Abstract
Fourteen people with human immunodeficiency virus type 1 had longitudinal measurements of intact, defective, and total proviral DNA over the course of two decades of antiretroviral therapy. Three patterns of intact proviral DNA decay were revealed: (1) biphasic decline with markedly slower second-phase decline, (2) initial decline that transitions to a zero-slope plateau, and (3) initial decline followed by later increases in intact proviral DNA. Defective proviral DNA levels were essentially stable. Mechanisms of slowing or reversal of second-phase decay of intact proviral DNA may include the inability to clear cells with intact but transcriptionally silent proviruses and clonal expansion of cells with intact proviruses.
Keywords: HIV, antiretroviral therapy, intact proviruses, reservoirs
In people with Human Immunodeficiency Virus and viral suppression by antiretroviral therapy, intact proviral DNA levels initially decay rapidly, followed by a marked slowing of the decay and, in some individuals, a late increase in proviral DNA levels.
Human immunodeficiency virus type 1 (HIV-1) infection cannot be cured in most people because of indefinite persistence of a latent reservoir during antiretroviral therapy (ART). An intact proviral DNA (IPD) assay has been developed to estimate the HIV-1 reservoir [1]. This assay separately quantifies intact proviruses, which are potentially replication competent, and defective proviruses, which are not. Intact provirus levels correlate with measures of inducible infectious virus outgrowth [1–3], the standard method of quantifying the HIV-1 reservoir. However, the IPD assay has several advantages over virus outgrowth assays: (1) it is a simpler, more feasible method of measurement; and (2) it detects HIV-1 proviruses that may replicate in vivo but cannot be activated to replicate in ex vivo cultures.
Several studies have shown that intact HIV-1 proviruses selectively decay during ART, compared with defective proviruses [4, 5], and that there is a biphasic decay in intact proviruses, with a more rapid initial decline followed by slower subsequent decay [6]. These previous studies, however, evaluated a limited number of time points during the first decade of ART and even fewer during the second decade of treatment. As a result, there is incomplete understanding of the longitudinal decay patterns of intact proviruses over the course of very long-term ART. Such an understanding is crucial for efforts to better characterize and eliminate persistent intact proviruses and achieve long-term ART-free remission (“HIV-1 cure”). In the current study, we performed longitudinal measurements of intact, defective and total proviral DNA in people with HIV-1 over the course of 2 decades of well-documented virally suppressive ART, thereby revealing new patterns of IPD decay.
METHODS
We evaluated a longitudinal cohort of participants with chronic HIV-1 infection who started ART in AIDS Clinical Trials Group (ACTG) trials for treatment-naive persons and were subsequently followed up while continuing to receive ART (ACTG studies A5001 and A5321) [7]. Commercial assays showed plasma HIV-1 RNA levels <50 copies/mL starting at week 48 of ART and at all subsequent time points, with no reported ART interruptions.
We performed the following measurements on CD4+ T cells isolated from peripheral blood mononuclear cells: IPD, 5′ or 3′ defective proviral DNA, and total proviral DNA (sum of defective, hypermutated, and intact proviruses), as reported elsewhere [1]. The number of cell equivalents assayed was determined for each sample, and proviral DNA measurements were normalized per million CD4+ T cells. The limit of detection was dependent on the number of CD4+ T cells available for the assay. Samples from 16 participants were initially included, but IPD was not analyzable in 2 participants (13%) because of signal failure (amplification or detection failure); results from the remaining 14 participants are the focus of this report.
Biexponential statistical modeling [8, 9] of IPD frequencies was performed using nonlinear regression, separately by participant. This model provides decay rate estimates for both a first and a second exponential phase. The transition time between phases was the estimated number of years of ART when the contributions to the IPD frequency were the same from both phases.
RESULTS
Fourteen participants who started ART during chronic HIV-1 infection were included in this study. All participants had documented suppression of viremia (HIV-1 RNA <50 copies/mL) for >15 years of ART and no known episodes of treatment interruption or viral rebound during those years. Blood samples had been collected from participants longitudinally from year 1 to years 17–23 after ART initiation (median ART duration, 20 years; 8–10 time points). CD4+ T cells were isolated from cryopreserved peripheral blood mononuclear cells and assayed for IPD, 5′ or 3′ defective proviral DNA, and total proviral DNA [1].
Five participants reported female sex assigned at birth, and the remainder endorsed male sex assigned at birth. The median pre-ART age (range) was 44 ( 23–56) years); the median pre-ART plasma HIV-1 RNA level, 4.2 (2.3–5.5) log10 copies/mL; and the median pre-ART CD4+ T-cell count, 377/µL (27–762/µL). Additional participant characteristics appear in Supplementary Table 1.
