Abstract
Lung cancer is prone to bone metastasis, and osteopontin (OPN) has an important significance in maintaining bone homeostasis. The goal of this study was to explore the impact of OPN level on bone metabolism and the molecular mechanism of inhibiting bone metastasis in non-small cell lung cancer (NSCLC). The expression of OPN in NSCLC was ascertained by Western blot and immunohistochemistry, and the correlation between the expression level of OPN and survival of patients was analyzed. Then the shRNA technology was applied to reduce the expression of OPN in NSCLC cells, and CCK-8 assay was carried out to investigate the effect of low expression of OPN on the proliferation of NSCLC cells. In addition, the effects of low expression of OPN on osteoclast differentiation, osteoblast generation and mineralization were studied using osteoclast precursor RAW264.7 and human osteoblast SaOS-2 cells, and whether OPN could regulate miR-34c/ Notch pathway to affect bone metabolism was further explored. The findings showed that the high level of OPN in NSCLC was closely related to the poor prognosis of patients and the abnormal proliferation of NSCLC cell lines. The suppression of OPN was beneficial to inhibit the differentiation of osteoclasts and promote the mineralization of osteoblasts. Besides, this study confirmed that the deletion of OPN can regulate bone metabolism through the regulation of miR-34c/Notch1 pathway, which will contribute to inhibiting the occurrence of osteolytic bone metastasis in NSCLC.
Key words: steopontin, non-small cell lung cancer, bone metastasis, miR-34c, Notch pathway
Introduction
According to the latest global cancer burden data in 2020, lung cancer is currently the world’s leading cause of cancer death.1 On the basis of histological classification, lung cancer can be divided into small cell lung cancer and non-small cell lung cancer (NSCLC), of which NSCLC constitutes about 85%.2 Bone metastasis is a common symptom in patients with advanced lung cancer, and similar to bone metastasis of other tumors, a series of bonerelated events will occur, such as pathological fractures, bone pain, and hypercalcemia.3 Normal bone metabolism is a homeostatic process of osteoblast (OB) -mediated bone formation and osteoclast (OC) -mediated bone resorption. When tumor cells metastasize to bone, they interact with cells and matrix in the bone microenvironment to break the dynamic balance of bone metabolism, resulting in osteolytic bone destruction or osteogenic bone destruction.3 Researches have indicated that bone metastases in NSCLC are generally osteolytic bone metastases.4 Limited by the current treatments, once bone metastases occur, it is usually difficult to cure, which seriously damages the survival quality of patients and reduces their survival time.5
Osteopontin (OPN) is a phosphorylated secreted glycoprotein with a specific ARG-Gly-ASP sequence in its structure, which can promote cell chemotaxis, adhesion and migration, and play a key part in the mineralization of bone matrix, bone resorption and bone remodeling.6 Since Sanger et al. in 1979 first reported that malignant transformed epithelial cells can secrete OPN, more and more evidence has shown that OPN is a crucial mediator in tumor invasion and metastasis.7 Researches have found that OPN is highly expressed in serum of patients with breast cancer, lung cancer, liver cancer and other tumors, and its level is correlated to the undesirable prognosis and survival of patients.8-10 Although there are many research findings on the mechanism of OPN involved in the regulation of tumor metastasis, such as the regulation of epithelial- mesenchymal plasticity, upregulation of chemokine receptors, activation of JAK1/STAT1 and other pathways,11-13 the mechanism through which OPN regulates tumor bone metastasis still needs to be further studied.
MicroRNAs (miRNAs) are small endogenous non-coding RNAs that regulate gene expression by targeting protein-coding mRNAs.14 MiRNAs are key regulators in bone homeostasis, miR- 34c, miR-466 and miR-7-5p have been confirmed to be involved in the bone metastasis of cancer.15-17 Whether OPN could regulate bone metabolism and then affect bone metastasis of NSCLC by regulating the expression of these miRNAs needs to be explored. It has been reported that miR-34c regulates Notch signaling to induce defects in OB mineralization and proliferation and promote OC proliferation.18 MiR-34c has also been proven to inhibit osteosarcoma progression by targeting the Notch pathway.19 We hypothesized that there may be some correlation between OPN, miR-34c and Notch pathway, which jointly regulates bone metabolism, and then affect the bone metastasis of NSCLC.
This study aims to investigate the mechanism of OPN regulating bone metastasis in NSCLC. Firstly, the expression level of OPN in NSCLC and its correlation with the survival rate were clarified, and the effect of knocking down of OPN on the proliferation of NSCLC cells was also tested. After that, the effect of OPN deletion on the differentiation of OCs and the activity and mineralization of OBs was studied using the osteoclast precursor RAW264.7 and human osteoblast SaOS-2 cells, and whether OPN could regulate the level of miR-34c and Notch pathway to affect bone metabolism was further investigated. The results of our investigation may provide a basis for new therapeutic strategies for inhibiting bone metastasis in NSCLC.
