Highlights
-
•
Investigate the application value of deep learning tech based on CE-MRI in the differential diagnosis of spinal metastases.
-
•
Study involves a deep learning algorithms and CE-MRI parameter maps as the input of CNN.
-
•
CE-MRI based deep learning are feasible in the differential diagnosis of spinal metastases from bone cancer.
-
•
Inception-ResNet model with CE-MRI image achieve high accuracy similar to the diagnostic accuracy of radiomics.
Keywords: Radiomics, Deep learning, Spinal tumor, CE-MRI, improved U-Net, Inception-ResNet
Abstract
Objective
The objective of this study was to investigate the use of contrast-enhanced magnetic resonance imaging (CE-MRI) combined with radiomics and deep learning technology for the identification of spinal metastases and primary malignant spinal bone tumor.
Methods
The region growing algorithm was utilized to segment the lesions, and two parameters were defined based on the region of interest (ROI). Deep learning algorithms were employed: improved U-Net, which utilized CE-MRI parameter maps as input, and used 10 layers of CE images as input. Inception-ResNet model was used to extract relevant features for disease identification and construct a diagnosis classifier.
Results
The diagnostic accuracy of radiomics was 0.74, while the average diagnostic accuracy of improved U-Net was 0.98, respectively. the PA of our model is as high as 98.001%. The findings indicate that CE-MRI based radiomics and deep learning have the potential to assist in the differential diagnosis of spinal metastases and primary malignant spinal bone tumor.
Conclusion
CE-MRI combined with radiomics and deep learning technology can potentially assist in the differential diagnosis of spinal metastases and primary malignant spinal bone tumor, providing a promising approach for clinical diagnosis.
1. Introduction
Cancer stands as the predominant cause of mortality on a global scale. Among the various sites prone to metastasis, the liver emerges as the most common, followed by the lungs and bones [1]. Specifically, spinal metastases account for approximately 68% of bone metastases, with a substantial portion of patients experiencing pathological fractures that lead to nerve damage and spinal nerve compression. These complications significantly compromise both the patients' quality of life and their chances of survival, while also incurring substantial medical resource consumption and costs [2]. Unfortunately, the overall effectiveness of interventions for spinal metastases continues to decline. Once a pathological compression fracture in the spine occurs, the risk of paralysis escalates, inevitably resulting in a drastic reduction in patients' quality of life. From a clinical perspective, the ability to predict the vertebral segment at risk of suffering from a pathological fracture enables the implementation of effective preventive measures to avert, mitigate, or delay such detrimental events. Given these challenges, accurate and prompt assessment of spinal metastases is crucial in preventing the progression of spinal metastases to pathological compression fractures. It allows for the selection of an appropriate treatment strategy prior to the patient's health deterioration and serves as a solid foundation for informed decision-making regarding interventional treatments [3], [4].
In this context, the study of radiographic risk factors assumes paramount significance. The sensitivity and accuracy of magnetic resonance imaging (MRI) in detecting spinal metastases surpass those of computed tomography (CT) and X-ray. This distinction arises from the physiological and pathological basis of the spine itself, coupled with the diverse imaging techniques employed in MRI [5]. The typical vertebral body predominantly comprises bone tissue, red marrow, and yellow marrow. In adults, the average fat content of red bone marrow within the vertebral body ranges from 25% to 30%. However, as individuals age, red bone marrow gradually diminishes, giving way to an increase in yellow bone marrow. On T1-weighted imaging (T1WI), yellow bone marrow exhibits high signal intensity, while on T2-weighted imaging (T2WI), it demonstrates slightly elevated signal intensity when compared to normal bone tissue associated with aging. In contrast, metastases display an affinity for red bone marrow. Consequently, upon the entry of tumor thrombi into the vertebral body, metabolic changes occur in the early stages, followed by morphological and structural alterations that ultimately lead to pathological fractures [6], [7].
Upon infiltration of the vertebral body by tumor thrombi, the initial damage manifests as the invasion of yellow bone marrow within the vascular wall or the interstitial space between cells, thereby giving rise to tumor tissue without disrupting the morphological structure of the vertebral body. In comparison to vertebral yellow bone marrow, tumor tissue exhibits a significant increase in water content, resulting in low signal intensity on T1WI, high signal intensity on T2WI, and high signal intensity on the short τ inversion recovery (STIR) sequence. These distinctions distinctly differentiate tumor tissue from normal yellow bone marrow, exemplifying the fat replacement sign. As the disease progresses, further structural damage ensues within the vertebral body, with compressive changes gradually extending to the surrounding soft tissues. Therefore, the present study aims to explore the potential of deep learning technology in augmenting the diagnostic accuracy of spinal metastases in CE-MRI scans [8].
2. Materials and methods
2.1. Retrospective analysis of research objects
From July 2020 to March 2022, the medical history of patients with spinal metastases admitted to Department of Orthopedics, the First Affiliated Hospital, Fujian Medical University included 81 patients with spinal metastases or primary malignant spinal bone tumor confirmed by pathology or clinical follow-up. CE-MRI was performed in each patient. Inclusion criteria: Patients with suspected spinal tumors before MRI, who did not undergo chemotherapy, radiotherapy, surgery, or needle biopsy, and who were diagnosed with spinal metastasis or primary malignant spinal bone tumor by pathological biopsy or clinical follow-up after MRI. Exclusion criteria: patients who had received treatment or needle biopsy prior to MRI examination, were unable to perform or refused enhanced MRI scan. The study included 81 patients, with a mean age of (60.2±11.4) years (range, 31–80 years), consisting of 42 males and 39 females. Among the patients, 36 had primary malignant spinal bone tumor and 45 had metastasis in other sites, including 14 cases of lung cancer, 9 cases of breast cancer, 7 cases of prostate cancer, 6 cases of thyroid cancer, 5 cases of liver cancer, and 4 cases of renal cancer.