After year 1 of ART, the median (interquartile range) IPD was 204 (99–876) copies/million CD4+ T cells, and the median intact provirus percentage (intact divided by total proviruses) was 66% (41%–83%). By the last time point (median ART duration, 20 years), the median (interquartile range) IPD had fallen to 16 (3–120) copies/million CD4+ T cells, and the median intact provirus percentage had fallen to 7% (4%–10%). We observed decay of intact proviruses but not defective proviruses over the course of time on ART: intact proviruses declined by 13-fold from the first to the last time point, whereas total proviruses (including intact provirus) declined by only 3-fold and defective provirus levels were generally stable (median reduction, 0.7-fold) (Table 1).
Table 1.
Decreases in Intact, Defective and Total Proviral DNA From First to Last Time Point
| Proviral DNA | Fold Reduction From First to Last Time Point (n = 14)a |
|
|---|---|---|
| Median (IQR) | Range | |
| Intactb | 13.4 (4.6–46.0) | 1.8–91 |
| Defective | 0.7 (0.3–1.7) | 0.1–4.5 |
| Total | 2.7 (1.2–4.9) | 0.3–6.7 |
Abbreviation: IQR, interquartile range.
The first time point was year 1 of antiretroviral therapy (ART); the last time point, years 17–23.
For 2 participants with censored results, the change in intact proviral DNA was calculated using the estimated assay lower limit.
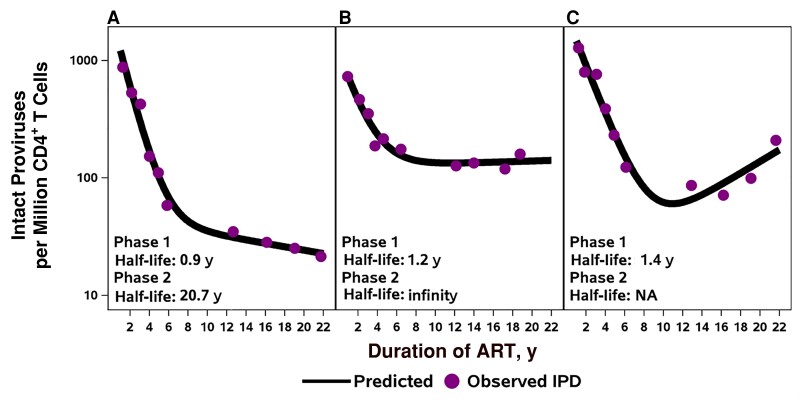
The longitudinal measurements revealed several patterns of IPD change during the 2-decade period of observation. Five participants had biphasic decay of IPD levels, 3 had biphasic decay with a second-phase plateau (slope effectively zero), and 2 showed evidence of increased IPD levels during the second decade of ART (see Figure 1 for examples; see Supplementary Figures 1–3 for changes in IPD, percentage IPD, and CD4+ T-cell counts for all 14 participants). The inflection or transition of decay occurred a median of 5 years after ART initiation (range, 2–13 years). For 8 of the 10 participants who were modeled, the estimated transition time to slower or reversal of decay was before year 10 of ART (range, 2–9 years of ART). Four participants with substantial censored DNA levels or other longitudinal patterns were not included in the modeling.
Figure 1.
Examples of decay patterns in intact proviral DNA (IPD) for participants on long-term antiretroviral therapy (ART). A, Five participants had a similar pattern with slowing decay of intact proviruses during the second decade of ART. B, Three participants showed plateauing of decay (second phase slope effectively zero). C, Two participants had patterns of late increases. Abbreviation: NA, not applicable.
The median IPD first-phase half-life was 1 year (n = 10), and the median IPD second-phase half-life, >25 years (n = 8). For the 2 participants with late IPD increases, the second-phase half-life could not be estimated. The changes in IPD from nadir value to final value for these 2 participants were 2.9-fold and 6.3-fold increases. In the 4 other participants, there was a variable pattern of IPD decay, in part owing to fewer cells assayed or low IPD levels. When the ratio of intact to total proviruses was evaluated, the 10 participants with an evaluable pattern showed rapid decline, followed by a plateau (Supplementary Figure 2).
DISCUSSION
In people with HIV-1 infection on very long-term ART with continual suppression of viremia, 3 patterns of IPD decay were revealed by multiple longitudinal measurements: (1) biphasic decline with markedly slower second-phase decline, (2) initial decline that transitions to a zero-slope plateau, and (3) initial decline followed by late increases in IPD. The slowing or reversal of IPD decay suggests that the mechanisms of infected cell clearance and persistence are very different during the first phase of decay (median half-life, 1 year) compared with the second phase (median half-life, >25 years).
