Summary
With a critical need for more complete in vitro models of human development and disease, organoids hold immense potential. Their complex cellular composition makes single-cell sequencing of great utility; however, the limitation of current technologies to a handful of treatment conditions restricts their use in screens or studies of organoid heterogeneity. Here, we apply sci-Plex, a single-cell combinatorial indexing (sci)-based RNA sequencing (RNA-seq) multiplexing method to retinal organoids. We demonstrate that sci-Plex and 10× methods produce highly concordant cell-class compositions and then expand sci-Plex to analyze the cell-class composition of 410 organoids upon modulation of critical developmental pathways. Leveraging individual organoid data, we develop a method to measure organoid heterogeneity, and we identify that activation of Wnt signaling early in retinal organoid cultures increases retinal cell classes up to 6 weeks later. Our data show sci-Plex’s potential to dramatically scale up the analysis of treatment conditions on relevant human models.
Keywords: single-cell sequencing, retina, organoids, cell signaling, neuron
Graphical abstract
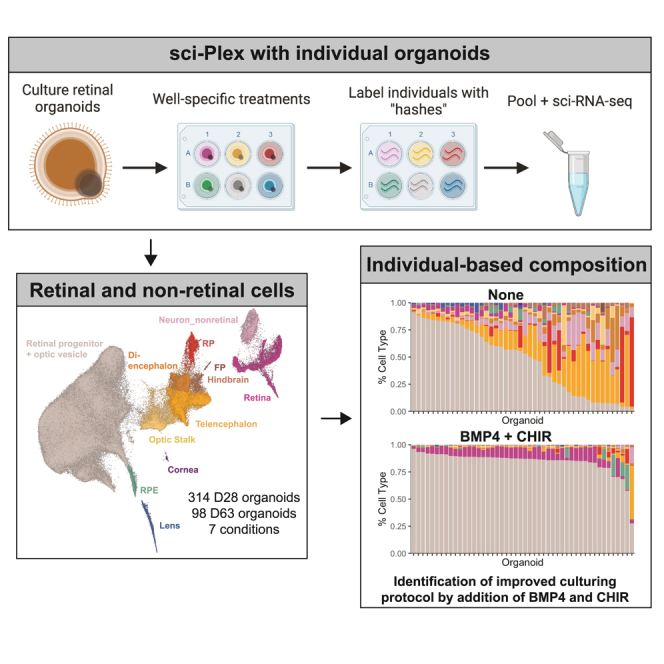
Highlights
-
•
Hundreds of individual retinal organoids sequenced at single-cell resolution
-
•
Detection of significant changes in cell-type abundance and heterogeneity
-
•
Wnt and BMP signaling activation increases retinal cell abundance
Motivation
The gaining popularity of organoids for the study of human development and disease will require technologies that advance the scale and depth by which they can be analyzed. In current retinal organoid protocols, high organoid heterogeneity is a critical but poorly understood reality. By adapting the single-nucleus RNA-seq multiplexing technique sci-Plex to assay the cell-type composition and heterogeneity of hundreds of individual retinal organoids, our method provides an unparalleled look at the factors driving heterogeneity and serves as a tool to screen for better culturing protocols.
Tresenrider et al. adapt the single-cell sequencing-based multiplexing strategy sci-Plex to assay the composition and heterogeneity of 410 individual retinal organoids after the perturbation of developmental signaling pathways. They find that the activation of BMP and Wnt signaling improves retinal organoid composition.
Introduction
There has been good progress developing in vitro systems that recapitulate the structure and function of a variety of organs.1,2,3,4,5,6,7,8,9 Retinal organoids, for example, contain all the major retinal neuronal and glial (Müller glia) cell classes and follow the developmental timing and laminar organization of the retina.10 Importantly, retinal organoids represent an accessible and renewable in vitro system for studies of development11 and disease progression. When generated from patient induced pluripotent stem cells (iPSCs), retinal organoids model patient-specific disease phenotypes,12,13 with applications in personalized medicine or for cell replacement in degenerative diseases.14,15,16
However, several roadblocks limit the full potential of organoids, including organoid-to-organoid heterogeneity, batch effects, and high costs.17 While several well-established protocols exist for retinal organoid generation, most protocols require manual selection or dissection of organoids.18,19 A protocol has been developed that eliminates the need for dissection, but it relies on specialized micro-well plates that have not been widely adopted, and it still only produces ∼80% retinal organoids.20 For this reason, the presence of non-retinal cells in retinal organoid cultures, in an unpredictable and heterogeneous manner, limits their reproducibility, especially for large-scale experiments.18,19,21
To advance the standardization of retinal organoids, we developed a high-throughput single-cell sequencing-based screening pipeline. Current techniques for screening in organoids rely on microscopy-based approaches that implement measurements of size and morphology, use viability stains, and/or perform immunostaining/fluorescence for a small number of marker genes.22,23,24,25 Single-cell sequencing reveals the full array of cell classes, permits the discovery of unpredictable cellular states, and allows for a treatment’s effect to be measured across all cell types. However, the high per-sample cost cannot practically scale past a handful of conditions (Table S1). sci-Plex, which builds upon single-cell combinatorial indexing-based RNA sequencing (sci-RNA-seq), facilitates the labeling and simultaneous processing of thousands of conditions with a single-cell output.26 Originally performed in culture, sci-Plex was recently applied to thousands of individual zebrafish embryos exposed to genetic or temperature-based perturbations.27,28 A handful of studies have sequenced individual organoids, but their experimental designs and level of replication are sharply constrained by the cost and throughput of droplet-based single-cell sequencing technologies.29,30 By contrast, sci-Plex increases the capacity to process individual organoids at single-cell resolution and enables the development of protocols that better recapitulate in vivo development.
Here, we demonstrate the first application of sci-Plex in organoids. To confirm the validity of the method, we show strikingly similar cell-class compositions of matched bulk retinal organoids prepared by sci-Plex compared with commercially available droplet-based single-cell sequencing methods. We then perform sci-Plex on 410 individual organoids while modulating key developmental signaling pathways. We find that activation of BMP followed by Wnt signaling produces organoids with a greater proportion of retinal cells compared with BMP activation alone without increasing organoid-to-organoid heterogeneity. With the depth of information obtained across hundreds of individuals, we vastly expand the types of analyses possible in single-cell sequencing-based methods in organoids, and we identify a culturing protocol that improves the composition of retinal organoids.
Results
Successful multiplexing of retinal organoids using sci-Plex
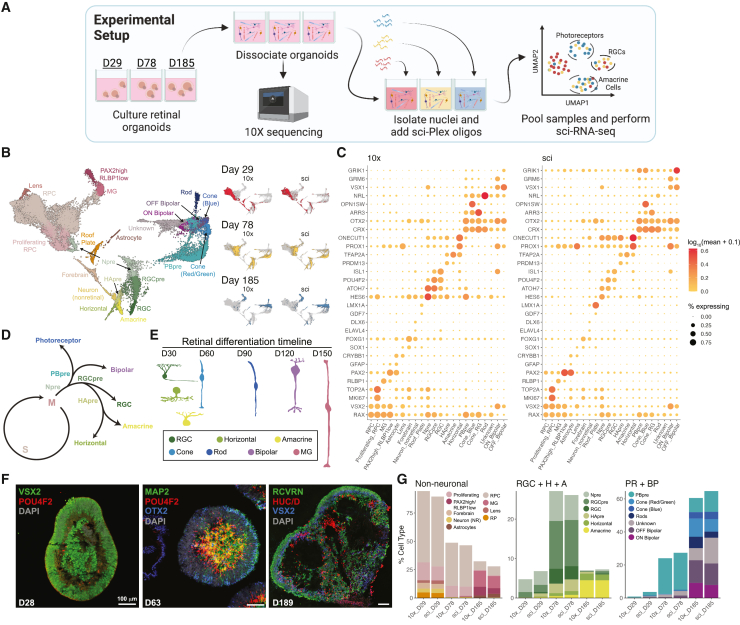
To demonstrate that multiplexed samples from sci-Plex have cell-class compositions (1) comparable to those recovered using commercial droplet-based techniques and (2) that would be expected developmentally, retinal organoids cultured for 29, 78, or 185 days were dissociated into single-cell suspensions and split for processing by either the 10× Chromium platform or sci-Plex (Figures 1A and S1A). sci-Plex recovered 12,867 cells (median unique molecular identifiers [UMIs] for day 29: 5,512, day 78: 5,797, day 185: 5,695.5), and 10× recovered 16,914 (median UMI for day 29: 6,335, day 78: 4,118, day 185: 3,950) (Figures S1B–S1H). Using the hash barcodes, the sample from which a cell originated was determined for 95% of the cells (Figures S1D and S1E).
Figure 1.
Comparison between sci-Plex and 10×
(A) Retinal organoids were cultured for 29, 78, or 185 days. The cells were split and either processed by a 10× RNA-seq pipeline or prepared by sci-Plex.
(B) Integrated UMAP of sci-Plex and 10× datasets. Left: cells colored by cell type. RPC, retinal progenitor cell; MG, Müller glia; Npre, retinal neuronal precursor; RGC, retinal ganglion cell; RGCpre, RGC precursor; HApre, horizontal/amacrine precursor; PBpre, photoreceptor/bipolar precursor. Right: cells are faceted by organoid age and technology. Cells are colored by age.
(C) Dot plot of genes used to define cell types in (B). Dot size indicates the percentage of cells that express the gene of interest. Color indicates the log10 mean UMIs per cell.
(D) Representation of retinal neuron developmental trajectories.
(E) Timeline of retinal cell differentiation in human organoids.
(F) Immunostaining of day 28, 63, and 189 organoids for progenitor (VSX2-day 28), RGC (POU4F2), photoreceptor/bipolar (OTX2), neuronal (MAP2), amacrine/horizontal/RGC (HuC/D), bipolar cell (VSX2-day 189), and photoreceptor (RCVRN) markers. Scale bar represents 100 μm.