2.2. Instrument and scanning parameters
A superconducting whole-body MR Scanner was used: SiemensTrio3T and GEMR750.Sequences included sagittal T1WI, axial T2WI, sagittal T2WI, and fat-suppressed T2WI. When found on sagittal position lesions, using three-dimensional volume plaque gas check sequence (three-dimensional volumeinter polated breath- hold examination) to CE-axis a MRI lesions, Parameters: TR4. 1 ms, TE1. 4 ms, turning angle 10°, matrix 256×192, field of view 250 mm × 250 mm, layer thickness 3 mm, time resolution 10–14 s, 12-layer image. Gadolinium speed-meglumine salt (Gd-DTPA) was injected with 0.1 mmol/kg through a high-pressure syringe at a flow rate of 2 ml/s, followed by 20 ml saline at the same flow rate.
2.3. Lesion segmentation
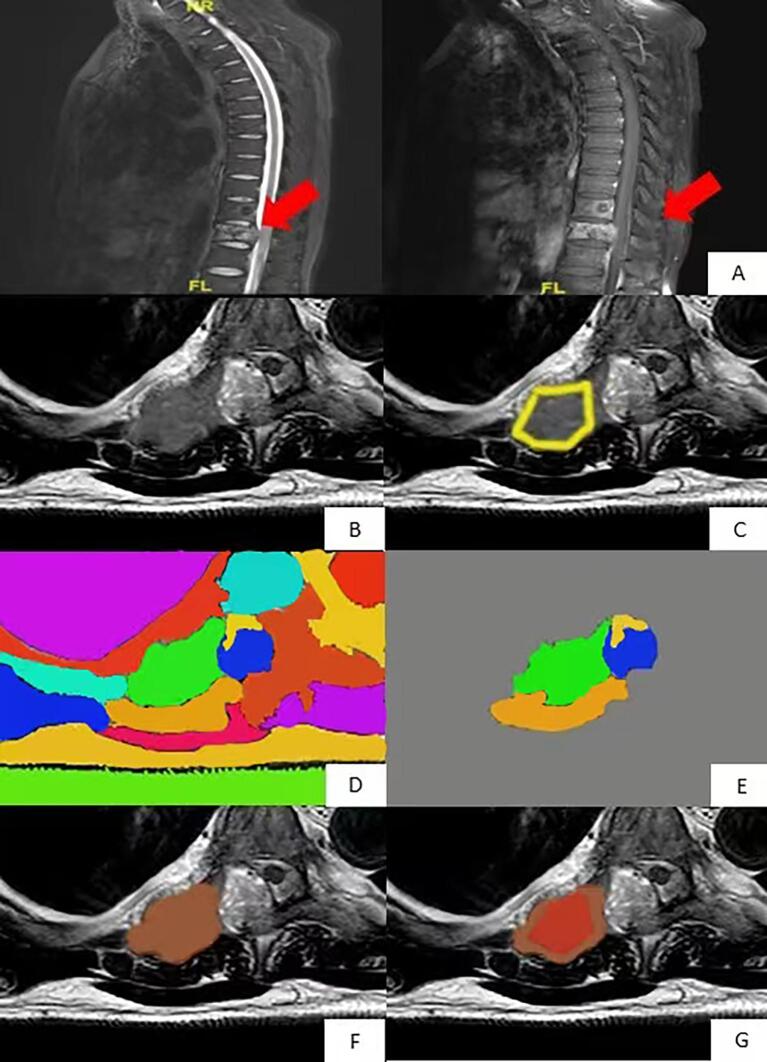
Region growing algorithm was used as a standard to perform segmentation [9], and the purpose of segmenting lesions was to further classify lesions by radiomics. Abnormal areas were found in sagittal view, and axial CE-MRI was performed, as shown in Fig. 1.
Fig. 1.
Region growth algorithm based on sagittal T2WI images for normalized axial CE-MRI segmentation. The lesion was found in the (A) sagittal position and was manually divided. Axial CE scan was performed on the (B) focal vertebra. The focal region outlined in the sagittal position (C) was transformed into the focal region in the axial position, and the red line region was used as the initial region of the region growth algorithm for segmentation. As shown in (D), in order to cover all lesions, the region growth algorithm expanded the initial region by five times and divided the image into several different partitions represented by different colors. (E) is reserved for and shows the partition that overlaps the red line area. (F) is a 3D area where the remaining partitions in all layers are fused. The pixel with the highest degree of enhancement in the (G) 3D region is the result shown by the seed region growth algorithm of the region, and the red region is the segmented tumor region. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. Radiomics differential diagnosis
Radiomics analysis was employed to extract and examine the texture features of contrast-enhanced magnetic resonance imaging (CE-MRI) parameters. The CE-MRI parameters were subjected to analysis based on the research conducted by Haralick and colleagues [10]. In total, 1316 radiomic features were computed using a gray-level co-occurrence matrix (GLCM). To eliminate confounding factors, the maximum relevance minimum redundancy (mRMR) algorithm was employed. This algorithm aided in selecting the most informative features while minimizing redundancy. The relevance-redundancy indexes were used to index the extracted features accordingly. From the pool of features, the top ten were retained for further analysis.
Subsequently, the least absolute shrinkage and selection operator (LASSO) logistic regression method was applied to identify the optimal features for constructing the radiomic signature. Through a ten-fold cross-validation process, features with nonzero coefficients were selected. These selected features were determined based on their predictive relevance. To generate the radiomics score (Rad-score), a weighted sum of the chosen features was calculated. The Rad-score facilitated the assessment of the probability of bone metastasis. The radiomic signature was developed using the training cohort and subsequently evaluated in the validation cohort.
After the most relevant features was selected, a logistic regression classifier was then constructed based on these selected features. The accuracy of the classifier was evaluated using a 10-fold cross-validation method and the receiver operating characteristic (ROC) curve. After completing 10 tests and training the final diagnostic classifier, all case results were used to test the accuracy of the classifier.
