Abstract
CMG (Cdc45‐MCM‐GINS) helicase assembly at the replication origin is the culmination of eukaryotic DNA replication initiation. This process can be reconstructed in vitro using defined factors in Saccharomyces cerevisiae; however, in vertebrates, origin‐dependent CMG formation has not yet been achieved partly due to the lack of a complete set of known initiator proteins. Since a microcephaly gene product, DONSON, was reported to remodel the CMG helicase under replication stress, we analyzed its role in DNA replication using a Xenopus cell‐free system. We found that DONSON was essential for the replisome assembly. In vertebrates, DONSON physically interacted with GINS and Polε via its conserved N‐terminal PGY and NPF motifs, and the DONSON‐GINS interaction contributed to the replisome assembly. DONSON's chromatin association during replication initiation required the pre‐replicative complex, TopBP1, and kinase activities of S‐CDK and DDK. Both S‐CDK and DDK required DONSON to trigger replication initiation. Moreover, human DONSON could substitute for the Xenopus protein in a cell‐free system. These findings indicate that vertebrate DONSON is a novel initiator protein essential for CMG helicase assembly.
Keywords: CMG helicase, DNA replication, DONSON, replisome assembly, Xenopus cell‐free replication system
Subject Categories: DNA Replication, Recombination & Repair
The microcephaly gene product DONSON emerges as a missing factor contributing to higher eukaryotic replisome formation, by interacting with GINS and DNA polymerase epsilon.
Introduction
Drosophila humpty dumpty, or DONSON in vertebrates, is an essential gene required for cellular proliferation, which is named after its mutant phenotype that produces thin eggshells (Bandura et al, 2005). The structural components of eggshells are encoded by clusters of chorion genes, whose copy number is amplified through endo‐replication in the ovarian follicle cells during early development to support eggshell biosynthesis (Calvi et al, 1998). Mutations in the humpty dumpty gene cause a defect in the amplification of the chorion genes, resulting in the reduced availability of eggshell proteins (Bandura et al, 2005; Lesly et al, 2017). The human DONSON gene is a humpty dumpty homolog that was identified using advanced next‐generation sequencing. Its mutation causes microcephalic primordial dwarfism in patients with Meier–Gorlin syndrome, microcephaly–micromelia syndrome, as well as microcephaly and short stature with limb abnormalities (Evrony et al, 2017; Reynolds et al, 2017; Schulz et al, 2018; Knapp et al, 2020). Interestingly, most of the underlying genes of Meier–Gorlin syndrome are categorized as initiation factors for DNA replication, such as ORC1, ORC4, ORC6, CDT1, CDC6, GMININ, CDC45, and MCM5 (de Munnik et al, 2015; Knapp et al, 2020). Therefore, it is possible that DONSON is involved in DNA replication initiation; however, there is currently no direct evidence substantiating this.
The basic mechanism of DNA replication initiation is highly conserved among eukaryotes (Parker et al, 2017; Moiseeva & Bakkenist, 2018; Lin & Prasanth, 2021; Costa & Diffley, 2022; Gillespie & Blow, 2022). The origin recognition complex (ORC) and mini chromosome maintenance (MCM) complex are each composed of six subunits, ORC1–6 and MCM2–7, respectively. The gene products ORC1/4/6 and MCM5 are the components of the ORC and MCM hetero‐hexamers, respectively. The ORC can recognize and demarcate potential replication initiation sites (origins) on chromosomal DNA (Fragkos et al, 2015). During the G1 phase of the cell cycle, the origin‐bound ORC, together with Cdc6 and Cdt1, promotes the sequential recruitment of two MCM single hexamers to create a stable MCM double hexamer at the origin, called the pre‐replicative complex (pre‐RC). The MCM has potential helicase activity, but it is inactive as a single or double hexamer. At the onset of the S‐phase of the cell cycle, one MCM double hexamer is converted into two active replicative helicases, called CMG (Cdc45‐MCM‐GINS) helicases (Li & O'Donnell, 2018). GINS is a hetero‐tetramer consisting of Sld5, Psf1, Psf2, and Psf3 (Kubota et al, 2003; Takayama et al, 2003). CMG helicase formation is coupled with initial origin melting, from where the two CMG helicases travel apart in a 3′–5′ direction on each leading strand, thereby producing two replication forks that proceed in opposite directions (Burgers & Kunkel, 2017; Attali et al, 2021). At the replication forks, replicative DNA polymerases α/δ/ε and other factors, such as Claspin and Tipin‐Tim1, are further assembled around the CMG to establish the replisome complex (Bellelli & Boulton, 2021).
In Saccharomyces cerevisiae, the conversion process from pre‐RC to CMG requires several chaperone‐like initiator proteins, such as Sld2, Sld3, Sld7, and Dpb11, that are regulated by the S‐phase cyclin‐dependent kinase CDK (S‐CDK) and Dbf4/Drf1‐dependent Cdc7 kinase (DDK) (Tanaka & Araki, 2013; Zegerman, 2015). Sld2 and Sld3 are the only two S‐CDK targets that are essential for replication initiation (Tanaka et al, 2007; Zegerman & Diffley, 2007). In the absence of S‐CDK, Sld3 forms a sub‐complex with Sld7 and Cdc45 (Heller et al, 2011; Tanaka et al, 2011; Yeeles et al, 2015). On the other hand, DDK binds the double hexamer MCM in the pre‐RC and phosphorylating the MCM2/4/6 subunits (Cho et al, 2006; Masai et al, 2006; Sheu & Stillman, 2006, 2010; Tsuji et al, 2006; Randell et al, 2010; Ramer et al, 2013), which attracts the Sld3‐Sld7‐Cdc45 complex via interaction between the MCM and Sld3 (Deegan et al, 2016; Fang et al, 2016). Upon S‐CDK activation, the phosphorylated Sld2 forms a sub‐complex with Dpb11, GINS, and Polε (the pre‐loading complex, pre‐LC), while the phosphorylated Sld3 recruits pre‐LC via the interaction with Dpb11 to form the pre‐initiation complex (pre‐IC; Muramatsu et al, 2010; Heller et al, 2011; Tanaka et al, 2013). The pre‐IC is then converted into the inactive CMG complex, whose activation presumably requires MCM10 (van Deursen et al, 2012; Douglas & Diffley, 2016). These chaperone‐like initiator proteins appear to have divergent primary structures among eukaryotes. Vertebrate Treslin, MTBP, and TopBP1 are the functional counterparts of yeast Sld3, Sld7, and Dpb11, respectively (Van Hatten et al, 2002; Kumagai et al, 2010; Sansam et al, 2010; Boos et al, 2013). In a Xenopus cell‐free replication system, Treslin‐MTBP and TopBP1 associate with chromatin independently of each other, Cdc45, or GINS (Hashimoto & Takisawa, 2003; Kubota et al, 2003; Kumagai & Dunphy, 2017; Volpi et al, 2021), but the phosphorylation of Treslin by S‐CDK promotes their interaction with TopBP1 on chromatin, which is essential for the formation of CMG (Kumagai et al, 2011). In contrast, there is no clear Sld2 functional equivalent in vertebrates. Vertebrate RecQL4 is a member of the RECQ family of helicases that are essential for cell viability; however, its N‐terminal region has limited sequence homology with Sld2 (Mann et al, 2005; Abe et al, 2011; Xu et al, 2021). MCM10 mediates the RecQL4 association with CMG helicase (Xu et al, 2009), and RecQL4 is required for Polα loading during replication initiation (Sangrithi et al, 2005; Matsuno et al, 2006). However, there is no direct evidence that RecQL4 is necessary for the assembly of CMG. The CMG helicase assembly and activation can be reproduced in vitro using defined factors in S. cerevisae (Yeeles et al, 2015; Douglas et al, 2018); however, in vertebrates, the origin‐dependent CMG formation has not yet been achieved partly due to the lack of a complete set of initiator proteins.
Once assembled, CMG helicase is highly stable until replication termination, after which CMG is disassembled by the p97 (also known as Cdc48/VCP) complex that recognizes poly‐ubiquitylated MCM7 by Cullin E3 ligase upon the convergence of two replication forks (Dewar & Walter, 2017; Moreno & Gambus, 2020). We previously reported that S1 nuclease (single strand‐specific endonuclease) treatment induces the dissociation of GINS from CMG during DNA replication in the absence of Rad51 recombinase and proposed a model in which Rad51 is required for the restoration of CMG integrity after GINS dissociation upon fork collapse (Hashimoto et al, 2011). Recently, a similar phenomenon entailing the GINS dissociation from CMG was reported. When replication forks encounter inter‐strand crosslink (ICL) sites, most forks proceed to the replication traverse pathway and few forks await the arrival of the opposite fork followed by the replication‐coupled ICL repair pathway (Huang et al, 2013; Zhang et al, 2015, 2021; Semlow & Walter, 2021). According to the replication traverse model, DONSON constitutively associates with CMG to form a CMG‐D complex in euchromatin, and CMG‐D transiently loses GINS to transform into the CM‐D complex, which can bypass the ICL site (Zhang et al, 2020). In heterochromatin, FANCM performs a similar function to DONSON in the traverse replication by transiently restructuring CMG to CM‐F (Huang et al, 2019; Zhang et al, 2020). Thus, DONSON and FANCM appear to be profoundly involved in the regulation of CMG under specific replication stress conditions.
In this study, we aimed to elucidate the functional relationship between DONSON and CMG using a Xenopus cell‐free replication system, which allows for a precise biochemical assessment of the DONSON requirements during each DNA replication regulatory process. We found that DONSON was required for DNA replication initiation, particularly for CMG helicase formation, and that DONSON directly interacted with Cdc45, GINS, and Polε via its N‐terminal disordered region independent of replication initiation. These findings indicate that DONSON is a novel factor which is essential for DNA replication initiation in vertebrates and suggest that DONSON may be a functional counterpart of S. cerevisiae's Sld2.
Results
DONSON's chromatin association increases during replication initiation and persists until replication termination
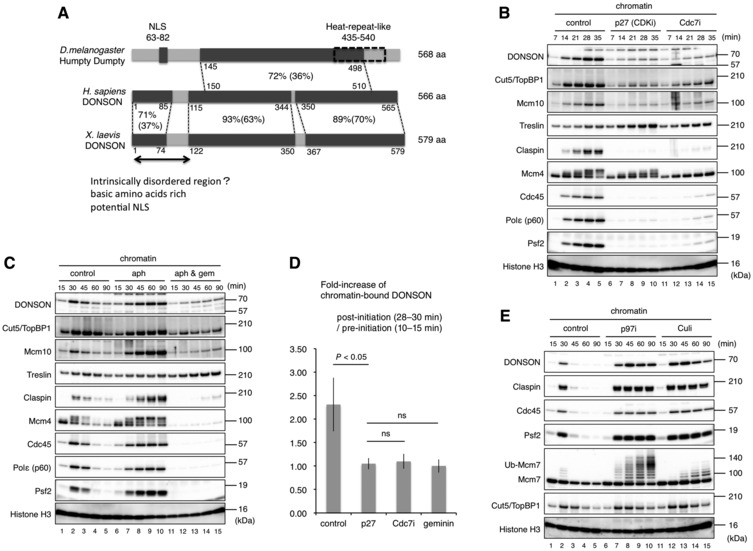
We first compared the primary structure of the DONSON homologs between Drosophila melanogaster, Homo sapiens, and Xenopus leavis (Fig 1A). Drosophila Humpty Dumpty (HD) and human DONSON share 72% similarity and 36% identity within their middle regions, whereas Xenopus DONSON is more closely related to human DONSON with 83% similarity and 57% identity overall. The local regions of the N‐terminal, middle, and C‐terminal share 71, 93, and 89% similarity and 37, 63, and 70% identity, respectively. The N‐terminal region is rich in basic amino acids (aa 1–122, the isoelectric point 10.69) and contains a potential nuclear localization signal. According to the AlphaFold protein structure database (https://alphafold.ebi.ac.uk), the N‐terminal region is intrinsically disordered and surrounds the core domain which is composed of the middle and C‐terminal regions (Fig EV1A).
Figure 1. DONSON's chromatin association increases upon replication initiation and persists until the end of replication termination.
-
AComparison of primary structures of DONSON orthologs in Drosophila melanogaster, Homo sapiens, and Xenopus leavis. The percentages of sequence similarity and identity in each region are indicated outside and within parenthesis, respectively. NLS: Nuclear Localization Signal. aa: amino acids.
