balagura G, Baynam G, Brower AM, et al. The human phenotype ontology in 2021. Nucleic Acids Res. 2021;49(D1):D1207–D1217. doi: 10.1093/nar/gkaa1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwitek AE, Tonellato PJ, Chen D, Gullings-Handley J, Cheng YS, Twigger S, Scheetz TE, Casavant TL, Stoll M, Nobrega MA, et al. Automated construction of high-density comparative maps between rat, human, and mouse. Genome Res. 2001;11(11):1935–1943. doi: 10.1101/gr.173701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, Hoffman D, Jang W, Kaur K, Liu C, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–D844. doi: 10.1093/nar/gkz972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulederkind SFJ, et al. Phenominer: quantitative phenotype curation at the Rat Genome Database. Database (Oxford). 2013;2013:bat015. doi: 10.1093/database/bat015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulederkind SJF, Hayman GT, Wang SJ, Hoffman MJ, Smith JR, Bolton ER, De Pons J, Tutaj MA, Tutaj M, Thota J. Rat Genome Databases, repositories, and tools. Methods Mol Biol. 2019;2018:71–96. doi: 10.1007/978-1-4939-9581-3_3. [DOI] [PubMed] [Google Scholar]
- Laulederkind SJF, Shimoyama M, Hayman GT, Lowry TF, Nigam R, Petri V, Smith JR, Wang SJ, de Pons J, Kowalski G, et al. The Rat Genome Database curation tool suite: a set of optimized software tools enabling efficient acquisition, organization, and presentation of biological data. Database (Oxford). 2011;2011(0):bar002. doi: 10.1093/database/bar002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BT, Barber GP, Benet-Pagès A, Casper J, Clawson H, Diekhans M, Fischer C, Gonzalez JN, Hinrichs AS, Lee CM, et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50(D1):D1115–D1122. doi: 10.1093/nar/gkab959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K. Three's company. Nature. 2004;428(6982):475–476. doi: 10.1038/428475a. [DOI] [PubMed] [Google Scholar]
- Liu W, et al. Ontomate: a text-mining tool aiding curation at the Rat Genome Database. Database (Oxford). 2015;2015:bau129. doi: 10.1093/database/bau129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P, Fexova S, George N, Manning JR, Miao Z, Mohammed S, Muñoz-Pomer A, Fullgrabe A, Bi Y, Bush N, et al. Expression Atlas update: gene and protein expression in multiple species. Nucleic Acids Res. 2022;50(D1):D129–D140. doi: 10.1093/nar/gkab1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas FW. Online Mendelian Inheritance in Animals (OMIA): a record of advances in animal genetics, freely available on the internet for 25 years. Anim Genet. 2021;52(1):3–9. doi: 10.1111/age.13010. [DOI] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. The complete sequence of a human genome. Science. 2022;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Wang GD, Larson G, vonHoldt BM, Davis BW, Jagannathan V, Hitte C, Wayne RK, Zhang YP; Dog10K Consortium . Dog10K: an international sequencing effort to advance studies of canine domestication, phenotypes and health. Natl Sci Rev. 2019;6(4):810–824. doi: 10.1093/nsr/nwz049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Z, Yang J, Liu Y, Chen X, Ma Z, Wu S, Huang Y, Song Y, Gu Y, Zhao S, et al. Gliomarker: an integrated database for knowledge exploration of diagnostic biomarkers in gliomas. Front Oncol. 2022;12:792055. doi: 10.3389/fonc.2022.792055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P, et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40(5):560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(suppl_1):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikawa T, Mashimo T, Takizawa A, Okajima R, Maedomari N, Kumafuji K, Tagami F, Neoda Y, Otsuki M, Nakanishi S, et al. National BioResource Project-Rat and related activities. Exp Anim. 2009;58(4):333–341. doi: 10.1538/expanim.58.333. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang SJ, et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43(D1):D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama M, Smith JR, De Pons J, Tutaj M, Khampang P, Hong W, Erbe CB, Ehrlich GD, Bakaletz LO, Kerschner JE. The Chinchilla Research Resource Database: resource for an otolaryngology disease model. Database (Oxford). 2016;2016:baw073. doi: 10.1093/database/baw073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19(9):D1630–D1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Bolton ER, Dwinell MR. The rat: a model used in biomedical research. Methods Mol Biol. 2019;2018:1–41. doi: 10.1007/978-1-4939-9581-3_1. [DOI] [PubMed] [Google Scholar]
