Summary
Over 200 genes are known to underlie human congenital hearing loss (CHL). Although transcriptomic approaches have identified candidate regulators of otic development, little is known about the abundance of their protein products. We used a multiplexed quantitative mass spectrometry-based proteomic approach to determine protein abundances over key stages of Xenopus otic morphogenesis to reveal a dynamic expression of cytoskeletal, integrin signaling, and extracellular matrix proteins. We correlated these dynamically expressed proteins to previously published lists of putative downstream targets of human syndromic hearing loss genes: SIX1 (BOR syndrome), CHD7 (CHARGE syndrome), and SOX10 (Waardenburg syndrome). We identified transforming growth factor beta-induced (Tgfbi), an extracellular integrin-interacting protein, as a putative target of Six1 that is required for normal otic vesicle formation. Our findings demonstrate the application of this Xenopus dataset to understanding the dynamic regulation of proteins during otic development and to discovery of additional candidates for human CHL.
Subject areas: Molecular network, Developmental biology, Proteomics, Model organism
Graphical abstract
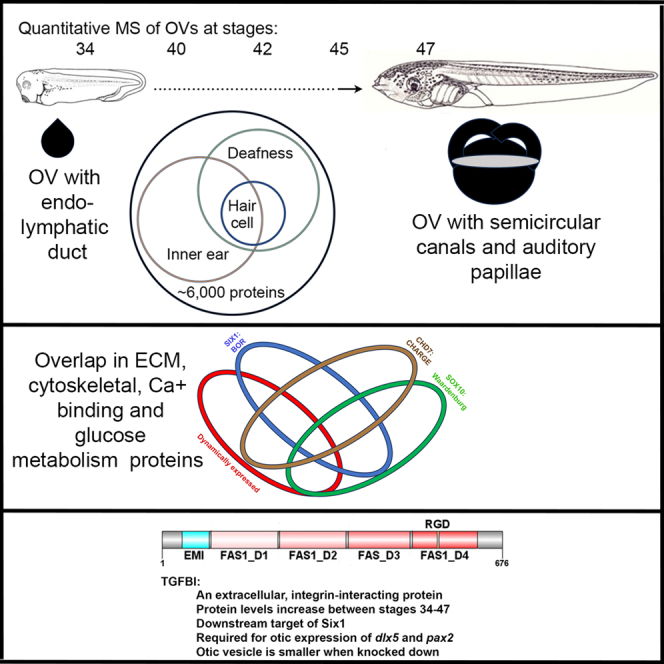
Highlights
-
•
Quantitative MS was used to determine protein abundances during otic morphogenesis
-
•
Cytoskeletal, integrin signaling, and ECM proteins are dynamically expressed
-
•
Comparison to congenital hearing loss datasets reveals potential disease targets
-
•
Tgfbi is a Six1 target that is required for otic gene expression and morphogenesis
Molecular network; Developmental biology; Proteomics; Model organism
Introduction
The vertebrate inner ear is an intricate sensory organ comprising a fluid filled labyrinth containing sensory hair cells that detect sound (hearing), rotation (spatial orientation), and acceleration (balance). Defects in the development of the inner ear commonly lead to congenital hearing loss (CHL), a prevalent birth defect in which about 20–30% of children have inner ear malformations. Although more than 200 genes are already implicated as causative for CHL, it is estimated that hundreds more remain to be identified.1 Recent studies in mammals, have primarily focused on the development of inner ear sensory patches and hair cell differentiation because these structures are essential for audition.2,3,4,5,6,7 However, advancing our understanding of the molecular programs that orchestrate earlier aspects of inner ear development, such as otic morphogenesis and patterning, is likely to reveal additional candidates that underlie unexplained causes of human CHL.
An important step toward understanding how the inner ear develops is to evaluate how its transcriptome and proteome change over time as it develops from a fluid-filled otic vesicle (OV) into a complex, three-dimensional, labyrinth housing both vestibular and auditory sensory organs. Early studies cataloged a number of protein-coding genes expressed in different regions of the fetal and adult inner ear, many of which caused an otic defect when mutated.8,9,10 These studies assisted in identifying genes that are essential for inner ear function but lack information on their dynamic expression during otic development. More recent transcriptomic analyses of purified inner ear cell types provided important insights into inner ear and hair cell developmental processes,2,6,11,12 but similar reports during early otic morphogenesis are not yet available. In addition, although transcriptomic studies provide a sensitive measure of gene expression, proteomic analyses represent gene product maturation and a measure of functioning pathways at a given time. Since there are several examples in developing systems in which mRNA and protein levels do not correlate,13,14 we chose to directly measure protein abundance levels over developmental time to provide information regarding the molecular dynamics involved in the early steps of otic morphogenesis.
To accomplish this, we performed a time-resolved quantitative proteomic analysis for five stages of otic development of Xenopus: from a simple, larval OV to a complex tadpole inner ear that contains both vestibular and auditory domains, semicircular canals, and differentiated hair cells. Xenopus was chosen as an excellent model in which to conduct this analysis because, as a tetrapod, its inner ear elements are highly homologous to human, its genome and proteome are well defined, and its transparency at the stages chosen allows a simple, non-protease–based dissection of the OV. We report the dynamic expression and enrichment of extracellular matrix (ECM) and cytoskeletal proteins as the OV undergoes morphogenetic patterning to form a complex labyrinth.
To identify potential candidates for human CHL, our comprehensive Xenopus protein dataset was mined for putative downstream targets of causative genes for branchiootorenal (BOR), CHARGE, and Waardenburg syndrome, leveraging publicly available differential datasets. We show that one such candidate, transforming growth factor beta-induced (Tgfbi), is: an ECM integrin-interacting protein whose expression is upregulated over the course of otic morphogenesis; a putative downstream target of the BOR gene Six1; and is required for normal OV gene expression and size. These results demonstrate the application of our Xenopus dataset to understanding the dynamic expression of proteins during otic morphogenesis and to the discovery of additional candidates for understanding the causes of human CHL.
Results
Time-resolved high-resolution mass spectrometry based proteomic analysis over the course of early otic morphogenesis
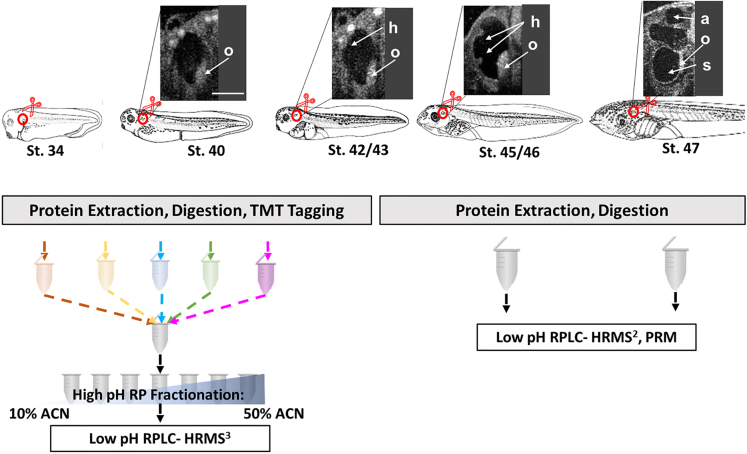
To assess proteome dynamics during the course of Xenopus otic morphogenesis, we manually dissected the entire OV, including its internal fluid, from the surrounding head mesoderm with sharpened forceps. Because larvae and tadpoles are nearly transparent and the OVs are encapsulated in a reflective ECM, no protease treatment was necessary to manually dissect OVs cleanly from the surrounding tissue. We collected samples from 5 distinct stages of otic morphogenesis and hair cell differentiation: (1) simple, fluid-filled OVs with a nascent endolymphatic duct starting to emerge (stage 3415); (2) polarized OVs with distinguishable pars superior (vestibular) and pars inferior (auditory) (stage 40); (3) OVs containing pouches in the pars superior that presage the horizontal, anterior, and posterior semicircular canals (stage 42); (4) OVs in which the protrusions that will form the horizontal canal grow further inward and stereocilia bundles on hair cells are very distinct (∼5 μm in length) (stage 45); and (5) OVs in which the axial protrusions are fused to form the horizontal, anterior, and posterior semicircular canals and the sensory patches in the sacculus, utricle, semicircular canals, and auditory lagena are recognizable and stereocilia bundles are established (stage 47)16,17 (Figure 1). These stages were chosen because they include the simplest structure displaying onset of tubular duct growth (st 34), the initial polarization and compartmentalization of the OV into distinct functional regions and the appearance of otoliths (st 40), the onset of semicircular canal formation (st 42), the onset of sensory hair cell formation (st 45) and the completion of semicircular canal formation, sensory patch formation and hair cell differentiation (st 47). Five biological replicates from these 5 distinct stages were subjected to high-resolution mass spectrometry (HRMS) based proteomic analysis.
Figure 1.
Workflow for time-resolved proteome analysis of otic morphogenesis
Xenopus larvae and tadpoles from stages 34–47 were selected for otic proteome analysis. These stages were chosen because they include the simplest structure displaying onset of tubular duct growth (st 34), the initial polarization and compartmentalization of the OV into distinct functional regions and the appearance of otoliths (st 40), the onset of semicircular canal formation (st 42/43), the onset of sensory hair cell formation (st 45) and the completion of semicircular canal formation, sensory patch formation and hair cell differentiation (st 47). Insets show stage associated optical coherence tomography-based images of the otic vesicle. Otolith (o), pouch and protrusions for the horizontal canal (h), anterior canal (a), sacculus (s). Bar indicates 100 μm. Otic vesicles were dissected and processed using a bottom-up proteomic workflow. Peptides were tagged with isobaric mass tags to quantify relative changes in protein abundances during the course of otic morphogenesis. Tagged peptides were fractionated offline using RP spin columns at a high pH and then analyzed by LC-HRMS. Quantification of select proteins was verified using targeted (PRM) HRMS.
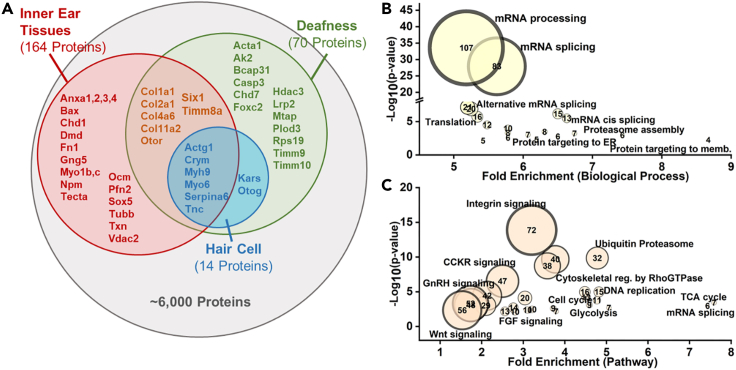
Overall, combining the results from all 5 stages of otic development, about 6,000 proteins were identified. To assess the depth of coverage, we compared our Xenopus dataset to previously published transcriptomic data from Xenopus inner ears as well as proteomic data from dissected postnatal mouse inner ear tissues.4,18 From our dataset of ∼6000 proteins, 178 proteins were previously shown to be present in inner ear tissues (n = 164) and hair cells (n = 14) (Figure 2A), including proteins that play important roles in proper functioning of the inner ear such as alpha-tectorin (Tecta), a protein present in the tectorial membrane that connects the stereocilia bundles of hair cells (Table S1). We also compared our Xenopus dataset to known human deafness genes reported on OMIM and matched >70 proteins to genes that are associated with syndromic CHL such as SIX1 (associated with BOR syndrome, OMIM entry 601205) and CHD7 (associated with CHARGE syndrome, OMIM entry 608892). The identification of a large number of proteins known to have functional roles in inner ear and hair cell development, as well as proteins that are associated with human CHL syndromes indicates that the depth of coverage in our analysis is sufficient to gain meaningful insights from our temporal dataset into pathways that regulate otic morphogenesis.
