Abstract
Background:
The eradication rate of Helicobacter pylori infection with empirical therapy has decreased due to increased drug resistance. The latest guidelines recommend genotypic resistance-guided therapy, but its clinical efficacy remains unclear.
Objectives:
The purpose of our study was to evaluate whether tailored therapy based on genotypic resistance is superior to empirical therapy for H. pylori infection.
Design:
A systematic review and meta-analysis of randomized controlled trials (RCTs) comparing tailored therapy based on genotypic resistance with empirical therapy was performed.
Sources and methods:
We retrieved relevant studies from PubMed, Embase, and the Cochrane Library. The primary outcome was H. pylori eradication rate and the adverse events (AEs) was the secondary outcome. A random-effect model was applied to compare pooled risk ratios (RRs) with related 95% confidence intervals (CIs).
Results:
A total of 12 qualified RCTs containing 3940 patients were identified in our systematic review and meta-analysis. The pooled eradication rates of tailored therapy based on the detection of genotypic resistance were consistently higher than those in the empirical treatment group, with no statistical significance. In triple therapy, the eradication rate was significantly higher in the tailored group than in the empirical group by intention-to-treat analysis (ITT) and per-protocol analysis (PP) analysis (p < 0.0001, RR: 1.20; 95% CI: 1.12–1.29; p < 0.0001, RR: 1.20; 95% CI: 1.15–1.25). In quadruple therapy, the eradication rate was higher in the empirical group (p = 0.001, RR: 0.93; 95% CI: 0.89–0.97; p = 0.009, RR: 0.95; 95% CI: 0.92–0.99). And this result was true for both bismuth quadruple therapy (BQT) and non-BQT. Regarding total AEs, the pooled rate was 34% in the tailored group and 37% in the empirical group, and no difference between the two groups was found (p = 0.17, RR: 0.88; 95% CI: 0.74–1.06).
Conclusion:
In conclusion, tailored therapy based on molecular methods may offer better efficacy than empirical triple therapy, but it may not be superior to empirical quadruple therapy in eradicating H. pylori infection. Larger and more individualized RCTs are needed to aid clinical decision-making.
Registration (PROSPERO):
CRD42023408688
Keywords: empirical, eradication, genotypic resistance, Helicobacter pylori, meta-analysis, tailored
Background
Helicobacter pylori is a major pathogenic factor of gastritis, peptic ulcer, and gastric adenocarcinoma.1,2 Compared to other infectious diseases, the eradication success rate for H. pylori remains poor. Common reasons for treatment failure include antibiotic resistance, rapid drug metabolism, poor compliance, and insufficient treatment duration.3,4 A previous systematic review and meta-analysis assessing the distribution of H. pylori resistance to commonly used antibiotics found that primary resistance rates to clarithromycin and levofloxacin were more than 15% in most regions. 5 The eradication rate of empirical therapy has declined dramatically due to increasing antibiotic resistance. In areas with high rates of clarithromycin resistance, bismuth quadruple therapy (BQT) has been recommended as a first-line empiric treatment. 6 High-dose dual therapy (HDDT) also offers an effective regimen with less use of antibiotics for H. pylori infection.1,7 Both regimens are recommended to be used for primary and secondary eradication of H. pylori in China. 7
However, there are still some patients who fail to eradicate H. pylori twice or more and are considered refractory H. pylori infections. With the increase in antibiotic resistance, individualized treatment has attracted great attention in recent years. As stated in the Maastricht VI/Florence Consensus Report, it is reasonable to recommend that susceptibility tests are routinely performed in terms of antibiotic stewardship, even before prescribing first-line treatment. 1 And if available, clarithromycin susceptibility testing should be performed before prescribing any clarithromycin-containing therapy to ensure satisfactory eradication rates. 1 Tailored treatments based on culture-based susceptibility testing have been confirmed to have guiding value for H. pylori eradication.8,9 However, due to the time-consuming and labor-intensive of the culture technique, its clinical application is limited. Recently, tailored therapy based on molecular detection of antibiotic resistance seems to provide a promising approach with its superior characteristics of easy operation and fast detection. The most widely used technique is polymerase chain reaction (PCR), which enables detecting point mutations associated with antibiotic resistance, such as A2142G and A2143G point mutations in 23S ribosomal RNA (rRNA) that are related to clarithromycin resistance, with high efficiency and accuracy.10,11
However, the concordance of antibiotic susceptibility testing based on genotype and phenotype remains uncertain. Although it is generally believed that the detection of antibiotic resistance by molecular method and E-test method is highly consistent, some samples have been reported to be phenotypically resistant but genotypically sensitive, suggesting that some rare mutations associated with phenotypic resistance may be missed by molecular method. 12 A recent study demonstrated that the molecular testing-guided therapy was comparable to susceptibility testing-guided therapy in first-line treatment and non-inferior to susceptibility testing-guided therapy in third-line treatment, supporting the use of molecular testing-guided therapy for H. pylori eradication. 13 But the evidence for the use of genotypic resistance-guided therapy in eradicating H. pylori infection compared to conventional treatment regimens remains to be evaluated. Hence, this systematic review and meta-analysis was conducted to determine whether tailored therapy based on genotypic detection of antibiotic resistance was superior to empirical therapy for H. pylori eradication, to evaluate the clinical value of molecular detection of genotypic resistance.
Materials and methods
This systematic review and meta-analysis was registered in PROSPERO (registration no.: CRD42023408688) and conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Supplemental Table S1).
Search strategy
We mainly conducted a systematic literature search through the following databases: (1) PubMed, (2) the Cochrane Library, and (3) Embase. Potentially relevant RCTs, published up to February 2023, were retrieved by the following mesh terms: ‘Helicobacter pylori or H. pylori’ and ‘tailored or susceptibility or personalized or resistance or mutation or individualized’. English was the only language searched in all databases. The detailed search strategies and results are shown in Supplemental Table S2. In addition, references retrieved from relevant meta-analyses, articles, and reviews were also manually screened to further incorporate eligible studies.
Selection criteria
All studies enrolled in our systematic review and meta-analysis were screened on the basis of following PICOS principles: (1) P (population): adult patients infected with H. pylori; (2) I and C (intervention and comparison): articles comparing tailored therapy based on genotypic resistance detection with empiric treatment recommended by the current consensus or guidelines, and having specific explanation of the drugs used in both regimens; (3) O (outcomes): end points included the eradication rate at least; and (4) S (study): randomized controlled trials (RCTs) published in English.