-
B–E(B, C, E) Characterization of DONSON's chromatin association. Sperm nuclei (5,000/μl) were incubated for the indicated times in 20 μl of interphase egg extract. Chromatin fractions were isolated and analyzed by immunoblotting. His‐p27 (100 μg/ml, p27 (CDKi)), PHA‐767491 (50 μM, Cdc7i), aphidicolin (10 μg/ml, aph), His‐geminin (50 μg/ml, gem), NMS‐873 (100 μM, p97i), and MLN‐4924 (20 μM, Culi) were supplemented as indicated. (D) Fold increase of the chromatin‐bound DONSON. Similar experiments with (B) and (C) were performed in triplicate and DONSON signal intensity was quantified by ImageJ. The mean ratio of the post‐initiation (28 or 30 min) to the pre‐initiation (14 or 15 min) DONSON intensities is shown in the bar graph. Error bar, mean ± standard deviation (SD). P‐values were calculated using the unpaired t‐test (two‐tailed). ns, not significant.
Source data are available online for this figure.
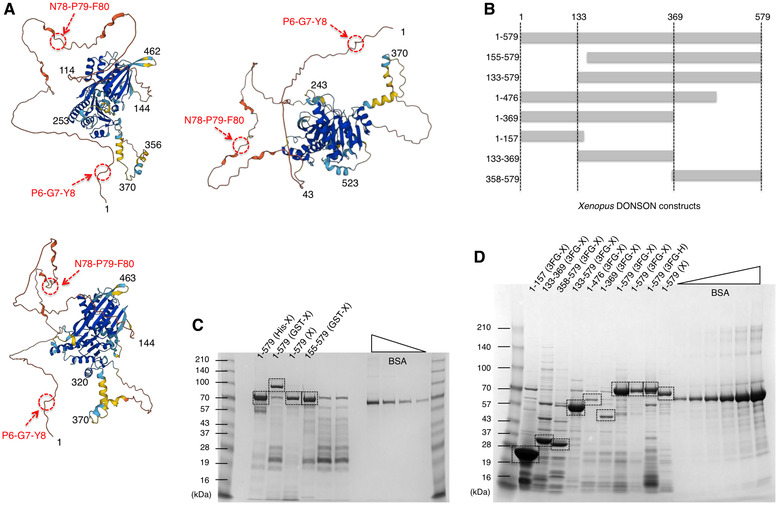
Figure EV1. The structure of DONSON and its deletion construct.
-
AThe three‐dimensional structure of human DONSON was predicted by the AlphaFold database. The views from three different directions are shown. The amino acid numbers are indicated in the structure.
-
BA schematic diagram for the deletion constructs of Xenopus DONSON. Each number indicates the residues number of the protein.
-
C, DPurified recombinant DONSON proteins were subjected to SDS–PAGE and stained with Coomassie Brilliant Blue. His‐X, 8 × His‐tagged Xenopus DONSON. GST‐X, GST‐tagged Xenopus DONSON. X, untagged Xenopus DONSON. 3FG‐X, 3 × FLAG‐tagged Xenopus DONSON. 3FG‐H, 3 × FLAG‐tagged human DONSON. The proper size of each product is indicated with dotted lines.
To study the role of DONSON in DNA replication, we utilized a Xenopus cell‐free replication system in which sperm nuclei synchronously initiate and terminate DNA replication within approximately 90 min. We examined the chromatin association of DONSON before, during, and after DNA replication through immunoblotting (Fig 1B–D). When sperm DNA was added to the untreated interphase extracts, the amount of associated DONSON gradually increased, peaking at around 30 min, when most of the nuclei had initiated DNA replication, and decreased between 45 and 90 min toward the end of replication (Fig 1B and C, lanes 1–5). This behavior was similar to that of typical replisome factors such as Claspin, Cdc45, Psf2 (a subunit of GINS), and Polε. In the presence of a kinase inhibitor to either CDK or Cdc7, which suppresses replication initiation, the amount of associated DONSON did not increase and remained at the basal level (Fig 1B, lanes 6–15 and D). This basal binding was not inhibited by geminin, a pre‐RC formation inhibitor (McGarry & Kirschner, 1998; Tada et al, 2001) (Fig 1C, lanes 11–15 and D). These features resembled those of TopBP1 and Mcm10, both of which are implicated in replication initiation. When aphidicolin, a DNA polymerase α/δ/ε inhibitor, impeded the replication fork progression, DONSON did not dissociate from the chromatin even after 90 min as with the other replisome factors Claspin, Cdc45, Psf2, and Polε (Fig 1C, lanes 6–10). Replisome disassembly during replication termination is dependent on the poly‐ubiquitylation of Mcm7 by Cullin E3 ligase and the subsequent recognition by p97 resolvase (Maric et al, 2014; Moreno et al, 2014). Thus, the addition of a p97 inhibitor resulted in the accumulation of poly‐ubiquitylated Mcm7 while a Cullin E3 inhibitor suppressed ubiquitylation (Fig 1E, lanes 6–15). In the presence of either of these inhibitors, DONSON remained on the chromatin for 90 min as Claspin, Cdc45, and Psf2, indicating that DONSON's chromatin association persists until replication termination. These results suggest that DONSON is an authentic component of the replisome complex.
DONSON is required for replisome assembly during DNA replication initiation
To examine whether DONSON was required for replisome assembly, we performed immunodepletion using two types of anti‐DONSON antibodies to reduce the endogenous DONSON protein levels by > 95% (Fig 2A). The concentration of endogenous DONSON was estimated as 50–100 nM through the comparison of the signal intensities of the endogenous and recombinant DONSON immunoblots (Figs 2A, lanes 1–8 and EV1C). This depletion procedure also reduced the concentrations of TopBP1 and Claspin by −50 and −70%, respectively (Fig 2A), which may have affected the initiation efficiency in the DONSON‐depleted extract. To allow for sufficient time for initiation and avoid replisome disassembly due to termination, we assessed the chromatin fractions at 60 min in the presence of aphidicolin. Interestingly, the chromatin binding of Claspin, Cdc45, Psf2, Polε, and PCNA was severely impaired in the DONSON‐depleted extract (Fig 2B, lane 5); a deficit that was rescued by adding various concentrations of recombinant DONSON (untagged; Fig 2B, lanes 6–8). The chromatin binding of Treslin and Mcm4 was not affected by the DONSON depletion, while that of TopBP1 and Claspin decreased in proportion to their concentrations in the DONSON‐depleted extract. These results suggest that DONSON was required for replisome assembly during DNA replication initiation without affecting the origin licensing.
Figure 2. DONSON is required for replisome assembly during DNA replication initiation.
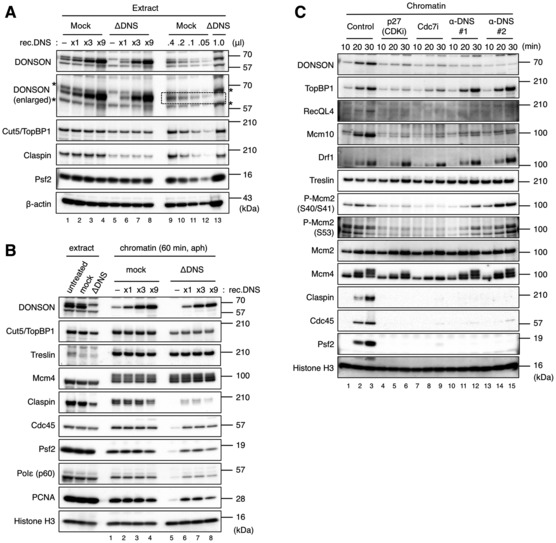
- Immunodepletion from the interphase egg extract was performed using control IgG beads (mock) or anti‐DONSON antibody beads (ΔDNS). Recombinant Xenopus DONSON (untagged) was added to the mock‐ and DONSON‐depleted extracts at 0 μM (−), 0.06 μM (×1), 0.18 (×3), and 0.36 μM (×9) as indicated. The extracts (1 μl) were analyzed by immunoblotting, except lanes 9–11, where 0.4 (0.4), 0.2 (0.2), and 0.1 (0.1) μl were used. Since the anti‐DONSON antibody was recognized by multiple bands, the presumable DONSON band was indicated by dotted lines in a vertically expanded blot. Asterisk (*), non‐specific band.
- DONSON requirement for the replisome assembly. Sperm nuclei (5,000/μl) were incubated in 20 μl of the mock‐ and DONDON‐depleted extracts as prepared in (A) for 60 min in the presence of 10 μg/ml aphidicolin. The extracts (0.5 μl) and isolated chromatin fractions were analyzed by immunoblotting.
- DONSON requirement for replisome assembly. Sperm nuclei were incubated for the indicated times and conditions in the egg extract and the chromatin fractions were analyzed by immunoblotting. Each of two types of anti‐DONSON antibodies (α‐DNS #1, α‐DNS #2) was added at 14 μg/ml to suppress DONSON function.
Source data are available online for this figure.
We also attempted to neutralize the function of endogenous DONSON by adding two kinds of anti‐DONSON antibodies (α‐DNS #1, #2) separately to the egg extract (Fig 2C). Since the concentrations of TopBP1 and Claspin should be unaffected in this setting, we examined the chromatin fractions at earlier times without aphidicolin. Both anti‐DONSON antibodies almost completely inhibited the chromatin binding of Claspin, Cdc45, and Psf2 at 20–30 min, which is the timing of replication initiation (Fig 2C, lanes 10–15). Assuming that the anti‐DONSON antibodies successfully neutralized the endogenous DONSON, these results confirm that DONSON is required for replisome assembly.
The amount of chromatin‐bound TopBP1, RecQL4, Mcm10, and Drf1 (a main partner of the Cdc7 kinase in Xenopus egg extract; Takahashi & Walter, 2005) increased during replication initiation (Fig 2C, lanes 1–3). Interestingly, the increase of TopBP1 was suppressed by CDK or Cdc7 inhibitors, but not by the anti‐DONSON antibodies (Fig 2C, lanes 4–15). These treatments all suppressed the increase of RecQL4 and Mcm10, while none of them affected the Drf1 binding. Treslin was consistently associated with chromatin under every condition. The Mcm4 migration on SDS–PAGE stalled over time possibly due to the sequential phosphorylation by Cdc7 and CDK (Fig 2C, lanes 1–3 vs. lanes 4–9). Consistent with the Drf1 binding, anti‐DONSON antibodies did not suppress the migration shift of Mcm4 or Mcm2 phosphorylation at Ser40/Ser41 and Ser53, which are known Cdc7‐dependent phosphorylation sites (Montagnoli et al, 2008; Fig 2C, lanes 10–15). These results suggest that DONSON is not required for origin licensing and that CDK and DDK can act on TopBP1, Mcm4, and Mcm2 in the absence of functional DONSON before replisome assembly.
DONSON is required for DNA replication
We next examined the replication activity at 60 min (the middle timing of DNA replication) in the extracts supplemented with anti‐DONSON antibodies by observing the incorporation of Cy3‐dCTP into the nuclei under fluorescence microscopy (Fig 3A), and in the DONSON‐depleted extracts by monitoring the incorporation of Cy5‐dUTP into the sperm genome DNA via agarose gel electrophoresis (Fig 3B). We found that both the addition of anti‐DONSON antibodies and DONSON depletion caused a severe reduction in Cy3 and Cy5 intensities, which suggests that DONSON is required for DNA replication which is consistent with the role of DONSON in replisome assembly. Moreover, the addition of GST‐ or His‐tagged or untagged versions of the recombinant DONSON proteins could partially but significantly restore this activity (Figs 3B, lanes 1, 2, 3, 6 and EV1B and C). Interestingly, deletion mutants lacking the N‐terminal 154 amino acids could not restore these activities (Fig 3B, lanes 4, 5), demonstrating that the N‐terminal region is essential for replication initiation and confirms the significance of the rescue achieved by full‐length DONSON.
Figure 3. DONSON is required for DNA replication.