Materials and Methods
Immunohistochemical staining
NSCLC clinical tissue samples were obtained from 85 patients who underwent radical excision with informed consent in 2017. This study was approved by the Ethics Committee on Human Research of the Ningbo First Hospital (approval number: 2022-201-5). Patients receiving neoadjuvant therapy or dying during perioperative period were excluded. All paraffin-embedded tumor tissue sections were examined by 2 experienced pathologists without knowledge of patients’ information or the study purpose. The paraffin sections were baked in a thermostatic oven for 30 min, then dewaxed and rehydrated. The endogenous peroxidase was blocked by 3% H2O2 solution. Then the sections were subjected to antigen recovery with 10mM, pH 6.0 citrate buffer (Wuhan Boster Bio-engineering Co. Ltd., Wuhan, China). After washing, the sections were added with anti-OPN (sc-21742, 1:200, Santa Cruz Biotechnology, Dallas, TX, USA), anti-VEGF (abs155843, 1:300, Absin Bioscience, Shanghai, China), anti-E-cadherin (ab231303, 1:300, Abcam, Cambridge, UK) and reacted overnight at 4°C. Sections incubated with PBS instead of primary antibody were set as the negative control. The known positive tissue sections were used as the positive control. The next morning, the sections were washed with PBS, and reacted with the secondary antibody (ab6728, 1:1000; Abcam) at 37°C for 1 h. Afterward, DAB substrate (Wuhan Chemstan Biotechnology Co., Ltd, Wuhan, China) was added, after color reaction, the sections were rinsed and counterstained with hematoxylin (Shanghai Uteam Biotechnology Co., Ltd, Shanghai, China) for 5 min. At last, the sections were dehydrated, transparent and sealed, and the degree of staining was observed and analyzed with a microscope (Ts2-FC; Nikon, Tokyo, Japan). The expression levels were judged according to the degree of staining and analyzed with ImageJ software.
Cell culture and treatment
Four kinds of NSCLC cell lines A549, H1355, H1299 and H460, human bronchial epithelial cell line 16HBE, OC precursor RAW264.7 cells and human OB SaOS-2 cells were obtained from ATCC. After thawing, the cells were kept in DMEM high glucose medium (Hyclone Laboratories, San Angelo, TX, USA) containing 10% fetal bovine serum (Tianhang Biological Technology, Huzhou, China), 100 U/mL penicillin (Biosharp, China) and 0.1 mg/mL streptomycin (BioSharp, Hefei, China). The cultivation environment was 37°C and 5% CO2. For the induction of RAW264.7 cells, the Transwell chambers were used. Specifically, 1.104/mL of the transfected A549 cells and 5.103/mL of the RAW264.7 cells were inoculated in the upper and lower chamber, respectively. They were co-cultured in 37°C and 5% CO2 for 7 days, then the differentiation of RAW264.7 cells into OCs was detected. In addition, to elucidate the effect of OPN on bone metabolism through Notch pathway, RAW264.7 cells and SaOS-2 cells were treated with Notch pathway inhibitor LY3039478 (final concentration was 1 nM).
Cell transfection
Plasmids sh-OPN and sh-NC, as well as miR-34c and NCmimics were designed and synthesized by Wuhan Viraltherapy Technologies Co., Ltd. A549 and H460 cells were inoculated into Petri dishes one day in advance. On the second day, 1 μg of shRNA or miRNA mimics and 3 μL of Entranster-R4000 (EnGreen Biosystem, Beijing, China) were diluted into 50 μL with the serum-free medium. After standing for 10 min at indoor temperature, they were mixed into transfection complexes and added to cells that had been replaced with fresh medium. The transfection efficiency was assessed by Western blot. Subsequent experiments were performed 48 h after transfection.
Detection of cell viability
The cell viability was detected to assess the proliferation capacity of A549 and H460 cells. After cells transfected with sh-OPN or sh-NC were completely adherent, CCK-8 reagent (MedChem Express, Monmouth Junction, NJ, USA) was added and reacted with cells for another 4 h. Five replicates were determined in parallel for each group. The absorbance of each well was monitored by the microplate reader (CMaxPlus; Molecular Devices, San Jose, CA, USA) and then the viability was figured out.
Western blotting
About 1.106 of A549 and H460 cells, RAW264.7 and SAOS-2 cells were gathered and added with RIPA (Beyotime Biotechnology, Shanghai, China), then the cells were lysed for 30 min on ice. The mixture was centrifuged at 12000 g for 5 min at 4°C, and the protein concentration of the supernatant was measured using BCA protein assay kit (Solarbio, Beijing, China). Then the denatured sample was injected into the loading well of the precast gels. At the end of electrophoresis, the gel was removed and then the transmembrane was performed. After that, the transferred PVDF membrane (GE, US) was rinsed and blocked with 1% BSA (BioFRoxx, Einhausen, Germany). Then the PVDF film was incubated with primary and secondary antibody solutions sequentially. Finally, ECL luminescence reagent (Yeasen Biotechnology, Shanghai, China) was dropped onto the membrane for reaction, and the chemiluminescence instrument (610020-9Q; Clinx Science Instruments Co. Ltd., Shanghai, China) was used for development. The antibodies used in this study were purchased from Affinity Biosciences (Cincinnati, OH, USA), including anti-OPN (AF0227, 1:1000 dilution), anti-Cathepsin K (DF6614, 1:500 dilution), anti- CTR (DF10202, 1:2000 dilution), anti-RANKL (AF0313, 1:500 dilution), anti-M-CSF (DF12536, 1:300 dilution), anti-OPG (DF6824, 1:500 dilution), anti-Cleaved-Notch 1 (AF5307, 1:100 dilution), and anti-β-actin (AF7018, 1:1000 dilution).