Studies were excluded based on the following criteria: case reports, meta-analyses, reviews, summary only, and literature with incomplete data, tailored treatment based on minimum inhibitory concentration detection of H. pylori culture.
Data extraction
The data from the included studies were independently extracted by two members using a form specially designed for this meta-analysis: year of publication, first author, nation, total numbers of patients, baseline characteristics of patients (age and previous experience in H. pylori eradication), drug regimen, origin of specimen, methods to confirm H. pylori infection and eradication, molecular methods and results for the detection of antibiotic resistance, the eradication rate (intention-to-treat analysis, ITT; per-protocol analysis, PP), and adverse events (AEs). The third reviewer checked all collected data for any discrepancies or errors.
Quality assessment
The quality of all eligible RCTs was assessed by the guidelines of the Cochrane Handbook for systematic reviews, and the main evaluation indicators were as follows: random sequence generation, allocation concealment, blinding of patients and personnel, incomplete outcome data, selective reporting, and other bias. 14 We considered studies with a score of 3 or more to be of high quality. The authors assessed the quality of each study separately and reached a consensus on the 12 studies included.
Statistical analysis
In this systematic review and meta-analysis, the following two data management software packages were used for statistical analysis: STATA 14.0 (Stata Corp) and Review Manager 5.3 (The Nordic Cochrane Centre). The risk ratios (RRs) and the corresponding 95% confidence intervals (CIs) were used for the eradication rates and adverse reactions. Quantitative analyses were performed on ITT and PP basis. Heterogeneity in the combined results was assessed by the I2 statistic and χ2 test. To reduce the potential bias due to heterogeneity, a random-effect model was used. Obvious heterogeneity was indicated when p < 0.10, and I2 > 50% in the χ2 test. Moreover, we also explored the publication bias through the funnel plot and Egger et al.’s test. 15 Statistical significance was defined as p < 0.05.
Results
Search results and quality assessment
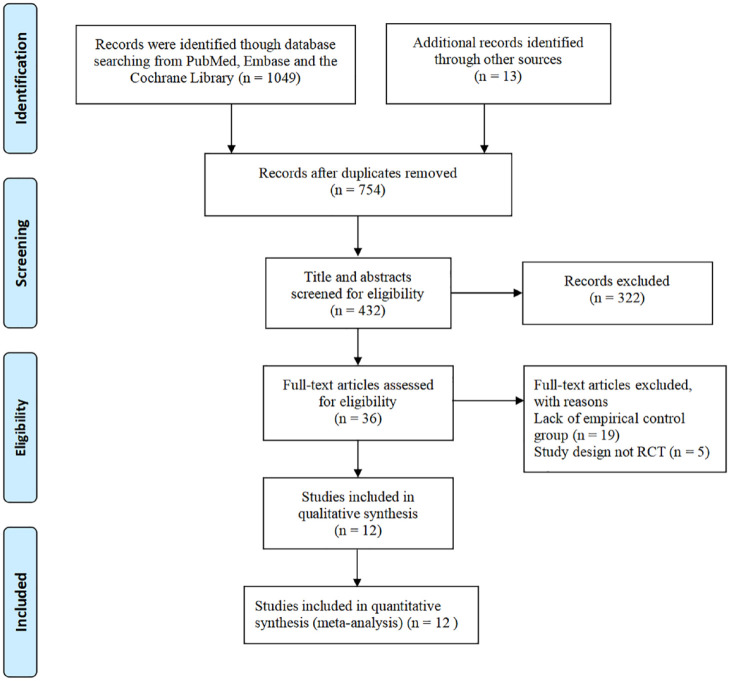
As shown in Figure 1, 1049 records were identified through careful literature screening. A total of 12 qualified RCTs containing 3940 patients (tailored group, 1780 patients; empirical group, 2160 patients) were finally included in our meta-analysis.16–27 One of the studies included two eligible control groups that were compared to the tailored group. We combined the data into one control group for analysis. 25 Only one study containing patients with refractory H. pylori infection (failure after two or more eradication therapies). In all, 11 studies were conducted in Asia and one in Europe. The main characteristics of the enrolled studies are listed in Table 1. Furthermore, all of the studies were of high quality and scored 3–5 points (Supplemental Table S3 and Figure S1).
Figure 1.
Flow chart of studies included in the systematic review and meta-analysis.
Table 1.
Characteristics of the studies included in the systematic review and meta-analysis.
| First authors | Year | Country | Study design | Treatment line | Tailored determinant | Patients (tailored/empiric) | Eradication regime | Eradication rate of TT | Eradication rate of ET | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tailored regimens | Empiric regimens | ITT | PP | ITT | PP | |||||||
| Cha | 2021 | Korea | RCT | 1 | DPO-PCR | 182/178 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 7 days CLA-r: BIS 300 mg QID + MET 500 mg TID + TET 500 mg QID + RAB 20 mg BID, 7 days |
BIS 300 mg QID + MET 500 mg TID + TET 500 mg QID + RAB 20 mg BID, 7 days | 66.3% (118/178) | 81.4% (118/145) | 78.0% (142/182) | 88.8% (142/160) |
| Choi | 2021 | Korea | RCT | 1 | DPO-PCR | 110/107 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + LAN 30 mg BID, 14 days CLA-r: BIS 300 mg QID + MET 500 mg BID + TET 500 mg QID + LAN 30 mg BID, 10 days |
AMO 1000 mg BID + MET 500 mg BID + CLA 500 mg BID + LAN 30 mg BID, 10 days | 82.7% (91/110) | 90.1% (91/101) | 82.2% (88/107) | 91.6% (87/95) |
| Cho | 2021 | Korea | RCT | 1 | DPO-PCR | 141/141 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + PAN 40 mg BID, 7 days CLA-r: BIS 300 mg QID + MET 500 mg TID + TET 500 mg QID + PAN 40 mg BID, 7 days |