Prior studies have shown that, after initiation of ART, the dynamics of plasma HIV-1 RNA and IPD decay are markedly different. The first phase of plasma HIV-1 RNA decay has a half-life of approximately 1 day, and its second phase a half-life of approximately 14 days [10]. By contrast, a 2022 study found that, during the first few months after ART initiation, IPD has an initial phase of decay with a half-life of about 14 days, but after approximately 3 months of ART, the IPD decay rate slows to a half-life of 19 months [8], similar to the half-life we describe over the first few years of ART. That study, however, evaluated only IPD dynamics during the first year of ART rather than long-term decay over decades of ART, as in the current study.
We found that IPD decays steadily during the first 2–10 years of ART, with a half-life of 1 year. After that relatively rapid decay, however, there is marked slowing in the IPD decay rate, to a half-life of >25 years. Compared with previous work, the frequent measurements allowed more precise modeling of decay and of the transition from relatively rapid decay to much slower or no further decay. The time of transition varied considerably and, in some participants, occurred after only 2–3 years of ART, which differs from a prior estimate of approximately 7 years in an analysis implementing the same transition timing for all individuals [6]. Of note, in 2 participants, both women, the transition to slower decay was followed by late increases in IPD, which has not been reported elsewhere to our knowledge. Thus, IPD does not monotonically decline over decades of ART, and there are varied patterns of change in IPD.
The mechanisms of the marked slowing or reversal of IPD decay are uncertain. One possibility is that during the first few years of ART, transcriptionally active intact proviruses are selectively cleared, perhaps through immune-mediated mechanisms or viral cytopathic effects. Once those transcriptionally active intact proviruses have been cleared, the decay rate may slow because remaining intact proviruses are less transcriptionally active or are silent and thus invisible to the immune system. This model is consistent with the rapid decay followed by a plateau in the ratio of intact to total proviruses observed in individual participants (Supplementary Figure 2) and other published results showing that transcriptionally active proviruses are selected against during long-term ART [11].
Another possible explanation for slowing or reversal of IPD decay is clonal expansion of cells with intact proviruses that offsets or exceeds clearance of cells with intact proviruses. Previous studies have demonstrated clonal expansion of infected cells, including those containing intact proviruses. Indeed, most of the HIV-1 reservoir is found in clonally expanded cells, and clonal proliferation is a major mechanism of reservoir persistence [12, 13]. Clonal expansion is largely driven by cognate antigen–induced cell proliferation and is rarely related to the specific site of proviral integration [14]. Cytomegalovirus and HIV-1 antigens drive expansion of some clones [15], but many other antigens may be involved. High scientific priorities include further identification of drivers of clonal expansion of cells with intact proviruses, including potential differences between women and men, and strategies to selectively block proliferation of infected cells.
Our study had several limitations. The number of participants was relatively small, mainly because it is very difficult to find individuals with documented suppression of plasma viremia for 20 years of ART and with no episodes of treatment interruption or viral rebound and frequent longitudinal sampling during those years. In the future, it will be important to study more participants to define the frequency of different patterns of IPD decay and provide insights into correlates of patterns of decay. Another limitation of our study is that the mechanisms behind different patterns of decay observed were not defined; longitudinal integration site and proviral sequencing are necessary to determine why decay in IPD levels slows over time and why some individuals appear to have late increases in IPD levels.
Our findings also have several important implications. First, over the course of 2 decades of ART, there is selective decay of intact proviruses and relatively little to no decay in defective or total proviral DNA. Cells harboring intact proviruses may be shorter lived because viral protein expression may trigger cell death or immune-mediated killing. We and others have seen similar proviral decay patterns during the first decade of ART [4–6], but the data in the current study extend into the second decade of treatment.
Second, the observation of a marked slowing or, at times, reversal of IPD decay during the second decade of ART indicates that the mechanism by which cells carrying intact proviruses are eliminated changes during the course of long-term ART, perhaps because (1) proviruses that persist are in silent, nongenic regions of the human genome ; (2) there is less HIV-specific immune effector function ; and/or (3) there is increasing proliferation of infected cell clones. If the decay rate during the second phase (median half-life, >25 years) could be accelerated to approximate the decay rate more closely during the first phase (median half-life, 1 year), it may be possible to reduce the viral reservoir to the degree needed to achieve HIV-1 remission. To achieve this goal, a more detailed understanding of the host and viral mechanisms involved in the varied decay of cells with intact proviruses is needed.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Rajesh T Gandhi, Infectious Disease Division, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Ronald J Bosch, Center for Biostatistics in AIDS Research, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Hanna Mar, Center for Biostatistics in AIDS Research, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Gregory M Laird, Accelevir Diagnostics, Baltimore, Maryland, USA.
Elias K Halvas, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Laura Hovind, Frontier Science and Technology Research Foundation, Amherst, New York, USA.