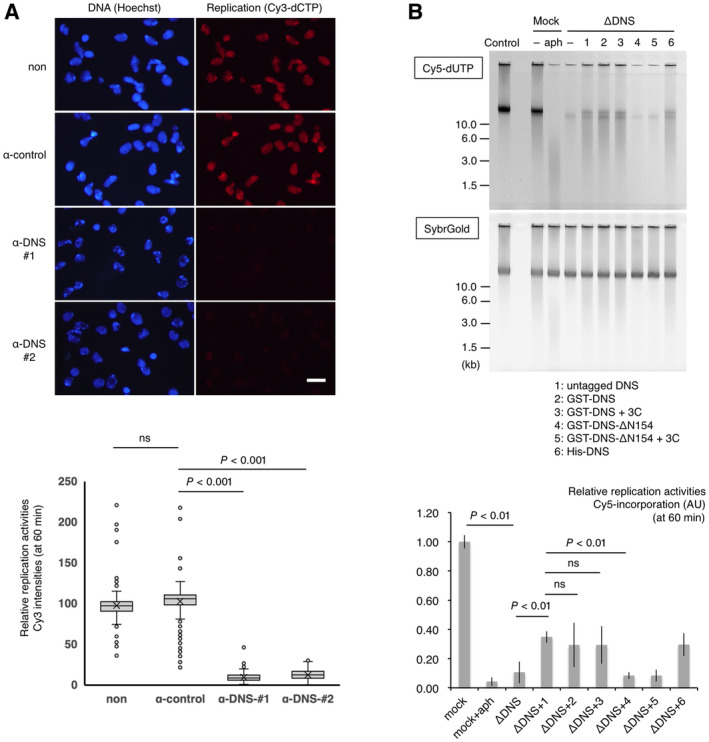
- DONSON requirement for DNA replication. Sperm nuclei were incubated for 60 min in egg extract with no IgG (non), control IgG (α‐control), or one of the two kinds of anti‐DONSON antibodies (α‐DNS #1, α‐DNS #2) in a final concentration of 14 μg/ml. Cy3‐dCTP was added to all of the reactions at 1 μΜ to visualize the DNA replication. After incubation, the nuclei were fixed, stained with Hoechst 33258, and observed under a fluorescent microscope. Representative images are shown above. Scale bar; 50 μm. The relative Cy3 intensity of > 100 nuclei was quantified using ImageJ and is shown in the box plot below. The boxes define the upper and lower quartiles; the whiskers define max to min values; the X‐mark indicates each mean value; the central band indicates each median; the circles indicate outliers. P‐values were calculated using the Mann–Whitney test. ns, not significant.
- DONSON requirement for DNA replication. Sperm nuclei (5,000/μl) were incubated for 60 min in 10 μl of the untreated (control), mock‐ (mock), and DONDON‐depleted (ΔDNS) extracts with 2 μM Cy5‐dUTP. Aphidicolin (aph) was added to the mock‐depleted extract (2.5 μg/ml) as a negative control. Various versions of the recombinant Xenopus DONSON (1–6) were added to the DONSON‐depleted extracts at about 0.06 μM. 1; untagged DONSON, 2 and 3; GST‐DONSON, 4 and 5; GST‐DONSON (residues 155–579) (ΔN154), 6; His‐DONSON. In the reactions corresponding to lanes 3 and 5, 3C protease was added to separate the GST from DONSON. After incubation, the genomic DNA was isolated and subjected to 0.8% TAE agarose gel electrophoresis, followed by SYBR Gold staining. The Cy5 and SYBR Gold fluorescent signals show replicated DNA and total DNA, respectively. The same experiment was performed in triplicate; the relative Cy5 intensities to the control reaction were quantified using Image J, as shown in the bar graph below. Error bar, mean ± standard deviation (SD). P‐values were calculated using the unpaired t‐test (two‐tailed). ns, not significant.
Source data are available online for this figure.
We failed to fully rescue the replication activity through the addition of our recombinant DONSON in the DONSON‐depleted extract. This could be due to several reasons: a reduction in the initiation and replisome factors such as TopBP1 and Claspin (and possibly other factors), abnormal activation of checkpoint signaling, insufficient quality of recombinant DONSON, or the multiple roles of DONSON in DNA replication (e.g., initiation as well as progression). Although the TopBP1 level was reduced in the DONSON‐depleted extract, we previously demonstrated that a small amount of TopBP1 was sufficient for replication initiation (Hashimoto & Takisawa, 2003). To determine whether a Claspin shortage may be the reason, we added recombinant Claspin and DONSON to the DONSON‐depleted extracts at a level similar to that of the endogenous proteins (Fig EV2A). However, the activity was not fully rescued (Fig EV2B and C). Therefore, it is unlikely that a reduction in TopBP1 and Claspin was responsible for the incomplete rescue.
Figure EV2. Requirement of Claspin for DNA replication in the DONSON‐depleted extract.
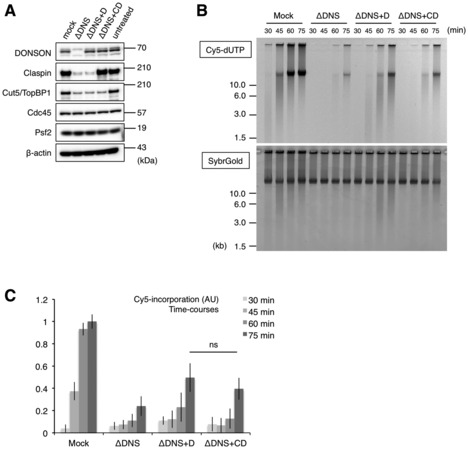
- The immunoblotting of the extracts used is shown.
- The isolated genomic DNA was analyzed by agarose gel electrophoresis.
- The relative Cy5 intensities in three independent experiments are shown in the bar graph. Error bar, mean ± SD. P‐values were calculated using the unpaired t‐test (two‐tailed). ns, not significant.
We then addressed whether DONSON depletion could suppress the origin firing by activating the ATM/ATR checkpoint signaling pathways. Immunoblotting of the nuclear fractions showed that DONSON depletion did not cause the phosphorylation of Chk1, an ATR effector kinase, in the absence of aphidicolin (Fig EV3A, nuclei, lanes 1–6), which indicates that the ATR‐Chk1 pathway was not activated. The addition of aphidicolin to the mock‐depletion extract induced Chk1 phosphorylation and was nullified by the addition of caffeine, an ATM/ATR inhibitor (Fig EV3A, nuclei, lanes 7, 8). In contrast, Chk1 phosphorylation scarcely occurred in the DONSON depletion extract with aphidicolin, and the addition of recombinant DONSON partially induced Chk1 phosphorylation, which was equivalent to the level of replication activities under each condition without aphidicolin (Figs EV3A, nuclei, lanes 9–12 and 3B). The immunoblotting of the chromatin fractions showed that caffeine treatment increased the amount of the replisome factors Claspin, Cdc45, and Psf2 in the mock depletion in both the absence and presence of aphidicolin (Fig EV3A, chromatin, lanes 1, 2, 7, 8). This indicates that the suppression of checkpoint signaling increased the replisome number irrespective of replication stress under control conditions. The caffeine treatment also increased the replisome number in DONSON depletion extract with recombinant DONSON in the presence of aphidicolin (Fig EV3A, chromatin, lanes 11, 12), but the treatment did not increase the replisome number in the absence of aphidicolin (Fig EV3A, chromatin, lanes 5, 6). Moreover, the replication activity was not restored to the level of the mock depletion by caffeine treatment in DONSON depletion extract with recombinant DONSON (Fig EV3B). These results suggest that the ATM/ATR checkpoint pathway was not active in the DONSON depletion extract without aphidicolin. Therefore, partial rescue by the recombinant DONSON was not due to the abnormal activation of checkpoint signaling.
Figure EV3. Involvement of checkpoint signaling in DNA replication in the DONSON‐depleted extract.
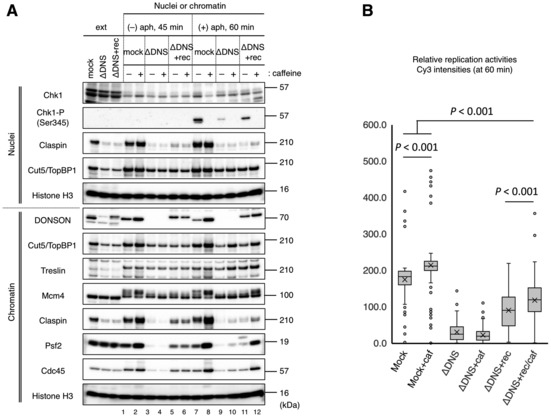
- Involvement of checkpoint signaling in the replisome assembly. Sperm nuclei (5,000/μl) were incubated in 20 μl of the mock‐ and DONDON‐depleted extracts for 45 or 60 min in the absence (−) or presence (+) of 10 μg/ml aphidicolin (aph). 3 × FLAG‐tagged recombinant Xenopus DONSON (rec) and caffeine (caf) were added at 0.06 μM and 5 mM, respectively. The nuclear and chromatin fractions were isolated and analyzed by immunoblotting with 0.5 μl of the extracts (ext).
- Involvement of checkpoint signaling in the replication activity. This was the same as the experiment in Fig 3A and was performed using the mock‐ and DONSON‐depleted extracts under aphidicolin‐free conditions as in (A). The relative Cy3 intensity of > 100 nuclei was quantified using ImageJ and is shown in the box plot. The boxes define the upper and lower quartiles; the whiskers define max to min values; the X‐mark indicates each mean value; the central band indicates each median; the circles indicates outliers. P‐values were calculated using the Mann–Whitney test. ns, not significant.
DONSON is dispensable for replication restart and progression
To clarify whether DONSON was required for replication progression, we examined the salt‐sensitivity of DONSON's chromatin binding by increasing the NaCl concentration in the chromatin isolation buffer, which originally contained 100 mM KCl. The addition of 0.2 M NaCl treatment almost completely dissociated DONSON as well as TopBP1, Treslin, Polε, and Claspin from the chromatin under various conditions (Fig 4A, lanes 3, 6, 9, 12). In contrast, Mcm7, Cdc45, Psf2, and PCNA were resistant to this treatment (Fig 4A, lanes 3, 12), suggesting that DONSON can be eliminated from the replisome complex without disrupting the integrity of the CMG helicase. Accordingly, we examined the requirement of DONSON for replication restart and progression after fork stalling as shown in Fig 4B. In this procedure, DNA replication was first initiated in the presence of aphidicolin, and nuclear fractions containing stalled replication forks were isolated and transferred to an aphidicolin‐free extract in the presence of CDK inhibitors, in which replication restart and progression were permissible but without a new origin firing. Under these conditions, we examined whether the presence of a control IgG or anti‐DONSON antibody cause a difference in the chromatin association of the replication‐replated proteins (Fig 4C) and DNA replication activity (Fig 4D and E). When analyzing the chromatin fractions, we added aphidicolin to the second extract to suppress the replication termination. Although the amount of all the examined proteins were slightly decreased by the addition of the NaCl treatment (Fig 4C, lanes 1–4 vs. 5–8), the chromatin binding of the salt‐sensitive proteins (TopBP1, Claspin, Polε, and DONSON) was restored in the second extract with the control IgG (Fig 4C, lanes 1, 2, 4, 5). However, the anti‐DONSON antibody specifically suppressed DONSON binding without affecting the other replication‐related proteins in both the NaCl‐treated and untreated samples (Fig 4C, lanes 3, 4, 7, 8). These results suggest that the re‐association of TopBP1, Claspin, and Polε with the replisome at the stalled forks does not require DONSON once DNA replication has been initiated. Importantly, DONSON inhibition did not cause any significant difference in the observed DNA replication activities without aphidicolin under both NaCl‐treated and untreated conditions (Fig 4D, Normal/α‐C or α‐DNS vs. 0.2 M NaCl/α‐C or α‐DNS). Taken alone, the second extracts did not support the de novo DNA replication initiation due to the presence of CDK inhibitors (Fig 4D, 2nd extract/α‐C, α‐DNS). Similar results were obtained when analyzing the incorporation of Cy5‐dUTP into the sperm genome DNA (Fig 4E). These results suggest that DONSON is dispensable for replication restart and progression after fork stalling. Therefore, the reason for the incomplete rescue in the DONSON depletion extract may be either because the recombinant DONSON was not fully functional for replication initiation or because other limiting factor were missing in the depleted extract.
Figure 4. DONSON is dispensable for replication restart and progression after fork stalling.