2.5. Deep learning for differential diagnosis
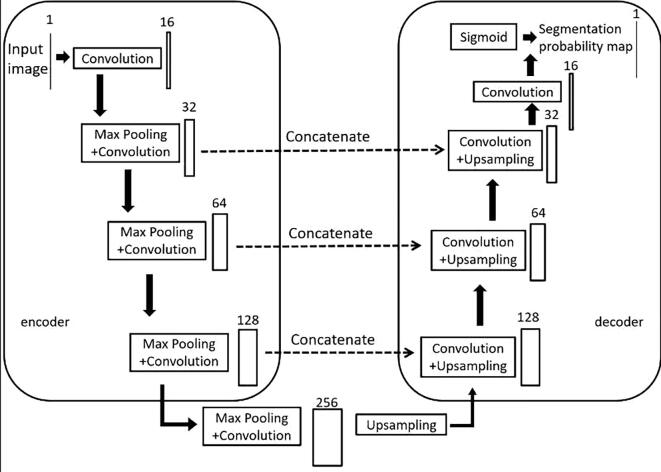
This paper proposes a modification to the U-Net architecture by replacing two 3×3 convolutions with residual blocks [8] and adding an attention gate module to the skip connections (see Fig. 2). The model consists of an encoder on the left half, which extracts features of different dimensions of MRI images using residuals and pooling layers, and a decoder on the right half, which restores the high-level semantic feature map to the resolution of the original MRI image using residuals and up-sampling. The gate module combines the feature maps obtained from the encoder and decoder, allowing for the combination of more abstract features extracted through multiple convolutional layers with less abstract but higher resolution features. This results in a feature map with a higher resolution while remaining abstract. The final output is obtained through convolution and classification. Overall, this modification improves the U-Net architecture for better segmentation performance in medical image analysis.
Fig. 2.
Model structure diagram of the convolutional neural network.
This model takes grayscale images of size 512×512 as input with one channel. The MRI images were preprocessed with bilinear interpolation method to 512×512.
The number of channels is increased from 1 to 16 after passing through a residual block. Then, a maximum pooling layer is used to halve the size of the feature map to 256×256. The decoder section of the model uses up-sampling to restore the feature map size from 32×32 back to the original size of 512×512.
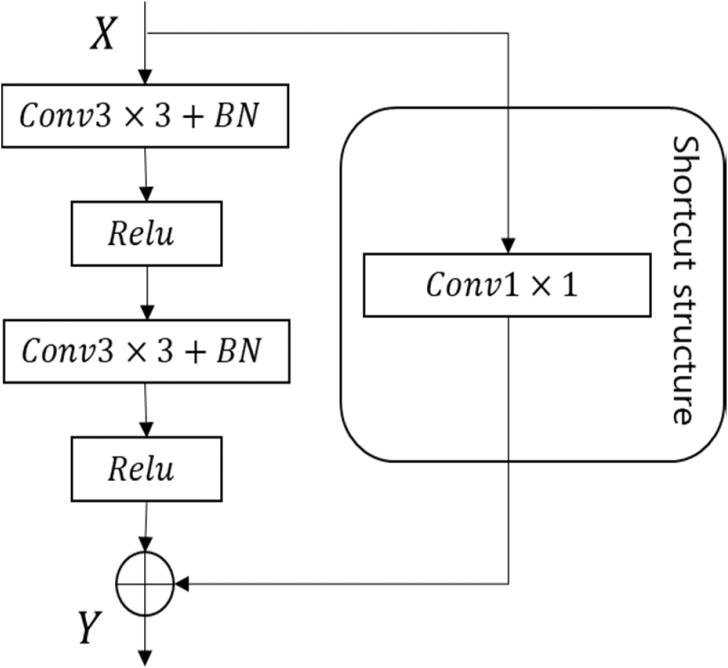
The residual block used in this model (see Fig. 3) employs a shortcut connection that directly maps the input to the output, allowing data to flow between different layers without negatively impacting the model's learning ability. This avoids the problem of decreased prediction accuracy due to gradient vanishing, while also avoiding an increase in computational complexity that can occur with the addition of 1×1 convolution layers. Specifically, the input X to the residual block is the feature map obtained after maximum pooling or up-sampling at each stage of the model structure. The input X is then transformed into a residual map through two convolution layers with Batch Norm and ReLU activation. The shortcut connection uses a 1×1 convolution layer to match the feature dimensions, and the input X and output Y are added together for feature fusion.
Fig. 3.
Residual block of the deep learning model.
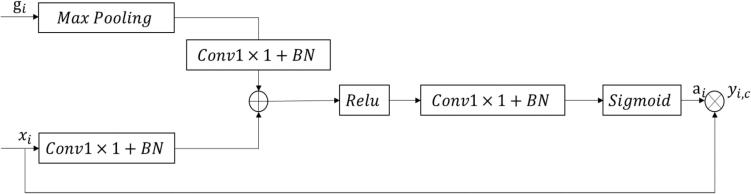
The gate module [11] used in this model is designed to enhance the feature maps obtained from low-level and high-level layers (see Fig. 4). As the low-level feature maps contain more location information and the high-level feature maps contain rich category information, the module uses the semantic information from the high-level feature maps to reinforce the feature weights of the brain tumor regions in the low-level feature maps. This helps to incorporate more detailed information into the low-level feature maps, thereby improving the segmentation accuracy of the model. The module uses a calculation method as follows:
| (2) |
Fig. 4.
Attention gate block.
In the formula, i is the pixel space size, c is the channel size, xi and gi are low-level and high-level feature maps respectively, Wx, Wg and W are linear transformation parameters, m is maximum poolization, b1 and b2 are offset terms, σ1 and σ2 are Relu activation function and Sigmoid activation function respectively. Since the sizes of xi and gi are inconsistent, gi needs to go through a maximum pooling layer first to obtain more detailed information. The obtained results and xi are converted to the same size by a simple convolutional network respectively. The two matrices obtained after conversion are added to obtain intermediate output ti, which is then compressed by Relu and convolutional neural network. Then add nonlinearity using Sigmoid[12] to obtain attention weight αi(0 αi I 1). Finally, multiply the attention weight with the encoding matrix to obtain the weighted feature graph yi(c). Attention weight can play a role in the selection of low-level feature maps, so that low-level feature maps can have rich category information as well as more accurate location information.[13].