- Smith JR, Hayman GT, Wang SJ, Laulederkind SJF, Hoffman MJ, Kaldunski ML, Tutaj M, Thota J, Nalabolu HS, Ellanki SLR. The year of the rat: the Rat Genome Database at 20: a multi-species knowledgebase and analysis platform. Nucleic Acids Res. 2020;48(D1):D731–D742. doi: 10.1093/nar/gkz1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Park CA, Nigam R, Laulederkind SJ, Hayman GT, Wang SJ, Lowry TF, Petri V, Pons JD, Tutaj M, et al. The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications. J Biomed Semantics. 2013;4(1):26. doi: 10.1186/2041-1480-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg Woods LC, Stelloh C, Regner KR, Schwabe T, Eisenhauer J, Garrett MR. Heterogeneous stock rats: a new model to study the genetics of renal phenotypes. Am J Physiol Renal Physiol. 2010;298(6):F1484–F1491. doi: 10.1152/ajprenal.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollis E, Mosaku A, Abid A, Buniello A, Cerezo M, Gil L, Groza T, Güneş O, Hall P, Hayhurst J, et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2022;51(D1):D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer C. Rat models of human diseases and related phenotypes: a systematic inventory of the causative genes. J Biomed Sci. 2020;27(1):84. doi: 10.1186/s12929-020-00673-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Smith H, Vanderlinden LA, Hoffman PL, Saba LM. Networking in biology: the hybrid rat diversity panel. Methods Mol Biol. 2019;2018:213–231. doi: 10.1007/978-1-4939-9581-3_10. [DOI] [PubMed] [Google Scholar]
- Twigger S, Lu J, Shimoyama M, Chen D, Pasko D, Long H, Ginster J, Chen CF, Nigam R, Kwitek A, et al. Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res. 2002;30(1):125–128. doi: 10.1093/nar/30.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedi M, Nalabolu HS, Lin CW, Hoffman MJ, Smith JR, Brodie K, De Pons JL, Demos WM, Gibson AC, Hayman GT, et al. MOET: a web-based gene set enrichment tool at the Rat Genome Database for multiontology and multispecies analyses. Genetics. 2022;220(4):iyac005. doi: 10.1093/genetics/iyac005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Brodie KC, De Pons JL, Demos WM, Gibson AC, Hayman GT, Hill ML, Kaldunski ML, Lamers L, Laulederkind SJF, et al. Ontological analysis of coronavirus associated human genes at the COVID-19 disease portal. Genes (Basel). 2022;13(12):2304. doi: 10.3390/genes13122304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Laulederkind SJ, Hayman GT, Petri V, Liu W, Smith JR, Nigam R, Dwinell MR, Shimoyama M. Phenominer: a quantitative phenotype database for the laboratory rat, Rattus norvegicus. Application in hypertension and renal disease. Database (Oxford). 2015;2015:bau128. doi: 10.1093/database/bau128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3(1):160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalaratne SM, Juty N, Kunze J, Janée G, McMurry JA, Beard N, Jimenez R, Grethe JS, Hermjakob H, Martone ME, et al. Uniform resolution of compact identifiers for biomedical data. Sci Data. 2018;5(1):180029. doi: 10.1038/sdata.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Sternberg PW, Lipshitz HD. Making biological knowledge useful for humans and machines. Genetics. 2022;220(4):iyac001. doi: 10.1093/genetics/iyac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley KC, Weinstock GM, Gibbs RA. Rats in the genomic era. Physiol Genomics. 2008;32(3):273–282. doi: 10.1152/physiolgenomics.00208.2007. [DOI] [PubMed] [Google Scholar]
- Ye W, Wu Z, Gao P, Kang J, Xu Y, Wei C, Zhang M, Zhu X. Identified gefitinib metabolism-related lncRNAs can be applied to predict prognosis, tumor microenvironment, and drug sensitivity in non-small cell lung cancer. Front Oncol. 2022;12:939021. doi: 10.3389/fonc.2022.939021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Smith JR, Wang SJ, Dwinell MR, Shimoyama M. Quantitative phenotype analysis to identify, validate and compare rat disease models. Database (Oxford). 2019;2019:baz037. doi: 10.1093/database/baz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The data (https://rgd.mcw.edu/wg/data-menu/) and tools (https://rgd.mcw.edu/wg/tool-menu/) described in the article are available from the RGD website (https://rgd.mcw.edu), REST APIs (https://rest.rgd.mcw.edu/rgdws/swagger-ui.html), and file download site (https://download.rgd.mcw.edu/data_release), and the software is available on the GitHub (https://github.com/rat-genome-database) link at the top right of the RGD homepage. For more information on the RGD webpages and tools, there are help pages (https://rgd.mcw.edu/wg/help3/) and video tutorials (https://rgd.mcw.edu/wg/home/rgd_rat_community_videos/rgd-tutorials/). RGD also offers virtual office hours that are available by appointment using the “Contact” link on the homepage (https://rgd.mcw.edu/rgdweb/contact/contactus.html).