(G) Using the cell-type assignments from (B), cells were counted, and the percentage of cell type was determined for each technology and organoid age.
The retina develops in a highly coordinated way with all retinal neurons and the Müller glia (MG) arising from a shared retinal progenitor cell (RPC) population31(Figure 1D) with conserved developmental timing between human retinal organoids and the fetal retina10 (Figure 1E). Staining of retinal organoids supported previous observations indicating that the layered organization of the retina is maintained10 (Figure 1F). We used the knowledge of developmental timing to determine how well sci-Plex segregated samples of differing ages (Figures 1B, 1G, and S1K). As expected, day 29 organoids included mainly RPCs, differentiating retinal ganglion cells (RGCs), other retinal neuron precursors, and brain progenitor cells (Figures 1C, 1G, S1J, and S1K; see Table S2 for marker genes). Day 78 organoids had far more RGCs and horizontal, amacrine, and photoreceptor precursor cells (Figures 1C, 1G, and S1K). By day 185, rods, cones, and bipolar cells were dominant (Figures 1C, 1G, S1J, and S1K). MG and astrocyte clusters were also defined (Figures 1B, 1C, 1G, S1J, and S1K). Comparing directly between 10× and sci-Plex, the cell-class distribution was highly concordant, although some low-abundance cell classes were detected more frequently in sci-Plex, possibly due to differences between nuclear and whole-cell transcript recovery (Figure 1G). We also found that processing just the sci-Plex data using Monocle3 resulted in a comparable cell-class classification (Figures S1L–S1N). These analyses demonstrated that sci-Plex can both identify a cell’s well of origin and correctly assign its cell type.
Detection of cell-class abundance changes across treatments through multiplexing of individual organoids
We next aimed to use the multiplexing ability of sci-Plex to screen conditions that modulate signaling pathways to (1) better understand the effect signaling pathways have on cell-fate decisions in young retinal organoids and (2) use this information to design better organoid differentiation protocols. The BMP, Sonic hedgehog, Wnt, and fibroblast growth factor (FGF) signaling pathways are all key regulators of eye and brain development.32,33 Modulators of these pathways, BMP4 (BMP protein), SAG (Sonic hedgehog agonist), CHIR99021 (Wnt agonist), and SU5402 (FGF inhibitor), have all been used in published retinal organoid protocols.19 However, there is no agreed upon “best” combination of these factors.
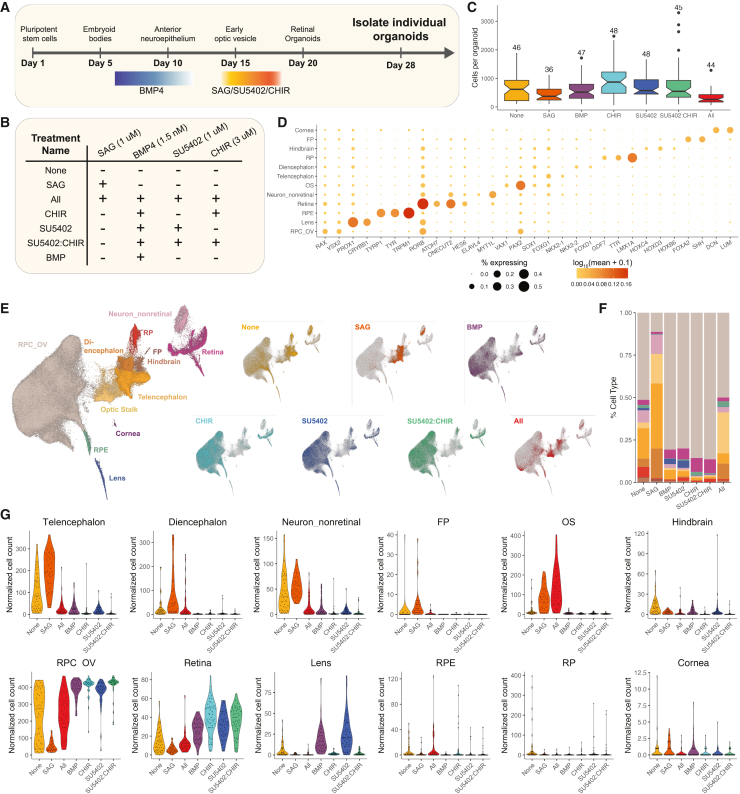
To perform a systematic and quantitative analysis, sci-Plex was used to label 314 individual organoids from seven different treatment conditions. Organoids were prepared for sci-Plex 28 days after initiation of differentiation (Figures 2A and 2B). No selection of organoids that morphologically appeared retinal was applied, as we aimed to understand the factors driving the heterogeneity that obligates such selection steps. An early time point was chosen to better understand the effects of signaling modulation on the choice between retinal and non-retinal fates. We distributed the organoids into 96-well plates with each well harboring a single organoid, permitting the hash-based barcoding to be performed on individuals. After hashing, cells were subjected to sci-RNA-seq3, a version of combinatorial indexing that greatly increases the throughput of the method.34 After filtering, 203,511 cells (median UMI: 350) were recovered (Figures S2A–S2F) with a median of 505 cells per individual (Figure 2C). Our successful recovery of cells from hundreds of individual organoids set us up to perform statistical analyses relating to cell-class abundance and organoid-to-organoid heterogeneity in an unprecedented manner.
Figure 2.
sci-Plex applied to individual organoids after signaling pathway modulation
(A) Overview of the retinal organoid differentiation protocol.
(B) A summary of the seven treatments performed.
(C) Boxplot of the number of cells recovered per organoid for each treatment. Number above the boxplot indicates the number of individual organoids in which >50 cells were recovered for that treatment. The notch of the boxplots represents the median cells per organoid. The colored region indicates the values from the lower 25th to the upper 75th quartile of cell counts. The whiskers span the range of the cell counts within 1.5× the distance between the 25th–75th percentiles. Points outside of that range are displayed as outliers.
(D) Dot plot of the genes used to define cell types for day 28 organoids. Dot size indicates the percentage of cells expressing the gene of interest. Color indicates the log10 mean UMIs per cell.
(E) Left: UMAP of the recovered cells with cell-type annotations. Right: for each UMAP, cells from the specified treatment are colored. RPC_OV, retinal progenitor cell/optic vesicle; RPE, retinal pigmented epithelium; OS, optic stalk; RP, roof plate; FP, floor plate.
(F) Stacked bar plot with mean cell-type composition of day 28 organoids displayed for each treatment.
(G) Violin plots of size-factor-normalized cell counts for each cell type from day 28 organoids. Colored by treatment.
Despite the lower UMI counts with sci-RNA-seq3, cell classes were readily defined with clustering followed by the analysis of marker gene expression (Figures 2D, 2E, and S2H; Table S2). The largest population of cells was the RPC/optic vesicle (RPC_OV), but we also recovered retinal neuron precursors and non-retinal cells (Figures 2D, 2E, and S2G). RPC_OV cells dramatically decreased in the sonic hedgehog (SHH) activation (SAG) treatment when compared with the “None” treatment (no exposure to any of the signaling modulators), yet they increased in the BMP, SU5402, CHIR, and SU5402:CHIR treatments (Figures 2E–2G). At this time point, large changes in cell composition were apparent, but smaller changes in cell-class abundance were not.
We next aimed to use statistical methods to assess treatment-dependent changes across all cell classes. We harnessed the power of our individual organoid data to fit a statistical model using the beta-binomial distribution (STAR Methods).27,28 Our model found that the organoids from treatments that include addition of BMP4, with the exception of “All” (exposure to all of the signaling modulators), produced significantly more RPC_OV and retinal cells compared with None (Figures 2G and 3A). This supports previous reports indicating that the addition of BMP4 improves the efficiency of retinal differentiation from stem cells.18 We also saw that SHH activation decreased RPC_OV cell counts and significantly increased diencephalon, telencephalon, optic stalk, and non-retinal neurons (Figure 3A). Interestingly, the optic stalk was significantly increased in the All treatment (Figure 3A). Thus, with our well-powered experiment and statistical approach, we were able to interpret cell-class-level effects from each treatment.
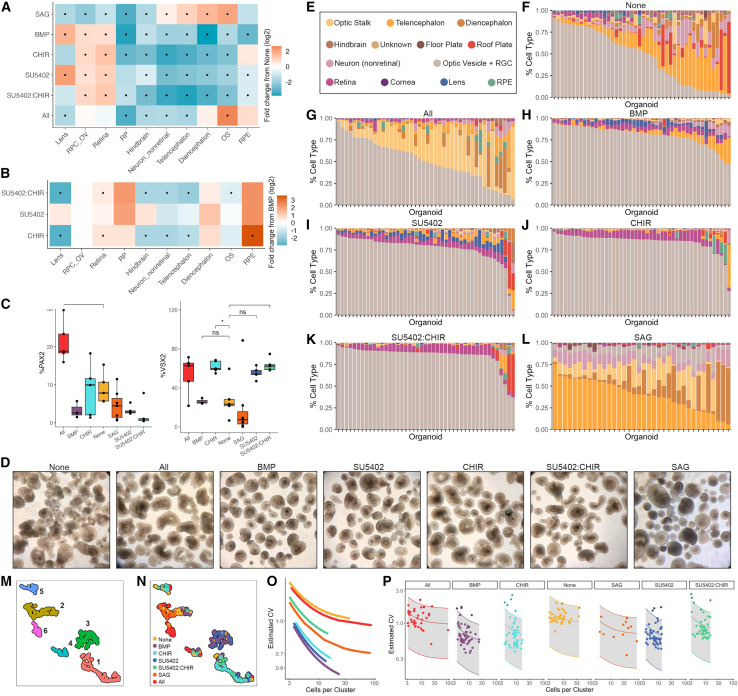
Figure 3.
sci-Plex allows the detection of significant changes in cell-type abundance and heterogeneity
(A) Heatmap of fold change in cell-type abundance compared to “None” for day 28 organoids. The data was modeled using a beta-binomial distribution and significance was determined by a Wald test. An asterisk (∗) in the center of the box indicates a q value of less than 0.05 after Benjamini-Hochberg correction.