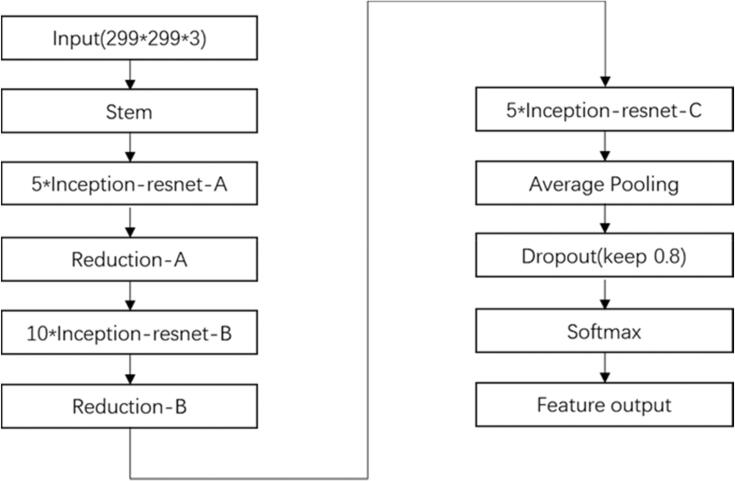
Following the fully connected layer, the Softmax activation function is employed for the classification of the input images [14]. To train our model, we utilized the Inception-ResNet-v2 convolutional neural network (CNN) architecture, employing the vast ImageNet dataset, which encompasses more than 1 million images. With a total of 164 layers, this network demonstrates its ability to classify images into approximately 1000 object categories [29]. Consequently, the model becomes proficient in learning comprehensive attribute representations across a diverse range of images. Our selection of the architecture was based on thorough experimental evaluations and comparative analyses conducted with other prominent deep learning models.
The initial ResNet block of our model incorporates convolutional filters and residual connections of varying sizes [30]. We specifically opted for the Inception-ResNet-v2 model due to its exceptional balance between model performance (accuracy) and resource requirements. Given the intended deployment of this model in edge environments, we deliberately avoided selecting extensive and bulky models that would hinder practical implementation. For a visual representation, refer to Fig. 5, illustrating the architecture of our custom model. The final structure of the Inception-ResNet-v2 network is depicted within the same figure [31].
Fig. 5.
Structure of Inception-ResNet-v2 network.
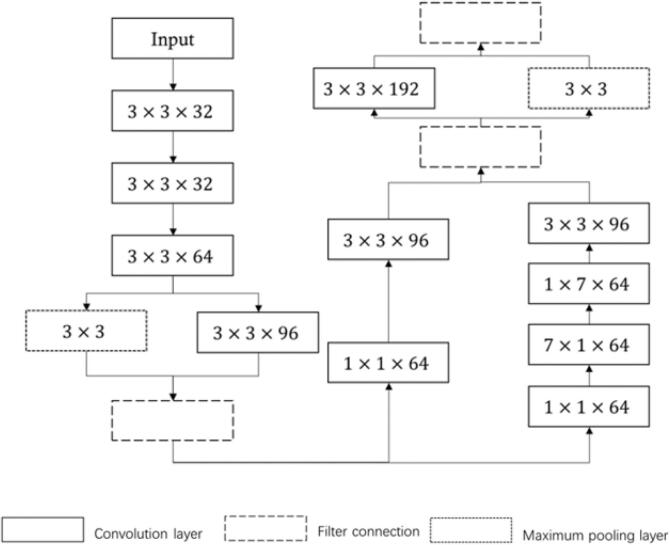
Within the combined network, the stem network assumes the role of a shallow feature extraction module. It incorporates various types of convolutions in parallel, employing an asymmetric approach that involves splitting large convolution kernels into multiple smaller ones, such as 1×7 and 7×1 kernels. This unique configuration enables enhanced extraction of multi-level structural features, thereby augmenting their diversity. Fig. 6 visually depicts the architecture of the stem network, providing a clear representation of its structure.
Fig. 6.
Structure of stem.
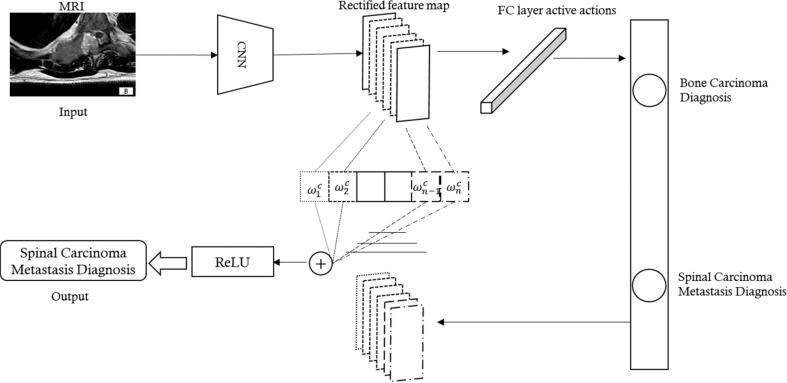
Our workflow comprises image acquisition, data preparation, DL model building, and diagnostic feedback. Data preparation is a crucial step that influences the accuracy of the results. It involves operations such as equalizing the number of images in each class, simple filtering, and denoising. The datasets are divided into ten groups, for 10-fold cross-validation. The allocation of training set and test set was consistent with that of the previous radiomics to ensure the scientific validity of performance comparison later. Tuning experiments are conducted during the training process to optimize the network parameters. As the model learns, its classification output becomes more accurate. Finally, after aggregating and deploying the experiments, the DL network is tested on the unseen images in the test set. Fig. 7 provides a more detailed overview of the proposed system network.
Fig. 7.
The total model architecture.
To enhance Inception-ResNet's classification ability, a self-attention mechanism is introduced. To achieve this, a process known as channel attention is employed. This entails compressing each channel of the output feature map by performing global average pooling, resulting in a feature vector. Subsequently, the feature weight of each channel is learned through an excitation mechanism. This learned feature weight is then utilized to generate a weighted feature map, effectively adjusting the relative strengths of different channels within the feature map. This process enables the network to emphasize important channels and suppress less relevant ones, enhancing the discriminative power of the model. The self-attention mechanism considers inter-channel dependencies, leading to improved performance. Experimental results show that the computational overhead is acceptable compared to the performance improvement. [34], [35].