Figure 2.
Characterization of the global proteome
(A) Approximately 6,000 proteins were identified from otic tissues across all analyzed developmental stages and the five biological replicates. Proteins correlated with publicly available proteomic and transcriptomic datasets from inner ear tissues. A number of proteins previously reported in inner ear tissues (red) and hair cells (blue) were included in our dataset. Additional proteins correlated with deafness genes reported on OMIM (green).
(B) Over-representation analysis (PantherDB) for biological processes demonstrating a fold enrichment >5 indicated an enrichment of processes such as mRNA processing and splicing in the otic proteome.
(C) Over-representation analysis (PantherDB) for pathways demonstrated an enrichment in pathways such as Integrin, Wnt, and FGF signaling.
Identified proteins were annotated for function using the PantherDB gene list analysis, which showed an over-representation (>5-fold enrichment) of biological processes relating to mRNA splicing, processing, translation and protein transport to the membrane across all stages (Figure 2B). This indicated active transcription, mRNA processing, and protein expression during otic morphogenesis, as would be expected for a tissue undergoing dramatic growth and differentiation. Pathway enrichment analysis indicated a significant over-representation (>5-fold enrichment) of important metabolic and signaling pathways (Figure 2C), including proteins related to: (1) energy metabolism such as glycolysis and the TCA cycle; (2) ubiquitin proteasome and proteasome assembly proteins that play key roles during mouse hair cell development19,20,21; (3) Wnt and FGF pathways that are essential for proper patterning of the OV19,22; (4) integrin signaling proteins; and (5) cytoskeleton regulation pathways, many of which are important for the specialized differentiation of cell types in the inner ear.23
The otic proteome is remodeled over developmental time
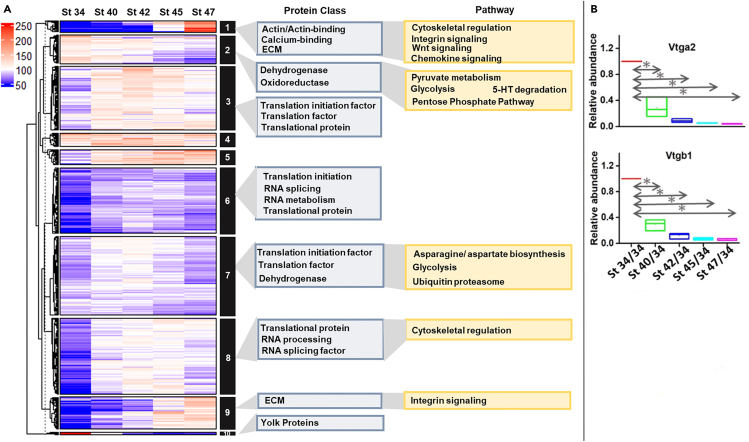
To assess the dynamics of protein expression across the 5 distinct stages of otic morphogenesis and hair cell differentiation, we performed a hierarchal cluster analysis (with K means clustering, k = 10), and generated 10 clusters that represented distinct protein abundance patterns as otic development temporally progressed (Figure 3). Cluster 10 primarily represented yolk proteins that decrease in abundance from stage 34 to stage 47; the representative results for two yolk protein (vitellogenin) fragments, Vtga2 and Vtgb1, are shown in Figure 3B. In Xenopus, intracellular yolk proteins are depleted as an energy source supporting embryonic metabolism until tadpoles begin to feed (after stage 45). Therefore, the expected decrease in the abundance of yolk proteins represented in our dataset confirmed the quantitative accuracy of our time-resolved analysis.
Figure 3.
Temporal proteome dynamics during otic development
(A) Hierarchical clustering analysis supplemented by k-means clustering, cluster numbers indicated in adjoining black bars. Overrepresented GO categories for dynamic clusters for protein class (blue) and pathway (yellow) are shown.
(B) Relative quantification of yolk protein fragments Vtga2 and Vtgb1 over developmental stages when maternal yolk proteins are metabolized. ∗, p < 0.05 (ANOVA). See also Table S1.
Cluster 2 also represented proteins that decrease in abundance between stages 34 and 47. They were stable or showed a modest decrease in abundance (∼1.5-fold) between stages 34 and 43 and a larger decrease (>3-fold) by stage 47. The proteins in Cluster 2 were over-represented in glycolysis and pyruvate metabolism. Experiments using human embryonic stem cells have demonstrated that dynamic regulation of glycolytic metabolism is critical for neural differentiation.24,25 These experiments primarily indicate a modification of the glycolytic pathway such that pyruvate is shunted away from mitochondrial respiration in favor of lactate synthesis even in the presence of free oxygen. In addition, aerobic glycolysis and lactate synthesis have been shown to promote neurosensory development in the OV via activation of FGF signaling.26 For example, phosphoglycerate kinase 1 (Pgk1) is co-expressed with FGF ligands in the mouse OV and promotes FGF signaling by promoting synthesis and secretion of lactate.26 Lactate in turn activates MAPK and basal levels of Etv5b, an important effector of FGF signaling, and promotes cell responsiveness to dynamic FGF signaling during otic development. It is interesting in our dataset that Pgk1 abundance increased between stages 34 and 40 and decreased at later stages, suggesting an important developmental window for FGF signaling. An enrichment of glycolysis and pyruvate metabolism proteins in earlier stages (34–40, 40–42) over the course of otic development could indicate a metabolic switch from aerobic respiration (TCA cycle) to lactate synthesis and metabolism as the inner ear develops.
Clusters 4–6 represent proteins that were stably expressed across all stages of otic development with a <1.5-fold change in abundance between stages 34 and 47. These proteins included ribosomal proteins and other proteins such as Sod1 and Prdx2 that are associated with regulating cellular oxidative state. Housekeeping proteins such as Gapdh were also represented in these Clusters.
Clusters 3 and 8 represented proteins that became enriched at intermediate stages (stage 40, 42, 43, 45), when OV wall protrusions form and grow centrally, and decreased by stage 47 when the protrusions have fused and all compartments of the inner ear fully form.16 Proteins involved in mRNA processing and translation increased during these intermediate stages of otic morphogenesis. For example, mRNA splicing proteins such as Prpf3 and Snrpa were more abundant at stages 42 and 45 relative to stage 34. Translation initiation and elongation factors (e.g., Eif5a, Eif3b, Etf1, Eef2), ribosomal proteins (e.g., Rps20, Rplp1, Rplp2, Rpl7, Rpl9), as well as tRNA ligases (e.g., Sars2, Iars2, Farsb) showed a similar dynamic pattern. These data suggest upregulation in translational activity as development progresses from stage 34–45 during which the size of the otic tissue dramatically increases, OV wall protrusions form and grow centrally, and sensory patches form.
Clusters 1 and 9 represent proteins that increased in abundance by later stages of otic development with a >3-fold increase in abundance in the inner ear (stages 45 and 47) compared to stage 34. Proteins grouped in Cluster 1 showed a 5-fold increase in abundance at stage 47 compared to stage 34 and included proteins that regulate the cytoskeleton, Wnt signaling, integrin signaling, chemokine signaling, actin binding, calcium binding, and contribute to the ECM. Cluster 1 included actin proteins (Acta1, Actb) and actin-binding proteins. β-actin is important for stereocilia maintenance and mutations in human β-actin are associated with deafness.27,28,29 Our quantitative analysis also revealed that skeletal and cardiac myosin proteins (Myh1, Myh6) increase during hair cell differentiation and stereocilia growth from stage 34 to stage 47 otic tissues. These dynamic changes are expected since these cytoskeletal proteins are involved in tissue growth as well as inner ear cell type differentiation.
Cluster 1 also included proteins regulated by Wnt signaling (e.g., Acta1, Actb, Myh3). An increase in abundance over developmental time is consistent with the observations that canonical Wnt signaling is required for the fusion of the protrusions from the OV wall to form the semicircular canals,30 regulates proliferation in the prosensory domain and maintains hair cell numbers.31 In addition, cytoskeletal remodeling through the Wnt-PCP pathway is required for proper orientation of hair cell bundles in the mouse cochlea.32 However, in our dataset the abundance of β-catenin was similar across all stages of otic development, the secreted frizzled related protein 1 (Sfrp1), an antagonistic inhibitor of Wnt signaling, was highest during intermediate stages (stages 40–45), and Wnt ligands were not detected.
Cluster 9 (∼4-fold increase at stages 45 and 47 compared to stage 34) also included integrin signaling and ECM proteins (Figure 3A). Notable amongst the ECM proteins are: (1) a number of collagens (Col2a1, Col3a1, Col9a2, Col9a3, Col11a1, Col11a2, Col12a) that were significantly more abundant at stages 45 and 47 compared to stage 34, some of which are associated with CHL syndromes and (2) proteins found in cochlear sensory epithelia (Aggrecan (Acan), hyaluronan and proteoglycan link proteins 1 (Hapln1)), that were significantly more abundant at stage 47 (Figure 3A).
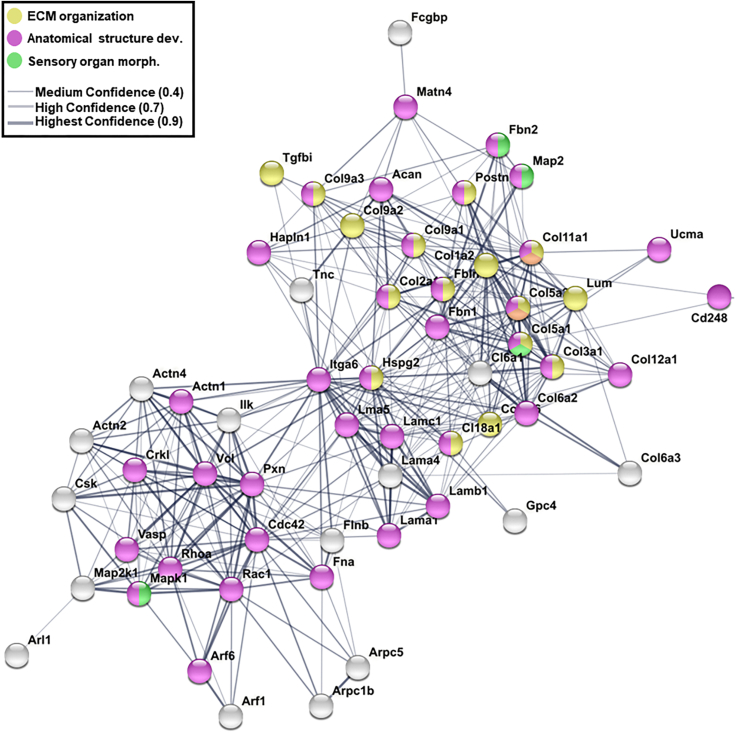
STRING analysis of ECM and integrin-interacting proteins present within Cluster 1 and Cluster 9 suggest a functional interaction network with enrichment of biological processes associated with ECM organization (e.g., Tgfbi, Lum, Col1a2), anatomical structure formation (e.g., Acan, Hapln1, Fbn2, Pxn), as well as sensory organ morphogenesis (e.g., Fbn2, Mapk1, Map2) (Figure 4). These results would be expected for a tissue undergoing significant morphological changes in which ECM proteins function as adhesive substrates to guide cell migration, control the spatial distribution of growth factors and cytokines and define tissue boundaries.33 Therefore, it is likely that ECM proteins have important roles in regulating the functional compartmentalization of the OV, the rapid growth of its wall protrusions that contribute to the complex labyrinthine morphology, and formation of sensory patches during stage 45 and 47.