(B) Heatmap of fold change in cell-type abundance compared with BMP for day 28 organoids. Statistical analysis was performed as in (A).
(C) Quantification of day 27 immunostained organoids. Significance was determined by performing t tests between the treatment of interest and None. Benjamini-Hochberg correction was performed to adjust the p value. ∗p < 0.05. The line of the boxplots represents the median cells per organoid. The colored region indicates the values from the lower 25th to the upper 75th quartile of cell counts. The whiskers span the range of the cell counts within 1.5× the distance between the 25th–75th percentiles.
(D) Bright-phase images of organoids the day before sci-Plex (day 27). Scale bars represent 200 μM.
(E) Cell-type legend for (F)–(L).
(F) Stacked bar plot, each bar represents an individual day 28 organoid exposed to the None treatment. Bars colored by cell-class composition.
(G–L) Same as (F) but for (G) All, (H) BMP, (I) SU5402, (J) CHIR, (K) SU5402:CHIR, and (L) SAG.
(M) UMAP generated using the size-factor-normalized cell type by organoid matrix generated from day 28 individual organoids. Colored by cluster.
(N) UMAP from (M) with the organoids colored by treatment.
(O) The mean and coefficient of variation (CV) were modeled for each cluster and each treatment. Displayed are the modeled relationships colored by treatment.
(P) Model from (O) faceted by treatment. The shaded area marks the range within 2 standard deviations from the mean. Points represent the individual cluster values.
To better investigate whether SU5402, CHIR, or SU5402:CHIR led to improvements over BMP4 alone, we trained a new model comparing these groups. In contrast to the addition of BMP4 alone, the addition of BMP4 combined with Wnt activation (CHIR, SU5402:CHIR) caused a striking loss of lens cells (Figure 3B). Both treatments with Wnt activation also robustly decreased most other non-retinal cell classes (Figure 3B). Of the conditions with high RPC_OV cell counts, CHIR treatment led to the highest relative numbers of retinal pigmented epithelium (RPE) cells in the organoids, consistent with prior evidence that Wnt activation steers cells toward the RPE fate both in vivo and in vitro.35,36 Unexpectedly, both the CHIR and SU5402:CHIR treatments increased neural retinal cell counts (Figure 3B). Overall, our results indicated that the addition of CHIR99021 to the culturing protocol increased the retinal content of organoids compared with the addition of BMP4 only.
To determine whether any of the treatments affected the production of specific retinal cell classes, we selected only the RPC_OV and retinal cells and carried out dimensionality reduction, clustering, and cell-type annotation using marker genes (Figures S3A–S3C; Table S2). We found that CHIR, SU5402, and SU5402:CHIR treatments increased photoreceptor/bipolar cell precursors (PBpres) (Figures S3D and S3E). CHIR and SU5402:CHIR treatments also significantly increased neurogenic precursor (Npre) cells (Figures S3D and S3E). Overall, the results of this experiment showed that exposure to CHIR99021 reduced non-retinal cell classes, increased retinal cells, and may have specifically enhanced photoreceptor/bipolar cell production.
Cell abundance changes detected by sci-Plex are concordant with immunostaining
To further investigate our observations from sci-Plex, we performed immunostaining on cryosections from matched day 27 organoids (Figure S3F). Consistent with the sci-Plex results, we observed a significant increase in PAX2+ optic stalk-like regions in the All treatment (Figure 3C). Also consistent with sci-Plex, we found significant increases in VSX2+ regions (RPCs and retinal neuron precursors) in CHIR- and SU5402:CHIR-treated organoids (Figure 3C). However, the immunolabeling results and sci-Plex did not agree when comparing the BMP and None treatments: sci-Plex showed an increase in VSX2+ regions, yet this was not apparent in the cryosectioned organoids. This suggests that the reduced number of organoids used for immunostaining hindered our ability to find some of the significant changes in cell composition that sci-Plex identified. Overall, we did not detect the same breadth of cell-class abundance changes by immunostaining; however, all significant changes detected with immunostaining were also observed by sci-Plex.
Organoid subtypes are identified within treatment groups
In order to quantify heterogeneity across organoids, we looked at the cell-class compositions of each individual. Recent innovations have increased the percentage of organoids in a given culture that are retinal, but the definition of retinal is based on qualitative bright-field images where variation is observable, and the molecular source of variation is not defined18,19 (Figure 3D). Our individual organoid sci-Plex dataset provided a more quantitative picture of the variation that exists within and across treatments (Figures 3E–3L). Without exposure to any of the signaling modulators (None), organoids ranged from being primarily RPCs to almost completely non-retinal (Figure 3F). While BMP4 treatment markedly reduced the heterogeneity of the organoids (BMP, CHIR, SU5402, SU5402:CHIR) (Figures 3H–3K), a subset of the organoids demonstrated atypical cell-class distributions.
To define organoids within a culture that are divergent in their cell-class compositions, we developed a method to categorize organoids into “archetypes.” We counted the number of cells of each type in each organoid and used the resulting normalized cell class × organoid matrix (see STAR Methods) to create a uniform manifold approximation and projection (UMAP) in which each of the points represented an individual organoid. We performed clustering and found that organoids of a given treatment fell predominantly into a single archetype (cluster) (Figures 3M, 3N, and S3G). We also became interested in why a specific secondary archetype may appear for one treatment and not another. For example, archetype 2, defined by high counts of hindbrain cells and non-retinal neurons, was higher for BMP than it was for the other high-retinal-cell-content treatments that had more organoids from archetype 5 (high RPE, high roof plate) (Figures 3M, 3N, and S3H). These cell classes (i.e., hindbrain, non-retinal neurons, RPE, and roof plate) were not seen as significantly increased compared to the BMP treatment (except for RPE in CHIR), yet they were compellingly present in high numbers for a small set of individuals (Figures 3B and 3H–3K). Together, these results show that sci-Plex can define the typical cellular composition of organoids and detect the way by which a minority of organoids within a given treatment deviate from the established typical cell-class composition.
Statistical measurements of organoid heterogeneity identify treatments that decrease homogeneity
We next sought to quantify the variability in cell composition across treatments. We used a method that accounts for the mean-variance relationship in cell abundances across individuals as previously described.27,28,37 Cells were divided into 67 clusters (as used to define cell classes) (Figure S3I). The variance and mean among individuals were calculated for each treatment within each cluster (see STAR Methods). After modeling, we plotted the relationship between the number of cells per cluster and the expected coefficient of variation (CV) (Figure 3O). Lines lower on the plot have a lower CV and thus less heterogeneity across clusters. Comparing BMP with each of the other treatments in a pairwise fashion, we used a likelihood ratio test on the models of variance with and without a treatment term in order to assess whether other treatments affected organoid heterogeneity. We found that CHIR and SU5402 treatments did not significantly increase heterogeneity compared with BMP (Table S3). Our modeling also enabled us to identify specific clusters of cells that have a higher CV than statistically expected (Figures 3P, S3J, and S3K). This most commonly included RPE and roof plate cells, followed by other non-retinal cell classes (Figures S3J and S3K). The analysis used a statistical method in a novel manner to find that the additional activation of Wnt or the inhibition of FGF signaling does not change a culture’s homogeneity compared with treatment with BMP4 alone, but that all other treatments increase heterogeneity.
Early modulation of signaling pathways affects long-term cellular composition
To see whether the effects of signaling modulation carry forward as the cells differentiate into more mature fates, we performed sci-Plex on 96 individual organoids cultured for 63 days. After filtering, 194,335 hashed cells were recovered (median UMIs: 341, median features: 298) with a median of 1841.5 cells per organoid (Figures S4A–S4G). Cell classes were defined using marker genes (Figures 4A and S4H). Although RPCs and retinal neurons made up the majority of the cells, cells with brain characteristics persisted. Again, Wnt activation (CHIR, SU5402:CHIR) led to significant decreases in mean cell counts for non-retinal/cortical neurons and undefined cells (Figures 4B and 4C). In addition, there was a significant increase in RPE cells upon FGF inhibition and Wnt activation (SU5402:CHIR) (Figures 4B and 4C). At day 63, many retinal neuron cell classes, including PBpres, horizontal/amacrine precursors (HApres), proliferating RPCs (RPCprolifs), and proneural RPCs (RPCproneurals), were broadly increased in the CHIR and SU5402:CHIR treatments, when compared with BMP, but not to the level of significance (Figures 4B and S4I). Plotting the individual organoid cell composition (Figure 4D) also highlighted the fact that Wnt activation (CHIR) reduced the number of organoids containing high representation of cortical neurons when compared with BMP treatment. However, overall heterogeneity was not significantly different in any of the tested treatments (Figure S4K; Table S3), and non-retinal cell classes tended to be more variable than retinal cell classes (Figures S4J and S4L). Immunolabeling of cryosections of organoids from the same batch used for sci-Plex confirmed the representation of non-retinal cells in these organoids (Figure 4E). Taken together, this suggested that the early modulation of signaling pathways established durable shifts in retinal cell-class representation.