In order to enhance the classification accuracy of primary malignant spinal bone tumor using contrast-enhanced magnetic resonance imaging (CE-MRI), a novel approach is employed. Instead of using 2D images, 3D cubes are utilized as input, as CE-MRI inherently consist of natural 3D image sequences. Previous research has demonstrated that utilizing 3D cubes yields improved classification performance, primarily due to the presence of dependencies between consecutive slices within the CE-MRI. To extract image features at a deeper level, a 3D convolutional neural network (CNN) is employed. This deep learning architecture enables the extraction of intricate and discriminative features from the 3D cubes, ultimately leading to enhanced classification accuracy. By considering the spatial information across the 3D data, the 3D CNN can effectively capture and utilize the contextual relationships between slices, resulting in more accurate classification of primary malignant spinal bone tumor in CE-MRI. The Inception-ResNet model is a decision tree-based ensemble model that, in this study, is intended to be used for the final classification. In order to control overfitting, a bootstrapping technique with replacement is applied to the data set, and each tree in the forest is trained on a randomly selected subset of features [15]. The output results were represented by spinal metastases and primary malignant spinal bone tumor, the test results were cross-validated by 10- fold, and the accuracy was represented by the average value and range.[16].
3. Results
3.1. Radiomics
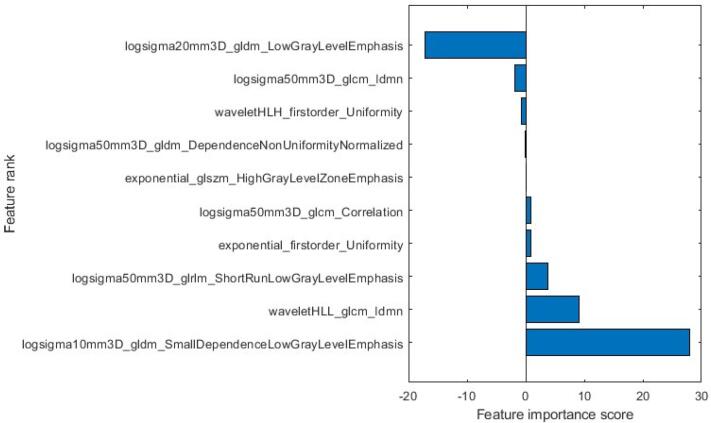
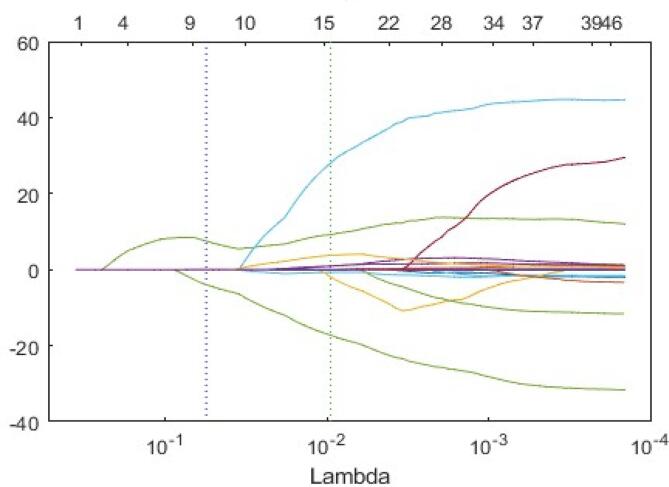
After applying the LASSO algorithm, the ten most significant features were selected to construct the radiomic model. The names of these features, along with their corresponding coefficients, are presented in Fig. 8. Moreover, Fig. 9 illustrates how the LASSO algorithm eliminated radiomics signatures in higher dimensions, ultimately retaining the ten most relevant features for further analysis. [17], [18].
Fig. 8.
Selected radiomic features and their corresponding coefficients.
Fig. 9.
Trace plot of coefficients of LASSO.
3.2. Deep learning
To evaluate the performance of the proposed Inception-ResNet, we use several commonly used classification evaluation metrics, including accuracy (ACC), precision (PRE), recall (REC), F1 score and ROC. The evaluation process involves labeling and classification, and the indicators are calculated based on true positives (TP), false negatives (FN), false positives (FP), and true negatives (TN). The equations used to calculate ACC, PRE, REC, F1 score are shown in Eqs. (6) to (9). [19], [20]
| (6) |
| (7) |
| (8) |
| (9) |
Table 1 contains additional information regarding the performance of the radiomic signature. In the training cohort, the radiomic signature achieved an area under the receiver operating characteristic curve (AUC) of 0.78, indicating a reasonably good discriminative ability. Similarly, in the test cohort, the radiomic signature exhibited an AUC of 0.76, suggesting consistent performance in an independent dataset. The performance metrics used for segmentation model evaluation include pixel precision (PA), Intersection-Over-Union (IoU) and Dice. The related formula as follows in Eqs. (10) to (12).
| (10) |
| (11) |
| (12) |
Table 1.
Prediction accuracy of Radiomics model.
| AUC (%) | ACC (%) | PRE (%) | REC (%) | F1 score (%) | |
|---|---|---|---|---|---|
| Radiomics | 0.76 | 0.74 | 0.71 | 0.70 | 0.72 |
TP, TN, FP and FN denote the number of true positives, true negatives, false positives and false negatives, respectively. The average precision is the area enclosed by the recall-precision curve and the coordinate axis, and n represents the number of categories in the classification. The calculation of average precision is complex, and single-valued limits for precision and recall can be set.
3.3. Performance comparison of segmented MRI
Table 2 presents the performance comparison of the two models. From Fig. 3, we can see that our model has high PA, IoU, and Dice values of 98.001%, 96.819%, and 98.384%, respectively, which is a better performance compared to egion growing algorithm.
Table 2.
Shows the performance comparison of the two model segmentations.
| Model | PA (%) | IoU (%) | Dice (%) |
|---|---|---|---|
| Improved U-Net method | 98.001 | 96.819 | 98.384 |
| Region growing algorithm | 91.445 | 87.007 | 93.052 |
In Fig. 10, the red curve indicates the ROC curve of Improved U-Net method and the blue indicates the ROC curve of Radiomics model. The area between the curve and the lower right corner is the AUC, and the value of AUC represents the diagnostic performance. From Fig. 10, we can see that our method has superior diagnostic performance than Radiomics model.