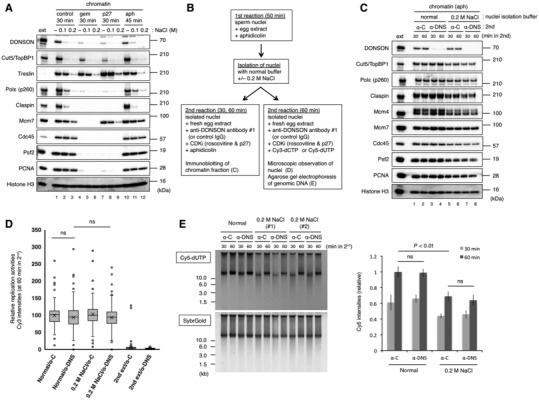
- Salt‐sensitivity of chromatin‐bound DONSON. Sperm nuclei (5,000/μl) were incubated in 20 μl of egg extract under each of the conditions as indicated, and the chromatin fractions were isolated with normal buffer plus 0 (−), 0.1, or 0.2 M NaCl, and analyzed by immunoblotting. In addition, 0.5 μl of each extract (ext) was analyzed for comparison.
- Experimental procedure to investigate the requirement of DONSON in replication restart and progression after fork stalling. In this procedure, DNA replication was first initiated in the presence of aphidicolin, and the nuclear fractions containing the stalled replication forks were isolated and transferred to an aphidicolin‐free extract in the presence of CDK inhibitors, in which replication restart and progression are allowed without a new origin firing.
- The chromatin fractions were isolated and analyzed by immunoblotting.
- The nuclei were observed under fluorescent microscopy as in Fig 3A. The relative Cy3 intensity of > 100 nuclei was quantified using ImageJ and is shown in the box plot. The boxes define the upper and lower quartiles; the whiskers define max to min values; the X‐mark indicates each mean value; the central band indicates each median; the circles indicates outliers. α‐C; control IgG. α‐DNS; anti‐DONSON antibody. Negative controls were prepared by incubating the sperm nuclei in the second extract without undergoing the first reaction (2nd ext/α‐C, 2nd ext/α‐DNS). P‐values were calculated using the Mann–Whitney test. ns, not significant.
- (left) The isolated genomic DNA was analyzed in the same manner as in Fig 3B. (right) The same experiment was repeated six times. The relative Cy5 intensities to the control reaction (normal α‐C) were quantified using Image J, as shown in the bar graph. Error bar, mean ± standard deviation (SD). P‐values were calculated using the unpaired t‐test (two‐tailed). ns, not significant.
Source data are available online for this figure.
N‐terminal region of DONSON physically interacts with Cdc45, GINS, and Polε
The N‐terminal 154 amino acid deletion mutant could not rescue the DNA replication (Fig 3B). Thus, we further prepared a series of N‐terminal and C‐terminal deletion mutants (Figs EV1B and D, and 5A, 1–157, 133–579, 1–476, 1–369) to evaluate their ability to support the replisome assembly; however, none of those deletion mutants could restore the chromatin binding of the replisome factors in the DONSON‐depleted extract (Fig 5A). These results suggest that the entire DONSON protein contains essential sections for replisome assembly.
Figure 5. Identification of GINS‐ and Polε‐interacting motifs within DONSON.
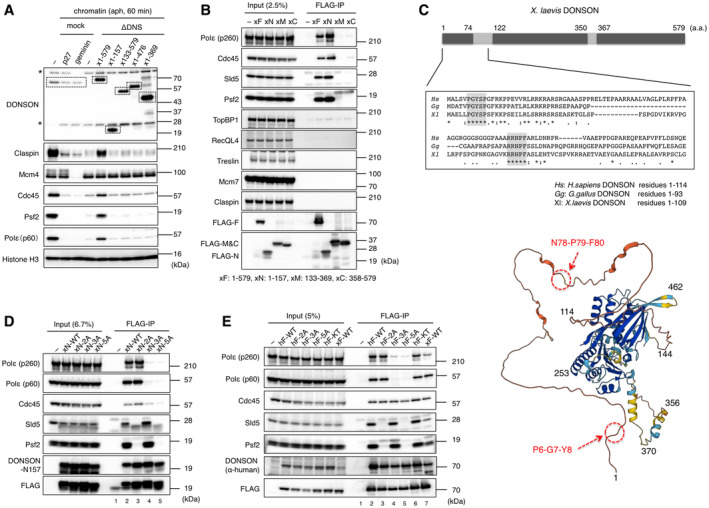
- Ability of DONSON deletions to support replisome assembly. The DONSON depletion experiment, as that in Fig 2B, was performed using 3 × FLAG‐tagged versions of the full length (amino acid residues 1–579) or N‐terminal/C‐terminal deletions (amino acid residues 1–157, 133–579, 1–476, 1–369) of Xenopus (x) DONSON, whose final concentrations were about 0.06 μM. The signals derived from DONSON are indicated with dotted lines. His‐p27 and His‐geminin were used as negative controls. Asterisk (*), non‐specific bands.
- Interaction of DONSON with the replication factors. 3 × FLAG‐tagged full length (xF), N‐terminal region residue 1–157 (xN), middle region residue 133–369 (xM), and C‐terminal region residue 358–579 (xC) versions of Xenopus DONSON were added to an interphase egg extract at about 8.8 μM. After incubation for 15 min, the whole extract was subjected to a pull‐down assay using FLAG‐M2 beads, and the precipitated proteins were analyzed by immunoblotting (FLAG‐IP). The extracts (2.5%) were analyzed for comparison (Input).
- (above) CLUSTALW alignment of the N‐terminal amino acid sequences of DONSON in Homo sapiens (Hs) (residues 1–114), Gallus gallus (Gg) (residues 1–93) and Xenopus laevis (Xl) (residues 1–109). Identical amino acid (*), amino acid with identical nature (:), amino acid with similar nature (.). Two highly conserved amino acid stretches (PGYSP, RRNPF) are shadowed. (below) A three‐dimensional structure of human DONSON predicted by the AlphaFold database. The amino acid numbers are indicated in the structure.
- Identification of GINS‐ and Polε‐interacting motifs in the N‐terminal region of DONSON. The FLAG‐IP experiment, as in (B), was performed with 2 μM of 3 × FLAG‐tagged versions of wild‐type (xN‐WT), P6A/Y8A (xN‐2A), N67A/P68A/F69A (xN‐3A), and P6A/Y8A/N67A/P68A/F69A (xN‐5A) of Xenopus DONSON each with an N‐terminal with 157 amino acids. The extracts (6.7%) were analyzed for comparison (Input).
- Interaction of human DONSON with Xenopus GINS and Polε in egg extract. The FLAG‐IP experiment, as in (B), was performed with 1 μM of 3 × FLAG‐tagged versions of wild‐type (hF‐WT), P6A/Y8A (hF‐2A), N78A/P79A/F80A (hF‐3A), P6A/Y8A/N78A/P79A/F80A (hF‐5A), K489T (hF‐KT) of full‐length human DONSON each as well as 3 × FLAG‐tagged wild‐type Xenopus DONSON (xF‐WT). The extracts (5%) were analyzed for comparison (Input).
Source data are available online for this figure.
We next examined whether DONSON interacts with other replication proteins using a pull‐down assay with FLAG‐tagged DONSON fragments in the absence of sperm nuclei (Fig 5B). The full length and N‐terminal 1–157 amino acid fragments co‐precipitated strongly with Polε, Cdc45, and GINS (Sld5/Psf2), but not with TopBP1, RecQL4, Treslin, Mcm7, and Claspin. No strong interaction was detected with the middle (amino acids 133–369) or C‐terminal regions (amino acids 358–579). Two short amino acid sections were conserved in the N‐terminal disordered region of the vertebrate DONSON (H. sapiens, G. gallus, and X. laevis): the first was PGYSP (residues 6–10 in Xenopus, the PGY motif) and the second was RRNPF (residues 65–69 in Xenpus, the NPF motif) (Figs 5C and EV1A). We mutated the PGY and NPF to AGA (PY‐2A) and AAA (NPF‐3A), respectively, or both to AGA/AAA (PY/NPF‐5A). The PY‐2A and NPF‐3A mutation in the N‐terminal and full‐length Xenopus DONSON fragments lost their affinity for GINS and Polε, respectively, and the PY/NPF‐5A mutants lost affinity for both (Figs 5D, lanes 1–5 and EV4A, lanes 1–5), indicates that the PGY and NPF motifs physically interact with GINS and Polε, respectively. The interaction with Cdc45 was weakened and lost by the NPF‐3A and PY/NPF‐5A mutations, respectively (Figs 5D, lanes 4, 5 and EV4A, lanes 4, 5). Since these motifs are conserved in human DONSON (P6GYSP10, R76RNPF80), we examined whether it also interacted with GINS, Polε, and Cdc45 in Xenopus egg extract. As with the Xenopus protein, the full‐length human DONSON co‐precipitated with GINS and Polε in a PGY and NPF motif‐dependent manner (Fig 5E, lanes 1–5, 7). Although we observed an interaction with Cdc45, it was not affected by the PY/NPF‐5A mutations in contrast to the Xenopus protein (Fig 5E, lane 5). The C‐terminal point mutation K489T, which has been identified in patients with microcephaly (Reynolds et al, 2017), did not affect the interaction with GINS, Polε, and Cdc45 as was predicted from its position (Fig 5E, lane 6). These results indicate that DONSON forms a sub‐complex with GINS, Polε, and Cdc45 in solution independent of replication initiation and that the interaction of DONSON with GINS and Polε is mediated by its conserved PGY and NPF motifs in the N‐terminal region of vertebrate DONSON.
Figure EV4. Interaction of DONSON with GINS is important for DNA replication initiation.
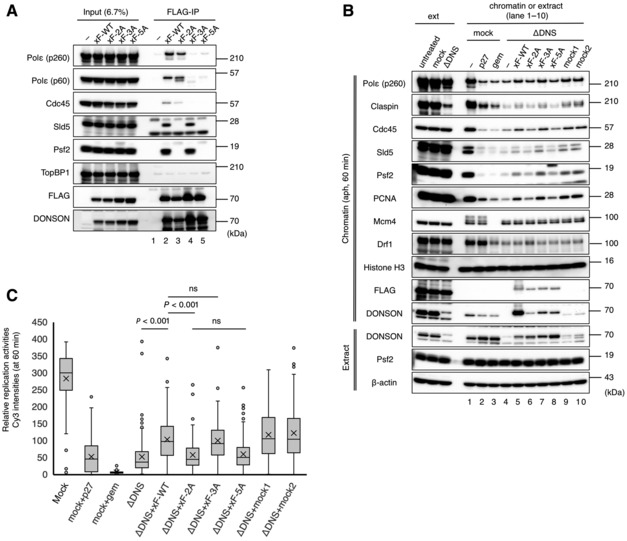
- This FLAG‐IP experiment was performed in the same manner as in Fig 5B using 1.2 μM of 3 × FLAG‐tagged versions of the wild‐type (xF‐WT), P6A/Y8A (xF‐2A), N67A/P68A/F69A (xF‐3A), and P6A/Y8A/N67A/P68A/F69A (xF‐5A) of full‐length Xenopus DONSON. The extract (6.7%) was analyzed for comparison (Input).
- The DONSON depletion experiment, as in Fig 2B, was performed using the same recombinant proteins as in (A) (final concentration of each, 0.06 μM). His‐p27 (27) and His‐geminin (gem) were used as negative controls. The DONSON‐depleted (ΔDNS) and mock‐depleted extracts were mixed at 9:1 (mock1, lane 9) and 8:2 (mock2, lane 10), respectively.
- Under the same conditions as in (B) except that it was aphidicolin‐free, the replication activity was analyzed in the same manner as Fig 3A. The relative Cy3 intensity of > 100 nuclei was quantified using ImageJ and is shown in the box plot. The boxes define the upper and lower quartiles; the whiskers define max to min values; the X‐mark indicates each mean value; the central band indicates each median; the circles indicates outliers. P‐values were calculated using the Mann–Whitney test. ns, not significant.