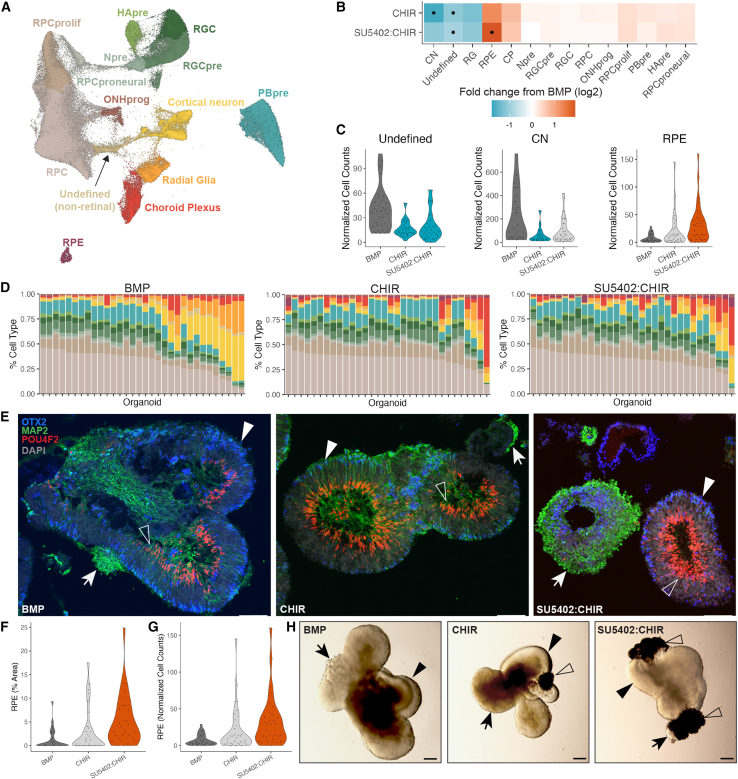
Figure 4.
Day 63 organoids demonstrate continued perturbation by signaling modulation
(A) UMAP with cell-type annotations from individually hashed D63 organoids exposed to BMP, CHIR, and SU5402:CHIR treatments. RPCprolif, proliferating RPC; RPCproneural, proneural RPC; ONHprog, optic nerve head progenitor; CN, cortical neuron; RG, radial glia; CP, choroid plexus.
(B)Heatmap of fold change in cell-type abundance compared with BMP. The data was modeled using a beta-binomial distribution and significance was determined by a Wald test. An asterisk (∗) in the center of the box indicates a q value of less than 0.05 after Benjamini-Hochberg correction.
(C) Violin plots of size-factor-normalized cell counts for day 63 organoids. Dark gray, control treatment in (B); light gray, no significant change; teal, significant decrease in abundance (p < 0.05); orange, significant increase in abundance (p < 0.05). Significance was determined as in (B).
(D) Stacked bar plots, each bar represents an individual organoid. Bars are colored by the cell-type composition. Right: cell-type color legend.
(E) Immunostaining of cryosectioned day 63 organoids with OTX2 (photoreceptors/RPE/CP, blue), MAP2 (retinal and cortical neurons, green), POU4F2 (RGC, red), and DAPI (gray). Arrow, non-retinal neurons; filled arrowhead, Otx2+ retinal tissue; empty arrowhead, Pou4f2+ retinal tissue. Scale bar represents 100 μm.
(F) Violin plots of percentage of area measurements for RPE as calculated from bright-phase microscopy images of day 62 organoids. Paired t tests followed by Bonferroni correction were used to determine significance (p < 0.05). Plots colored as in (C).
(G) Violin plots of size-factor-normalized RPE cell counts for day 63 organoids. Paired t tests followed by Bonferroni correction were used to determine significance (p < 0.05). Plots colored as in (C).
(H) Representative bright-field images of day 62 organoids from each group quantified in (F) and (G). Arrow, non-retinal tissue; filled arrowhead, retinal tissue; empty arrowhead, RPE. Scale bar represents 200 μm.
Lastly, we compared the sci-Plex cell counts to measurements of organoid composition obtained by bright-field microscopy. Prior to dissociation, each organoid was imaged in a 96-well plate such that the imaged organoid could be matched to a specific hash barcode. Areas of predicted retina, RPE, and non-retinal tissue were quantified (Figures 4F–4H, S4M, and S4N). Despite differences in the modalities, trends were conserved between the imaging and sci-Plex datasets. Notably, RPE was significantly increased in SU5402:CHIR by both sci-Plex and bright-field microscopy (Figures 4B and 4F). The cell count and area-based measurements from each individual also correlated, although sci-Plex detected a higher retinal composition and the bright-field images detected higher RPE and non-retinal composition (Figure S4O). Overall, we have shown agreement between two highly divergent methods. However, our analysis by bright-field microscopy was limited to a quantification based on broad cell-class domains using 2D images of a 3D object. The ability to quantify all cell classes across hundreds of individual organoids in a manner that agrees with and surpasses the sensitivity and breadth of other methods sets sci-Plex apart as a critical new method for the study of individual organoid cell-class composition.
Discussion
By assaying 410 individual organoids using sci-Plex, we show here that previously intractable analyses are now possible. We identify significant changes in cell-class abundance with a statistically principled methodology. We are also able to, for the first time, comprehensively measure the extensive heterogeneity of retinal organoids, a known but poorly quantified feature of current culturing protocols. Recent publications emphasize the utility of single-cell sequencing to investigate individual organoids and organoid heterogeneity29,30; however, no other study has profiled cell-class composition in comparable sample sizes across multiple treatments (32–48 individuals per treatment). Our work represents the most complete investigation of cell-class composition and heterogeneity in any organoid system to date.
Reassuringly, we find that the effect of treatments on cell-class abundance expected from the literature are readily detected. BMP4 treatment increased retinal and lens character in organoids,18,36,38 Wnt activation in the OV promoted RPE fate determination,32 and SAG treatment reduced OV cells, reflecting the role of SHH signaling in defining the ventral forebrain.39,40
Additionally, our results suggest that retinal organoid protocols might be optimized to generate adjacent neural tissues as well. Tissue that appears non-retinal by bright-field microscopy is routinely dissected away to improve the homogeneity of the cultures. However, when we add a combination of signaling modulators, we generate organoids with optic stalk-like cells, a cell type that is of clinical relevance but for which an established 3D culturing system does not exist.41 Optic stalk cells would have been discarded as non-retinal by conventional bright-field microscopy; however, they are captured in our study by the unbiased survey of the entire composition of organoids at scale. Further use of the technology in other developmental contexts has the potential to uncover novel culturing systems for additional disease-relevant cell types that may form unexpectedly or that may be difficult to detect by traditional methods.
Another advantage of surveying all cell classes within an organoid, specifically when it can be performed across individuals, is the ability to clarify the sources underlying organoid heterogeneity. We show that it is possible to separate individual organoids into subtypes based on their cell-class composition. With CHIR treatment, >75% of organoids fall into a single subtype defined by high RPC and retinal cell counts, but a secondary subtype defined by high RPE and roof plate content persists. Additional developmental signals that specifically regulate roof plate development could be incorporated in the protocol to reduce heterogeneity.42
Lastly, our most surprising result is the CHIR-dependent increase in neural retina at day 28 and the decrease in non-retinal neurons through day 63. Active canonical Wnt signaling is associated with decreased telencephalic identity and increased diencephalic identity, but the role of Wnt in determining the eye field is less well understood.43,44 Future studies will be needed to determine whether Wnt activation directly promotes the retinal fate or, alternatively, stimulates other non-retinal cells to indirectly drive the retinal identity.
Ultimately, the methodological advancement of sci-Plex and our analytical pipeline facilitates the expansion of perturbation studies in organoids. Sci-Plex can readily be expanded to thousands of individuals across as many conditions as can be cultured. We imagine this will lead to continued improvements to organoid culturing protocols, as well as an expansion in the use of organoids in chemical screening platforms. Although there are many applications for sci-Plex, we envision its utility in the near future to identify ganglion cell survival molecules and to screen for molecules that aid in cone survival as potential treatments for macular degeneration. Outside the retina, this technology holds promise for use with the constantly expanding number of 3D culturing systems.