Fig. 10.
Comparison of the ROC curves of Improved U-Net and Radiomics model.
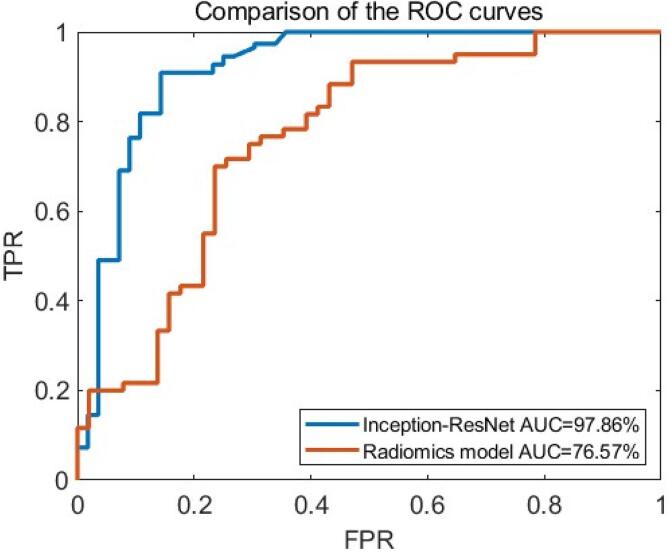
The experimental findings are exhibited in Table 3, delineating the Area Under the Curve (AUC), accuracy, precision, recall, and F1 score of the enhanced U-Net approach for the dataset, aiming to facilitate an exhaustive evaluation of its performance.
Table 3.
Performance of Inception-ResNet:
| Model | AUC (%) | ACC (%) | PRE (%) | REC (%) | F1 score (%) |
|---|---|---|---|---|---|
| Inception-ResNet | 97.86 | 98.56 | 97.41 | 94.34 | 95.85 |
| Radiomics model | 76.57 | 74.45 | 71.51 | 70.31 | 72.54 |
4. Discussion
4.1. Significance of this study
Due to metastasis or complications, a large number of patients died. Early detection of the primary lesion can lead to active treatment. The most common place for cancer to metastasize is the liver, followed by the lungs and the bones. The spine is the most common site of bone metastasis, accounting for about 68% of bone metastases. Some studies have shown that 86% of cancer patients have found spinal metastases in autopsy [1], [2], [3]. Although CT-guided spinal puncture can be used to perform pathological biopsy of metastases, this high-risk invasive procedure is a last resort. Under the condition of the patients with no clear primary focal history, usually as the preferred imaging examination project, but spinal metastases lack of specificity in morphology, can show the osseous or dissolve into osseous changes, also can be single or multiple, identify a lot of difficulties, so often clinically for possible primary focal place for screening a variety of imaging methods one by one, this includes expensive PET-CT. The search for primary lesions is time-consuming and time-consuming. If CE-MRI, a low-risk noninvasive means of spinal metastasis, can be used alone to predict the most likely source of primary lesions, and certain sites can be examined preferentially, the cost and time will be greatly reduced compared with the screening system one by one. CE-MRI is a highly useful diagnostic tool for evaluating tumor angiogenesis, and has been extensively utilized in the diagnosis and preoperative staging of various types of cancers. Moreover, CE-MRI has been increasingly used in the diagnosis of spinal diseases, including primary spinal tumors such as myeloma, chordoma, and lymphoma [16], [17], [18], [19]. In addition, it has also been found to be effective in diagnosing benign lesions such as spinal tuberculosis and giant cell tumor of bone [16], [20], as well as in comparing spinal metastases between different cancer types, such as renal cancer with rich blood supply and prostate cancer with poor blood supply [21], and spinal metastases from other sources [22], [23].
Radiomics can extract massive information in high-throughput images and excavate high-level and deep-level features, which provide far more value for differential diagnosis of spinal tumors than morphological manifestations, and have been widely used to identify cancer subtypes and predict treatment effects and prognosis [24]. Radiomics requires the segmentation of tumors to extract features based on pixel histograms and advanced texture features.
These parameters are combined for feature selection to construct the optimal diagnostic and predictive classifiers. Radiomics is usually used to analyze some conventional magnetic resonance sequence images, such as T1WI, T2WI, DWI, etc., but there are many studies on CE-MRI. For example, Wu et al. [25] used CE-MRI-based radiomics to judge breast cancer classification. There are also studies exploring the correlation between radiomics features based on CE-MRI and Ki-67 antigen expression in other forms of cancer [26], [27]. There are no other studies on spinal metastasis at present.
In this study, features were extracted and analyzed from CE parameter maps: inflow, outflow, and peak. The region growing algorithm was used to standardize the 3D segmentation of lesions. Histogram features were analyzed first, and texture features were added to observe whether the diagnosis results were improved. The rapid development of deep learning depends on the breakthrough of algorithm theory in recent years, the expansion of data samples and powerful image processing hardware equipment. Deep learning has the ability to extract complex features from data, without requiring manual feature engineering, and can be trained using input data to achieve the goal of diagnosis. However, there is a limited amount of research on using CE-MRI as input for deep learning models. One notable study on this topic involves the classification of breast cancer as benign or malignant using deep learning techniques [28].
In this study, 12 layers of CE-MRI images were collected and 12 groups of images were uniformly planned. U-Net segments the image, and then evaluates it with the corresponding evaluation method, such as DICE.
4.2. Analysis of research results
In this study, CE-MRI parameters were calculated to obtain the CE-MRI parameter map. The diagnostic accuracy of radiomics is 0.7445, and the results showed that adding texture features did not have much value in improving the diagnostic accuracy. In order to compare deep learning model with radiomics model, same CE-MRI data were used to validate the deep learning performance. The diagnostic accuracy of deep learning model was 0.9856. The results show that using deep learning model and CE-MRI can achieve high accuracy.
Machine learning technology [29] for AI based medical image diagnosis [30] and physiological assessment of spinal metastases can be implemented to enhance medical analysis. It may be worthwhile noting that medical image segmentation using deep neural networks [31], [32] is also vital in medical image analysis. Furthermore, modelling of spine related cells coupled with an understanding of the cellular interactions [33] may be of high medical informatics value in understanding tumor development and how it affects spinal structures, and other spinal related issues in orthopedics [34].