Interaction of DONSON with GINS is required for efficient DNA replication initiation
To clarify the significance of the interaction between DONSON and GINS and Polε for the replisome assembly, we examined whether recombinant human and Xenopus DONSONs with the PY‐2A, NPF‐3A, and PY/NPF‐5A mutations could restore the replisome assembly in the DONSON‐depleted extracts (Figs 6A and EV4B). Wild‐type human DONSON was as efficient as Xenopus DONSON for the loading of replisome factors Polε, Cdc45, GINS, PCNA, and Claspin onto chromatin (Fig 6A, lanes 6 and 11). This indicates that human DONSON is functional for the replisome assembly in Xenopus egg extract and implies that the DONSON function as an initiation factor is conserved in mammalian cells. Although equivalent amounts of recombinant proteins were added to each extract (Fig 6A, extract, lanes 6–10), PY‐2A and PY/NPF‐5A were less efficient than the wild‐type protein for the loading of replisome factors (Fig 6A, chromatin, lanes 7, 9); however, NPF‐3A and K489T were as efficient as the wild‐type proteins (Fig 6A, chromatin, lanes 8, 10). Similar results were obtained with the Xenopus PY‐2A, NPF‐3A, and PY/NPF‐5A mutants (Fig EV4B, lanes 5–8). These results suggest that the interaction of DONSON with GINS is important for the replisome assembly in replication initiation while the interaction with Polε is dispensable. However, both NPF‐3A and K489T mutants did not associate with chromatin as abundantly as the wild‐type protein did (Figs 6A, chromatin, lanes 6, 8, 10, and EV4B, chromatin, lanes 5, 7), suggesting that the interaction with Polε and integrity of the C‐terminal region may be required to stably maintain DONSON as part of the replisome.
Figure 6. Interaction of DONSON with GINS is required for efficient DNA replication.
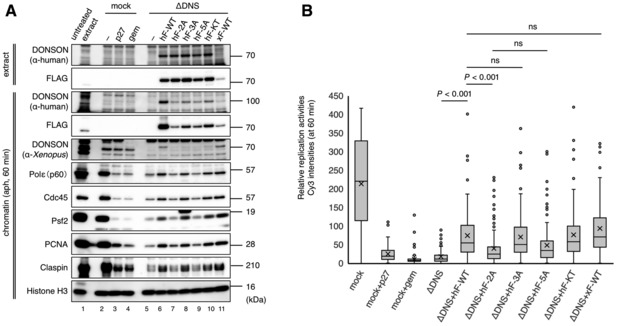
- Requirement of GINS‐ and Polε‐interacting motifs within DONSON for replisome assembly. This DONSON depletion experiment, as that in Fig 2B, was performed using the same recombinant proteins as in Fig 5E. Human proteins were added at 0.3 μM each and Xenopus protein at 0.06 μM. His‐p27 (p27) and His‐geminin (gem) were used as negative controls.
- Requirement of GINS‐ and Polε‐interacting motifs within DONSON for replication activity. The same experiment procedure was used as in Fig 3A for the mock‐ and DONSON‐depleted extracts with the same recombinant proteins as in Fig 5E. Xenopus proteins were added at 0.06 μM each while human proteins were added at 0.3 μM. The relative Cy3 intensity of > 100 nuclei was quantified using ImageJ and is shown in the box plot. The boxes define the upper and lower quartiles; the whiskers define max to min values; the X‐mark indicates each mean value; the central band indicates each median; the circles indicates outliers. P‐values were calculated using the Mann–Whitney test. ns, not significant.
Source data are available online for this figure.
We also examined whether these recombinant DONSON proteins could restore the replication activity in the DONSON‐depleted extracts by monitoring the incorporation of Cy3‐dCTP (Figs 6B and EV4C). Both the human and Xenopus wild‐type DONSON could partially restore the replication activities at similar levels, and comparable activities were obtained by the NPF‐3A (human or Xenopus) or K489T (human) mutants (Figs 6B and EV4C). In contrast, the PY‐2A and PY/NPF‐5A mutants (human or Xenopus) were substantially less efficient than the wild‐type in restoring the replication activities (Figs 6B and EV4C). These results emphasize the importance of the DONSON‐GINS interaction in the DNA replication initiation.
DONSON's chromatin association requires TopBP1
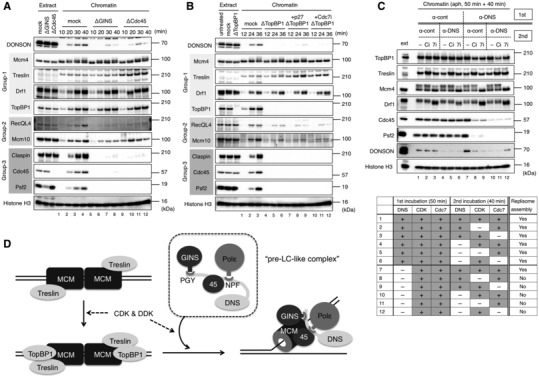
To gain further insight into the mechanism of replisome assembly, we examined whether the chromatin binding of DONSON and the other initiation factors were dependent on GINS, Cdc45, and TopBP1 (Fig 7A and B). These analyses categorized the initiation factors into three major groups. Group‐1 included MCM, Treslin, Drf1, and TopBP1, which were independent of GINS and Cdc45. In addition, MCM, Treslin, and Drf1 were independent of TopBP1. Group‐2 included RecQL4 and Mcm10, whose increased and stable bindings were dependent on GINS, Cdc45, and TopBP1. Group‐3 included Claspin, Cdc45, and GINS, which were fully dependent on GINS, Cdc45, and TopBP1. Cdc45 and GINS were dependent on each other as previously demonstrated (Kubota et al, 2003). DONSON's behavior was different from those of the other initiation factors. In the absence of GINS or Cdc45, DONSON chromatin binding transiently and slightly increased at 30 min and declined at 40 min (Fig 7A, lanes 5–12), which suggests that GINS and Cdc45 are necessary for stable DONSON chromatin association. Interestingly, DONSON chromatin binding was entirely dependent on TopBP1 (Fig 7B, lanes 5–12). Thus, DONSON appears to be situated between Group‐2 and ‐3. Since DONSON forms a sub‐complex with GINS, Cdc45, and Polε independent of replication initiation (Figs 5 and EV4A), these results suggest that chromatin‐bound TopBP1 recruits the sub‐complex via DONSON under the control of CDK and DDK.
Figure 7. CDK and DDK cannot accomplish their functions in the replisome assembly without DONSON.
-
A, BRequirement of GINS, Cdc45, and TopBP1 for the DONSON chromatin association. Sperm nuclei (5,000/μl) were incubated in 20 μl of mock‐, GINS‐, Cdc45‐ or TopBP1‐ depleted egg extract (ΔGINS, ΔCdc45, ΔTopBP1) for the indicated times and conditions, and the chromatin fractions were isolated and analyzed by immunoblotting. The extracts (0.5 μl) were also analyzed. Replication proteins were categorized into three groups (Group‐1, −2, −3) based on their dependency on GINS, Cdc45, and TopBP1.
-
C(above) Requirement of DONSON for CDK and DDK function in the replisome assembly. Sperm nuclei (5,000/μl) were incubated for 50 min in 120 μl of egg extract with control IgG (α‐cont) or anti‐DONSON antibody #1 (α‐DNS). Each part of this first reaction (1st) was divided equally into six parts, and the nuclear fractions were isolated and transferred to 20 μl of flesh extract with control IgG (α‐cont) or anti‐DONSON antibody #1 (α‐DNS) in the presence or absence of CDK inhibitors (Ci: 100 μM roscovitine, 100 μg/ml His‐p27) and Cdc7 inhibitor (7i: 50 μM PHA‐767491). Aphidicolin was added to the first and second extracts at 10 μg/ml to suppress replication termination. After incubation for 40 min in the second reaction (2nd), the chromatin fractions were isolated and analyzed by immunoblotting. In addition, the extracts (0.5 μl) were analyzed (ext). (below) It is summarized whether or not replisome assembly was allowed under each condition, where DONSON, CDK, and Cdc7 were either functional (+) or non‐functional (−) in the 1st and 2nd incubations.
-
DModel of DONSON function in the replisome assembly based on our observations.
Source data are available online for this figure.
CDK and DDK require DONSON to promote replisome assembly
The chromatin binding of Drf1 and the phosphorylation‐dependent migration shift of Mcm4 were not affected by the presence or absence of DONSON, GINS, Cdc45, and TopBP1 (Figs 2C and 7A and B), which suggests that CDK and DDK may regulate MCM, Treslin, and TopBP1 for replication initiation in the absence of DONSON. To examine whether the actions of CDK and DDK, before the loading of DONSON onto the chromatin, were sufficient for replisome assembly, we performed a nuclear transfer experiment (Fig 7C). The sperm nuclei were first incubated with control IgG or anti‐DONSON antibody, and then, the nuclei were isolated and incubated with control IgG or anti‐DONSON antibody in the presence or absence of CDK and Cdc7 inhibitors. Since the replisome assembly had already occurred in the first extract with the control IgG, similar amounts of Cdc45 and Psf2 were detected on the chromatin irrespective of the presence of anti‐DONSON antibody, or CDK and Cdc7 inhibitors in the second extract (Fig 7C, lanes 1–6). Although the anti‐DONSON antibody should have inhibited the replisome assembly in the first extract, Cdc45 and Psf2 loading were allowed in the second extract with the control IgG (Fig 7C, lane 7). This demonstrates that the effects of DONSON inhibition were successfully negated so that the replisome assembly was restored in the second extract. Nevertheless, Cdc45 and Psf2 loading was inhibited by the addition of either a CDK or Cdc7 inhibitor in the second extract with the control IgG (Fig 7C, lanes 8, 9), indicating that CDK and DDK had not accomplished their functions in the first extract with the anti‐DONSON antibody. When the anti‐DONSON antibody was included in both the first and second extracts, DONSON did not allow for replication initiation, thus resulting in no replisome assembly (Fig 7C, lanes 10–12). To exclude the possibility that CDK‐ and DDK‐dependent phosphorylations were lost before DONSON could generate the replisome assembly in the second extract, we performed the same experiment in the presence of phosphatase inhibitors (Fig EV5). When okadaic acid, a protein phosphatase 1 and 2A inhibitor, was added at > 1.5 μΜ, the DNA replication activity was severely inhibited (Fig EV5A). Therefore, we used okadaic acid at lower concentrations (0.4–1.6 μΜ) in both the first and second extracts in the nuclear transfer experiment (Fig EV5B). In addition, we used a nuclear isolation buffer supplemented with a phosphatase inhibitor cocktail. However, insufficient replisome assembly was observed under every condition (Fig EV5B, lanes 2–12) other than in the control (Fig EV5B, lane 1). These results suggest that both CDK and DDK require DONSON for DNA replication initiation and that CDK and DDK without DONSON are not sufficient for replisome assembly when DONSON is not present at the same time as the kinases.
Figure EV5. Effect of phosphatase inhibitors on DNA replication.
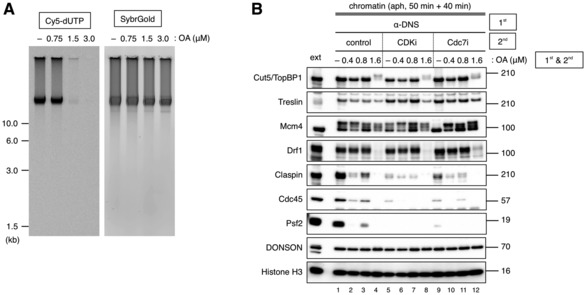
- The effect of okadaic acid (OA) on the replication activity. Sperm nuclei (5,000/μl) were incubated for 60 min in 10 μl of extract in the presence of 0 (−), 0.75, 1.5, and 3.0 μM OA. DNA replication activity was detected in the same manner as Fig 3B.
- The effect of phosphatase inhibitors on the replisome assembly after nuclei transfer. The same experiment as in Fig 7C was performed in the presence of phosphatase inhibitors. The indicated concentrations of OA were added to the 1st and 2nd extracts. After the first incubation, nuclei were isolated using a buffer containing a phosphatase inhibitor cocktail.