BIS 600 mg BID + AMO 1000 mg BID + MET 750 mg BID + PAN 40 mg BID, 14 days | 80.9% (114/141) | 89.0% (113/127) | 85.8% (121/141) | 93.5% (116/124) |
| Delchier | 2020 | France | RCT | 1 and 2 | GenoType HelicoDR® PCR | 266/260 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + PPI BID, 7 days CLA-r + LEV-s: AMO 1000 mg BID + LEV 250 mg BID + PPI BID, 10 days CLA-r + LEV-r: AMO 1000 mg BID + MET 500 mg BID + PPI BID, 14 days (OME 20 mg BID or ESO 20 mg BID or LAN 30 mg BID or PAN 40 mg BID or RAB 20 mg BID) |
First line: AMO 1000 mg BID + CLA 500 mg BID + PPI BID, 7 days Second line: AMO 1000 mg BID + MET 500 mg BID + PPI BID, 14 days (OME 20 mg BID or ESO 20 mg BID or LAN 30 mg BID or PAN 40 mg BID or RAB 20 mg BID) |
85.5% (177/207) | 86.5% (173/200) | 73.1% (152/208) | 74.4% (151/203) |
| Furuta | 2007 | Japan | RCT | 1 | PCR | 150/150 | CLA-s: AMO 500 mg TID + CLA 200 mg TID + LAN 15 mg/30 mg BID (According to CYP2C19), 7 days CLA-r: AMO 500 mg QID + LAN 15 mg/30 mg BID (According to CYP2C19), 14 days |
AMO 750 mg BID + CLA 400 mg BID + LAN 30 mg BID, 7 days | 96.0% (144/150) | 96.6% (144/149) | 70.0% (105/150) | 72.9% (105/144) |
| Hsieh | 2022 | China | RCT | 1 | PCR-RLFP | 91/91 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 7 days CLA-r, LEV-s: AMO 1000 mg BID + LEV 500 mg QD + RAB 20 mg BID, 7 days CLA-r, LEV-r: AMO 1000 mg BID (14 days) + CLA 500 mg BID (7 days) + MET 500 mg BID (7 days) or AMO 1000 mg QID + RAB 20 mg QID, 14 days |
AMO 750 mg BID + CLA 500 mg BID + RAB 20 mg BID, 7 days | 89.0% (81/99) | 91.0% (81/89) | 75.8% (69/91) | 79.3% (69/87) |
| Kawai | 2008 | Japan | RCT | 1 | PCR | 35/35 | CLA-s: AMO 750 mg BID + CAM 400 mg BID + LAP 30 mg BID, 7 days CLA-r/mixed infections: AMO 750 mg BID + MET 250 mg BID + RAB 10 mg BID, 7 days |
AMO 750 mg BID + CAM 400 mg BID + LAN 30 mg BID, 7 days | 94.3% (33/35) | 94.3% (33/35) | 71.4% (25/35) | 78.1% (25/32) |
| Kim | 2020 | Korea | RCT | 1 | DPO-PCR | 36/36 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + ESO 40 mg BID, 10 days CLA-r: BIS 300 mg BID + TET 500 mg QID + MET 500 mg TID + ESO 40 mg BID, 10 days |
AMO 1000 mg BID + CLA 500 mg BID + ESO 40 mg BID, 10 days | 88.9% (32/36) | 97.0% (32/33) | 75.0% (27/36) | 81.8% (27/33) |
| Kim | 2022 | Korea | RCT | 1 | DPO-PCR | 145/145 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 14 days CLA-r: TET 1000 mg QID + MET 500 mg TID + BIS 120 mg QID + RAB 20 mg BID, 14 days |
AMO 1000 mg BID + CLA 500 mg BID + MET 500 mg + RAB 20 mg BID, 14 days | 85.5% (124/145) | 94.6% (122/129) | 82.8% (120/145) | 88.6% (117/132) |
| Lee | 2013 | Korea | RCT | 1 | DPO-PCR | 218/616 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 7 days CLA-r: AMO 1000 mg BID + MET 500 mg TID + RAB 20 mg BID, 7 days |
APC: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 7 days APM: AMO 1000 mg BID + MET TID + RAB 20 mg BID, 7 days |
80.7% (176/218) | 91.2% (176/193) | 70.3% (433/616) | 77.5% (433/559) |
| Liou | 2018 | China | RCT | 3 and more | PCR | 205/205 | (EPZ 40 mg BID + AMP 1000 mg BID), 7 days + (EPZ 40 mg BID + MET 500 mg BID + LEV 250 mg/CLA 500 mg/TET 500 mg BID) (According to genotypic resistance), 7 days | (EPZ 40 mg BID + AMP 1000 mg BID), 7 days + (EPZ 40 mg BID + MET 500 mg BID + LEV 250 mg/CLA 500 mg/TET 500 mg BID), 7 days | 78.0% (160/205) | 78.4% (156/199) | 72.2% (148/205) | 74.4% (145/195) |
| Ong | 2019 | Korea | RCT | 1 | PCR | 201/196 | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + LAN 30 mg BID, 14 days CLA-r: AMO 1000 mg BID + MET 500 mg BID + LAN 30 mg BID, 14 days |
AMO 1000 mg BID + CLA 500 mg BID + MET 500 mg BID + LAN 30 mg BID, 14 days | 81.6% (164/201) | 86.5% (154/178) | 86.2% (169/196) | 90.2% (157/174) |
AMO, amoxicillin; APM, amoxicillin + rabeprazole + metronidazole; APM, amoxicillin + rabeprazole + clarithromycin; BID, two times a day; BIS, bismuth; CLA, clarithromycin; CLA-r, clarithromycin resistant; CLA-s, clarithromycin sensitive; DPO-PCR, dual priming oligonucleotide polymerase chain reaction; EPZ, esomeprazole; ET, empirical therapy; FUR, furazolidone; LAN, lansoprazole; LEV, levofloxacin; LEV-r, levofloxacin resistant; LEV-s, levofloxacin sensitive; MET, metronidazole; MET-r, metronidazole resistant; MET-s, metronidazole sensitive; PCR-RLFP, polymerase chain reaction-restriction fragment length polymorphism; PPI, proton-pump inhibitor; QD, once a day; QID, four times a day; RAB, rabeprazole; RCT, randomized control trial; TET, tetracycline; TID, three times a day; TT, tailored therapy.