The use of advanced deep learning segmentation techniques has the potential to enhance medical diagnosis, particularly in the evaluation of spinal metastases. This in turn could have a significant impact on the effectiveness of deep learning for physiological analysis in this area of research.
Using machine intelligence applied to advanced medical diagnostics [35], [36] specifically based on biomedical imaging [37], the accuracy of expert diagnosis can be enhanced, which can help save lives.
The main limitations of the study were the size and diversity of the sample. The study only included 81 patients, 36 of whom were primary malignant spinal bone tumor and 45 had metastasis, and the small sample size may affect the stability and reliability of the study results. In addition, the diversity of the sample is also limited and may not fully reflect the situation of different populations. Therefore, the conclusions of this study need to be further studied and verified.
5. Conclusion
The use of CE-MRI in diagnosing spinal metastases is crucial due to its ability to provide detailed information about tumor angiogenesis and enhance the visualization of lesions. The developed segmentation model demonstrated its potential in assisting radiologists and clinicians in identifying and delineating metastatic lesions, which is essential for treatment planning and monitoring disease progression., this study focused on contrast-enhanced magnetic resonance image (CE-MRI) segmentation for the diagnosis of spinal metastases using an improved U-Net and Inception-ResNet architecture. In conclusion, the results demonstrated the effectiveness of the proposed method in accurately segmenting metastatic lesions in the spine. By incorporating the U-Net and Inception-ResNet networks, the model achieved improved performance in terms of segmentation accuracy and efficiency. The application of an improved U-Net and Inception-ResNet for CE-MRI segmentation in the diagnosis of spinal metastases shows promising results and potential for further advancements in computer-aided diagnosis and treatment planning for patients with spinal metastases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was sponsored by Joint Funds for the Innovation of Science and Technology, Fujian Province (No.2021Y9091) and Fujian Provincial Health Technology Project (2020CXA045).
References
- 1.Piccioli A., Maccauro G., Spinelli M.S., Biagini R., Rossi B. Bone metastases of unknown origin: epidemiology and principles of management [J] Journal of Orthopaedics and Traumatology. 2015;16(2):81–86. doi: 10.1007/s10195-015-0344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ugras N., Yalcinkaya U., Akesen B., et al. Solitary bone metastases of unknown origin [J] Acta Orthopaedica Belgica. 2014;80:139–143. [PubMed] [Google Scholar]
- 3.Zacharia B., Subramaniam D., Joy J. Skeletal Metastasis-an Epidemiological Study [J] Indian J Surg Oncol. 2018;9(1):46–51. doi: 10.1007/s13193-017-0706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar V., Gu Y., Basu S., Berglund A., Eschrich S.A., Schabath M.B., Forster K., Aerts H.J.W.L., Dekker A., Fenstermacher D., Goldgof D.B., Hall L.O., Lambin P., Balagurunathan Y., Gatenby R.A., Gillies R.J. Radiomics: the process and the challenges [J] Magnetic Resonance Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X., Yang W., Weinreb J., Han J., Li Q., Kong X., Yan Y., Ke Z., Luo B.o., Liu T., Wang L. Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning [J] Scientific Reports. 2017;7(1) doi: 10.1038/s41598-017-15720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masood A., Sheng B., Li P., Hou X., Wei X., Qin J., Feng D. Computer-Assisted Decision Support System in Pulmonary Cancer detection and stage classification on CT images [J] Journal of Biomedical Informatics. 2018;79:117–128. doi: 10.1016/j.jbi.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 7.van der Burgh H.K., Schmidt R., Westeneng H.-J., de Reus M.A., van den Berg L.H., van den Heuvel M.P. Deep learning predictions of survival based on MRI in amyotrophic lateral sclerosis [J] Neuroimage Clin. 2017;13:361–369. doi: 10.1016/j.nicl.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang N., Zhang E.L., Su M.Y., et al. Dynamic contrast-enhanced MRI in differential diagnosis of spinal myeloma and primary non-Hodgkin's lymphoma [J] Chinese Journal of Medical Imaging. 2018;26:135–139. [Google Scholar]
- 9.Huang S.H., Chu Y.H., Lai S.H., et al. Learning-based vertebra detection and iterative normalized-cut segmentation for spinal MRI [J] IEEE Transactions on Medical Imaging. 2009;28:1595–1605. doi: 10.1109/TMI.2009.2023362. [DOI] [PubMed] [Google Scholar]
- 10.Haralick R.M., Shanmugam K., Dinstein I. Textural Features for Image Classification [J] IEEE Transactions on Systems, Man, and Cybernetics. 1973;SMC-3(6):610–621. [Google Scholar]
- 11.Liaw A., Wiener M. Classification and Regression by Inception-ResNet [J] R News. 2002;2:18–22. [Google Scholar]
- 12.Chang P., Grinband J., Weinberg B.D., Bardis M., Khy M., Cadena G., Su M.-Y., Cha S., Filippi C.G., Bota D., Baldi P., Poisson L.M., Jain R., Chow D. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas [J] American Journal of Neuroradiology. 2018;39(7):1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochreiter S., Schmidhuber J. Long short-term memory [J] Neural Computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 14.Nair V., Hinton G.E. Omnipress; Haifa, Israel: 2010. Rectified linear units improve restricted boltzmann machines [C] [Google Scholar]