Limitations of the study
We designed this study in favor of a high number of individual organoids to sample the organoid-to-organoid variation and included only a single organoid culturing technique. Additionally, our analyses focused on cell-class abundances instead of gene expression, namely because the treatments were performed weeks before the organoids were collected. Thus, the effects on gene expression caused at the time of treatment were not captured. Lastly, in our sci-RNA-seq3 hashing experiments, our hash rates were only ∼50%. Furthermore, the original sci-RNA-seq3 protocol has a low cell recovery rate, so this protocol is not ideal for rare cell types.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| POU4F2 | Santa Cruz Biotechnology | Cat# SC-6026; RRID: AB_673441 |
| PAX2 | BioLegend | Cat# 901001; RRID: AB_2565001 |
| VSX2 | Exalpha Biologicals | Cat# X1180P; RRID: AB_2314191 |
| VSX2 | SantaCruz | Cat# Sc-365519; RRID:AB_10842442 |
| MAP2 | Sigma-Aldrich | Cat# M9942; RRID:AB_477256 |
| OTX2 | Abcam | Cat# ab21990; RRID:AB_776930 |
| ELAVL3/4 | Thermo Fisher Scientific | Cat# A-21271; RRID:AB_221448 |
| RCVRN | MIllipore | Cat# AB5585; RRID:AB_2253622 |
| Donkey anti-goat 488 (1:250 dilution) | Thermo Fisher Scientific | Cat# A-11055; RRID:AB_2534102 |
| Donkey anti-rabbit 647 (1:250 dilution) | Thermo Fisher Scientific | Cat# A-31573; RRID:AB_2536183 |
| Donkey anti-sheep 647 (1:300 dilution) | Thermo Fisher Scientific | Cat# A-21448; RRID:AB_2535865 |
| Donkey anti-mouse 488 (1:250 dilution) | Jackson ImmunoResearch Labs | Cat# 715-546-151; RRID:AB_2340850 |
| Donkey anti-mouse 568 (1:250 dilution) | Thermo Fisher Scientific | Cat# A10037; RRID:AB_2534013 |
| Donkey anti-goat 568 (1:250 dilution) | Thermo Fisher Scientific | Cat# A-11057; RRID:AB_2534104 |
| Donkey anti-rabbit 647 (1:250 dilution) | Thermo Fisher Scientific | Cat# A-31573; RRID:AB_2536183 |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel | Corning | CLS354277 |
| ReLeSR | StemCell Technologies | 100–0484 |
| BMP4 | R&D Systems | 314-BP-050 |
| CHIR99021 | Cedarlane Laboratories | 04-0004-02 |
| SU5402 | Cayman Chemical Co | 13182 |
| SAG | Calbiochem | 566660 |
| DPBS (no calcium, no magnesium) | ThermoFisher | 14190144 |
| Accutase | Sigma | A6964-100ML |
| FBS | Corning | 35-011-CV |
| Papain and Ovomucoid | Worthington Biochemical | LK003150 |
| Fluromount-G | SouthernBiotech | 0100–01 |
| IGEPAL | Sigma-Aldrich | I8896 |
| SuperaseIn RNase Inhibitor | Ambion | AM2694 |
| BSA | NEB | B9000S |
| 5% paraformaldehyde | EMS | 50-980-493 |
| Triton X-100 | ThermoFisher | A16046.AP |
| 4′,6-diamidino-2-phenylindole (FACS) | Invitrogen | D1306 |
| 4′,6-diamidino-2-phenylindole (Immunostaining) | Sigma | D9542 |
| EB buffer | Qiagen | 19086 |
| mRNA Second Strand Synthesis buffer | NEB | E6111L |
| N7 loaded custom TDE1 enzyme | MacroLab, UC Berkeley | N/A |
| Dimethyl Formamide | Sigma-Aldrich | 227056-100ML |
| DNA binding buffer | Zymo Research | D4004-1-L |
| D1000 Screen Tape | Agilent Technologies | 5067–5583 |
| DNA purification | Zymo Research | D4014 |
| AMPure XP Reagent | Beckman Coulter Life Sciences | A63881 |
| Critical commercial assays | ||
| Single Cell 3′ v3.1 Dual Index Gene Expression kit (Dual Index Kit TT Set A 96 rxns) | 10x Genomics | 1000215 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE220661 |
| Experimental models: Cell lines | ||
| Human: H9 hESCs NRL+/eGFP | Phillips38 | N/A |
| Oligonucleotides | ||
| Hashing oligo: 5′-GTCTCGTGGGCTCGGAGATG TGTATAAGAGACAG-[10bp-barcode]-BAAAAAA AAAAAAAAAAAAAAAAAAAAAAAAAA-3′ where B is G, C or T |
Srivatsan et al.26 | N/A |
| Indexed 2-level RT oligo: oligo-dT primer 5ʹ-ACGA CGCTCTTCCGATCTNNNNNNNN-[10bp index]-TT TTTTTTTTTTTTTTTTTTTTTTTTTTTTVN-3ʹ, where N is any base and V is either A, C or G |
Cao et al.45 | N/A |
| Indexed 3-level RT oligo: (5’-[PHO]- CAGAGCNNN NNNNN-[10-bp barcode]-TTTTTTTTTTTTTTTTTTT TTTTTTTTTTT-3′ where N is any base |
Cao et al.34 | N/A |
| Indexed 3-level ligation oligo: 5′-GCTCTG-[9-bp or 10-bp barcode A]-/dideoxyU/ACGACGCTCTT CCGATCT-[reverse complement of barcode A]-3′ |
Cao et al.34 | N/A |
| Indexed P5 PCR oligo: 5′-AATGATACGGCGACC ACCGAGATCTACAC-[i5]-ACACTCTTTCCCTACA CGACGCTCTTCCGATCT-3′ |
Cao et al.45 | N/A |
| Indexed P7 PCR oligo: 5′-CAAGCAGAAGACGGC ATACGAGAT-[i7]-GTCTCGTGGGCTCGG-3′ |
Cao et al.45 | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al.46 | https://imagej.nih.gov/ij/ |
| 10x Genomics Cell Ranger | Zheng et al.47 | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Monocle3 | Cao et al.34 | https://cole-trapnell-lab.github.io/monocle3/ |
| BBI pipeline is available at the bbi-lab github page under the bbi-dmux and bbi-sci repositories | NA | https://github.com/bbi-lab |
| Seurat | Stuart et al.48 | https://satijalab.org/seurat/ |
| VGAM | Yee49 | https://CRAN.R-project.org/package=VGAM |
| Code related to this paper | This paper | https://zenodo.org/badge/latestdoi/654269063 |
| Other | ||
| Keyence BZ-X810 microscope | Keyence | N/A |
| Zeiss Axio Observer D1 | Zeiss | N/A |
| Zeiss LSM 990 | Zeiss | N/A |
| 1.5 mL LoBind microcentrifuge tube | Eppendorf | Z666491 |
| twin.tec™ 96 Well LoBind PCR Plates | Eppendorf | 0030129512 |
| Bioruptor Plus | diagenode | B01020001 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas A. Reh (tomreh@uw.edu).
Materials availability
No unique materials were generated by this study.
Experimental model and study participant details
The parent cell line used for all experiments was the female human embryonic stem cell line (hESC) H9 (WiCell, Madison, WI, https://www.wicell.org). H9 hESCs contain an NRL+/eGFP reporter38 contain. All organoids of the same age were from the same batch, but each age was from a different starting batch of organoids. The time matched sci-Plex and immunostaining experiments used organoids from the same batch.
| Experiment | Age | Line | Organoids Used |
|---|---|---|---|
| 10x match | D78 | H9 | 5 pooled |
| 10x match | D185 | H9 | 5 pooled |
| 10x match | D29 | H9 | 10 pooled |
| D28 | D28 | H9 | 314 individuals |
| D63 | D63 | H9 | 96 individuals |
Method details
Stem cell and retinal organoid culture
hESCs were maintained by dissociation and propagation in StemFlex media (ThermoFisher, A3349401) on Matrigel (Corning, CLS354277) coated plates at 37°C in 5% CO2. hESC colonies were passaged by dissociation with ReLeSR (StemCell Technologies, 100–0484). Retinal organoids were grown as described in18 with minor alterations mentioned herein. The cell culture medias used were Neural induction media, or NIM: 484.5mL DMEM/F12 (Life Technologies, Catalog #11330-057), 5mL N2 supplement (Life Technologies, Catalog #17502048), 5mL MEM NEAA (Life Technologies, Catalog# 11140050), 5mL penicillin-streptomycin (Life Technologies, Catalog# 15240062). Retinal differentiation media, or RDM: 240mL DMEM/F12 (Life Technologies, Catalog #11330-057), 240mL DMEM (Life Technologies, Catalog #12430062), 10mL B27 supplement (Life Technologies, Catalog #17504001), 5mL MEM NEAA (Life Technologies, Catalog #11140050), 5mL penicillin-streptomycin (Life Technologies, Catalog #15240062) + 1%, 3%, or 5% FBS (Corning, Catalog #35-011-CV) based on protocol needs.
In brief, confluent stem cell colonies were lifted from the plate using dispase (2 mg/mL, Life Technologies, Catalog #17105041) for 5–10 min, then forcefully removed by pipetting 2 mL/well of DMEM (Life Technologies, Catalog #12430062) directly onto cells using a 1 mL pipette. Colonies were removed from the 6 well plates and allowed to settle to the bottom of a 15 mL collection tube by gravity. The supernatant was removed, and colonies were transferred to a T25 tissue culture flask in a 3:1 StemFlex:NIM mix for a total of 10 mL per flask. Full media change was performed the following day (Day 1) with 10 mL of 1:2 StemFlex:NIM. Day 2 a full media change was performed and replaced with 10 mL of NIM. Day 3 a full media change was performed and replaced with 10 mL of NIM. Day 5 a full media change was performed and replaced with 10 mL of NIM. Day 7 a full media change was performed and replaced with 10 mL of NIM, and BMP4 BMP4 (1.5 nM, R&D Systems, 314-BP-050) was added to the media to a final concentration of 1.5nM. Day 8 EBs were then evenly distributed between the 6 wells of a 6 well plate. The media was not changed, and EBs were moved and cultured in the BMP4 containing NIM media they were in. 200 μL of FBS was added to allow for EBs to stick to the bottom of the plate. Days 10–20, every other day, a full media change was performed with fresh NIM. Days 14 and 16 a full media change was performed with NIM, and the signaling factor modulators CHIR99021 (3 μM, Cedarlane Laboratories, 04-0004-02), SU5402 (1 μM, Cayman Chemical Co, 13182), and/or SAG (1 μM, Calbiochem, 566660) were added depending on the treatment group. For the None treatment, DMSO was added at the same volume and time in which SU5402 was added. EBs were then manually lifted from the plates on day 20 or 21 by forcefully pipetting 2 mL/well of DMEM directly onto EBs using a 1 mL pipette. EBs were removed from the 6 well plates and allowed to settle to the bottom of a 15 mL collection tube by gravity. From this point on we consider the cells retinal organoids. The supernatant was removed, then organoids were resuspended in RDM with 1% FBS. Two days later media was exchanged with RDM +3% FBS, and two days after that with RDM +5% FBS. From this point on, cells were maintained with a full media change of RDM +5% FBS every two to three days.