Eradication rates
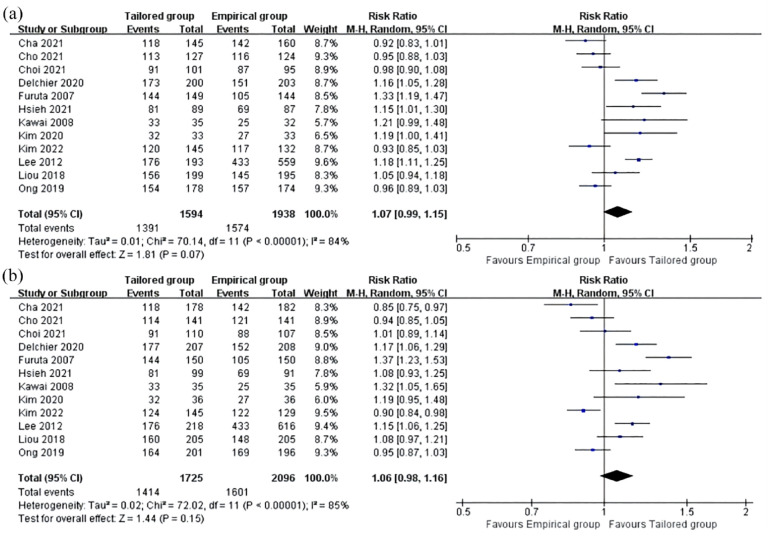
The pooled H. pylori eradication rates, obtained from ITT and PP analyses, were compiled and presented in Table 1. The pooled eradication rates of tailored therapy were consistently higher than those in the empirical treatment group, although the difference did not reach statistical significance [p = 0.07, RR: 1.07; 95% CI: 0.99–1.15, Figure 2(a); p = 0.15, RR: 1.06; 95% CI: 0.98–1.16, Figure 2(b)]. In the ITT analysis, the overall eradication rates of tailored therapy and empirical treatment were 82% and 76%, respectively. In the PP analysis, the eradication rates were 87% for tailored therapy and 81% for empirical treatment. But high cure rates (>90%) were not achievable in both the tailored group and the empirical group. Meanwhile, the level of heterogeneity between the enrolled studies in both analyses was substantial (ITT: p < 0.00001, I2 = 84%; PP: p < 0.00001, I2 = 85%).
Figure 2.
Forest plots of the pooled H. pylori eradication rates with PP (a) and ITT (b) analysis for the comparison of tailored therapy versus empirical therapy.
ITT, intention-to-treat; PP, per-protocol.
Advert events
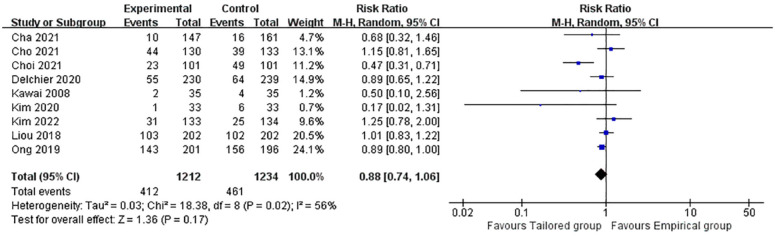
Of all the studies included, nine reported total AEs in both therapeutic regimens (heterogeneity: p = 0.02, I2 = 56%). Overall, the pooled rate was 34% in the tailored group and 37% in the empirical group. No significantly increased risk between the two groups was observed (p = 0.17, RR: 0.88; 95% CI: 0.74–1.06, Figure 3). The symptoms reported were mild, with a bitter taste, nausea, or vomiting and diarrhea being the most common.
Figure 3.
Forest plot of the adverse events comparing tailored treatment with empirical treatment.
Subgroup analysis
The enrolled studies in our meta-analysis were heterogeneous and varied in design. To verify the accordance of the results, subgroup analyses were conducted through the following categories of variables: country, duration of therapy, empirical regimen (triple versus quadruple), treatment lines, the types of antibiotics tested, and origin of specimen. Moreover, we performed subgroup analyses of BQT and non-BQT. The results are shown in Table 2.
Table 2.
Subgroup analysis for the eradication rate of H. pylori.
| Group | No. studies | ITT | PP | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pooled estimate | Test of heterogeneity | Pooled estimate | Test of heterogeneity | ||||||
| RR (95% CI) | p | I2 (%) | p value | RR (95% CI) | p | I2 (%) | p value | ||
| Total | 12 | 1.06 (0.98–1.16) | 0.15 | 84 | <0.00001 | 1.07 (0.99–1.15) | 0.07 | 84 | <0.00001 |
| Nation | |||||||||
| Korea | 7 | 0.98 (0.90–1.07) | 0.67 | 78 | 0.0001 | 1.00 (0.92–1.09) | 0.92 | 84 | <0.00001 |
| China | 2 | 1.08 (0.99–1.18) | 0.09 | 0 | 0.98 | 1.09 (1.01–1.19) | 0.04 | 3 | 0.31 |
| Japan | 2 | 1.36 (1.23–1.50) | <0.00001 | 0 | 0.76 | 1.30 (1.18–1.43) | <0.0001 | 0 | 0.42 |
| France | 1 | 1.17 (1.06–1.29) | – | – | – | 1.16 (1.05–1.28) | – | – | – |
| Empirical therapy | |||||||||
| Triple therapy | 6 | 1.20 (1.12–1.29) | <0.00001 | 48 | 0.08 | 1.20 (1.15–1.25) | <0.00001 | 0 | 0.45 |
| Quadruple therapy | 5 | 0.93 (0.89–0.97) | 0.001 | 4 | 0.39 | 0.95 (0.92–0.99) | 0.009 | 0 | 0.85 |
| BQT | 2 | 0.90 (0.81–1.00) | 0.05 | 37 | 0.21 | 0.94 (0.88–1.00) | 0.03 | 0 | 0.54 |
| Non-BQT | 3 | 0.94 (0.89–0.99) | 0.03 | 10 | 0.33 | 0.96 (0.91–1.01) | 0.10 | 0 | 0.73 |
| Sequential therapy | 1 | 1.08 (0.97–1.21) | – | – | – | 1.05 (0.94–1.18) | – | – | – |
| Treatment duration | |||||||||
| 7 days | 4 | 1.11 (1.02–1.22) | 0.02 | 46 | 0.14 | 1.12 (1.01–1.24) | 0.04 | 74 | 0.09 |
| 14 days | 3 | 0.97 (0.87–1.07) | 0.54 | 73 | 0.02 | 0.97 (0.91–1.04) | 0.45 | 36 | 0.21 |
| Mixed | 5 | 1.11 (0.97–1.26) | 0.15 | 85 | <0.0001 | 1.10 (0.97–1.25) | 0.14 | 89 | <0.00001 |
| Treatment lines | |||||||||
| First | 10 | 1.05 (0.95–1.16) | 0.32 | 86 | <0.00001 | 1.06 (0.98–1.15) | 0.16 | 86 | <0.00001 |
| First and second | 1 | 1.17 (1.06–1.29) | – | – | – | 1.16 (1.05–1.28) | – | – | – |
| Third and more | 1 | 1.08 (0.97–1.21) | – | – | – | 1.05 (0.94–1.18) | – | – | – |
| Antibiotics tested | |||||||||
| CLA | 9 | 1.05 (0.94–1.17) | 0.38 | 88 | <0.00001 | 1.05 (0.96–1.15) | 0.26 | 87 | <0.00001 |
| CLA + LEV | 3 | 1.12 (1.05–1.20) | 0.0009 | 0 | 0.50 | 1.12 (1.05–1.20) | 0.0003 | 0 | 0.39 |
| Specimen origin | |||||||||
| Gastric biopsy | 10 | 1.04 (0.95–1.14) | 0.35 | 81 | <0.00001 | 1.04 (0.97–1.11) | 0.31 | 81 | <0.00001 |
| Other | 2 | 1.17 (0.96–1.42) | 0.11 | 54 | 0.14 | 1.16 (1.05–1.29) | 0.005 | 0 | 0.68 |
BQT, bismuth quadruple therapy; CI, confidence interval; CLA, clarithromycin; ITT, intention-to-treat; LEV, levofloxacin; PP, per-protocol; RR, relative risk; –, not available.