- 15.Baldi P., Sadowski P. The Dropout Learning Algorithm [J] Artificial Intelligence. 2014;210:78–122. doi: 10.1016/j.artint.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang N., Su M.-Y., Xing X., Yu H.J., Yuan H. Morphological and dynamic contrast enhanced MR imaging features for the differentiation of chordoma and giant cell tumors in the Axial Skeleton [J] Journal of Magnetic Resonance Imaging. 2017;45(4):1068–1075. doi: 10.1002/jmri.25414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang N., Yuan H., Yu H.J., Su M.-Y. Diagnosis of Spinal Lesions Using Heuristic and Pharmacokinetic Parameters Measured by Dynamic Contrast-Enhanced MRI [J] Academic Radiology. 2017;24(7):867–875. doi: 10.1016/j.acra.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang P., Honda G., Roberts T., Vahlensieck M., Johnston J.O., Rosenau W., Mathur A., Peterfy C., Gooding C.A., Genant H.K. Musculoskeletal neoplasm: perineoplastic edema versus tumor on dynamic postcontrast MR images with spatial mapping of instantaneous enhancement rates [J] Radiology. 1995;197(3):831–839. doi: 10.1148/radiology.197.3.7480764. [DOI] [PubMed] [Google Scholar]
- 19.Moulopoulos L.A., Maris T.G., Papanikolaou N., Panagi G., Vlahos L., Dimopoulos M.A. Detection of malignant bone marrow involvement with dynamic contrast-enhanced magnetic resonance imaging [J] Annals of Oncology. 2003;14(1):152–158. doi: 10.1093/annonc/mdg007. [DOI] [PubMed] [Google Scholar]
- 20.Lang N., Su M.-Y., Yu H.J., Yuan H. Differentiation of tuberculosis and metastatic cancer in the spine using dynamic contrast-enhanced MRI [J] European Spine Journal. 2015;24(8):1729–1737. doi: 10.1007/s00586-015-3851-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha A, Peck KK, Lis E, et al. Magnetic resonance perfusion characteristics of hypervascular renal and hypovascular prostate spinal metastases: clinical utilities and implications [J]. Spine (Phila Pa 1976). 2014. 39. E1433-E1440. [DOI] [PMC free article] [PubMed]
- 22.Khadem N.R., Karimi S., Peck K.K., Yamada Y., Lis E., Lyo J., Bilsky M., Vargas H.A., Holodny A.I. Characterizing hypervascular and hypovascular metastases and normal bone marrow of the spine using dynamic contrast-enhanced MR imaging [J] American Journal of Neuroradiology. 2012;33(11):2178–2185. doi: 10.3174/ajnr.A3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang N., Su M.-Y., Yu H.J., Lin M., Hamamura M.J., Yuan H. Differentiation of myeloma and metastatic cancer in the spine using dynamic contrast-enhanced MRI [J] Magnetic Resonance Imaging. 2013;31(8):1285–1291. doi: 10.1016/j.mri.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Zhu Y., Burnside E.S., Drukker K., Hoadley K.A., Fan C., Conzen S.D., Whitman G.J., Sutton E.J., Net J.M., Ganott M., Huang E., Morris E.A., Perou C.M., Ji Y., Giger M.L. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays [J] Radiology. 2016;281(2):382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Peiqi, Zhao Ke, Wu Lei. et al. Relationship between radiomics features and molecular classification of breast cancer based on diffusion weighted imaging and dynamic contrast-enhanced MRI [J]. Chin J Radiology. 2018. 52. 338-343.
- 26.Ma W., Ji Y., Qi L., Guo X., Jian X., Liu P. Breast cancer Ki67 expression prediction by DCE-MRI radiomics features [J] Clinical Radiology. 2018;73(10):909.e1–909.e5. doi: 10.1016/j.crad.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 27.Juan M., Yu J.i., Peng G., Jun L., Feng S., Fang L. Correlation between DCE-MRI radiomics features and Ki-67 expression in invasive breast cancer [J] Oncology Letters. 2018 doi: 10.3892/ol.2018.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antropova N., Abe H., Giger M.L. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks [J] J Med Imaging (Bellingham) 2018;5(01):1. doi: 10.1117/1.JMI.5.1.014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong K.K.L., Fortino G., Abbott D. Deep learning-based cardiovascular image diagnosis: A promising challenge. Future Generation Computer Systems. 2020;110:802–811. [Google Scholar]
- 30.Wong K.K.L., Wang D., Ko J.K.L., Mazumdar J., Le T.-T., Ghista D. Computational Medical Imaging and Hemodynamics Framework for Functional Analysis and Assessment of Cardiovascular Structures. BioMedical Engineering OnLine. 2017;16(1) doi: 10.1186/s12938-017-0326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong K.K.L., Zhang A.n., Yang K.e., Wu S., Ghista D.N. GCW-UNet segmentation of cardiac magnetic resonance images for evaluation of left atrial enlargement[J] Computer Methods and Programs in Biomedicine. 2022;221:106915. doi: 10.1016/j.cmpb.2022.106915. [DOI] [PubMed] [Google Scholar]
- 32.Zhao M., Wei Y., Lu Y.u., Wong K.K.L. A novel U-Net approach to segment the cardiac chamber in magnetic resonance images with ghost artifacts. Computer Methods and Programs in Biomedicine. 2020;196:105623. doi: 10.1016/j.cmpb.2020.105623. [DOI] [PubMed] [Google Scholar]
- 33.Wong K.K.L. Three-dimensional discrete element method for the prediction of protoplasmic seepage through membrane in a biological cell. Journal of Biomechanics. 2017;8(65):115–124. doi: 10.1016/j.jbiomech.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 34.Deng X., Zhu Y., Wang S., Zhang Y., Han H., Zheng D., Ding Z., Wong K.K.L. CT and MRI Determination of Intermuscular Space within Lumbar Paraspinal Muscles at Different Intervertebral Disc Level. PLoS One1. 2015;10(10):e0140315. doi: 10.1371/journal.pone.0140315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu S., Zhang Z., Yan Z., Wang Y., Cheng T., Zhou R., Yang G. Mutually aided uncertainty incorporated Dual Consistency Regularization with Pseudo Label for Semi-Supervised Medical Image Segmentation. Neurocomputing. 2023;126411 [Google Scholar]
- 36.K.K.L. Wong, Cybernetical Intelligence: Engineering Cybernetics with Machine Intelligence, John Wiley & Sons Limited, England, U.K., ISBN: 9781394217489, 2024.
- 37.Wong K.K.L., Kelso R.M., Worthley S.G., Sanders P., Mazumdar J. Medical imaging and processing methods for cardiac flow reconstruction. Journal of Mechanics in Medicine and Biology. 2009;9(1):1–20. [Google Scholar]