Discussion
Almost all essential factors for replication initiation in yeast have functional counterparts in higher eukaryotes including vertebrates (Moiseeva & Bakkenist, 2018; Gillespie & Blow, 2022). To date, Sld2 is the sole exception. Regarding its role in DNA replication initiation, DONSON appears more closely related to Sld2 than RecQL4, despite the lack of obvious sequence homology between DONSON and Sld2. We demonstrated that DONSON forms a sub‐complex with GINS, Polε, and Cdc45 independent of replication initiation (Figs 5 and EV4), which somewhat resembles the pre‐LC (Dpb11‐Sld2‐GINS‐Polε) in S. cerevisiae (Muramatsu et al, 2010) except that the sub‐complex contains Cdc45 instead of TopBP1 (a Dpb11 ortholog). Here we called this sub‐complex (DONSON‐GINS‐Polε‐Cdc45) the pre‐LC‐like complex (Fig 7D). Considering that the Dpb11–Sld2 interaction is essential for the replication initiation and CMG assembly in S. cerevisiae (Kamimura et al, 1998; Takayama et al, 2003; Tak et al, 2006; Muramatsu et al, 2010), and assuming that DONSON performs a Sld2‐like function in vertebrates, DONSON may interact with TopBP1 before CMG assembly. In contrast to the yeast Dpb11, we found that TopBP1 stably binds to the chromatin independent of GINS, Cdc45, and DONSON in Xenopus egg extract (Figs 2C and 7A). Therefore, TopBP1 may not need to join with the pre‐LC‐like complex in solution before being loaded onto the chromatin. Instead, TopBP1 together with Treslin and MCM may create a platform on the chromatin for the loading of the pre‐LC‐like complex (Fig 7D). Moreover, we observed that the chromatin binding of DONSON was entirely dependent on TopBP1 (Fig 7B). In S. cerevisiae, Cdc45 is recruited as a complex with Sld3‐Sld7, is exclusively depending on MCM and DDK and does not require CDK and pre‐LC (Heller et al, 2011; Yeeles et al, 2015). In contrast, in Xenopus egg extracts, Cdc45 requires not only MCM, DDK, and Treslin‐MTBP but also S‐CDK, GINS, DONSON, and TopBP1 (Figs 2B and C, and 7A and B; Mimura & Takisawa, 1998; Jares & Blow, 2000; Walter, 2000; Van Hatten et al, 2002; Hashimoto & Takisawa, 2003; Kubota et al, 2003; Kumagai et al, 2010; Kumagai & Dunphy, 2017). Therefore, Cdc45 may be recruited as a member of the pre‐LC‐like complex. However, whether this pre‐LC‐like complex exists as a stoichiometric complex in egg extract is currently unknown. Since DONSON depletion from egg extract did not cause any obvious reduction in the amount of GINS, Polε, and Cdc45 (Figs 2A and B, and EV4B), it is possible that only a small portion of DONSON interacts with these proteins. We considered the concentrations of endogenous DONSON, GINS, and Cdc45 to be 50–100, 120, and 160 nM, respectively (Fig 2A; unpublished data) (Mimura & Takisawa, 1998). In the future, the stoichiometry of the interactions between DONSON, GINS, Polε, and Cdc45 requires further investigation.
We demonstrated that both S‐CDK and DDK kinase activities are required for CMG assembly after phosphorylating MCM (and possibly Treslin and TopBP1) in the absence of functional DONSON (Fig 7C). The formation of the pre‐LC‐like complex is likely independent of S‐CDK and DDK as it occurred in a Xenopus egg extract without sperm nuclei (Fig 5). Therefore, at least one substrate in the pre‐LC‐like complex may need to be phosphorylated by S‐CDK and DDK before the complex is loaded onto the chromatin platform made of MCM‐Treslin‐TopBP1. This scenario differs from that in S. cerevisiae, where the pre‐LC formation itself is dependent on S‐CDK (Muramatsu et al, 2010). MCM4 is presumably the only essential DDK substrate required for replication initiation, since deletion of the auto‐inhibitory domain of MCM4's N‐terminus bypassed the DDK requirement (Sheu & Stillman, 2006, 2010). In the future, it is important to clarify which substrates are required to be phosphorylated for loading of the pre‐LC‐like complex and how those phosphorylation events are coordinated with CMG assembly.
We identified the PGY and NPF motifs in DONSON as the GINS and Polε interaction sites, respectively (Fig 5). Both motifs reside within the N‐terminal region, which is predicted to be intrinsically disordered (AlphaFold database) and may function as a flexible interaction arm (Fig EV1A). The PGY and NPF motifs are not unique to DONSON and their function as protein–protein interaction motifs has been previously described (de Beer et al, 2000; Morgan et al, 2003; Bonnon et al, 2007). Paranodin, a neuronal paranodal membrane glycoprotein, has a motif with PGY repeats which mediates Paranodin trafficking through the interaction with contactin (Bonnon et al, 2007). The Esp15 homology (EH) domain is often found in proteins associated with endocytosis and vesicle trafficking, and EH domains bind to their target proteins via NPF motifs (de Beer et al, 2000; Morgan et al, 2003). Therefore, similar structural interfaces may be used for these interactions.
Since the DONSON protein with the PY‐2A mutation was substantially less efficient in DNA replication (Figs EV4 and 6), it can be deduced that the interaction via the PGY motif between DONSON and GINS contributes to CMG assembly. However, the PGY motif is not conserved in Drosophila HD and C. elegans DONSON‐like protein (Gene symbol: dnsn‐1, Gene ID: 173431). In these organisms, DONSON orthologs may function differently from that of vertebrates and may possibly be more similarly to that of yeast. On the other hand, the NPF motif is conserved in both Drosophila and C. elegans. Although this motif was apparently dispensable for replication initiation under the control condition, the DONSON protein with the NPF‐3A mutation did not bind to chromatin as stably as the wild‐type DONSON (Figs EV4 and 6). Therefore, the NPF motif may allow DONSON retention as a replisome component rather than Polε recruitment. In the future studies, it will be of interest to explore the physiological function of the DONSON‐Polε interaction.
We found that human DONSON was functional for DNA replication in Xenopus egg extract (Figs 5 and 6). This suggests that the role of DONSON is conserved in mammalian cells, which allowed us to test the biochemical activity of human DONSON proteins with the various mutations identified in patients with microcephaly. In human cells, DONSON depletion by siRNA causes increased rates of spontaneous fork stalling, which can be rescued by the overexpression of wild‐type GFP‐DONSON but not by the K489T mutant GFP‐DONSON (Reynolds et al, 2017). Other mutants such as Y282C, F292L, and M446T show no or reduced nuclear localization but K489T has normal nuclear localization (Reynolds et al, 2017). We examined the replication ability of K489T and observed no significant difference from the wild‐type in replication initiation in Xenopus egg extract (Fig 6B). However, we observed that the K489T mutant, as the NPF‐3A mutant, was unstable on the chromatin (Fig 6A); thus, this defect may cause microcephaly symptoms in patients. In DONSON‐depleted cells, spontaneous chromosomal breakage and the fragmentation of mitotic chromosomes increased, which was suppressed with the co‐depletion of MUS81 or XPF (structure specific fork targeting nucleases) (Reynolds et al, 2017). Taken together, DONSON appears to play multiple roles in DNA replication: CMG assembly, replication traverse of ICL, and protection of stalled forks from nuclease attack. Future research should address which function is defective in each DONSON mutation identified in patients with Meier–Gorlin syndrome, microcephaly–micromelia syndrome, and microcephaly and short stature with limb abnormalities.
In conclusion, we identified DONSON as a novel initiator protein that is essential for CMG helicase assembly in vertebrates. These findings will contribute to the understanding of the control of replication initiation and help the establishment of the in vitro reconstitution system of vertebrate DNA replication using defined factors.
Materials and Methods
cDNA cloning, mutagenesis, and primers
We inserted the annealed DNA oligo (5′‐BglII‐3 × FLAG‐BamHI‐3′‐sense and 5′‐BamHI‐3 × FLAG‐BglII‐3′‐anti) that encodes for 3 × FLAG into the BamHI restriction site of the pGEX 6P‐1 vector (Cytiva) to create a modified pGEX 6P‐1 vector, which we named pGFEX 6P‐1. This vector was used to produce a GST‐3 × FLAG‐tagged recombinant protein that retained the PreScission Protease target sequence (Cytiva) between the GST and 3 × FLAG.
The cDNA encoding for the wild‐type X. laevis DONSON (xDONSON) was amplified by PCR with the primers 5′‐EcoRI‐xDONSON and xDONSON‐SalI‐3′ using X. laevis oocyte cDNA as a template and cloned it into pGEX 6P‐1, pGFEX 6P‐1, and pHEX‐1 between the EcoRI and SalI restriction sites (xDONSON/pGEX 6P‐1, xDONSON/pGFEX 6P‐1, xDONSON/pHEX‐1). The pHEX‐1 vector is a modified pGEX 6P‐1 in which the GST‐encoding sequence was replaced by the His8‐encoding sequence (provided by Dr S Mochida, Kumamoto University). The xDONSON/pGEX 6P‐1 vector was digested with BamHI, which produced two fragments: one, the cDNA encoding xDONSON residues 155–579 with pGEX6P‐1 and the other, encoding the xDONSON residues 1–154 with a large portion of the vector's multi cloning sites. The former fragment was isolated and self‐ligated to create xDONSON 155‐579/pGEX 6P‐1. Each of a series of the cDNA sequences encoding the xDONSON residues 1–157, 133–579, 1–467, 1–369, 133–369, and 358–579 was amplified by PCR using the xDONSON/pGEX 6P‐1 vector as a template with each of the following primer sets: 5′‐EcoRI‐xDONSON and xDONSON‐V157‐SalI‐3′ (for 1–157), 5′‐EcoRI‐xDONSON‐D133 and xDONSON‐SalI‐3′ (for 133–579), 5′‐EcoRI‐xDONSON and xDONSON‐K467‐SalI‐3′ (for 1–467), 5′‐EcoRI‐xDONSON and xDONSON‐A369‐SalI‐3′ (for 1–369), 5′‐EcoRI‐xDONSON‐D133 and xDONSON‐A369‐SalI‐3′ (for 133–369), and 5′‐EcoRI‐xDONSON‐E358 and xDONSON‐SalI‐3′ (for 358–579). These PCR products were cloned into pGFEX 6P‐1 between the EcoRI and SalI restriction sites to create xDONSON 1‐157/pGFEX 6P‐1, xDONSON 133‐579/pGFEX 6P‐1, xDONSON 1‐467/pGFEX 6P‐1, xDONSON 1‐369/pGFEX 6P‐1, xDONSON 133‐369/pGFEX 6P‐1, and xDONSON 358‐579/pGFEX 6P‐1.
The cDNA sequences encoding for the xDONSON full‐length and residues 1–157 with the P6A/Y8A mutations were each amplified by PCR using the xDONSON/pGEX 6P‐1 vector as a template with each of the following primer sets: 5′‐EcoRI‐xDONSON‐P6A/Y8A and xDONSON‐SalI‐3′ (for full length), and 5′‐EcoRI‐xDONSON‐P6A/Y8A and xDONSON‐V157‐SalI‐3′ (for 1–157). These PCR products were cloned into pGFEX6P‐1 between the EcoRI and SalI restriction sites to create xDONSON P6A/Y8A (PY‐2A)/pGFEX 6P‐1 and xDONSON 1–157 P6A/Y8A (PY‐2A)/pGFEX 6P‐1. The entire plasmid sequences were amplified using xDONSON/pGFEX 6P‐1 and xDONSON 1‐157/pGFEX 6P‐1 as the template DNA with two complementary primers containing the N67A/P68A/F69A mutations (xDONSON‐N67A/P68A/F69A‐sense and xDONSON‐N67A/P68A/F69A‐anti). The PCR mixture was treated with DpnI to digest the template DNA, and the remaining PCR product was transformed into E. coli to create xDONSON N67A/P68A/F69A (NPF‐3A)/pGFEX 6P‐1 and xDONSON 1–157 N67A/P68A/F69A (NPF‐3A)/pGFEX 6P‐1. These vectors were used as the template DNA to create xDONSON P6A/Y8A/N67A/P68A/F69A (PY/NPF‐5A)/pGFEX 6P‐1 and xDONSON 1–157 P6A/Y8A/N67A/P68A/F69A (PY/NPF‐5A)/pGFEX 6P‐1 in the same manner as for the P6A/Y8A mutations.
The cDNA encoding for the wild‐type H. sapiens DONSON (hDONSON) was amplified by PCR with the primers 5′‐EcoRI‐hDONSON and hDONSON‐SalI‐3′ using the HeLa cell cDNA as a template and cloned into pGFEX 6P‐1 between the EcoRI and SalI restriction sites to create hDONSON/pGFEX 6P‐1. This vector was used as the template DNA to create hDONSON P6A/Y8A (PY‐2A)/pGFEX 6P‐1, hDONSON N78A/P79A/F80A (NPF‐3A)/pGFEX 6P‐1, hDONSON P6A/Y8A/N78A/P79A/F80A (PY/NPF‐5A)/pGFEX 6P‐1, and hDONSON K489T/pGFEX 6P‐1 as with the xDONSON mutants using the primers: 5′‐EcoRI‐hDONSON‐P6A/Y8A, hDONSON‐SalI‐3′, hDONSON‐N78A/P79A/F80A‐sense, hDONSON‐N78A/P79A/F80A‐anti, hDONSON‐K489T‐sense, and hDONSON‐K489T‐antisense.