As shown in Table 2, the tailored therapy had a superior eradication efficacy over empirical treatment in Japan with both ITT and PP analysis, while a higher eradication rate was only shown in PP analysis in China. In Korea, there was no significant difference. In triple therapy, the eradication rate was significantly higher in a tailored group than in an empirical group by ITT and PP analyses. Conversely, in quadruple therapy, the eradication rate was higher in the empirical group (p = 0.001, RR: 0.93; 95% CI: 0.89–0.97; p = 0.009, RR: 0.95; 95% CI: 0.92–0.99). The pooled eradication rates of tailored therapy and empirical quadruple therapy were 79% and 85% in the ITT analysis. And in the PP analysis, the eradication rates were 86% for tailored therapy and 90% for empirical quadruple treatment. This result was true for both BQT and non-BQT (Table 2). The detailed characteristics of the included studies of genotypic resistance-guided therapy and empirical quadruple therapy are shown in Table 3. Under the guidance of genotypic resistance testing, all the studies used a triple regimen for sensitive strains in the tailored group, and the duration of treatment was 7 days in some studies.16–18,24,27 Regarding treatment duration, the tailored group had significantly increased eradication rates compared with the empirical group in a 7-day regimen (p = 0.02, RR: 1.11; 95% CI: 1.02–1.22; p = 0.04, RR: 1.12; 95% CI: 1.01–1.24). But the pooled results in the 14-day regimen between the two groups showed no significant difference (p = 0.54, RR: 0.97; 95% CI: 0.87–1.07; p = 0.45, RR: 0.97; 95% CI: 0.91–1.04). Concerning genotypic resistance of antibiotic tested, the tailored group proved superior to empiric treatment by ITT and PP analysis for both clarithromycin and levofloxacin tested subgroups (RR: 1.12; 95% CI: 1.05–1.20, Table 2). When antibiotics were selected through clarithromycin resistance only, we found no statistically significant difference between the two groups. In addition, 10 RCTs compared tailored therapy with empirical treatment as first-line treatment, which showed no significant difference between them. We also performed a subgroup analysis according to the origin of the specimen, only two studies used specimens other than gastric biopsy. One was gastric juice, and another was stool. No significant results were found in the subgroup of gastric biopsy studies.
Table 3.
Characteristics of the included studies of genotypic resistance-guided therapy and empirical quadruple therapy.
| Group | Antibiotics tested determinant | Eradication regime | Eradication rate of TT | Eradication rate of ET | ITT | PP | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tailored regimens | Empiric regimens | ITT | PP | ITT | PP | RR (95% CI) | p | RR (95% CI) | p | ||
| BQT | 72.7% (232/319) | 84.9% (231/272) | 81.4% (263/323) | 90.8% (258/284) | 0.90 (0.81–1.00) | 0.05 | 0.94 (0.88–1.00) | 0.03 | |||
| Cha 2021 | CLA | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 7 days CLA-r: BIS 300 mg QID + MET 500 mg TID + TET 500 mg QID + RAB 20 mg BID, 7 days |
BIS 300 mg QID + MET 500 mg TID + TET 500 mg QID + RAB 20 mg BID, 7 days | 66.3% (118/178) | 81.4% (118/145) | 78.0% (142/182) | 88.8% (142/160) | 0.85 (0.75–0.97) | 0.92 (0.83–1.01) | ||
| Cho 2021 | CLA | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + PAN 40 mg BID, 7 days CLA-r: BIS 300 mg QID + MET 500 mg TID + TET 500 mg QID + PAN 40 mg BID, 7 days |
BIS 600 mg BID + AMO 1000 mg BID + MET 750 mg BID + PAN 40 mg BID, 14 days | 80.9% (114/141) | 89.0% (113/127) | 85.8% (121/141) | 93.5% (116/124) | 0.94 (0.85–1.05) | 0.95 (0.88–1.03) | ||
| Non-BQT | 83.1% (379/456) | 86.1% (365/424) | 87.7% (379/432) | 90.2% (361/401) | 0.94 (0.89–0.99) | 0.03 | 0.96 (0.91–1.01) | 0.1 | |||
| Choi 2021 | CLA | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + LAN 30 mg BID, 14 days CLA-r: BIS 300 mg QID + MET 500 mg BID + TET 500 mg QID + LAN 30 mg BID, 10 days |
AMO 1000 mg BID + MET 500 mg BID + CLA 500 mg BID + LAN 30 mg BID, 10 days | 82.7% (91/110) | 90.1% (91/101) | 82.2% (88/107) | 91.6% (87/95) | 1.01 (0.89–1.14) | 0.98 (0.90–1.08) | ||
| Kim 2022 | CLA | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + RAB 20 mg BID, 14 days CLA-r: TET 1000 mg QID + MET 500 mg TID + BIS 120 mg QID + RAB 20 mg BID, 14 days |
AMO 1000 mg BID + CLA 500 mg BID + MET 500 mg + RAB 20 mg BID, 14 days | 85.5% (124/145) | 94.6% (122/129) | 82.8% (120/145) | 88.6% (117/132) | 0.90 (0.84–0.98) | 0.94 (0.88–1.00) | ||
| Ong 2019 | CLA | CLA-s: AMO 1000 mg BID + CLA 500 mg BID + LAN 30 mg BID, 14 days CLA-r: AMO 1000 mg BID + MET 500 mg BID + LAN 30 mg BID, 14 days |
AMO 1000 mg BID + CLA 500 mg BID + MET 500 mg BID + LAN 30 mg BID, 14 days | 81.6% (164/201) | 86.5% (154/178) | 86.2% (169/196) | 90.2% (157/174) | 0.95 (0.87–1.03) | 0.93 (0.85–1.03) | ||
| Pooled analysis | 78.8% (611/775) | 85.6% (596/696) | 85.0% (642/755) | 90.4% (619/685) | 0.93 (0.89–0.97) | 0.001 | 0.95 (0.92–0.99) | 0.009 | |||
AMO, amoxicillin; BID, two times a day; BIS, bismuth; CLA, clarithromycin; CLA-r, clarithromycin resistant; CLA-s, clarithromycin sensitive; EPZ, esomeprazole; ET, empirical therapy; LAN, lansoprazole; LEV, levofloxacin; MET, metronidazole; PPI, proton-pump inhibitor; QD, once a day; QID, four times a day.; RAB, rabeprazole; TET, tetracycline; TID, three times a day; TT, tailored therapy.