Figure 4.
Integrin and ECM protein interaction networks
STRING analysis and pathway enrichment analysis using the Xenopus interaction database show enrichment of processes related to ECM organization (yellow), anatomical structure development (pink), and sensory organ development (green). Interactions among proteins are reported with medium to highest confidence represented by thickness of lines.
CHL candidate genes are dynamically expressed
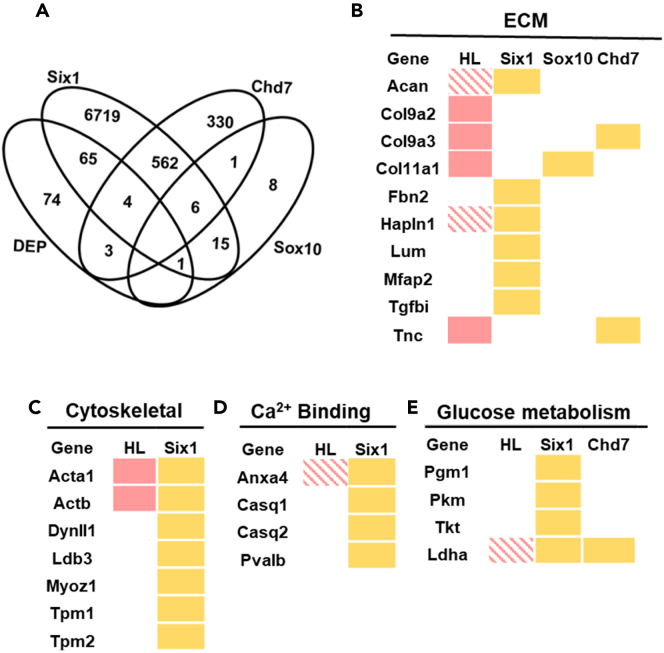
The genetic etiologies of many human syndromes that include hearing loss are not fully understood. To identify genes that might cause of CHL, we correlated the set of dynamically expressed proteins (DEPs = 206 (144 unique proteins)) in our Xenopus dataset to genes that are putative binding partners, downstream targets, effectors, and/or regulators of other genes that are known to be causative for three human CHL syndromes: BOR (SIX1), CHARGE (CHD7), and Waardenburg (SOX10) (Figure 5A; Table S2). To identify Six1-related genes, we manually curated lists of putative downstream targets of Six1 previously identified in the preplacodal ectoderm34,35 and the cochlear sensory epithelium.36 To identify Chd7-related genes, we compiled a list of proteins that are putatively regulated by CHD7 via epigenetic regulation of their enhancers in granule cell precursors and cerebellar morphogenesis.37 To identify genes related to Waardenburg syndrome, we compared our list of DEPs to putative downstream targets of two genes SOX10 and MITF in human iPSC derived neural crest cells and human melanoma cells.38,39 The majority of Six1-related genes (86%) did not overlap with Chd7-related or Sox10-related genes. There were ∼550 genes that overlapped between Six1- and Chd7-related genes, 22 genes that overlapped between Six1- and Sox10-related genes, and 7 genes that overlapped between Chd7- and Sox10-related genes. Our DEP dataset overlapped with 70 Six1-related, 7 Chd7-related and one Sox10-related proteins, indicating that the DEP dataset highly represents Six1-related genes.
Figure 5.
Dynamically expressed proteins overlap with putative downstream targets of CHL genes
(A–E) (A) Venn diagrams show overlap between dynamically expressed proteins (DEPs) and BOR (Six1), CHARGE (Chd7) and Waardenburg (Sox10) associated genes (B–E) Dynamically regulated ECM (B), cytoskeletal (C), calcium binding (D), and glucose metabolism (E) proteins that are putative downstream targets of CHL genes. Filled pink boxes are based on human data whereas the hatched pink boxes are based on mouse knockout data. Yellow boxes indicate genes in these categories found in the datasets analyzed in A. See also Table S2.
Mutations in SIX1 is one genetic cause of BOR, which is an autosomal dominant syndrome characterized by defects in the development of hyoid tissues, external, middle, and inner ear, and kidney.40 We found that of 7366 putative downstream targets of Six1, 70 overlapped with our DEP set (Figure 5A). These included ECM proteins such as Acan and Hapln1, which have been shown to be necessary for mouse otic and hair cell development.41 We then compared this list of 70 DEPs that overlap with Six1 to currently known human hearing loss genes to identify potential candidate genes that may be useful in identifying unexplained cases of BOR. This correlation indicated ECM proteins such as Fbn2, Lum, Mfap2 and Tgfbi, which have not yet been studied in the context of otic development and hearing loss, as potential BOR candidates (Figure 5B). This analysis also identified cytoskeletal proteins known to be hearing loss genes (Acta1, Actb) as Six1 targets in the DEP dataset, and Dynll1, Ldb3, Myoz1, Tpm1 and Tmp2 as potential BOR candidates (Figure 5C). Other putative BOR candidates were identified as proteins involved in calcium binding, e.g., Anxa4 that is expressed in auditory and vestibular hair cells42 (Figure 5D).
CHARGE syndrome is primarily caused by mutations in CHD7, which encodes an ATP-dependent chromatin remodeling protein. This is an autosomal dominant syndrome in which patients present with complex phenotypes, including coloboma, heart defects, choanal atresia, retardation in growth and development in addition to inner ear defects resulting in hearing loss and vestibular dysfunction. Of the 870 Chd7-related genes, only 7 overlapped with our DEP dataset, including ECM proteins (Col9a3, Tnc) that are associated with hearing loss in humans (Figure 5B). This list also included Ldha, which plays a role in lactate metabolism and also is a putative downstream target of Six1 (Figure 5E). There was no overlap between Chd7-related genes and our DEP dataset in the cytoskeletal or calcium binding categories (Figures 5C and 5D).
Waardenburg syndrome is caused by mutations in SOX10 and MITF, resulting in abnormalities in neural crest cell proliferation, migration, and differentiation.43 It is characterized by neurosensory hearing loss and pigmentation abnormalities, often presented in hair, eyes, skin, and the neural crest derived melanocytes of the cochlear stria vascularis. Comparison of our DEP dataset to putative downstream targets of Sox10 or MITF revealed only two overlapping proteins: Col11a1, an ECM protein that is a putative downstream target of Sox10 in human iPSCs (Figure 5B),39 and Apoe, an apolipoprotein that is a putative downstream target of MITF in human melanoma cells.38 No targets of Sox10 or Mitf overlapped with our DEP dataset in the other categories (Figures 5C–5E).
Transforming growth factor beta-induced (Tgfbi) is an ECM protein required for normal otic development
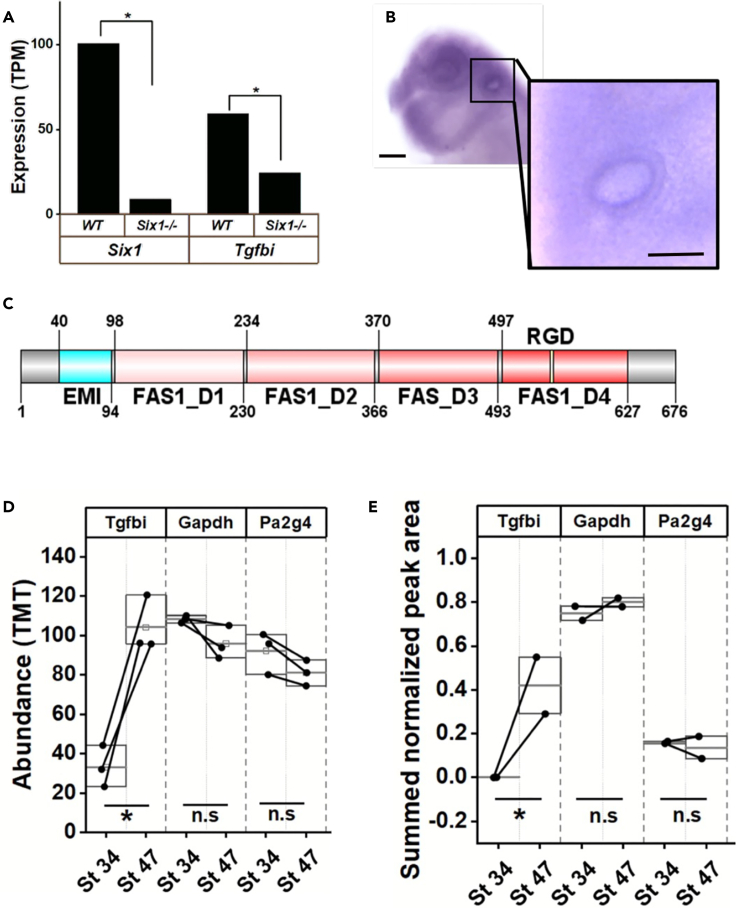
The correlation of upregulated ECM proteins with putative downstream targets of Six1 indicated four potential BOR candidates that are not well characterized in the context of inner ear development or function: Fbn2, Lum, Mfap2, and Tgfbi (Figure 5C). We chose to further analyze the expression of Tgfbi because several observations suggested a link to hearing loss. First, ChIP-sequencing experiments in mouse OVs showed that Six1 bound in close proximity to Tgfbi.36 Second, we find that at the transcript level Tgfbi is significantly reduced in OVs dissected from E10.5 Six1-null mice compared to OVs dissected from wild type littermates (Figure 6A). Together these findings indicate that Tgfbi is likely a direct target of Six1 in the developing inner ear. Third, tgfbi mRNA is expressed in the Xenopus larval OV44 (confirmed by us, Figure 6B). Fourth, Tgfbi is secreted into the perilymph of the human inner ear.45 Therefore, we characterized Tgfbi protein abundance over the course of Xenopus otic morphogenesis and determined whether it is required for normal OV development. Tgfbi is a secreted protein containing four fasciclin 1 (FAS1) domains and an RGD motif that facilitates cell adhesion and migration via interactions with integrins (e.g., αvβ3 and αvβ5)46,47 (Figure 6C).
Figure 6.
Characterization of Tgfbi expression
(A) Tgfbi is a putative downstream target of Six1. RNA-seq of E10.5 mouse OVs from Six1+/+ and Six1−/− embryos was done to evaluate gene expression changes in the absence of Six1. Expression of Tgfbi significantly decreased in Six1-null OVs compared to wild type (WT) OVs of littermates (∗, p < 0.05).