The cDNA encoding Xenopus Cdc45 (xCdc45) was amplified by PCR with the primers 5′‐SalI‐xCdc45 and xCdc45‐EagI‐3′ using X. laevis oocyte cDNA as a template and cloned into pGEX 6P‐1 between the SalI and NotI restriction sites to create xCdc45/pGEX 6P‐1.
The sequences of 3 × FLAG‐encoding oligonucleotides and PCR primers are as follows (single and double underline indicates the restriction and mutation sites, respectively):
5′‐BglII‐3 × FLAG‐BamHI‐3′‐sense (5′‐GATCTGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGG‐3′).
5′‐BamHI‐3 × FLAG‐BglII‐3′‐anti (5′‐GATCCCTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCA‐3′).
5′‐EcoRI‐xDONSON (5′‐TATGAATTCATGGCCGAGCTACTGCCCGGATATTC‐3′).
xDONSON‐SalI‐3′ (5′‐TATGTCGACTTATTTCCAGGTGTACCTGTAGTCGG‐3′).
xDONSON‐V157‐SalI‐3′ (TATGTCGACTTACACGGATCCCTTAGGTATCTC).
5′‐EcoRI‐xDONSON‐D133 (TATGAATTCGATGCCGATCCACTTGTAAAGACC).
xDONSON‐K467‐SalI‐3′ (TATGTCGACTTACTTAAGCGCGTGCATTGTTGC).
xDONSON‐A369‐SalI‐3′ (TATGTCGACTTATGCCTCATCTGCTTCTGCTGTGC).
5′‐EcoRI‐xDONSON‐E358 (TATGAATTCGAGAAAGAAAATGGCACAGCAGAAGC).
5′‐EcoRI‐xDONSON‐P6A/Y8A (TATGAATTCATGGCCGAGCTACTGGCCGGAGCTTCCCC).
xDONSON‐N67A/P68A/F69A‐sense (GTAAAGCGGCGGGCCGCAGCTGCCAGCCTAGAG).
xDONSON‐N67A/P68A/F69A‐anti (CTCTAGGCTGGCAGCTGCGGCCCGCCGCTTTAC).
5′‐EcoRI‐hDONSON (TATGAATTCATGGCCCTTTCGGTGCCCGGCTAC).
hDONSON‐SalI‐3′ (TATGTCGACTCAGGATCTCCAATTATAAATGTAG).
5′‐EcoRI‐hDONSON‐P6A/Y8A (TATGAATTCATGGCCCTTTCGGTGGCCGGCGCCTCACC).
hDONSON‐N78A/P79A/F80A‐sense (GCTGCTCGGAGGGCCGCAGCCGCCCGCCTGGACAA).
hDONSON‐N78A/P79A/F80A‐anti.
(TTGTCCAGGCGGGCGGCTGCGGCCCTCCGAGCAGC)
hDONSON‐K489T‐sense (CTGACCATGCTGCTCACATCTTCACAGAGTGG).
hDONSON‐K489T‐antisense.
(CCACTCTGTGAAGATGTGAGCAGCATGGTCAG)
5′‐SalI‐xCdc45 (TATGTCGACTCATGTTTGTGAGTGATCTCCG).
xCdc45‐EagI‐3′ (ATCGGCCGTCATGACTTTAGAGAAATAAGAGC).
Recombinant proteins and antibody production
GST‐xDONSON (full length, residues 155–579), GST‐3 × FLAG‐xDONSON (full length, residues 1–157, 133–579, 1–467, 1–369, 133–369, and 358–579), GST‐3 × FLAG‐xDONSON‐P6A/Y8A (PY‐2A) (full length, residues 1–157), GST‐3 × FLAG‐xDONSON‐N67A/P68A/F69A (NPF‐3A) (full length, residues 1–157), GST‐3 × FLAG‐xDONSON‐P6A/Y8A/N67A/P68A/F69A (PY/NPF‐5A) (full length, residues 1–157), and GST‐3 × FLAG‐hDONSON full length (wild‐type, P6A/Y8A, N78A/P79A/F80A, P6A/Y8A/N78A/P79A/F80A, K489T) were expressed in Rossetta (DE3) pLys (Novagen) and purified with Glutathione Sepharose 4B (glutathione beads) (Cytiva) according to the manufacturer's protocol. To remove the GST tag, the glutathione beads bound to the GST‐tagged proteins were treated with PreScission Protease (Cytiva) and the flow‐through fractions were collected. His‐xDONSON (full length) was expressed in Rossetta (DE3) pLys and purified with Ni‐NTA agarose (QIAGEN) according to the manufacturer's instructions. The purified proteins were concentrated, and buffer exchanged with phosphate buffered saline (PBS) (−) using an Amicon Ultra‐0.5 10 K (or 30 K) ultrafiltration device (Merck). His‐xDONSON was used as an antigen to immunize rabbits (#1, #2). Anti‐DONSON antibodies (#1, #2) were affinity purified with the antigen immobilized on Affi‐Gel10 (Bio‐Rad).
GST‐xCdc45 was expressed in BL21 (DE3) codonplus RIL‐X (Agilent Technologies). The E. coli pellet was suspended in lysis buffer (PBS (−) plus 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1% NP40), sonicated, and centrifuged. The insoluble pellet was washed with lysis buffer and re‐suspended in SDS–PAGE sample buffer. The sample was subjected to SDS–PAGE, followed by Zinc staining (Thermo Fisher). A band corresponding to GST‐xCdc45 was excised, de‐stained, washed with PBS (−), sheered through a G18 needle, and used as an antigen to immunize rabbits. To create the antigen beads, the insoluble pellet was re‐suspended and lysed with the buffer (0.1 M NaCl, 0.5% SDS, 5 mM DTT, 20 mM Hepes‐NaOH [pH7.5]) and coupled to Affi‐Gel15 (Bio‐Rad). Anti‐xCdc45 antibodies were affinity purified using the antigen beads.
His‐p27 and His‐geminin were prepared as previously described (Hashimoto et al, 2010) and were used at 100 and 50 μg/ml to suppress CDK activity and replication licensing reaction, respectively.
Xenopus laevis egg extracts, chromatin fractions, nuclear fractions, replication activity, and chemicals
The S‐phase (interphase) egg extract and demembranated sperm nuclei were prepared as previously described (Kubota & Takisawa, 1993). To normalize the replication activity, we used a mixture of three different extract preparations for the experiments. The concentration of the sperm nuclei was 5,000 nuclei/μl and the reaction temperature was 23°C in all experiments using the egg extracts. The chromatin and nuclear fractions were prepared as previously described (Hashimoto & Tanaka, 2022).
To detect the total replication activity, 2 μM Cy5‐dUTP (Cytiva) was included in each reaction, and the genomic DNA was purified and analyzed by agarose gel electrophoresis as previously described (Hashimoto & Tanaka, 2021). To detect the replication activity in each nucleus, 1 μM Cy3‐dCTP (Cytiva) was included in each reaction, and nuclei were collected by centrifugation onto a poly‐L‐lysine coated coverslip (IWAKI) and observed under a fluorescence microscope (Olympus IX70) as previously described (Hashimoto & Tanaka, 2021).
The following chemicals were used at the indicated concentrations: Aphidicolin (Sigma‐Aldrich, 10 μg/ml), PHA‐767491 (Sigma‐Aldrich, 50 μM), NMS‐873 (Sigma‐Aldrich, 100 μM), MLN4924 (Sigma‐Aldrich, 20 μM), roscovitine (Sigma‐Aldrich, 100 μM), okadaic acid (Sigma‐Aldrich, 0.4–3.0 μM), and caffeine (Wako, 5 mM).
Nuclear transfer experiment
The nuclear transfer experiments (in Figs 4, 7, and EV5) comprised of three steps: the first reaction, nuclei isolation, and second reaction. In the first reaction, the sperm nuclei (5,000/μl) were incubated in 20 μl of egg extract with aphidicolin for 50 min, which is sufficient for the completion of replication initiation under normal conditions. To isolate the nuclei, the first reaction mixture was diluted with 400 μl of EB buffer (100 mM KCl, 2.5 mM MgCl2, 50 mM HEPES‐KOH, pH 7.5) and layered onto 200 μl of a 30% (w/v) sucrose cushion made with the EB buffer. These buffers, containing the additional 0.2 M NaCl or phosphatase inhibitor cocktail (PhosSTOP, Roche), were used for the removal of the chromatin‐bound DONSON in Fig 4 or to suppress the de‐phosphorylation during nuclei isolation in Fig EV5, respectively. The nuclei were centrifuged at 6,620 g for 3 min at 4°C, washed with 300 μl of EB buffer containing 15% (w/v) sucrose, and centrifuged at 6,620 g for 1 min. In the second reaction, the nuclei pellet was re‐suspended in 20 μl of egg extract and further incubated under various conditions. When analyzing the chromatin‐bound proteins by immunoblotting (Figs 4C, 7C, and EV5B), aphidicolin was added to the second reaction to avoid replisome dissociation due to replication termination. In the experiments in Fig 4C–E, roscovitine and His‐p27 were added to the second reaction to suppress de novo replication initiation. In Fig 4E, Cy3‐dCTP was added to the second reaction to detect the replication activity. In Figs 7C and EV5B, anti‐DONSON antibody (#1) was added to suppress the functioning of endogenous DONSON in the first and/or second reactions while control IgG was used as a control for the anti‐DONSON antibody.
Immunodepletion
The interphase egg extract was incubated twice (for DONSON, Cdc45, and GINS) or three times (for Cut5/TopBP1) with ProteinA Sepharose Fast Flow (Cytiva) (15 μl per 100 μl extract) and then conjugated with the control IgG or specific antibody (antibody beads) for 30 min each. The amount of antibody used per depletion round (for 100 μl of extract) were as follows: anti‐DONSON antibodies (#1 30 μg, #2 30 μg) for DONSON depletion, anti‐Cdc45 antibody (40 μg) for Cdc45 depletion, anti‐Psf2 antibody (12 μg), and anti‐Psf3 antibody (28 μg) for GINS depletion, anti‐Cut5/TopBP1 antibody (20 μg) for Cut5/TopBP1 depletion, and control IgG (60, 40, or 20 μg) for mock‐depletion. The flow‐through extract after two rounds of antibody beads treatment was used as the depleted extract for further experiments. The anti‐Psf3 antiserum and antigen peptide were provided by Dr K Kamada (RIKEN) (Kamada et al, 2007). Anti‐Psf3 antibody was affinity purified using the immobilized antigen on an NHS‐Activated Sepharose 4 Fast Flow (Cytiva).
Immunoprecipitation (FLAG‐IP)
Each 20 μl of the interphase egg extract was combined with each of the various recombinant DONSON proteins and incubated for 15 min at 23°C. The mixture was diluted with 200 μl of EB buffer plus 0.2% NP40, supplemented with 5 μl of FLAG M2 affinity gel (Sigma‐Aldrich) and incubated for 40 min at 6°C with constant gentle rotation. The M2 gel was then centrifuged, washed twice with 0.5 ml of EB buffer plus 0.2% NP40, and washed once with 0.5 ml of 50 mM Tris–HCl (pH 7.5). The washed gel was re‐suspended in SDS–PAGE sample buffer to elute the proteins bound to the gel, which were then analyzed by immunoblotting.