Sensitivity analysis
We found significant heterogeneity when we initially analyzed the pooled H. pylori eradication rates and total AEs. Thus, to assess the sensitivity and stability of the final results, apart from using a random-effects model, we also analyzed the influence of each study on the overall results. The analysis results demonstrated that the pooled eradication rate and total AEs were stable (Supplemental Figures S2 and S3).
Publication bias
Upon examination, there was no evidence of publication bias regarding the pooled eradication rate (ITT and PP) or AEs (Supplemental Figure S4).
Discussion
H. pylori is a human-specific pathogen, which is a cause of peptic ulcers and gastritis, as well as a main cause of gastric carcinogenesis. 28 With the increase in antibiotic resistance, rational and efficient first-line treatment should be adopted to eradicate H. pylori, to avoid secondary resistance as far as possible. 29 The Maastricht VI/Florence Consensus Report recommended the clinical application of susceptibility-guided therapy based on culture with susceptibility tests or molecular detection of genotype resistance. 1
Currently, some studies have compared the efficacy, safety, or cost-effectiveness of tailored therapy with empirical therapy in the eradication of H. pylori, but the conclusions are inconsistent. A meta-analysis conducted by Lopez-Gongora et al. (2015) revealed that sensitivity-guided triple therapy was more effective than standard triple therapy in the first-line treatment of H. pylori infection. However, there was a lack of evidence to compare tailored therapy with currently recommended quadruple therapy and the sensitivity testing method involved in their studies was culture. 30 Another study reported that the susceptibility-guided therapy may be slightly better than empirical first-line triple therapy but was not superior to empirical first-line quadruple therapy or empirical rescue therapy. 31 A third study that included both RCTs and non-RCTs demonstrated that tailored therapy was more effective in both the empirical triple and quadruple therapy subgroups and the conclusion was consistent in both culture and molecular detection subgroups. 32 For the above-tailored therapy, the methods used to test antimicrobial susceptibility included molecular detection or culture. Compared with the traditional culture method, genotype resistance detection through PCR has the advantages of time-saving and acceptable accuracy. It can use fresh or formalin-fixed gastric biopsies, gastric fluid, or stool samples and could rapidly provide data on multiple antibiotics.33,34 Nevertheless, few studies have specifically conducted detailed analysis of eradication regimens on the basis of molecular methods. Thus, we performed this review and meta-analysis. The pooled eradication rates based on both PP and ITT analyses revealed that the tailored group exhibited a tendency toward superiority over the empirical treatment group, despite the lack of statistical significance. Moreover, our study demonstrates that genotypic resistance-guided therapy has an advantage only in empirical triple therapy but is not superior to empirical quadruple therapy.
In our analysis, most included studies used standard triple therapy as the empirical triple treatment in areas with clarithromycin resistance rate greater than 15%,19–23 which was not recommended by current H. pylori management guidelines.1,6 As determined by molecular methods, if no clarithromycin-resistant mutation was present, use clarithromycin; otherwise, switched to another antibiotic. Thus, it is reasonable to find that sensitivity-guided therapy is superior to empirical triple therapy. And under the guidance of genotypic resistance detection, irrational use of antibiotics that are ineffective for H. pylori eradication and may induce resistance in gut bacteria can be avoided.
Five studies compared quadruple therapy with tailored therapy.16–18,24,27 Unexpectedly, the pooled eradication rate showed that empirical quadruple therapy was even superior to the tailored group. The outcomes in the sensitivity-guided group did not show a significant advantage over the empirical BQT group, especially in the 14-day treatment group.16,17 This was not consistent with the conclusions of some existing studies, such as the systematic review and meta-analysis from Ouyang et al., 35 which showed that susceptibility-guided therapy was superior to empirical BQT as a first-line treatment for H. pylori infection. The majority of studies included in that meta-analysis utilized culture-based methods for drug sensitivity testing, with only one RCT employing molecular detection methods. Therefore, further research is needed to validate this conclusion and assess the potential application value of genotype testing. BQT includes proton-pump inhibitors (PPIs), bismuth, and another two susceptible antibiotics, while non-BQT includes a PPI plus another three antibiotics. 36 They have been shown to achieve high efficacy in both susceptible and resistant H. pylori strains.1,37 But a three-antibiotic regimen may increase the risk of antibiotic resistance and make subsequent treatment difficult. In some clinical studies, although there was no advantage in the eradication rate under the guidance of genotypic resistance, the number of antibiotics or the duration of treatment was reduced compared with the empirical group.16–18,24,27 Thus, the tailored approach for H. pylori infection plays a crucial role in avoiding unnecessary antibiotic use and reducing the potential side effects associated with antibiotic overuse. Indeed, these findings should be interpreted with caution, and there is a need for additional large-scale and high-quality RCTs to provide more robust evidence for guiding clinical decision-making. In addition, only one study we included compared the genotype resistance-guided sequential therapy with empirical therapy in refractory populations with the eradication rate of 78% (tailored group) and 72% (empirical group), respectively. 26 It is worth mentioning that the empirical group was treated with medication history-guided therapy, which is strongly approved by the latest guidelines.1,7 This suggests that obtaining as much detailed medication history as possible of previous eradication treatments could be of great clinical benefit.