(B) Expression pattern of Tgfbi as assessed by in situ hybridization at stage 32 (lateral view). Bar indicates 200 μm. Inset shows Tgfbi expression in otic vesicle. Bar indicates 100 μm.
(C) Xenopus laevis Tgfbi contains one EMI domain, four FAS1 domains and the RGD motif. (source Uniprot). Domain visualization generated using DOG (version 2).48 Numbers indicate amino acid residues.
(D) Label based (TMT) abundance for Tgfbi, Gapdh, and Pa2g4 between stage 34 and 47 otic vesicles (paired Student’s t test for unequal variance, ∗, p < 0.05).
(E) Summed and normalized peak intensities for Tgfbi, Gapdh, and Pa2g4 between stage 34 and 47 otic vesicles (paired Student’s t test for unequal variance, ∗, p < 0.05). n.s., not significant.
Relative quantification based mass spectrometry measurements showed that Tgfbi protein increases in abundance between stages 34 and 47 (Figures 3 and 6D). This was confirmed by using parallel reaction monitoring mass spectrometry (PRM-MS/MS) wherein proteotypic peptides of proteins are assayed using a targeted approach and peptide quantity is assessed based on area under the curve of extracted ion chromatograms. Two independent peptides of Tgfbi from OVs dissected from stage 34 and stage 47 were assayed alongside peptides from the reference gene Gapdh. This approach also showed that Tgfbi increased in abundance between stages 34 and 47 OVs (Figures 6D and 6E). With both proteomic assays, the housekeeping protein Gapdh and Pa2g4, a Six1 co-factor expressed in the otic placode and OV,49 showed similar abundances at both stages (Figures 6D and 6E).
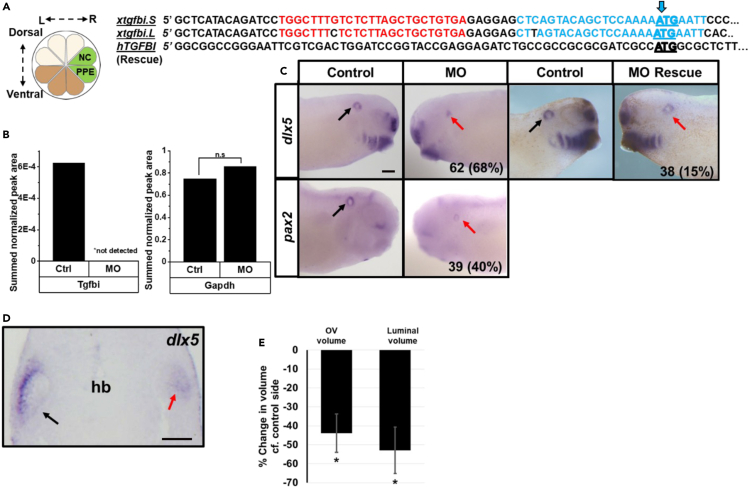
To determine whether Tgfbi is required during the early stages of Xenopus OV development, we knocked-down (KD) the levels of endogenous Tgfbi protein in neural crest and preplacodal ectoderm by microinjecting two different translation-blocking antisense morpholino oligonucleotides (MOs) into their precursor blastomeres (Figure 7A). The MOs were designed to target the 5′ UTR and the ATG translational start site of tgfbi mRNA encoded at loci on both the long (.L) and short (.S) Xenopus laevis chromosomes (Figure 7A). MOs were injected on one side of the embryo so that the uninjected side of the same embryo served as an internal control. KD of Tgfbi was confirmed using PRM-MS/MS: Tgfbi peptides were not detected in OVs dissected from the MO injected sides compared to the control sides of the same embryos, indicating the efficacy of the MOs, whereas there were no significant effects on the expression of Gapdh or Pa2g4, indicating the specificity of the MOs (Figure 7B). Next, we assessed the expression of two genes that are known to be expressed in larval OVs (dlx5, pax2) in Tgfbi morphants. Dlx5 plays a major role in specification of the endolymphatic duct and vestibular structures during otic development50 and pax2 deletion results in multiple inner ear defects.51,52,53 The OV expression domain of dlx5 was reduced in 68% (n = 62) of larvae (Figure 7C); this phenotype was reduced to 15% (n = 38), i.e., was rescued, by co-injecting mRNA encoding human TGFBI to which the MOs cannot bind (Figures 7A and 7C), further indicating that the MOs are specific to tgfbi. The OV expression domain of pax2 also was reduced by KD of Tgfbi (40%; n = 39; Figure 7C). To determine whether the phenotype was caused simply by gene expression changes or might include effects on morphogenesis, a subset of larvae stained for dlx5 expression was serial sectioned (n = 10). In addition to a smaller dlx5 expression domain (Figure 7D), the overall volume of the OV and the volume of the OV lumen were significantly decreased compared to paired control OVs (Figure 7E). These experiments demonstrate that Tgfbi is required for early stages of OV gene expression and initial morphogenesis.
Figure 7.
Loss-of-function experiments for tgfbi in the context of otic development
(A) Xenopus 16-cell blastomeres that are fated to primarily contribute to neural crest (NC) and pre-placodal ectoderm (PPE) are labeled in green, and were microinjected with antisense morpholino oligonucleotides (MO) with or without mRNA encoding human TGFBI to which the MOs do not bind. Left (L) and right (R) sides of embryo. Uninjected half of the embryo was used as control for phenotype assessment. In the right panel, sequences of the two alleles of Xenopus laevis tgfbi on the long (.L) and short (.S) chromosomes and human TGFBI are aligned. Red and blue regions indicate sites for MO binding, blue arrow marks the translation start site (ATG). hTGFBI 5′ sequence cannot bind the MOs and therefore could be used to rescue the effects of the MOs.
(B and C) Targeted MS analysis shows Tgfbi protein is not detected in OVs dissected from the MO injected sides of stage 34 larvae. Abundance of the reference protein Gapdh does not change significantly following Tgfbi KD (paired Student’s t test, p > 0.05) (C) MO injected embryos cultured to stage 32 and processed by ISH for dlx5 and pax2. While OV gene expression was normal on the control side (black arrow), it was greatly reduced on the MO-injected side (red arrow) of same larva in a large percentage of the cases (n = 63 larvae for dlx5, n = 39 larvae for pax2). Embryos injected with Tgfbi MOs plus the MO-insensitive human TGFBI mRNA showed a significantly reduced percentage of reduced dlx5 expression (p < 0.05, Chi-Squared test), demonstrating rescue of the morphant phenotype. MO: morpholino injection; MO Rescue: MOs + hTGFBI mRNA injection. Bar indicates 200 μm.
(D) Larvae were vibratome sectioned to measure OV and luminal volumes. In the image shown, dlx5 expression was reduced in the OV on the MO injected side (red arrow) compared to the control side (black arrow). hb, hindbrain.
(E) Otic and luminal volumes of MO injected sides represented as percent change in otic volume compared to control (uninjected) sides of the same larvae (paired Wilcoxon signed rank test, ∗, p < 0.05, Data are represented as mean ± SEM).
Discussion
CHL is a common birth defect worldwide with an estimated incidence of six cases per 1,000 live births.54 Of these, ∼30% are syndromic, i.e., are accompanied by additional clinical features such as craniofacial and renal anomalies.55,56 Because the genetic etiology of many CHL cases is not yet known, efforts to fully delineate the molecular programs that regulate otic morphogenesis are likely to identify additional CHL candidate genes. Although there has been significant progress in elucidating the transcriptome and proteome of the inner ear hair cells, because CHL often also involves structural defects in inner ears, we focused on early stages of otic development that entail key transitions in morphogenesis as well as the initial formation of the sensory patches and hair cells. Herein, we characterized proteome remodeling using a time-resolved quantitative proteomic analysis across five distinct stages of Xenopus otic morphogenesis and hair cell differentiation.
The otic proteome is remodeled over developmental time
Using our time-resolved quantitative Xenopus proteomic data, we mapped the dynamic expression of proteins across five critical stages of otic morphogenesis and hair cell differentiation. To further categorize our data, we used a k-means clustering approach that segregated the temporal data in the top ten primary patterns of protein expression across these stages. Gene ontology analysis of each cluster further helped classify the types of proteins that primarily increased, decreased, or remained stable over the five stages. We found that ECM and cytoskeletal proteins increased from stage 34 to stage 47 during which time the size of the OV increases and auditory and vestibular compartments of the inner ear are formed. While many ECM and cytoskeletal genes have already been linked to CHL, our data show that there is a rich complexity yet to be explored.
Mutations in collagen proteins are associated with several hearing loss syndromes. For example, mutations in COL2A1, COL9A1, and COL9A2 have been reported to contribute to hearing loss in Stickler’s syndrome.57 Many regions of the cochlea, such as the spiral limbus, basilar membrane, spiral ligament, and the stria vascularis, are enriched in ECM, and basket shaped ECM complexes surround the base of hair cells.41 Functional loss of these proteins disrupts ECM baskets and impairs the spatial coupling for pre- and postsynaptic elements leading to hearing impairment in mouse.41 It is well documented that collagens are essential for the integrity of ion transport systems in the spiral ligament. ECM proteins also increase during hair cell differentiation from human iPSCs.58 Our Xenopus data reveal a number of collagens and other ECM proteins that dynamically increase during the course of otic development, specifically at stages 45 and 47 with a >4-fold increase in abundance as compared to stage 34. These included ECM proteins such as Lum, Mfap2, and Hapln1 among others that are putative downstream targets of human CHL genes. It is therefore likely that there are additional ECM proteins that are not yet fully understood in the context of inner ear development but play important roles in otic development and thereby are CHL candidates.
As the OV undergoes morphogenesis, it is expected that proteins that play a role in cytoskeletal regulation are likely to be enriched. In inner ear hair cells, β- and γ-actins are important components of stereocilia and are necessary for hearing.28 Myosin proteins, actin-activated ATPases that move along actin filaments by hydrolyzing ATP, also are required for stereocilia development. Mutations in actin and myosin proteins are known to underlie human hearing loss. For instance, dominant mutations in ACTB are reported in Baraitser-Winter syndrome, patients of which are characterized by facial dysmorphologies, brain malformations and variable hearing loss.59 Our Xenopus data demonstrate a dynamic increase in the abundance of actin proteins such as Acta1, Actb, and Actn4 in otic tissues at stage 45 when protrusions of the semicircular canals grow centrally and at stage 47 when the protrusions have fused, and the inner ear compartments are fully formed. Proteins such as Actn1 and Actn2, on the other hand, show stable expression across the studied stages. Cytoskeletal proteins such as tropomyosins also are dynamically regulated during otic development and are putative downstream targets of Six1. These results motivate future studies of the cytoskeletal proteins we identified in the context of otic morphogenesis and hearing loss.
Syndromic hearing loss candidate genes are dynamically expressed
To place our Xenopus dataset in the context of human CHL and highlight potential candidate genes, we compared our dataset to putative downstream targets of known CHL genes. We focused on known CHL genes that have been previously studied to reveal their putative downstream targets or gene regulatory networks including SIX1, CHD7 and SOX10. We noted that many putative human CHL related genes that were also dynamically represented in our Xenopus dataset included proteins that have cytoskeletal and ECM function, as well as calcium binding and glucose metabolism functions. Each of these protein classes have been shown to play a role in normal development and functioning of the inner ear and are therefore meaningful candidates for CHL.