Single cell dissociation
Organoids were placed into 96-well v-bottom plates. For the 10x comparison, 5–10 organoids were added to each well. In the D28 and D63 experiments, a single organoid was added to each well. Media was removed, the organoids were washed in DPBS (no calcium, no magnesium) (ThermoFisher, 14190144). For the D28 and D63 organoids, 100 μL of accutase (Sigma, A6964-100ML) was added to the cultures. The cultures were incubated at 37°C on a nutator for 15 min. After 15 min, organoids were pipetted up and down as needed (every 5–10 min). The extent of dissociation was determined by bright-field microscopy. When clusters of cells could no longer be seen, the accutase was deactivated by the addition of 20 μl of FBS (Corning, 35-011-CV). This was done on an organoid-by-organoid basis as some dissociate faster than others. The dissociation was complete by 30–45 min after accutase addition. For the D78 and D185 organoids, the dissociation was performed with 100 μL of papain (Worthington Biochemical, LK003150). The dissociation was performed as above and was completed within 15–20 min. 100 μL of Ovomucoid (Worthington Biochemical, LK003150) was used to deactivate the reaction. After deactivation of accutase or papain cells were spun down at 700 g for 3 min, then resuspended in a single cell suspension in RDM media18 for sci-Plex processing.
Immunostaining
Organoids were fixed in 4% paraformaldehyde, 5% sucrose for 30–45 min followed by 3 washes in 1X PBS. All samples were then transitioned into 10%, then 20% and then 30% solutions of sucrose for at least 30 min each solution. Organoids were then embedded in OCT and cryosectioned at 14–16 mm. For immunostaining, slides were washed three times in 1X PBS, then blocked in blocking buffer (0.5%Triton and 10% horse serum) for 20–60 min, followed by addition of primary antibody diluted in blocking buffer and incubation at 4C overnight. The following day, slides were washed 3 times in 1X PBS, then the appropriate 488/568/647 fluorophore-conjugated secondary antibody, diluted in blocking buffer, was added and incubated for 1 h at room temperature. Slides were mounted using Fluromount-G (SouthernBiotech, 0100-01). Details of the antibodies used in this study are provided below. For the D28 organoids, between 3 and 7 organoids were stained per condition (All = 5, BMP = 3, CHIR = 5, None = 5, SAG = 7, SU5402 = 5, SU5402:CHIR = 5).
| Antibody | Source | Catalog # | Dilution |
|---|---|---|---|
| POU4F2 | Santa Cruz Biotechnology | SC-6026 | 1:300 |
| PAX2 | BioLegend | 901001 | 1:300 |
| VSX2 | Exalpha Biologicals | X1180P | 1:300 |
| VSX2 | SantaCruz | Sc-365519 | 1:300 |
| MAP2 | Sigma | M9942 | 1:200 |
| OTX2 | ABCAM | 21990 | 1:300 |
| ELAVL3/4 | Invitrogen | A-21271 | 1:100 |
| RCVRN | Chemicon | AB5585 | 1:200 |
Microscopy
For brightfield images, single organoids were placed into each well of a round-bottom 96-well plate and imaged at 4x magnification with a Keyence BZ-×810 microscope. Group bright-field images were obtained at 4x magnification using the Zeiss Axio Observer D1. Confocal images were obtained using the Zeiss LSM 990. Image processing was done in ImageJ.46 Area measurements were obtained by setting a standardized threshold per channel.
10x library preparation
Dissociated cells were subjected to the standard 10x workflow for Single Cell 3′ v3.1 Dual Index Gene Expression kit (Dual Index Kit TT Set A 96 rxns, 10x Genomics, 1000215).
Sci-plex (hashing)
Hashing of organoids uses a protocol adapted from the original sci-Plex publication.26 Single-cell suspensions in 96-well v-bottom plates were centrifuged at 600 x g for 5 min. This step and all following centrifugation steps were performed in a swinging bucket centrifuge that was chilled to 4°C. Media was removed by aspiration. Cells were washed with 200 μL 1 x dPBS (no calcium, no magnesium) in each well. After centrifugation of the v-bottom plate, the dPBS was removed from wells by aspiration, and 50 μL of CLB+hash solution (45 μL of Cold Lysis Buffer – 10mM Tris/HCl pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL, 1% (v/v, Sigma-Aldrich, I8896), SuperaseIn RNase Inhibitor (20 U/μL, Ambion, AM2694), 1% (v/v) BSA (20 mg/mL, NEB, B9000S) + 5μL of hash oligo (10 μM, IDT)) was added to the well. The cells were mixed with the CLB+hash by pipetting up and down for 5–10 strokes. The plate was allowed to sit on ice for 3 min. To fix the hashes to the nuclei, 200 μL of fixation buffer (5% paraformaldehyde (EMS, cat. no. 50-980-493), 1.25x dPBS) was mixed with the nuclei by pipetting up and down for 10 strokes. The nuclei were fixed on ice for 15 min and then all wells of a plate were pooled into a single 50 mL conical tube. The hashed nuclei were centrifuged for 5 min at 800xg. Pellets were resuspended in 1mL of NSB (Nuclei Buffer + SuperaseIn + BSA – 10mM Tris/HCl pH 7.4, 10mM NaCl, 3mM MgCl2, 1% (v/v) BSA, 1% (v/v) SuperaseIn RNase Inhibitor) and all 50 mL tubes were pooled into a single 50 mL conical. Nuclei were centrifuged for 5 min at 800 x g and resuspended again in 1 mL of cold NSB. Nuclei were transferred to a 1.5 mL LoBind microcentrifuge tube (Eppendorf, Z666491). The number of nuclei recovered was counted using a hemocytometer. In order to continue centrifuging in a swinging bucket centrifuge, the 1.5 mL tube was placed within a 15 mL conical. The nuclei were centrifuged one final time for 5 min at 800 x g and they were resuspended in 500 μL of NSB. Samples were flash frozen in liquid nitrogen and stored at −80C.
Sci-RNA-seq
The sci-RNA-seq experiment described in Figure 1 was prepared by the original sci-RNA-seq protocol45 with minor modifications. Briefly, fixed nuclei were thawed and centrifuged at 800 x g for 5 min. Nuclei were permeabilized with 500 μL NSB +0.2% Triton X-100 (ThermoFisher, A16046.AP) on ice for 3 min. Nuclei were centrifuged for 10 min at 800 x g and resuspended in 400 μL NSB. A final centrifugation step was performed at 800 x g for 10 min. The nuclei were diluted to ∼300 nuclei per uL across 2 x twin.tec 96 Well LoBind PCR Plates (Eppendorf, 0030129512). The reverse transcription reaction was set up as in Cao et al., and the RT reaction was carried out using an increasing temperature gradient (4°C 2 min, 10°C 2 min, 20°C 2 min, 30°C 2 min, 40°C 2 min, 50°C 2 min, 55°C 15 min) without the addition of any reagents to stop the reaction upon completion. All nuclei were pooled and stained with DAPI (4′,6-diamidino-2-phenylindole, 3 μM final) (Invitrogen, D1306). Using a FACSAria III cell sorter (BD Biosciences), 50 nuclei were sorted into each well of 4 twin.tec 96 Well LoBind PCR Plates that had 5 μL of EB buffer (Qiagen, 19086), 0.5 μL of 5 x mRNA Second Strand Synthesis buffer (New England Biolabs, E6111L), and 0.25 μL of mRNA Second Strand Synthesis enzyme (New England Biolabs, E6111L) in each well. The plates were incubated at 16°C for 3 h. To each well, 5.75 μL of Tagmentation mix (1.1 μL N7 loaded custom TDE1 enzyme (MacroLab, UC Berkeley) per 632.5 μL 2xTD buffer (20 mM Tris-HCl pH 7.6, 10 mM MgCl2, 20% v/v Dimethyl Formamide (Sigma-Aldrich, 227056-100ML)) was added and mixed by pipetting 10 x. Tagmentation was carried out at 55C for 5 min before being stopped by the addition of 12 μL of DNA binding buffer (Zymo Research, D4004-1-L) for 5 min at room temperature. Ampure-based bead purification, PCR, and final library purification was performed as published previously.45 The library was visualized using a D1000 Screen Tape (Agilent Technologies, 5067–5583).
Both of the individual organoid datasets were prepared following the sci-RNA-seq3 protocol from Srivatsan et al. with modifications noted below. As above, all centrifugation speeds were performed at 800 x g. Fixed nuclei were thawed and centrifuged for 5 min. Nuclei were permeabilized with 500 μL NSB +0.2% Triton X-100 on ice for 3 min. Nuclei were centrifuged for 10 min and resuspended in 400 μL NSB to wash away the Triton. The nuclei were sonicated with a Bioruptor Plus (diagenode) sonication device on low power for 12 s. A final centrifugation step was then performed for 10 min. Nuclei were suspended to a concentration of 1000–2500 nuclei/uL with 22,000–55,000 nuclei added to each well of 2 x twin.tec 96 Well LoBind PCR Plates. From here only minor modifications from the Srivatsan protocol were performed. Of note, before second strand synthesis 2500-3000 nuclei were distributed into each well, during tagmentation the 10 μL of tagmentation mix was composed of 0.05 μL of a custom TDE1 enzyme in 9.95μL of 2 x TD buffer, and when performing the well-based Ampure bead purification the volume of beads was increased to 60 μL per well. PCR was performed for 13 cycles and libraries were purified with column-based DNA purification (Zymo Research, D4014) followed by 0.8x Ampure bead cleanup. Libraries were visualized using D1000 or HS D1000 Screen Tapes.
In-depth protocols can be found on protocols.io under the following titles.
2-level: Single cell RNA sequencing library preparation (2-level sci-RNA-seq)
3-level: sci-RNA-seq3.
Processing of single cell RNA sequencing data
10x
The 10x libraries from D28, D78, and D185 organoids were sequenced on an Illumina NextSeq 2000 and a NextSeq 550 (Read 1: 28, Index 1: 10, Index 2: 10, Read 2: 75) and processed by CellRanger (3.1.0)47 using the default settings. The output was imported into Monocle3 (1.0.0),34 and data from the three runs were combined into a single object with the corresponding sci-Plex dataset.