For the treatment duration subgroup, the tailored group had significantly increased eradication rates compared with the empirical group in the 7-day regimen, while results in the 14-day regimen showed no significant difference. This was consistent with some previously published studies.28,31 Similarly, for first-line treatment, extended triple therapy with 14 days has been testified to be superior to 7 or 10 days of the same regimen. 38 This suggested that prolonging the duration of treatment may be one of the crucial factors in achieving a better cure rate. In addition, it was also confirmed that the effect of prolonging the duration to 14 days in sensitive strains was not significant. 39 However, there was limited data available to demonstrate whether tailored therapy can achieve efficacy comparable to that of the empirical group while reducing the duration of treatment. The uncertainty of drug regimens and the course of treatment weakened the concept of tailored therapy to some extent and limited its clinical application. In the future, more refined clinical trials will be needed to address this issue and demonstrate the advantages of individualized treatment guided by resistance gene testing.
Moreover, PPIs with different metabolic sensitivity to CYP2C19 polymorphism may also affect the eradication rate of H. pylori. 40 In our meta-analysis, three studies detected this gene and PPI was selected for patients according to CYP2C19 in two of them.20,21,26 Further analysis was not performed due to insufficient data. Regarding the AEs in our study, no significant decreased risk was found in the tailored group. And the data indicated that both strategies were relatively safe in clinical practice.
This review and meta-analysis is novel in the following two aspects: First, it was the first meta-analysis evaluating genotypic resistance-guided therapy for eradicating H. pylori infection and providing new insights into the application of molecular detection techniques to address antibiotic resistance. Second, we introduced a novel topic comparing the efficacy of personalized therapy with the currently widely recommended empirical BQT. This provided a new perspective for conducting high-quality clinical trials in the future. Several potential limitations should be noted in interpreting the results. First, the number of studies on quadruple therapy and rescue therapy was relatively limited, so the results were not extrapolated. Second, the baseline demographic data, study design, and evaluation criteria of each eligible study were different, which might increase the heterogeneity. We had taken this into account and used an appropriate random-effect model. Third, most of the included studies were conducted in Asia with high antibiotic resistance rates, which may limit the applicability of these findings in the European region. Fourth, the lack of economic benefit analysis of various treatments increased the difficulty of clinical feasibility.
Conclusion
In conclusion, tailored therapy based on molecular methods may offer better efficacy than empirical triple therapy, but may not be superior to empirical quadruple therapy in eradicating H. pylori infection. Larger and more individualized RCTs are needed to aid clinical decision-making.
Supplemental Material
Supplemental material, sj-doc-1-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-docx-2-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-docx-3-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-4-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-5-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-6-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-7-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Acknowledgments
The authors would like to express gratitude to the mentor for constant guidance and valuable insights throughout the entire process.
Footnotes
ORCID iD: Meng Li  https://orcid.org/0009-0001-0280-0111
https://orcid.org/0009-0001-0280-0111
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Meng Li, Department of Gastroenterology, Peking University First Hospital, Beijing, China.
Xiaolei Wang, Department of Gastroenterology, Peking University First Hospital, Beijing, China.
Wenting Meng, Department of Gastroenterology, Peking University First Hospital, Beijing, China.
Yun Dai, Department of Gastroenterology, Peking University First Hospital, Beijing, China.
Weihong Wang, Department of Gastroenterology, Peking University First Hospital, No. 8 Xishiku Street, Beijing 100034, China.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Meng Li: Writing – original draft; Writing – review & editing.
Xiaolei Wang: Data curation.
Wenting Meng: Data curation.
Yun Dai: Methodology.
Weihong Wang: Supervision.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
The authors declare that there is no conflict of interest.
Availability of data and materials: All data and materials involved in the article are available from the author.
References
- 1. Malfertheiner P, Megraud F, Rokkas T, et al. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut. Epub ahead of print August 2022. DOI: 10.1136/gutjnl-2022-327745 [DOI] [PubMed] [Google Scholar]
- 2. Hooi JKY, Lai WY, Ng WK, et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 2017; 153: 420–429. [DOI] [PubMed] [Google Scholar]
- 3. Liu DS, Wang YH, Zhu ZH, et al. Characteristics of Helicobacter pylori antibiotic resistance: data from four different populations. Antimicrob Resist Infect Control 2019; 8: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alba C, Blanco A, Alarcón T. Antibiotic resistance in Helicobacter pylori. Curr Opin Infect Dis 2017; 30: 489–497. [DOI] [PubMed] [Google Scholar]
- 5. Savoldi A, Carrara E, Graham DY, et al. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in world health organization regions. Gastroenterology 2018; 155: 1372–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shah SC, Iyer PG, Moss SF. AGA clinical practice update on the management of refractory Helicobacter pylori infection: expert review. Gastroenterology 2021; 160: 1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Helicobacter pylori Study Group, Chinese Society of Gastroenterology, Chinese Medical Association. Chinese national clinical practice guideline on Helicobacter pylori eradication therapy. Chin J Dig 2022; 42: 745–756. [Google Scholar]
- 8. Pan J, Shi Z, Lin D, et al. Is tailored therapy based on antibiotic susceptibility effective ? A multicenter, open-label, randomized trial. Front Med 2020; 14: 43–50. [DOI] [PubMed] [Google Scholar]
- 9. Chen Q, Long X, Ji Y, et al. Randomised controlled trial: susceptibility-guided therapy versus empiric bismuth quadruple therapy for first-line Helicobacter pylori treatment. Aliment Pharmacol Ther 2019; 49: 1385–1394. [DOI] [PubMed] [Google Scholar]
- 10. Oleastro M, Ménard A, Santos A, et al. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol 2003; 41: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerrits MM, van Vliet AH, Kuipers EJ, et al. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis 2006; 6: 699–709. [DOI] [PubMed] [Google Scholar]