Immunoblotting and antibodies
SDS–PAGE and immunoblotting were performed according to standard procedures. Chemiluminescent signals were detected utilizing a ChemiDoc XRS+ (Bio‐Rad) and quantified using ImageJ software. Primary antibodies or antisera were used at the indicated dilutions. Rabbit polyclonal antibodies to xDONSON (#1 51 μg/ml, 1:100) and xCdc45 (94.8 μg/ml, 1:500–1,000) were prepared in this study. Rabbit polyclonal anti‐xPsf2 antibody (62 μg/ml, 1:100) was prepared as previously described (Hashimoto & Tanaka, 2018). Rabbit antisera to xCut5/TopBP1 (1:3,000), xRecQL4 (1:600), xMcm10 (1:1,000), xClaspin (1:1,000), xPolε (p60) (1:1,000), xSld5 (1:300), and xMcm2 (1:2,000) were provided by Dr Y Kubota (Osaka University) and Dr H Takisawa (Osaka University). The rabbit antiserum to xDrf1 (1:3,000) was provided by Dr TS Takahashi (Kyusyu University). The following antibodies were obtained from the indicated companies: rabbit polyclonal antibodies to DONSON (1:500, Novus 38599), Treslin (1:500, abcam ab124268), MCM4 (1:2,000, abcam ab4459), Polε (p260) (1:1,000, Novus 57,240), phospho‐MCM2 (Ser53) (1:1,000, Bioss bs‐10608R), phospho‐MCM2 (Ser40/Ser41) (1:1,000, Bethyl A300‐788A), rabbit monoclonal anti‐Histone H3 (1:800, Cell Signaling 4499); mouse monoclonal antibodies to MCM7 (1:2,500, Santa Cruz sc‐9966), PCNA (1:1,000, Santa Cruz sc‐56), FLAG (1:1,000–3,000, Sigma‐Aldrich), and β‐actin (1:5,000, abcam ab8224).
Author contributions
Yoshitami Hashimoto: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing. Kota Sadano: Investigation. Nene Miyata: Investigation. Haruka Ito: Investigation. Hirofumi Tanaka: Supervision.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
The authors thank H Takisawa for the critical reading of the manuscript, Y Kubota for providing the antisera (to Cut5/TopBP1, RecQL4, Mcm10, Claspin, Polε (p60), Sld5, and Mcm2), K Kamada for providing the Psf3 antiserum and its antigen peptide, TS Takahashi for providing the Drf1 antiserum, and S Mochida for providing the pHEX‐1. This study was supported by JSPS KAKENHI grants to Y Hashimoto (19K06617) from the Ministry of Education, Cultures, Sports, Science and Technology (MEXT) in Japan.
The EMBO Journal (2023) 42: e114131
Data availability
No data have been deposited.
References
- Abe T, Yoshimura A, Hosono Y, Tada S, Seki M, Enomoto T (2011) The N‐terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells: role of the N‐terminal region of RECQL4 in cells. Biochim Biophys Acta 1813: 473–479 [DOI] [PubMed] [Google Scholar]
- Attali I, Botchan MR, Berger JM (2021) Structural mechanisms for replicating DNA in eukaryotes. Annu Rev Biochem 90: 77–106 [DOI] [PubMed] [Google Scholar]
- Bandura JL, Beall EL, Bell M, Silver HR, Botchan MR, Calvi BR (2005) humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila . Curr Biol 15: 755–759 [DOI] [PubMed] [Google Scholar]
- de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M (2000) Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol 7: 1018–1022 [DOI] [PubMed] [Google Scholar]
- Bellelli R, Boulton SJ (2021) Spotlight on the replisome: aetiology of DNA replication‐associated genetic diseases. Trends Genet 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Bonnon C, Bel C, Goutebroze L, Maigret B, Girault JA, Faivre‐Sarrailh C (2007) PGY repeats and N‐glycans govern the trafficking of paranodin and its selective association with contactin and neurofascin‐155. Mol Biol Cell 18: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Yekezare M, Diffley JF (2013) Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 340: 981–984 [DOI] [PubMed] [Google Scholar]
- Burgers PMJ, Kunkel TA (2017) Eukaryotic DNA replication fork. Annu Rev Biochem 86: 417–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC (1998) Cell cycle control of chorion gene amplification. Genes Dev 12: 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK (2006) CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A 103: 11521–11526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Diffley JFX (2022) The initiation of eukaryotic DNA replication. Annu Rev Biochem 91: 107–131 [DOI] [PubMed] [Google Scholar]
- Deegan TD, Yeeles JT, Diffley JF (2016) Phosphopeptide binding by Sld3 links Dbf4‐dependent kinase to MCM replicative helicase activation. EMBO J 35: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen F, Sengupta S, De Piccoli G, Sanchez‐Diaz A, Labib K (2012) Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J 31: 2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Walter JC (2017) Mechanisms of DNA replication termination. Nat Rev Mol Cell Biol 18: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Diffley JFX (2016) Recruitment of Mcm10 to sites of replication initiation requires direct binding to the Minichromosome maintenance (MCM) complex. J Biol Chem 291: 5879–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Ali FA, Costa A, Diffley JFX (2018) The mechanism of eukaryotic CMG helicase activation. Nature 555: 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cordero DR, Shen J, Partlow JN, Yu TW, Rodin RE, Hill RS, Coulter ME, Lam AN, Jayaraman D et al (2017) Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factor DONSON as the cause of microcephaly‐micromelia syndrome. Genome Res 27: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Cao Q, Lou H (2016) Sld3‐MCM interaction facilitated by Dbf4‐dependent kinase defines an essential step in eukaryotic DNA replication initiation. Front Microbiol 7: 885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos M, Ganier O, Coulombe P, Méchali M (2015) DNA replication origin activation in space and time. Nat Rev Mol Cell Biol 16: 360–374 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Blow JJ (2022) DDK: the outsourced kinase of chromosome maintenance. Biology (Basel) 11: 877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Takisawa H (2003) Xenopus Cut5 is essential for a CDK‐dependent process in the initiation of DNA replication. EMBO J 22: 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Tanaka H (2018) Mitotic entry drives replisome disassembly at stalled replication forks. Biochem Biophys Res Commun 506: 108–113 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Tanaka H (2021) Ongoing replication forks delay the nuclear envelope breakdown upon mitotic entry. J Biol Chem 296: 100033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Tanaka H (2022) Mre11 exonuclease activity promotes irreversible mitotic progression under replication stress. Life Sci Alliance 5: e202101249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V (2010) Rad51 protects nascent DNA from Mre11‐dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V (2011) RAD51‐ and MRE11‐dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 19: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP (2011) Eukaryotic origin‐dependent DNA replication in vitro reveals sequential action of DDK and S‐CDK kinases. Cell 146: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM (2013) The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell 52: 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhang J, Bellani MA, Pokharel D, Gichimu J, James RC, Gali H, Ling C, Yan Z, Xu D et al (2019) Remodeling of Interstrand crosslink proximal replisomes is dependent on ATR, FANCM, and FANCD2. Cell Rep 27: 1794–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P, Blow JJ (2000) Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev 14: 1528–1540 [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F (2007) Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol 14: 388–396 [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 18: 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp KM, Sullivan R, Murray J, Gimenez G, Arn P, D'Souza P, Gezdirici A, Wilson WG, Jackson AP, Ferreira C et al (2020) Linked‐read genome sequencing identifies biallelic pathogenic variants in DONSON as a novel cause of Meier‐Gorlin syndrome. J Med Genet 57: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Takisawa H (1993) Determination of initiation of DNA replication before and after nuclear formation in Xenopus egg cell free extracts. J Cell Biol 123: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H (2003) A novel ring‐like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (2017) MTBP, the partner of Treslin, contains a novel DNA‐binding domain that is essential for proper initiation of DNA replication. Mol Biol Cell 28: 2998–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2010) Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2011) Direct regulation of Treslin by cyclin‐dependent kinase is essential for the onset of DNA replication. J Cell Biol 193: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesly S, Bandura JL, Calvi BR (2017) Rapid DNA synthesis during early Drosophila embryogenesis is sensitive to maternal humpty dumpty protein function. Genetics 207: 935–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, O'Donnell ME (2018) The eukaryotic CMG helicase at the replication fork: emerging architecture reveals an unexpected mechanism. Bioessays 40: 10.1002/bies.201700208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Prasanth SG (2021) Replication initiation: implications in genome integrity. DNA Repair (Amst) 103: 103131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, Luo G (2005) Defective sister‐chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund‐Thomson syndrome. Hum Mol Genet 14: 813–825 [DOI] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K (2014) Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346: 1253596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T et al (2006) Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem 281: 39249–39261 [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H (2006) The N‐terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 26: 4843–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H (1998) Xenopus Cdc45‐dependent loading of DNA polymerase alpha onto chromatin under the control of S‐phase Cdk. EMBO J 17: 5699–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva TN, Bakkenist CJ (2018) Regulation of the initiation of DNA replication in human cells. DNA Repair (Amst) 72: 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C et al (2008) A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 4: 357–365 [DOI] [PubMed] [Google Scholar]
- Moreno SP, Gambus A (2020) Mechanisms of eukaryotic replisome disassembly. Biochem Soc Trans 48: 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A (2014) Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346: 477–481 [DOI] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Jin S, Augustine GJ, Lafer EM (2003) Eps15 homology domain‐NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J Biol Chem 278: 33583–33592 [DOI] [PubMed] [Google Scholar]
- de Munnik SA, Hoefsloot EH, Roukema J, Schoots J, Knoers NV, Brunner HG, Jackson AP, Bongers EM (2015) Meier‐Gorlin syndrome. Orphanet J Rare Dis 17: 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H (2010) CDK‐dependent complex formation between replication proteins Dpb11, Sld2, Polε, and GINS in budding yeast. Genes Dev 24: 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Botchan MR, Berger JM (2017) Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol 52: 107–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer MD, Suman ES, Richter H, Stanger K, Spranger M, Bieberstein N, Duncker BP (2013) Dbf4 and Cdc7 proteins promote DNA replication through interactions with distinct Mcm2‐7 protein subunits. J Biol Chem 288: 14926–14935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP (2010) Mec1 is one of multiple kinases that prime the Mcm2‐7 helicase for phosphorylation by Cdc7. Mol Cell 40: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JJ, Bicknell LS, Carroll P, Higgs MR, Shaheen R, Murray JE, Papadopoulos DK, Leitch A, Murina O, Tarnauskaitė Ž et al (2017) Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat Genet 49: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund‐Thomson syndrome. Cell 121: 887–898 [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA (2010) A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev 24: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S, Mensah MA, de Vries H, Fröber R, Romeike B, Schneider U, Borte S, Schindler D, Kentouche K (2018) Microcephaly, short stature, and limb abnormality disorder due to novel autosomal biallelic DONSON mutations in two German siblings. Eur J Hum Genet 26: 1282–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Walter JC (2021) Mechanisms of vertebrate DNA Interstrand cross‐link repair. Annu Rev Biochem 90: 107–135 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2006) Cdc7‐Dbf4 phosphorylates MCM proteins via a docking site‐mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2010) The Dbf4‐Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Méchali M, Blow JJ (2001) Repression of origin assembly in metaphase depends on inhibition of RLF‐B/Cdt1 by geminin. Nat Cell Biol 3: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H (2006) A CDK‐catalyzed regulatory phosphorylation for formation of the DNA replication complex Sld2‐Dpb11. EMBO J 25: 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Walter JC (2005) Cdc7‐Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev 19: 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H (2013) Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol 5: a010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK‐dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H (2011) Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin‐firing timing. Curr Biol 21: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Komeda Y, Umemori T, Kubota Y, Takisawa H, Araki H (2013) Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1‐GINS interaction. Mol Cell Biol 33: 2614–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Ficarro SB, Jiang W (2006) Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol Biol Cell 17: 4459–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM (2002) The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol 159: 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi I, Gillespie PJ, Chadha GS, Blow JJ (2021) The role of DDK and Treslin‐MTBP in coordinating replication licensing and pre‐initiation complex formation. Open Biol 11: 210121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JC (2000) Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem 275: 39773–39778 [DOI] [PubMed] [Google Scholar]
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y (2009) MCM10 mediates RECQ4 association with MCM2‐7 helicase complex during DNA replication. EMBO J 28: 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chang CW, Li M, Liu C, Liu Y (2021) Molecular mechanisms of the RECQ4 pathogenic mutations. Front Mol Biosci 8: 791194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF (2015) Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P (2015) Evolutionary conservation of the CDK targets in eukaryotic DNA replication initiation. Chromosoma 124: 309–321 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin‐dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC (2015) DNA interstrand cross‐link repair requires replication‐fork convergence. Nat Struct Mol Biol 22: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bellani MA, James RC, Pokharel D, Zhang Y, Reynolds JJ, McNee GS, Jackson AP, Stewart GS, Seidman MM (2020) DONSON and FANCM associate with different replisomes distinguished by replication timing and chromatin domain. Nat Commun 11: 3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bellani MA, Huang J, James RC, Pokharel D, Gichimu J, Gali H, Stewart G, Seidman MM (2021) Replication of the mammalian genome by replisomes specific for Euchromatin and heterochromatin. Front Cell Dev Biol 9: 729265 [DOI] [PMC free article] [PubMed] [Google Scholar]