Tgfbi is a secreted ECM protein required for normal otic development
Our analyses revealed Tgfbi to be dynamically regulated during early otic morphogenesis and that it was necessary for appropriate OV gene expression and size. Tgfbi is a secreted ECM protein that has FAS1 domains that interact with different integrin proteins. For instance, FAS1 and RGD domains of this protein mediate strong binding to αvβ3 integrin and inhibit tumor growth and angiogenesis in mice.47 It is of interest as a putative CHL gene because it is expressed in the OV, the organ of Corti and neurons of the spiral ganglion,44,45 and its expression is regulated by Six1 in the preplacodal ectoderm and the inner ear.34,36 TGFBI appears to be dysregulated in the perilymph from patients with vertigo.45 In other tissues, Tgfbi participates in branching morphogenesis,60,61,62,63 regulates microtubule functions via FAK and Rho-dependent integrin signaling,64 and plays a role in somite and craniofacial cartilage development through the regulation of canonical Wnt signaling.44 Our results uniquely demonstrate a requirement for Tgfbi for normal OV gene expression and size. Although the precise cellular mechanism by which this protein affects these early processes requires further investigation, the established role of Tgfbi in structural morphogenesis and its presence in neurosensory components of the inner ear mark this protein as an important putative candidate for CHL. This discovery highlights the usefulness of applying a time-resolved quantitative proteomic approach to characterize proteomic dynamics during early stages of otic morphogenesis. Curating proteins that are dynamically regulated during the course of Xenopus otic morphogenesis and are putatively regulated by other CHL genes can provide a list of potential candidates of human hearing loss.
Limitations of the study
It will be very important to test whether the many proteins revealed by the temporal proteomic analysis to be dynamically expressed play a role in otic development. Because we analyzed whole OVs, it will be important to discern whether these proteins have distinct spatial expression patterns. Although the developing frog inner ear is morphologically and functionally very similar to that of mammals, it will be important to confirm the reported dataset from a mammalian species to demonstrate which are conserved and which are unique. The interactions predicted by String analysis will need to be biochemically confirmed. The Six1 targets predicted by comparing our differentially expressed proteins to those implicated in CHL datasets should be analyzed for mutations in BOR patient samples that test negative for SIX1 and EYA1 variants. This analysis may reveal new causative genes. The developmental function of Tgfbi during otic morphogenesis needs to be elucidated in detail at the cellular and molecular levels.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Digoxigenin-AP, Fab fragments | Millipore Sigma | Cat#11093274910 |
| Chemicals, peptides, and recombinant proteins | ||
| Human Chorionic gonadotropin | Sigma | Cat#CG10 |
| C18 EASY-Spray HPLC column | Thermo | Cat#03-052-574 |
| TPCK-modified trypsin | Thermo | Cat#20233 |
| Pierce BCA Protein Assay kit | Thermo | Cat#23227 |
| Pierce Quantitative Colorimetric Peptide Assay kit | Thermo | Cat#PI23275 |
| Pierce High pH reverse phase peptide fractionation kit | Thermo | Cat#84868 |
| Deposited data | ||
| RNAseq data | NCBI | Submission ID: SUB13273105; BioProject ID: PRJNA967984 |
| Mass spectrometry proteomics data | ProteomeXchange Consortium via the PRIDE partner repository | PXD041991; https://doi.org/10.6019/PXD041991 |
| Experimental models: Organisms/strains | ||
| 129/S6 Six1-deleted mouse line | Dr. Kiyooshi Kawakami, Jichi Medical School | Ozaki et al., 200465 |
| Xenopus laevis, wild type | Nasco Education, Ft. Atkinson, WI, USA | No longer available |
| Oligonucleotides | ||
| 5′ AATTCATTTTTGGAGCTGTACTAAG 3′ | Gene-tools | custom |
| 5′ TCACAGCAGCTAAGAGAGAAAGCCA 3′ | Gene-tools | custom |
| Recombinant DNA | ||
| Xenopus laevis dlx5 cDNA | Dr. Thomas Sargent, NIH-NICHD | Luo et al., 200166 |
| Xenopus laevis pax2 cDNA | Dr. Andre Brandli, Ludwig-Maximilians Univ Munchen | Heller and Brandli 199767 |
| Human TGFBI | Origene | Cat#RC200411 |
| Software and algorithms | ||
| Skyline | SCIEX | https://sciex.com/products/software/skyline-software |
| Proteome Discovery V2.1 | ThermoFisher Scientific | Cat#OPTON-31103 for v3.0 |
| ComplexHeatmap package, 4.2 | In R | |
| PantherDB | Panther Classification System | https://pantherdb.org/ |
| OriginPro | OriginLab | https://www.originlab.com/ |
| STAR Aligner v2.5.2b | https://www.encodeproject.org/software/star/ | http://code.google.com/p/rna-star/ |
| featureCounts, subread package v.1.5.2 | Bioconductor Rsubread | https://subread.sourceforge.net/featureCounts.html |
| cellSense Entry | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/ |
| Prism 9.4.0 | GraphPad | https://www.graphpad.com/features |
| String | String database | https://string-db.org/ |
| Xenopus laevis proteome | UniProt58 | https://www.uniprot.org/ |
| PHROG database | Peshkin et al., 201514 | http://kirschner.med.harvard.edu/MADX.html |
| ThorImageOCT v.5.1.1 | Thorlabs | https://www.thorlabs.com/newgrouppage9.cfm?objectgroup_id=7982 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Sally A. Moody (samoody@gwu.edu).
Materials availability
This study did not generate any new reagents.
Experimental model and study participant details
Animals
Larvae and tadpoles at Nieuwkoop and Faber15 stages 34, 40, 43, 45 and 47 were obtained by natural mating of 4–5 year old adult males and females from our breeding colony of wild type Xenopus laevis, which were obtained from Nasco Education (Fort Atkinson, WI) and housed at the George Washington University. All procedures and experiments involving Xenopus were approved by the Institutional Animal Care and Use Committee (IACUC) at the George Washington University (A2022-020). The sex of the larvae and tadpoles was not determined but assumed to be represented in equal numbers; sex is not considered a biological variable in these studies.
The Six1-null mouse line, backcrossed onto the 129S6 background for 5 generations,68 was kindly provided by Dr. Kiyoshi Kawakami (Jichi Medical School, Japan). Sexually mature male and female heterozygous mice (∼10–15 months old) were mated to produce embryos. Embryos were collected at embryonic day 10.5. The sex of the embryos was not determined but assumed to be represented in equal numbers; sex is not considered a biological variable in these studies.
All experiments using these mice were approved by the IACUC at the George Washington University (A2022-019).
The George Washington University Animal Research Facility is certified by the Association for Assessment and Accreditation of Laboratory Animal Care.
Study participants
No human participants were involved in this study.
Method details
Obtaining Xenopus embryos and otic tissues
To obtain embryos, outbred male and female adult frogs were injected with human chorionic gonadotropin and placed in an aquarium for 12–15 h. Embryos were removed from the aquarium, treated with 2% cysteine to remove their jelly coats, and cultured to desired stages in Steinberg’s solution.65,69 Larvae and tadpoles were anesthetized in 0.05% benzocaine at Nieuwkoop and Faber15 stages 34, 40, 42, 45, and 47. The entire OV, including its internal fluid, was manually dissected from the surrounding head mesoderm with sharpened forceps. Because these stages are nearly transparent and the OVs are encapsulated in a reflective ECM, no protease treatment was necessary to manually dissect OVs cleanly from the surrounding tissue. Each replicate was dissected from the same clutch of embryos to control for natural size variations between clutches that can scale gene expression.70 For each stage, 5 OVs were pooled from up to 5 animals as one sample (one biological replicate) and immediately frozen on dry ice. A total of 5 biological replicates from 5 independent clutches were collected. The frozen samples were stored at −80°C until further processing.
Optical coherence tomography to visualize otic vesicles
Tadpoles (stages 40–47) were anesthetized in 0.01% benzocaine solution 5 min before OCT imaging. Animals were aligned in an agar-coated dish with their dorsal side facing the OCT imaging field and live imaged with an OCT Imaging System (Telesto series, Thorlabs) with a 1300 nm spectral domain using the 2-dimensional imaging mode.71 Images collected from living tadpoles were processed and visualized with the ThorImage OCT software (version 5.1.1). Sectional image stacks were collected every 2 μm and exported as tiff images into Free-D image reconstruction and modeling software.72
Sample processing for bottom-up proteomic analysis
For proteomic analysis, samples were subject to 3X cycles of freeze (−80 C) – thaw (RT) cycles. Following that, 100 mL of lysis buffer containing 150 mM NaCl, 20 mM Tris-HCl, 5 mM EDTA, and 1% sodium dodecyl sulfate was added. The samples were sonicated for 5 min or until all visible tissue debris were homogenized. Samples were then processed using a bottom up proteomic approach.73 Proteins were reduced in 0.5 M dithiothreitol at 60°C, then alkylated in 0.5M iodoacetamide at room temperature, and the reaction then quenched by the addition of 0.5 M dithiothreitol. Proteins were purified by overnight precipitation in −20°C acetone followed by centrifugation at 10,000 X g for 10 min at 4°C. The resulting pellets were suspended in 50mM ammonium bicarbonate and protein concentration determined by the BCA assay (Thermo). Proteins were digested with TPCK-treated trypsin (protein:enzyme = 1:50) and peptide concentration quantified using the Quantitative Colorimetric Peptide Assay (Thermo). After digestion of proteins to peptides, equal amounts of total pooled peptides from each of the five developmental stages were tagged with tandem mass tags (Thermo) to enable multiplexed relative quantification. A total of ∼100 μg TMT-labeled peptide mixture was fractionated under high pH in spin columns following vendor recommendations (Pierce High pH Reversed-Phase Peptide Fractionation Kit, Thermo). Peptides were eluted in 0.1% (v/v) trimethylamine containing acetonitrile at 10%, 12.5%, 15%, 17.5%, 20%, 25%, and 50% (v/v). Each fraction was dried in a vacuum concentrator and stored at −80°C until analysis.
Liquid chromatography-HRMS discovery quantitative proteomics
Dried peptides were thawed and dissolved in 5% acetonitrile containing 0.1% (v/v) formic acid to a concentration of ∼0.15 μg/μL. Finally, 10 μL of each reconstructed fraction (total ∼1.2 μg) was loaded onto the nanoLC for analysis.