Sci-Plex (10x comparison)
The sci-Plex library was sequenced on an Illumina Nextseq550 (High Output 75 cycle kit) with 18 cycles for Read1, 10 cycles for each index, and 52 cycles for Read2. The reads were processed using a pipeline developed by the Brotman Baty Institute (BBI). This BBI pipeline is available at the bbi-lab github page under the bbi-dmux and bbi-sci repositories (https://github.com/bbi-lab). The output was imported into Monocle3, and custom code (https://github.com/atresen/individual_retinal_organoids) was used to incorporate the hash information outputted by the pipeline into the Monocle3 cds object. Cells with fewer than 1000 UMIs and >20,000 UMIs were discarded. Hashing success was determined by the ratio of the most prevalent hash barcode for a cell compared to the second most prevalent hash barcode. If the top_to_second_best_ratio was >3 the cell was considered hashed. All cells below that cutoff were discarded. The filtered sci-Plex data was combined with the 10x data. The combined object was converted into a Seurat object for CCA integration following the “Introduction to scRNA-seq integration” vignette.48,50,51 The integration anchors were found using 100 dimensions and 100 anchor features. A 3D UMAP was constructed from 50 PCA dimensions with a mindist of 0.2 and 50 nearest neighbors. The UMAP coordinates outputted from the integration were transferred back to Monocle3 from which the remainder of the analysis and plotting was performed. Clusters were determined using cluster_cells() in Monocle3 using the default assigned resolution. Common marker genes for retinal cell classes compiled from previous single cell and bulk transcriptomic studies of retinal development in fetal human (from our group and others) were then plotted on to the UMAP to determine the clusters with which they were most associated (Table S2). For some clusters, common retinal markers were not sufficient to assign cell classes. In those cases, genes with high specificity to that cluster were determined using custom code (https://github.com/atresen/individual_retinal_organoids). These genes were then used to aid in a cell class determination (Table S2).
Sci-Plex (D28 individual organoids)
The sci-Plex library was sequenced on an Illumina NextSeq 2000 (P2 100 cycle kit) with 35 cycles for Read 1, 10 cycles for each index, and 70 cycles for Read2. Reads were processed using the BBI pipeline as described above. The output was imported into Monocle3. A preliminary filtering was performed for cells with >100 UMIs, less than 20,000 UMIs, and <15% mitochondrial reads. Cells with a top_to_second_best_ratio <3 were also removed from the analysis. Size factors were estimated, the data was preprocessed with 40 PCA dimensions, correction was performed to account for the number of UMIs per cell. UMAPs were projected in 2 dimensions with a mindist of 0.1 and 15 nearest neighbors. Cells were clustered with a resolution of 5 × 10−5. cell classes were assigned using well-established marker genes (Table S2). For the analysis of individual organoids, only organoids with at least 50 cells were used.
Sci-Plex (D63 individual organoids)
The sci-Plex library was sequenced on a Nextseq 2000 (P2 100 cycle kit) with 35 cycles for Read 1, 10 cycles for each index, and 70 cycles for Read2. Reads were processed using the BBI pipeline as described above. The output was imported into Monocle3. A preliminary filtering was performed for cells with >150 UMIs, less than 20,000 UMIs, and <15% mitochondrial reads. Cells with a top_to_second_best_ratio <2.5 were also removed from the analysis. Size factors were estimated, the data was preprocessed with 30 PCA dimensions, correction was performed to account for the number of UMIs per cell. UMAPs were projected in 2 dimensions with a mindist of 0.1 and 50 nearest neighbors. Cells were clustered with a resolution of 5 × 10−5. cell classes were assigned using well-established marker genes (Table S2). Only organoids with at least 50 cells were used in downstream analyses.
Quantification and statistical analysis
Calculation of size factor normalized cell class counts
Size factor normalized cell counts were calculated as in.27,28 Briefly, counts for cells of the same cell class within each organoid were added up and organized into a cell class by organoid matrix. A Monocle3 cds was created from the matrix, and size factors(), which takes the total number of cells in the organoid and divides it by the geometric mean of cell counts for all organoids, was run. The number of cells of each cell class in each organoid was divided by the size factor of that organoid. All values were rounded to the nearest whole number.
Detection of significant fold changes in cell class abundance
A generalized linear model was fit to the normalized data using a beta-binomial distribution52 as in.27,28
where yi represents the normalized cell count distribution for cell class i, μi is the mean of the distribution, and ρi is the overdispersion or “litter effect” associated with the beta-binomial distribution. The binary indicator.
variable Xt denotes which treatment the organoids were exposed to. All D28 treatments were first modeled with respect to the None treatment. The CHIR, SU5402, and SU5402:CHIR treatments were later modeled compared to the BMP treatment. At D63 CHIR and SU5402:CHIR treatments were modeled compared to BMP. The models were fit using the VGAM package, and p values were determined by a Wald test on βt.53,54 Abundance changes were determined significant if the q-value after Benjamini Hochberg correction was < 0.05.55 In D28 organoids, the model above was also used to determine significant abundances changes in specific retinal cell populations. When this was performed, all cell classes were included in the calculation even though only the retinal cell classes are displayed in the heatmap in Figure S3D.
Clustering of individual organoids
Counts for cells of the same cell class within each organoid were added up and organized into a cell class by organoid matrix. A Monocle3 cds was created from the matrix and size factors() was run. The cds was processed preprocess_cds(cds, method = “PCA”, norm_method = “size_only”, num_dim = 9) and then subjected to dimensionality reduction in two dimensions by UMAP reduce_dimension(cds, umap.n_neighbors = 30, umap.min_dist = 0.05). Clustering was performed with a resolution of 1 × 10−2.
Detection of treatments and cell clusters with high variance
A cluster by individual organoid matrix was calculated, size factors were determined, and a generalized linear model was fit to the normalized data using a beta-binomial distribution as above. The VGAM package doesn’t automatically model dispersions for the beta-binomial distribution, so simulate.vlm() was run 100 times to model the mean and standard deviation which was then used to calculate the coefficient of variation () for each cluster and each treatment (i.e., the normalized cell counts for the 48 organoids exposed to CHIR treatment were determined for cluster 1 and then the mean and variance were calculated. This was repeated for each of the treatments and then each of the clusters). Because variance scales with the mean, the coefficient of variation (CV) was calculated by taking the ratio of the standard deviation over the mean as a means to normalize the amount of variance such that we can compare the variance in clusters with high cell counts to clusters with low cells counts. As randomness is increased at smaller values, we expect that clusters with fewer cells will have higher CV and clusters with more cells will have lower CV values. A gamma-valued glm of the form pioneered by DESeq56 was then to capture the trend between the average number of cells in a cluster across organoids and that cluster’s CV using VGAM.53,54 To identify clusters with higher than expected CV, conditional upon treatment, we tested whether the CV for a given cluster exceeded this model’s 95% confidence interval’s upper limit.
To assess whether a treatment altered heterogeneity compared to the BMP treatment, a likelihood-ratio test was used to compare the output of the gamma distribution to a reduced model that excludes the treatment term. Treatments were considered more heterogeneous than BMP if the likelihood-ratio test produced a q-value after Benjamini Hochberg correction of <0.05 indicating that use of treatment as a factor improves the model and the sign of the treatment term was positive indicating that the treatment-dependent change led to an increase in heterogeneity.55
Acknowledgments
We thank members of the Trapnell and Reh labs for their comments and discussions. We thank Choli Lee for assistance in flow sorting, the Beliveau lab for use of their Keyence BZ-X810, and the Brotman Baty Institute for support with the data processing pipeline. This work was supported by the NIH (1R01HG010632 to C.T., R01EY021482-12 to T.A.R., and 1F32EY032331 to A.T.); the Paul G. Allen Family Foundation (Allen Discovery Center grant 12357 to C.T.); the CZI (CZF2019-002442 to C.T.); the Foundation Fighting Blindness (TA-RM-0620-0788-UWA to T.A.R.); the Damon Runyon Cancer Research Foundation (DRG-# 32-20 to K.C.E.); and the Hanna H. Gray Fellows Program Award from the HHMI (GT15994 to K.C.E.). Illustrations were created with BioRender.com.
Author contributions
Conceptualization, A.T., A.S., K.C.E., and T.A.R.; methodology, A.T. and C.T.; formal analysis, A.T., K.C.E., and A.S.; investigation, A.T., A.S., K.C.E., D.H., and S.C.; writing – original draft, A.T.; writing – review & editing, A.T., K.C.E., C.T., and T.A.R.; visualization, A.T., A.S., and K.C.E.; supervision, C.T. and T.A.R.; funding acquisition C.T. and T.A.R.
Declaration of interests
C.T. is a SAB member, consultant, and/or co-founder of Algen Biotechnologies, Altius Therapeutics, and Scale Biosciences. One or more embodiments of one or more patents and patent applications filed by the University of Washington may encompass methods, reagents, and the data disclosed in this manuscript. Some work in this study is related to technology described in patent applications.
Published: August 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100548.
Contributor Information
Cole Trapnell, Email: coletrap@uw.edu.
Thomas A. Reh, Email: tomreh@uw.edu.