Ann C Collier, Department of Medicine, University of Washington School of Medicine, Seattle, Washington, USA.
Sharon A Riddler, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Albine Martin, Accelevir Diagnostics, Baltimore, Maryland, USA.
Kristen Ritter, Accelevir Diagnostics, Baltimore, Maryland, USA.
Deborah K McMahon, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Joseph J Eron, Department of Medicine, University of North Carolina, Chapel Hill, North Carolina, USA.
Joshua C Cyktor, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
John W Mellors, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
AIDS Clinical Trials Group A5321 Team:
Evelyn Hogg, Rebecca LeBlanc, Christine Scello, David Palm, Monica Gandhi, Courtney Fletcher, Anthony Podany, Fran Aweeka, Jeymohan Joseph, Susan Pederson, Leah Rubin, Davey Smith, Serena Spudich, and Athe Tsibris
Notes
Acknowledgments. We acknowledge all members of the A5321 team, including Evelyn Hogg, Rebecca LeBlanc, Christine Scello, David Palm, Monica Gandhi, Courtney Fletcher, Anthony Podany, Fran Aweeka, Jeymohan Joseph, Susan Pederson, Leah Rubin, Davey Smith, Serena Spudich, and Athe Tsibris. We appreciate the site staff who enrolled participants into the AIDS Clinical Trials Group A5321 study and the participants who contributed so much to this study. We also thank Courtney Tern for all her help in preparing the manuscript.
Financial support . This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID; grants UM1AI068636, UM1AI106701, UM1AI068634, and UM1AI069481); the National Institutes of Health (NIH) Small Business Innovation Research grant program (grants R43AI142866 and R44AI124996 to G. M. L., A. M., and K. R.); the NIH Cooperative Agreement Grant (U24AI143502 to G. M. L.); the I4C Martin Delaney Collaboratory (NIH grant UM1 AI126603 to G. M. L.); the Beat-HIV Collaboratory (UM1 AI126620); the National Science Foundation Small Business Innovation Research grant program (grant 1738428 to G. M. L., A. M., and K. R.); the Harvard University Center for AIDS Research (funding to R. T. G. through NIH grant P30 AI060354); and the AIDS Clinical Trials Group (funding to R. T. G. through NIAID/NIH grant 2 UMAI069412).
References
- 1. Bruner KM, Wang Z, Simonetti FR, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Falcinelli SD, Kilpatrick KW, Read J, et al. Longitudinal dynamics of intact HIV proviral DNA and outgrowth virus frequencies in a cohort of individuals receiving antiretroviral therapy. J Infect Dis 2021; 224:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosch RJ, Gandhi RT, Mar H, et al. Associations between multiple measures of HIV-1 persistence in persons on suppressive antiretroviral therapy. J Infect Dis 2022; 225:2163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antar AAR, Jenike KM, Jang S, et al. Longitudinal study reveals HIV-1–infected CD4+ T cell dynamics during long-term antiretroviral therapy. J Clin Invest 2020; 130:3543–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gandhi RT, Cyktor JC, Bosch RJ, et al. Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis 2021; 223:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peluso MJ, Bacchetti P, Ritter KD, et al. Differential decay of intact and defective proviral DNA in HIV-1–infected individuals on suppressive antiretroviral therapy. JCI Insight 2020; 5:e132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gandhi RT, McMahon DK, Bosch RJ, et al. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 2017; 13:e1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White JA, Simonetti FR, Beg S, et al. Complex decay dynamics of HIV virions, intact and defective proviruses, and 2LTR circles following initiation of antiretroviral therapy. Proc Natl Acad Sci U S A 2022; 119:e2120326119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu H, Ding AA. Population HIV-1 dynamics in vivo: applicable models and inferential tools for virological data from AIDS clinical trials. Biometrics 1999; 55:410–8. [DOI] [PubMed] [Google Scholar]
- 10. Kuritzkes DR, Ribaudo HJ, Squires KE, et al. Plasma HIV-1 RNA dynamics in antiretroviral-naive subjects receiving either triple-nucleoside or efavirenz-containing regimens: ACTG A5166s. J Infect Dis 2007; 195:1169–76. [DOI] [PubMed] [Google Scholar]
- 11. Einkauf KB, Osborn MR, Gao C, et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 2022; 185:266–282.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bui JK, Sobolewski MD, Keele BF, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog 2017; 13:e1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hosmane NN, Kwon KJ, Bruner KM, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med 2017; 214:959–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coffin JM, Bale MJ, Wells D, et al. Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy. PLoS Pathog 2021; 17:e1009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simonetti FR, Zhang H, Soroosh GP, et al. Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J Clin Invest 2021; 131:145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.