A total of 1 μg of peptide mixture (confirmed by the Total Peptide Assay) was separated on a C18 EASY-Spray HPLC column at 200 nL/min (75 μm inner diameter, 2 μm particle size with 100 Å pores, 50 cm length, Acclaim PepMap 100, Thermo Cat#03-052-574). Separation used a nanoflow liquid chromatography system (Dionex Ultimate 3000 RSLCnano, Thermo) providing a 120 min gradient mixture from Buffer A (2% ACN, 0.1% FA) and Buffer B (100% ACN, 0.1% FA). Peptides were separated using a multistep gradient supplying Buffer B as follows: 3% for 10 min, then ramped to 40% in 110 min, to 80% in 5 min, returned to 3% in 10 min for equilibration over 15 min. Peptides were detected by nanoESI-HRMS. The electrospray was generated at 2.5 kV using a pulled fused silica capillary as emitter (10 μm tip, New Objective, Woburn, MA). Peptides were detected and sequenced using an Orbitrap-quadrupole-ion trap tribrid high-resolution mass spectrometer (Orbitrap Fusion Lumos, Thermo) executing data dependent acquisition. Ions were surveyed at 60,000 FWMH resolution in the orbitrap mass analyzer (MS1) with following settings: maximum IT, 50 ms; AGC, 2×105 counts; microscans, 1. Tandem MS was activated with collision-induced dissociation (CID) in helium at 35% NCE. Fragment ions were detected in the ion trap with rapid scan rate with the following settings: maximum IT, 50 ms; AGC, 5×104 counts; microscans, 1. Multinotch MS3 fragmentation was triggered on the top 10 MS2 fragments with HCD in nitrogen (65% NCE) and fragment ions were dynamically excluded for 60 s before being reconsidered for fragmentation. MS3 spectra were acquired at 15,000 FWMH in the Orbitrap (maximum IT, 120 ms; AGC, 1 × 105counts; microscans, 1).
Liquid chromatography-HRMS targeted quantification (parallel reaction monitoring)
Peptides (∼1 μg) were analyzed by LC-MS/MS using a Dionex Ultimate 3000 UPLC (Thermo Scientific) coupled online to a nanoESI source (details in the previous section) and the Fusion Lumos (Thermo Scientific). Peptides were separated on a C18 column (Thermo Cat#03-052-574) at a flow rate of 300 nL/min as follows: 2% B to 35% B over 70 min, then to 70% B over 5 min, held at 70% B for 3 min, then to 2% B over 2 min and held for 20 min to equilibrate the column. A peptide inclusion list was generated with retention time windows of 3–5 min and was used to isolate precursors for fragmentation. A scheduled PRM method was executed in which fragmentation was acquired using HCD and MS2 scans were acquired at a resolution of 30,000 and an isolation window of 1 m/z. Label-free targeted quantitation of peptides specific for a protein of interest and with identical sequences between stage 34 and stage 47 otic tissues was designed and analyzed using the Skyline software.74 Summed area under the curve of 2–3 transitions per peptide was used for quantitation. Peptide peak area values for each sample were scaled to the average of each peptide across a given analysis. Scaled peptide peak areas were normalized by peak area values for a reference (housekeeping) gene, Gapdh, which is expected to show stable expression across the stages and after Tgfbi knockdown. For each protein, 2 to 3 proteotypic peptide was considered for this portion of the study.
Data analysis for quantitative proteomics
Primary mass spectrometry data were processed in Proteome Discoverer v2.1 executing the SEQUEST-HT search engine. The MS−MS/MS data were searched against the SwissProt Xenopus laevis proteome (downloaded from UniProt58on 10/15/2017) supplemented with the mRNA-derived PHROG database (version 1.0) from Peshkin et al.14 The following search parameters were applied: digestion, tryptic; number of missed cleavages, maximum 2; minimum number of unique peptides, 1; fixed modification, cysteine carbamidomethylation; variable modification, methionine oxidation; maximum mass deviation for main search of precursor masses, 4.5 ppm; de novo mass tolerance for tandem mass spectra, 0.25 Da; sample type, reporter ion MS3; label, TMT; minimum score for modified peptides, 8; phosphorylation localization probability filtered to >0.9 and sites probability >0.9. Peptide and protein identifications were filtered to <1% false discovery rate (FDR) against a reversed-sequence database. Protein identifications were grouped by gene names based on the parsimony principle unless SwissProt or the mRNA-derived database provided evidence of isoforms.
Statistical analysis and temporal clustering
Three biological replicates out of five with the greatest number of protein identifications were used for further analysis. Hierarchal and k-means clustering were performed using the ComplexHeatmap package75 in R (version 4.2). Statistical overrepresentation tests of gene ontology (GO) terms were performed with the PANTHER gene analysis tools http://pantherdb.org76 using gene names as inputs and Bonferroni correction for multiple testing. Statistical tests including Student’s t-tests, ANOVA, Wilcoxon signed rank tests were performed in OriginPro and Microsoft Excel.
Obtaining mouse otic vesicles and RNA extraction
Embryonic day (E) 10.5 mouse embryos were harvested in PBS, OVs were manually dissected and stored in RNAlater (Thermo) until genotyping. Yolk sac DNA from each embryo was genotyped by PCR using primers that span exon 1 and the gfp gene insert.65 Once genotypes were established, OVs from 3 or more embryos were pooled to generate 2 biological replicates. Tissues were homogenized in TRI-reagent total RNA was extracted using the Direct-zol RNA microprep (Zymo Research) per vendor instructions.
RNA-sequencing and data analysis
Frozen extracted RNA was shipped to Azenta Life Sciences (South Plainfield, NJ) for library preparation and sequencing. RNA quality and RIN scores (>7) were determined. Paired-end libraries were constructed according to the Illumina protocol for the HiSeq2500 platform and subject to sequencing. Sequence reads were trimmed to remove adapter sequences and poor-quality nucleotides using Trimmomatic (v.0.36). Trimmed reads were aligned to the mouse reference genome (Mus musculus GRCm38) available on ENSEMBL using STAR aligner (v.2.5.2b). Unique hit counts were calculated using featureCounts (Subread package v.1.5.2). Counts were converted to TPM for further analysis.
Embryo microinjection
Two antisense oligo sequences (MO) were purchased (Gene-tools, Philomath, USA) and reconstituted per manufacturer’s instructions. MO1: 5′ TCACAGCAGCTAAGAGAGAAAGCCA 3′, was fluorescein conjugated and targeted the 5′ UTR of both the.L and.S tgfbi mRNAs and MO2: 5′ AATTCATTTTTGGAGCTGTACTAAG 3′, targeted the start site of both the.L and.S tgfbi mRNAs (Figure 7A). Embryos were obtained as above, and then chosen at the two-cell stage to accurately identify the dorsal and ventral animal blastomeres.69,77 Upon reaching the 16-cell stage, blastomeres on one side of the embryo that are the major precursors of the neural crest and cranial placodes (Figure 7A)78 were microinjected with 1 nL of an equimolar mixture of MO1 and MO2 at a total concentration of 4.5 ng/nL. In some experiments MOs were co-injected with an mRNA (200 pg) encoding human TGFBI whose sequence does not allow the MOs to bind (Figure 7A). For each injection, the uninjected side served as an internal control. Embryos were cultured in diluted Steinberg’s solution until fixation.
In vitro mRNA and antisense RNA probe synthesis
mRNA encoding human TGFBI was synthesized in vitro from a vector purchased from Origene (Cat# RC200411) according to the manufacturer’s protocols (mMessage mMachine kit, Invitrogen). Digoxigenin-labeled antisense RNA probes for Xenopus dlx5 and pax2 were synthesized in vitro according to manufacturer’s protocols (MEGAscript kit; Ambion).
In situ hybridization
Embryos were cultured to stages 32–34, screened for fluorescence to indicate MO localization to the OVs, fixed in MEMFA (MEM salts: MOPS, EGTA, and MgSO4 with formaldehyde), dehydrated to 100% ethanol and processed for wholemount in situ hybridization using digoxigenin-labeled RNA probes that detected by an anti-digoxigenin alkaline phosphatase labeled antibody (Millipore Sigma).79 The position, intensity and size of the expression domains of dlx566 and pax267 were compared between the injected, lineage-labeled side and the control, uninjected side of the same larva, thus controlling for inter-embryo variation in expression patterns. Embryos for each assay were derived from a minimum of three different sets of outbred parents. Gene expression changes were scored in three categories: 1) decreased expression, which was either a smaller domain or same sized domain with reduced intensity compared to the control side of the same embryo; 2) larger otic vesicle/broader expression domain, which was of the same intensity or increased intensity compared to the control side of the same larva; or 3) no change compared to the control side of the same larva. The frequency of phenotypes falling into each category were represented as a percentage of the total number of embryos analyzed for each gene.
Vibrotome sectioning
A subset of larvae that were processed by ISH for dlx5 expression were embedded in a gelatin-based medium (0.5% gelatin, 30% bovine serum albumin, 20% sucrose, hardened with glutaraldehyde (75 μL/mL)). Larvae were sectioned in the transverse plane with a thickness of 40 μm using a vibratome. Serial section images of the OV of 10 larvae were collected using an Olympus SZX 12 stereomicroscope. The area of both control and injected OV from every section per embryo was measured using the cellSense software (Olympus) to calculate the volumes of the OV and its lumen. Because the larvae were of different sizes, the volume of each OV on the MO injected side of the larva was expressed as a percentage of the control OV of the same larva and plotted as percent change from control: [(experimental−control)/control]×100. A two-tailed paired Wilcoxon signed rank test was used to determine if OV and luminal volumes were significantly different (p < 0.05).
Quantification and statistical analysis
For Hierarchal and k-means clustering three biological replicates out of five with the greatest number of protein identifications were used for further analysis. Hierarchal and k-means clustering were performed using the ComplexHeatmap package75 in R (version 4.2). Statistical overrepresentation tests of gene ontology (GO) terms were performed with the PANTHER gene analysis tools http://pantherdb.org76 using gene names as inputs and Bonferroni correction for multiple testing. Statistical tests including Student’s t-tests, ANOVA, Wilcoxon signed rank tests were performed in OriginPro and Microsoft Excel. The details can be found in the results, Figures 2 and 3 and legends for Figures 2 and 3.
For whole embryo ISH assays, embryos were derived from a minimum of three different sets of outbred parents. The number of embryos examined are found in the results and Figure 7. The percentage of the total number of embryos analyzed that showed reduced expression of the indicated genes can be found in Figure 7 and its legend. For otic volume measurements, a two-tailed paired Wilcoxon signed rank test was used to determine if OV and luminal volumes were significantly different (p < 0.05). Ten control and ten experimental OVd were analyzed. Statistics can be found in legend of Figure 7. Statistical tests were performed in GraphPad Prism 9.4.0.
For transcriptomic comparisons (Figure 6A), 3–5 OVs were included in each of two replicates per genotype; embryos were derived from multiple dams. Unique hit counts were calculated using featureCounts (Subread package v.1.5.2). Counts were converted to TPM for further analysis. TPMs from wild type and Six1-nulls were compared by a Student’s t test performed in GraphPad Prism 9.4.0. Details are presented in Figure 6 and the legend for Figure 6.
For parallel reaction monitoring mass spectrometry (Figures 6D, 6E, and 7B), 5 OV from each stage were included in each of three biological replicates. Peptides abundances were compared by a paired Student’s t test using the Skyline software. Details are presented in Figures 6D, 6E, and 7 and their legends.
For OV volume measurements (Figure 7E), the area of both control and injected OV from every section per larva (n = 10 larvae analyzed) was measured using the cellSense software (Olympus) to calculate the volumes of the OV and its lumen. Because the larvae were of different sizes, the volume of each OV on the MO injected side of the larva was expressed as a percentage of the control OV of the same larva and plotted as percent change from control: [(experimental−control)/control]×100. A two-tailed paired Wilcoxon signed rank test was used to determine if OV and luminal volumes were significantly different using GraphPad Prism software. Details are presented in Figure 7 and its legend.