Supplemental information
The coefficients and likelihood ratio test results obtained after modeling the CV vs. cluster size with a gamma distribution. The likelihood test compared CHIR or SU5402:CHIR to BMP treatments
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available at GEO: GSE220661. The accession number is also listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at Zenodo: https://doi.org/10.5281/zenodo.8088246 and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 2.Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., Saito K., Yonemura S., Eiraku M., Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia Y., Nivet E., Sancho-Martinez I., Gallegos T., Suzuki K., Okamura D., Wu M.-Z., Dubova I., Esteban C.R., Montserrat N., et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat. Cell Biol. 2013;15:1507–1515. doi: 10.1038/ncb2872. [DOI] [PubMed] [Google Scholar]
- 6.Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Takasato M., Er P.X., Becroft M., Vanslambrouck J.M., Stanley E.G., Elefanty A.G., Little M.H. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 8.Fordham R.P., Yui S., Hannan N.R.F., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T., et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannan N.R.F., Fordham R.P., Syed Y.A., Moignard V., Berry A., Bautista R., Hanley N.A., Jensen K.B., Vallier L. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Rep. 2013;1:293–306. doi: 10.1016/j.stemcr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sridhar A., Hoshino A., Finkbeiner C.R., Chitsazan A., Dai L., Haugan A.K., Eschenbacher K.M., Jackson D.L., Trapnell C., Bermingham-McDonogh O., et al. Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep. 2020;30:1644–1659.e4. doi: 10.1016/j.celrep.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eldred K.C., Hadyniak S.E., Hussey K.A., Brenerman B., Zhang P.-W., Chamling X., Sluch V.M., Welsbie D.S., Hattar S., Taylor J., et al. Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science. 2018;362 doi: 10.1126/science.aau6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayerl S.J., Bajgai S., Ludwig A.L., Jager L.D., Williams B.N., Bacig C., Stoddard C., Sinha D., Philpot B.D., Gamm D.M. Human retinal organoids harboring IMPG2 mutations exhibit a photoreceptor outer segment phenotype that models advanced retinitis pigmentosa. Stem Cell Rep. 2022;17:2409–2420. doi: 10.1016/j.stemcr.2022.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallman A., Capowski E.E., Wang J., Kaushik A.M., Jansen A.D., Edwards K.L., Chen L., Berlinicke C.A., Joseph Phillips M., Pierce E.A., et al. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Commun. Biol. 2020;3:82. doi: 10.1038/s42003-020-0808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aboualizadeh E., Phillips M.J., McGregor J.E., DiLoreto D.A., Jr., Strazzeri J.M., Dhakal K.R., Bateman B., Jager L.D., Nilles K.L., Stuedemann S.A., et al. Imaging transplanted photoreceptors in living nonhuman primates with single-cell resolution. Stem Cell Rep. 2020;15:482–497. doi: 10.1016/j.stemcr.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ripolles-Garcia A., Dolgova N., Phillips M.J., Savina S., Ludwig A.L., Stuedemann S.A., Nlebedum U., Wolfe J.H., Garden O.A., Maminishkis A., et al. Systemic immunosuppression promotes survival and integration of subretinally implanted human ESC-derived photoreceptor precursors in dogs. Stem Cell Rep. 2022;17:1824–1841. doi: 10.1016/j.stemcr.2022.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao J.R., Lamba D.A., Klesert T.R., Torre A.L., Hoshino A., Taylor R.J., Jayabalu A., Engel A.L., Khuu T.H., Wang R.K., et al. Transplantation of human embryonic stem cell-derived retinal cells into the subretinal space of a non-human primate. Transl. Vis. Sci. Technol. 2017;6:4. doi: 10.1167/tvst.6.3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eldred K.C., Reh T.A. Human retinal model systems: Strengths, weaknesses, and future directions. Dev. Biol. 2021;480:114–122. doi: 10.1016/j.ydbio.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Fligor C.M., Huang K.-C., Lavekar S.S., VanderWall K.B., Meyer J.S. Differentiation of retinal organoids from human pluripotent stem cells. Methods Cell Biol. 2020;159:279–302. doi: 10.1016/bs.mcb.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Wagstaff E.L., Heredero Berzal A., Boon C.J.F., Quinn P.M.J., Ten Asbroek A.L.M.A., Bergen A.A. The role of small molecules and their effect on the molecular mechanisms of early retinal organoid development. Int. J. Mol. Sci. 2021;22:7081. doi: 10.3390/ijms22137081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T.M., Krol J., Szikra T., et al. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020;182:1623–1640.e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Hara-Wright M., Gonzalez-Cordero A. Retinal organoids: a window into human retinal development. Development. 2020;147 doi: 10.1242/dev.189746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukonin I., Zinner M., Liberali P. Organoids in image-based phenotypic chemical screens. Exp. Mol. Med. 2021;53:1495–1502. doi: 10.1038/s12276-021-00641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeStefanis R.A., Kratz J.D., Olson A.M., Sunil A., DeZeeuw A.K., Gillette A.A., Sha G.C., Johnson K.A., Pasch C.A., Clipson L., et al. Impact of baseline culture conditions of cancer organoids when determining therapeutic response and tumor heterogeneity. Sci. Rep. 2022;12:5205. doi: 10.1038/s41598-022-08937-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renner H., Grabos M., Becker K.J., Kagermeier T.E., Wu J., Otto M., Peischard S., Zeuschner D., TsyTsyura Y., Disse P., et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife. 2020;9 doi: 10.7554/eLife.52904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fligor C.M., Langer K.B., Sridhar A., Ren Y., Shields P.K., Edler M.C., Ohlemacher S.K., Sluch V.M., Zack D.J., Zhang C., et al. Three-dimensional retinal organoids facilitate the investigation of retinal ganglion cell development, organization and neurite outgrowth from human pluripotent stem cells. Sci. Rep. 2018;8:14520. doi: 10.1038/s41598-018-32871-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivatsan S.R., McFaline-Figueroa J.L., Ramani V., Saunders L., Cao J., Packer J., Pliner H.A., Jackson D.L., Daza R.M., Christiansen L., et al. Massively multiplex chemical transcriptomics at single cell resolution. Science. 2019;6234:1–13. doi: 10.1126/science.aax6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorrity M.W., Saunders L.M., Duran M., Srivatsan S.R., Ewing B., Queitsch C., Shendure J., Raible D.W., Kimelman D., Trapnell C. Proteostasis governs differential temperature sensitivity across embryonic cell types. bioRxiv. 2022 doi: 10.1101/2022.08.04.502669. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders, L.M., Srivatsan, S.R., Duran, M., Dorrity, M.W., Ewing, B., Linbo, T., Shendure, J., Raible, D.W., Moens, C.B., Kimelman#, D., et al. Deep molecular, cellular and temporal phenotyping of developmental perturbations at whole organism scale. Preprint at bioRxiv. 10.1101/2022.08.04.502764. [DOI]
- 29.Uzquiano A., Kedaigle A.J., Pigoni M., Paulsen B., Adiconis X., Kim K., Faits T., Nagaraja S., Antón-Bolaños N., Gerhardinger C., et al. Single-cell multiomics atlas of organoid development uncovers longitudinal molecular programs of cellular diversification of the human cerebral cortex. bioRxiv. 2022 doi: 10.1101/2022.03.17.484798. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bues J., Biočanin M., Pezoldt J., Dainese R., Chrisnandy A., Rezakhani S., Saelens W., Gardeux V., Gupta R., Sarkis R., et al. Deterministic scRNA-seq captures variation in intestinal crypt and organoid composition. Nat. Methods. 2022;19:323–330. doi: 10.1038/s41592-021-01391-1. [DOI] [PubMed] [Google Scholar]
- 31.Reese B.E. Development of the retina and optic pathway. Vis. Res. 2011;51:613–632. doi: 10.1016/j.visres.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr. Top. Dev. Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diacou R., Nandigrami P., Fiser A., Liu W., Ashery-Padan R., Cvekl A. Cell fate decisions, transcription factors and signaling during early retinal development. Prog. Retin. Eye Res. 2022;91:101093. doi: 10.1016/j.preteyeres.2022.101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimura N., Taketo M.M., Mori M., Korinek V., Kozmik Z. Spatial and temporal regulation of Wnt/beta-catenin signaling is essential for development of the retinal pigment epithelium. Dev. Biol. 2009;334:31–45. doi: 10.1016/j.ydbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015;6:6286. doi: 10.1038/ncomms7286. [DOI] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips M.J., Capowski E.E., Petersen A., Jansen A.D., Barlow K., Edwards K.L., Gamm D.M. Generation of a rod-specific NRL reporter line in human pluripotent stem cells. Sci. Rep. 2018;8:2370. doi: 10.1038/s41598-018-20813-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 40.Rubenstein J.L., Beachy P.A. Patterning of the embryonic forebrain. Curr. Opin. Neurobiol. 1998;8:18–26. doi: 10.1016/s0959-4388(98)80004-4. [DOI] [PubMed] [Google Scholar]
- 41.Dutton G.N. Congenital disorders of the optic nerve: excavations and hypoplasia. Eye. 2004;18:1038–1048. doi: 10.1038/sj.eye.6701545. [DOI] [PubMed] [Google Scholar]
- 42.Le Dréau G., Martí E. Dorsal-ventral patterning of the neural tube: a tale of three signals. Dev. Neurobiol. 2012;72:1471–1481. doi: 10.1002/dneu.22015. [DOI] [PubMed] [Google Scholar]
- 43.Harrison-Uy S.J., Pleasure S.J. Wnt signaling and forebrain development. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson S.W., Houart C. Early steps in the development of the forebrain. Dev. Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yee T.W. The VGAM package for categorical data analysis. J. Stat. Software. 2010;32:1–34. [Google Scholar]
- 50.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ennis D.M., Bi J. The beta-binomial model: Accounting for inter-trial variation in replicated difference and preference tests. J. Sens. Stud. 1998;13:389–412. [Google Scholar]
- 53.Yee T.W., Wild C.J. Vector generalized additive models. J. R. Stat. Soc. Series B Stat. Methodol. 1996;58:481–493. [Google Scholar]
- 54.Yee T.W. Springer; 2015. Vector Generalized Linear and Additive Models: With an Implementation in R. [Google Scholar]
- 55.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 56.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The coefficients and likelihood ratio test results obtained after modeling the CV vs. cluster size with a gamma distribution. The likelihood test compared CHIR or SU5402:CHIR to BMP treatments
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available at GEO: GSE220661. The accession number is also listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at Zenodo: https://doi.org/10.5281/zenodo.8088246 and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.