- 12. Zhou J, Shen Y, Song X, et al. Evaluation of a molecular mosprie assay for detection of Helicobacter pylori and resistance to clarithromycin and levofloxacin. J Infect Dis 2022; 226: S503–S509. [DOI] [PubMed] [Google Scholar]
- 13. Chen MJ, Chen PY, Fang YJ, et al. Molecular testing-guided therapy versus susceptibility testing-guided therapy in first-line and third-line Helicobacter pylori eradication: two multicentre, open-label, randomised controlled, non-inferiority trials. Lancet Gastroenterol Hepatol 2023; 8: 623–634. [DOI] [PubMed] [Google Scholar]
- 14. Higgins JP, Altman DG, Gøtzsche PC, et al. The cochrane collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011; 343: d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cha B, Bang BW, Shin JB, et al. Bismuth containing quadruple therapy versus tailored therapy as first-line treatments for Helicobacter pylori infection in a high clarithromycin resistance area. Scand J Gastroenterol 2021; 56: 1017–1022. [DOI] [PubMed] [Google Scholar]
- 17. Cho JH, Jin SY, Park S. Comparison of tailored Helicobacter pylori eradication versus modified bismuth quadruple therapy in Korea: a randomized controlled trial. Expert Rev Anti Infect Ther 2022; 20: 923–929. [DOI] [PubMed] [Google Scholar]
- 18. Choi YI, Chung JW, Kim KO, et al. Tailored eradication strategy vs concomitant therapy for Helicobacter pylori eradication treatment in Korean patients. World J Gastroenterol 2021; 27: 5247–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delchier JC, Bastuji-Garin S, Raymond J, et al. Efficacy of a tailored PCR-guided triple therapy in the treatment of Helicobacter pylori infection. Med Mal Infect 2020; 50: 492–499. [DOI] [PubMed] [Google Scholar]
- 20. Furuta T, Shirai N, Kodaira M, et al. Pharmacogenomics-based tailored versus standard therapeutic regimen for eradication of H. pylori. Clin Pharmacol Ther 2007; 81: 521–528. [DOI] [PubMed] [Google Scholar]
- 21. Hsieh MS, Kuo FC, Wu MC, et al. Tailored susceptibility-guided therapy via gastric juice PCR for the first-line H. pylori eradication, a randomized controlled trial. J Formos Med Assoc 2022; 121: 1450–1457. [DOI] [PubMed] [Google Scholar]
- 22. Kawai T, Yamagishi T, Yagi K, et al. Tailored eradication therapy based on fecal Helicobacter pylori clarithromycin sensitivities. J Gastroenterol Hepatol 2008; 23(Suppl. 2): S171–S174. [DOI] [PubMed] [Google Scholar]
- 23. Kim JL, Cho SJ, Chung SJ, et al. Empiric versus clarithromycin resistance-guided therapy for Helicobacter pylori based on polymerase chain reaction results in patients with gastric neoplasms or gastric mucosa-associated lymphoid tissue lymphoma: a randomized controlled trial. Clin Transl Gastroenterol 2020; 11: e00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim SJ, Jee SR, Park MI, et al. A randomized controlled trial to compare Helicobacter pylori eradication rates between the empirical concomitant therapy and tailored therapy based on 23S rRNA point mutations. Medicine (Baltimore) 2022; 101: e30069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee HJ, Kim JI, Cheung DY, et al. Eradication of Helicobacter pylori according to 23S ribosomal RNA point mutations associated with clarithromycin resistance. J Infect Dis 2013; 208: 1123–1130. [DOI] [PubMed] [Google Scholar]
- 26. Liou JM, Chen PY, Luo JC, et al. Efficacies of genotypic resistance-guided vs empirical therapy for refractory Helicobacter pylori infection. Gastroenterology 2018; 155: 1109–1119. [DOI] [PubMed] [Google Scholar]
- 27. Ong S, Kim SE, Kim JH, et al. Helicobacter pylori eradication rates with concomitant and tailored therapy based on 23S rRNA point mutation: a multicenter randomized controlled trial. Helicobacter 2019; 24: e12654. [DOI] [PubMed] [Google Scholar]
- 28. Kotilea K, Bontems P, Touati E. Epidemiology, diagnosis and risk factors of Helicobacter pylori infection. Adv Exp Med Biol 2019; 1149: 17–33. [DOI] [PubMed] [Google Scholar]
- 29. Neri M, Milano A, Laterza F, et al. Role of antibiotic sensitivity testing before first-line Helicobacter pylori eradication treatments. Aliment Pharmacol Ther 2003; 18: 821–827. [DOI] [PubMed] [Google Scholar]
- 30. López-Góngora S, Puig I, Calvet X, et al. Systematic review and meta-analysis: susceptibility-guided versus empirical antibiotic treatment for Helicobacter pylori infection. J Antimicrob Chemother 2015; 70: 2447–2455. [DOI] [PubMed] [Google Scholar]
- 31. Gingold-Belfer R, Niv Y, Schmilovitz-Weiss H, et al. Susceptibility-guided versus empirical treatment for Helicobacter pylori infection: a systematic review and meta-analysis. J Gastroenterol Hepatol 2021; 36: 2649–2658. [DOI] [PubMed] [Google Scholar]
- 32. Rokkas T, Ekmektzoglou K, Graham DY. Current role of tailored therapy in treating Helicobacter pylori infections. A systematic review, meta-analysis and critical analysis. Helicobacter 2023; 28: e12936. [DOI] [PubMed] [Google Scholar]
- 33. Graham DY. Molecular-based Helicobacter pylori susceptibility testing is almost ready for prime time. Gastroenterology 2021; 160: 1936–1937. [DOI] [PubMed] [Google Scholar]
- 34. Pohl D, Keller PM, Bordier V, et al. Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing. World J Gastroenterol 2019; 25: 4629–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ouyang Y, Zhang W, He C, et al. Susceptibility-guided therapy vs. Bismuth-containing quadruple therapy as the first-line treatment for Helicobacter pylori infection: a systematic review and meta-analysis. Front Med (Lausanne) 2022; 9: 844915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding SZ, Du YQ, Lu H, et al. Chinese consensus report on family-based Helicobacter pylori infection control and management (2021 Edition). Gut 2022; 71: 238–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liou JM, Lee YC, Wu MS. Treatment of refractory Helicobacter pylori infection-tailored or empirical therapy. Gut Liver 2022; 16: 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan Y, Ford AC, Khan KJ, et al. Optimum duration of regimens for Helicobacter pylori eradication. Cochrane Database Syst Rev 2013; 2013: 1–205. [DOI] [PubMed] [Google Scholar]
- 39. Liou JM, Chen PY, Kuo YT, et al. Toward population specific and personalized treatment of Helicobacter pylori infection. J Biomed Sci 2018; 25: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwab M, Schaeffeler E, Klotz U, et al. CYP2C19 polymorphism is a major predictor of treatment failure in white patients by use of lansoprazole-based quadruple therapy for eradication of Helicobacter pylori. Clin Pharmacol Ther 2004; 76: 201–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-doc-1-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-docx-2-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-docx-3-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-4-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-5-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-6-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology
Supplemental material, sj-png-7-tag-10.1177_17562848231196357 for Empirical versus tailored therapy based on genotypic resistance detection for Helicobacter pylori eradication: a systematic review and meta-analysis by Meng Li, Xiaolei Wang, Wenting Meng, Yun Dai and Weihong Wang in Therapeutic Advances in Gastroenterology