Acknowledgments
This work was enabled by the support of the National Xenopus Resource (RRID:SCR_013731) and Xenbase (RRID:SCR_003280). Funding was provided by the National Institutes of Health grants F31 DC018742 (A.B.B.), R01 DE026434 (S.A.M.), and R35 GM124755 (P.N.). We thank Dr. Kiyoshi Kawakami (Jichi Medical School, Japan) for kindly providing the Six1-null mouse line.
Author contributions
Conceptualization, A.B.B., P.N., and S.A.M.; methodology, A.B.B., P.N., and S.A.M.; formal analysis, A.B.B. and S.A.M.; investigation, A.B.B. and S.A.M.; resources, A.B.B., S.A.M., and P.N.; data curation, A.B.B.; writing – original draft, A.B.B.; writing – review and editing, A.B.B., P.N., and S.A.M.; visualization, A.B.B. and S.A.M.; supervision, P.N. and S.A.M.; project administration, P.N. and S.A.M., funding acquisition, A.B.B., P.N., and S.A.M.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse and equitable conduct of research.
Published: August 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107665.
Supplemental information
Xenopus laevis otic vesicles (OV) dissected from stage 34, 40, 43, 45 and 47 larvae and tadpoles were collected; five OVs were pooled as one sample for each developmental stage and five replicates per stage were processed and analyzed. Peptides from each developmental stage were bar-coded with tandem mass tags. The dataset included in this table is the set of common proteins from the 3/5 most comprehensive biological replicates: the number of proteins reported = 2,353. Each category is color coded and can be accessed by clicking on the dropdown arrow next to the category of interest. In the dropdown, select the category of interest. Deafness and vestibular proteins are marked based on the list provided in Ramírez-Gordillo et al., 2015S1. Pathway protein and transcription factor annotations are based on PantherDB searches. Temporal change: proteins that increased over time are marked in yellow and proteins that decreased over time are marked in orange; threshold = log2 (Fold change): +/− 1 and p < 0.05. Gene ontology: proteins reported as deafness/vestibular are in red, transcription factors in green, pathways in blue, molecular function in purple. Abundance values: reported values are grouped abundances for isobaric mass tags use to tag each developmental stage.
Proteins that were dynamically expressed over OV morphogenesis stages (DEP) were compared to those known to be causative for hearing loss (HL) and those that are expressed downstream of causative genes for Branchio-oto-renal syndrome (SIX1, SIX5), Waardenburg syndrome (SOX10, MITF) and CHARGE syndrome (CHD7).
Data and code availability
-
•
Data: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD041991 and https://doi.org/10.6019/PXD041991; these data will be freely available upon publication. The RNA-seq data are submitted to NCBI. Submission ID: SUB13273105; BioProject ID: PRJNA967984. All data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Vona B., Nanda I., Hofrichter M.A.H., Shehata-Dieler W., Haaf T. Non-syndromic hearing loss gene identification: A brief history and glimpse into the future. Mol. Cell. Probes. 2015;29:260–270. doi: 10.1016/j.mcp.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Burns J.C., Kelly M.C., Hoa M., Morell R.J., Kelley M.W. Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat. Commun. 2015;6:8557. doi: 10.1038/ncomms9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darville L.N.F., Sokolowski B.H.A. In-depth proteomic analysis of mouse cochlear sensory epithelium by mass spectrometry. J. Proteome Res. 2013;12:3620–3630. doi: 10.1021/pr4001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickox A.E., Wong A.C.Y., Pak K., Strojny C., Ramirez M., Yates J.R., Ryan A.F., Savas J.N. Global analysis of protein expression of inner ear hair cells. J. Neurosci. 2017;37:1320–1339. doi: 10.1523/jneurosci.2267-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng H., Liu M., Pecka J., Beisel K.W., Ding S.J. Proteomic analysis of the Organ of Corti using nanoscale liquid chromatography coupled with tandem mass spectrometry. Int. J. Mol. Sci. 2012;13:8171–8188. doi: 10.3390/ijms13078171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheffer D.I., Shen J., Corey D.P., Chen Z.Y. Gene expression by mouse inner ear hair cells during development. J. Neurosci. 2015;35:6366–6380. doi: 10.1523/jneurosci.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Q.Y., Rozanas C.R., Thalmann I., Chance M.R., Alagramam K.N. Inner ear proteomics of mouse models for deafness, a discovery strategy. Brain Res. 2006;1091:113–121. doi: 10.1016/j.brainres.2006.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anagnostopoulos A.V. A compendium of mouse knockouts with inner ear defects. Trends Gen. 2002;18:499. doi: 10.1016/s0168-9525(02)02753-1. [DOI] [PubMed] [Google Scholar]
- 9.Beisel K.W., Shiraki T., Morris K.A., Pompeia C., Kachar B., Arakawa T., Bono H., Kawai J., Hayashizaki Y., Carninci P. Identification of unique transcripts from a mouse full-length, subtracted inner ear cDNA library. Genomics. 2004;83:1012–1023. doi: 10.1016/j.ygeno.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Resendes B.L., Robertson N.G., Szustakowski J.D., Resendes R.J., Weng Z., Morton C.C. Gene discovery in the auditory system: Characterization of additional cochleae-expressed sequences. J. Assoc. Res. Otolaryngol. 2002;3:45–53. doi: 10.1007/s101620020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai T., Jen H.I., Kang H., Klisch T.J., Zoghbi H.Y., Groves A.K. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J. Neurosci. 2015;35:5870–5883. doi: 10.1523/jneurosci.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkan-Miller T., Ulitsky I., Hertzano R., Rudnicki A., Dror A.A., Lenz D.R., Elkon R., Irmler M., Beckers J., Shamir R., Avraham K.B. Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear. PLoS One. 2011;6:e18195. doi: 10.1371/journal.pone.0018195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Peshkin L., Wühr M., Pearl E., Haas W., Freeman R.M., Gerhart J.C., Klein A.M., Horb M., Gygi S.P., Kirschner M.W. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell. 2015;35:383–394. doi: 10.1016/j.devcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieuwkoop P.D., Faber J. Garland Publishing; 1994. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis. [DOI] [Google Scholar]
- 16.Bever M.M., Jean Y.Y., Fekete D.M. Three-dimensional morphology of inner ear development in Xenopus laevis. Dev. Dynam. 2003;227:422–430. doi: 10.1002/dvdy.10316. [DOI] [PubMed] [Google Scholar]
- 17.Quick Q.A., Serrano E.E. Inner ear formation during the early larval development of Xenopus laevis. Dev. Dynam. 2005;234:791–801. doi: 10.1002/dvdy.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramírez-Gordillo D., Powers T.R., van Velkinburgh J.C., Trujillo-Provencio C., Schilkey F., Serrano E.E. RNA-Seq and microarray analysis of the Xenopus inner ear transcriptome discloses orthologous OMIM® genes for hereditary disorders of hearing and balance. BMC Res. Notes. 2015;8:1–9. doi: 10.1186/s13104-015-1485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Streit A. Induction of the inner ear: Stepwise specification of otic fate from multipotent progenitors. Hear. Res. 2013;297:3–12. doi: 10.1016/j.heares.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y.F., Tong M., Edge A.S.B. Destabilization of Atoh1 by E3 Ubiquitin Ligase Huwe1 and Casein Kinase 1 is essential for normal sensory hair cell development. J. Biol. Chem. 2016;291:21096–21109. doi: 10.1074/jbc.M116.722124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman S., Sanchez S.M., Pouyo R., Van Lerberghe P.B., Hanon K., Thelen N., Malgrange B. Proteostasis is essential during cochlear development for neuron survival and hair cell polarity. EMBO Rep. 2019;20:20. doi: 10.15252/embr.201847097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groves A.K., Fekete D.M. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X., Mahmood S.R., Gjorgjieva T., Percipalle P. Emerging roles of cytoskeletal proteins in regulating gene expression and genome organization during differentiation. Nucleus. 2020;11:53–65. doi: 10.1080/19491034.2020.1742066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata R., Vanderhaeghen P. Regulatory roles of mitochondria and metabolism in neurogenesis. Curr. Opin. Neurobiol. 2021;69:231–240. doi: 10.1016/j.conb.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F.H., Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:25. doi: 10.7754/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantarci H., Gou Y., Riley B.B. The Warburg Effect and lactate signaling augment Fgf-MAPK to promote sensory-neural development in the otic vesicle. Elife. 2020;9:e56301. doi: 10.7554/eLife.56301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond M.C., Belyantseva I.A., Friderici K.H., Friedman T.B. Actin in hair cells and hearing loss. Hear. Res. 2012;288:89–99. doi: 10.1016/j.heares.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrin B.J., Sonnemann K.J., Ervasti J.M. beta-actin and gamma-actin are each dispensable for auditory hair cell development but required for stereocilia maintenance. PLoS Genet. 2010;6:e1001158. doi: 10.1371/journal.pgen.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Procaccio V., Salazar G., Ono S., Styers M.L., Gearing M., Davila A., Jimenez R., Juncos J., Gutekunst C.A., Meroni G., Wainer B.H. A mutation of beta-actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am. J. Hum. Genet. 2006;78:947–960. doi: 10.1086/504271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noda T., Oki S., Kitajima K., Harada T., Komune S., Meno C. Restriction of Wnt signaling in the dorsal otocyst determines semicircular canal formation in the mouse embryo. Dev. Biol. 2012;362:83–93. doi: 10.1016/j.ydbio.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 31.Jacques B.E., Puligilla C., Weichert R.M., Ferrer-Vaquer A., Hadjantonakis A.K., Kelley M.W., Dabdoub A. A dual function for canonical Wnt/beta-catenin signaling in the developing mammalian cochlea. Development. 2012;139:4395–4404. doi: 10.1242/dev.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian D., Jones C., Rzadzinska A., Mark S., Zhang X., Steel K.P., Dai X., Chen P. Wnt5a functions in planar cell polarity regulation in mice. Dev. Biol. 2007;306:121–133. doi: 10.1016/j.ydbio.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozario T., DeSimone D.W. The extracellular matrix in development and morphogenesis: A dynamic view. Dev. Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riddiford N., Schlosser G. Dissecting the pre-placodal transcriptome to reveal presumptive direct targets of Six1 and Eya1 in cranial placodes. Elife. 2016;5:e17666. doi: 10.7554/eLife.17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan B., Neilson K.M., Ranganathan R., Maynard T., Streit A., Moody S.A. Microarray identification of novel genes downstream of Six1, a critical factor in cranial placode, somite, and kidney development. Dev. Dynam. 2015;244:181–210. doi: 10.1002/dvdy.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Zhang T., Ramakrishnan A., Fritzsch B., Xu J., Wong E.Y.M., Loh Y.-H.E., Ding J., Shen L., Xu P.X. Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. 2020;48:2880–2896. doi: 10.1093/nar/gkaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy N.C., Majidi S.P., Kong L., Nemera M., Ferguson C.J., Moore M., Goncalves T.M., Liu H.K., Fitzpatrick J.A.J., Zhao G., Gabel H.W. CHARGE syndrome protein CHD7 regulates epigenomic activation of enhancers in granule cell precursors and gyrification of the cerebellum. Nat. Commun. 2021;12:5702. doi: 10.1038/s41467-021-25846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoek K.S., Schlegel N.C., Eichhoff O.M., Widmer D.S., Praetorius C., Einarsson S.O., Valgeirsdottir S., Bergsteinsdottir K., Schepsky A., Dummer R., Steingrimsson E. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 39.Wen J., Song J., Bai Y., Liu Y., Cai X., Mei L., Ma L., He C., Feng Y. A model of Waardenburg Syndrome using patient-derived iPSCs with a SOX10 mutation displays compromised maturation and function of the neural crest that involves inner ear development. Front. Cell Dev. Biol. 2021;9:720858. doi: 10.3389/fcell.2021.720858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith R.J.H. In: GeneReviews. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K., Amemiya A., editors. 2018. Branchiootorenal spectrum disorders.https://www.ncbi.nlm.nih.gov/books/NBK1380/ [Google Scholar]
- 41.Sonntag M., Blosa M., Schmidt S., Reimann K., Blum K., Eckrich T., Seeger G., Hecker D., Schick B., Arendt T., Morawski M. Synaptic coupling of inner ear sensory cells is controlled by brevican-based extracellular matrix baskets resembling perineuronal nets. BMC Biol. 2018;16:99. doi: 10.1186/s12915-018-0566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N., Xi Y., Du H., Zhou H., Xu Z. Annexin A4 Is Dispensable for Hair Cell Development and Function. Front. Cell Dev. Biol. 2021;9:680155. doi: 10.3389/fcell.2021.680155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 44.Wang F., Hu W., Xian J., Ohnuma S.i., Brenton J.D. The Xenopus Tgfbi is required for embryogenesis through regulation of canonical Wnt signalling. Dev. Biol. 2013;379:16–27. doi: 10.1016/j.ydbio.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Lin H.C., Ren Y., Lysaght A.C., Kao S.Y., Stankovic K.M. Proteome of normal human perilymph and perilymph from people with disabling vertigo. PLoS One. 2019;14:e0218292. doi: 10.1371/journal.pone.0218292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J.E., Jeong H.W., Nam J.O., Lee B.H., Choi J.Y., Park R.W., Park J.Y., Kim I.S. Identification of motifs in the fasciclin domains of the transforming growth factor-beta-induced matrix protein beta ig-h3 that interact with the alpha v65 integrin. J. Biol. Chem. 2002;277:46159–46165. doi: 10.1074/jbc.M207055200. [DOI] [PubMed] [Google Scholar]
- 47.Kim J.E., Kim S.J., Lee B.H., Park R.W., Kim K.S., Kim I.S. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, beta ig-h3. J. Biol. Chem. 2000;275:30907–30915. doi: 10.1074/jbc.M002752200. [DOI] [PubMed] [Google Scholar]
- 48.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 49.Neilson K.M., Abbruzzesse G., Kenyon K., Bartolo V., Krohn P., Alfandari D., Moody S.A. Pa2G4 is a novel Sixl co-factor that is required for neural crest and otic development. Dev. Biol. 2017;421:171–182. doi: 10.1016/j.ydbio.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chatterjee S., Kraus P., Lufkin T. A symphony of inner ear developmental control genes. BMC Genet. 2010;11:68. doi: 10.1186/1471-2156-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou D., Silvius D., Rodrigo-Blomqvist S., Enerbäck S., Xu P.-X. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev. Biol. 2006;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grocott T., Tambalo M., Streit A. The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Dev. Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 53.Moody S.A., LaMantia A.S. Transcriptional regulation of cranial sensory placode development. Curr. Top. Dev. Biol. 2015;111:301–350. doi: 10.1016/bs.ctdb.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olusanya B.O. Neonatal hearing screening and intervention in resource-limited settings: an overview. Arch. Dis. Child. 2012;97:654–659. doi: 10.1136/archdischild-2012-301786. [DOI] [PubMed] [Google Scholar]
- 55.Kalatzis V., Petit C. The fundamental and medical impacts of recent progress in research on hereditary hearing loss. Hum. Mol. Genet. 1998;7:1589–1597. doi: 10.1093/hmg/7.10.1589. [DOI] [PubMed] [Google Scholar]
- 56.Koffler T., Ushakov K., Avraham K.B. Genetics of Hearing Loss - Syndromic. Otolaryngol. Clin. 2015;48:1041–1061. doi: 10.1016/j.otc.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoornaert K.P., Vereecke I., Dewinter C., Rosenberg T., Beemer F.A., Leroy J.G., Bendix L., Björck E., Bonduelle M., Boute O., Mortier G.R. Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur. J. Hum. Genet. 2010;18:872–880. doi: 10.1038/ejhg.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chacko L.J., Lahlou H., Steinacher C., Assou S., Messat Y., Dudas J., Edge A., Crespo B., Crosier M., Sergi C., et al. Transcriptome-wide analysis reveals a role for extracellular matrix and integrin receptor genes in otic neurosensory differentiation from human iPSCs. Interntl. J. Mol. Sci. 2021;22:23. doi: 10.3390/ijms221910849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Donato N., Rump A., Koenig R., Der Kaloustian V.M., Halal F., Sonntag K., Krause C., Hackmann K., Hahn G., Schrock E., Verloes A. Severe forms of Baraitser-Winter syndrome are caused by ACTB mutations rather than ACTG1 mutations. Eur. J. Hum. Genet. 2014;22:179–183. doi: 10.1038/ejhg.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan S.Y., Chan A.K.W., Cheung B.P.K., Liang Y., Leung M.P. Identification of genes expressed during myocardial development. Chin. Med. J. 2003;116:1329–1332. [PubMed] [Google Scholar]
- 61.Hirate Y., Okamoto H., Yamasu K. Structure of the zebrafish fasciclin I-related extracellular matrix protein (beta ig-h3) and its characteristic expression during embryogenesis. Gene Expr. Patterns. 2003;3:331–336. doi: 10.1016/s1567-133x(03)00035-8. [DOI] [PubMed] [Google Scholar]
- 62.Schorderet D.F., Menasche M., Morand S., Bonnel S., Büchillier V., Marchant D., Auderset K., Bonny C., Abitbol M., Munier F.L. Genomic characterization and embryonic expression of the mouse Bigh3 (Tgfbi) gene. Biochem. Biophys. Res. Commun. 2000;274:267–274. doi: 10.1006/bbrc.2000.3116. [DOI] [PubMed] [Google Scholar]
- 63.Schwab K., Patterson L.T., Aronow B.J., Luckas R., Liang H.C., Potter S.S. A catalogue of gene expression in the developing kidney. Kidney Int. 2003;64:1588–1604. doi: 10.1046/j.1523-1755.2003.00276.x. [DOI] [PubMed] [Google Scholar]
- 64.Ahmed A.A., Mills A.D., Ibrahim A.E.K., Temple J., Blenkiron C., Vias M., Massie C.E., Iyer N.G., McGeoch A., Crawford R., Brenton J.D. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moody S.A. Cell lineage analysis in Xenopus embryos. Methods Mol. Biol. 2000;135:331–347. doi: 10.1385/1-59259-685-1:331. [DOI] [PubMed] [Google Scholar]
- 66.Luo T., Matsuo-Takasaki M., Lim J.H., Sargent T.D. Differential regulation of Dlx gene expression by a BMP morphogenetic gradient. Int. J. Dev. Biol. 2001;45:681–684. [PubMed] [Google Scholar]
- 67.Heller N., Brändli A.W. Xenopus Pax-2 displays multiple splice forms during embryogenesis and pronephric kidney development. Mech. Dev. 1997;69:83–104. doi: 10.1016/s0925-4773(97)00158-5. [DOI] [PubMed] [Google Scholar]
- 68.Ozaki H., Nakamura K., Funahashi J.i., Ikeda K., Yamada G., Tokano H., Okamura H.O., Kitamura K., Muto S., Kotaki H., et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- 69.Moody S.A. Lineage tracing and fate mapping in Xenopus embryos. Cold Spring Harb. Protoc. 2018;2018:933–942. doi: 10.1101/pdb.prot097253. [DOI] [PubMed] [Google Scholar]
- 70.Leibovich A., Edri T., Klein S.L., Moody S.A., Fainsod A. Natural size variarion among embryos leads to the corresponding scaling in gene expression. Dev. Biol. 2020;462:165–179. doi: 10.1016/j.ydbio.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah A.M., Krohn P., Baxi A.B., Tavares A.L.P., Sullivan C.H., Chillakuru Y.R., Majumdar H.D., Neilson K.M., Moody S.A. Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis. Model. Mech. 2020;13:dmm043489. doi: 10.1242/dmm.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andrey P., Maurin Y. Free-D: an integrated environment for three-dimensional reconstruction from serial sections. J. Neurosci. Methods. 2005;145:233–244. doi: 10.1016/j.jneumeth.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 73.Baxi A.B., Lombard-Banek C., Moody S.A., Nemes P. Proteomic characterization of the neural ectoderm fated cell clones in the Xenopus laevis embryo by high-resolution mass spectrometry. ACS Chem. Neurosci. 2018;9:2064–2073. doi: 10.1021/acschemneuro.7b00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 76.Mi H., Ebert D., Muruganujan A., Mills C., Albou L.P., Mushayamaha T., Thomas P.D. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive api. Nucleic Acids Res. 2021;49:D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klein S.L. The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev. Biol. 1987;120:299–304. doi: 10.1016/0012-1606(87)90127-8. [DOI] [PubMed] [Google Scholar]
- 78.Moody S.A. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- 79.Sive H.L., Grainger R.M., Harland R.M. chapter 13. Cold Spring Harbor Laboratory Press; 2000. (Early Development of Xenopus laevis, a Laboratory Manual). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Xenopus laevis otic vesicles (OV) dissected from stage 34, 40, 43, 45 and 47 larvae and tadpoles were collected; five OVs were pooled as one sample for each developmental stage and five replicates per stage were processed and analyzed. Peptides from each developmental stage were bar-coded with tandem mass tags. The dataset included in this table is the set of common proteins from the 3/5 most comprehensive biological replicates: the number of proteins reported = 2,353. Each category is color coded and can be accessed by clicking on the dropdown arrow next to the category of interest. In the dropdown, select the category of interest. Deafness and vestibular proteins are marked based on the list provided in Ramírez-Gordillo et al., 2015S1. Pathway protein and transcription factor annotations are based on PantherDB searches. Temporal change: proteins that increased over time are marked in yellow and proteins that decreased over time are marked in orange; threshold = log2 (Fold change): +/− 1 and p < 0.05. Gene ontology: proteins reported as deafness/vestibular are in red, transcription factors in green, pathways in blue, molecular function in purple. Abundance values: reported values are grouped abundances for isobaric mass tags use to tag each developmental stage.
Proteins that were dynamically expressed over OV morphogenesis stages (DEP) were compared to those known to be causative for hearing loss (HL) and those that are expressed downstream of causative genes for Branchio-oto-renal syndrome (SIX1, SIX5), Waardenburg syndrome (SOX10, MITF) and CHARGE syndrome (CHD7).
Data Availability Statement
-
•
Data: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD041991 and https://doi.org/10.6019/PXD041991; these data will be freely available upon publication. The RNA-seq data are submitted to NCBI. Submission ID: SUB13273105; BioProject ID: PRJNA967984. All data reported in this paper will be shared by the lead contact upon request.
-
•
Code: This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.