Flow cytometry
Flow cytometry was applied to measure the positive rate of RANK in OCs and osteocalcin in OBs. The cells were collected and rinsed twice with pre-cooled PBS. PE anti-mouse CD265 (RANK) antibody (119805, 1:500 dilution; BioLegend, San Diego, CA, USA) or Human/Rat Osteocalcin Alexa Fluor®405-conjugated antibody (IC1419V, 1:200 dilution; R&D Systems, Minneapolis, MN, USA) was added according to the demand and co-incubated with cells for 30 min in dark. Then the unbound antibodies were washed off, and the fluorescence intensity of each sample was detected by flow cytometer (Accuri C6; Bd Biosciences, Franklin Lakes, NJ, USA).
Immunofluorescence staining
Immunofluorescence (IF) staining was conducted to monitor the area changes of F-actin ring in RAW264.7 cells and the expression of osteocalcin in SaOS-2 cells. RAW264.7 or SaOS-2 cells were seeded in plates pre-placed with sterile coverslips and treated with the medium of transfected A549 cells. After treatment, the cells were fixed with 1 mL of pre-cooled 4% paraformaldehyde (Shanghai Aladdin Biochemical Technology Co., Ltd., Shanghai, China) for 10 min, and the cell membrane was penetrated with 0.5% Triton X-100 (Shanghai Aladdin Biochemical Technology Co., Ltd.) for 2 min, then the cells were blocked with 3% BSA for 30-60 min. After that, cells were co-incubated with primary antibodies anti-osteocalcin (DF12303, 1:200 dilution; Affinity Biosciences) or anti-F-actin (ab205, 1:500 dilution; Abcam) and anti-β-actin (ab8224, 1:1000; Abcam), and secondary antibodies Alexa Fluor® 488-labeled goat anti-rabbit IgG H&L (ab150077, 1:1000 dilution; Abcam) or goat anti-mouse IgG H&L (ab150113, 1:1000 dilution; Abcam), successively. Cells in the negative control group were incubated with equal volume of PBS instead of primary antibody. Cells with known antigen to be tested were used as the positive control. Finally, the coverslips were carefully removed and sealed with DAPI-containing mounting medium. The images were observed and photographed using an inverted fluorescence microscope (TS2-FC; Nikon) and analyzed with ImageJ software.
Alizarin red S staining
This experiment was performed to assess the mineralization of SaOS-2 cells. SaOS-2 cells were fixed with 4% paraformaldehyde and the cell membrane was then penetrated with 0.3% Triton X- 100, respectively. Then 500 μL of Alizarin Red solution (0.2%, pH8.3) (Solarbio) was added to each well and stained for 20 min. After washing, the staining was observed and photographed under a microscope and analyzed with ImageJ software.
Dual-luciferase reporter assay
To verify the interaction of Notch1 and miR-34c, Notch1-3’UTR WT and MUT plasmids were constructed. The luciferase reporter plasmids and miR-34c mimics were transfected into cells 16 h after plating. Three duplicated wells were set for each sample. Fresh complete medium was replaced 6 h after transfection. After 48 h, the luciferase activity was measured using the Dualluciferase reporter assay kit (Solarbio) as indicated.
RT-qPCR
Cells were lysed by adding 1 mL Trizol reagent (Sangon Biotech, Shanghai, China) to each 1.107 cell. After centrifugation, the supernatant was added with 200 μL chloroform, the mixture was shaken and set for 2 min. After centrifugation again, 600 μL isopropanol was injected into the supernatant, the mixture was set aside for 15 min and centrifuged. Then, the supernatant was disposed and the precipitate was rinsed with 1 mL 75% ethanol and absolute ethanol successively, and the RNA was dissolved in 40 μL DEPC water after centrifugation. Subsequently, using the extracted RNA as template, reverse transcription reaction system was prepared by kit (Jiangsu CoWin Biotech Co., Ltd, Beijing, China). The reaction conditions were as follows: 42°C, 15 min; 85°C, 5 min. Then the qPCR reaction system was prepared by mixing specific primers and Ultra SYBR Mixture (Takara Bio Inc., Shiga, Otsu, Japan) with the synthesized template cDNA. Primer sequences used in this manuscript were shown in Table 1. Reactions were carried out using the real-time PCR device (CFX Connect, Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analysis
The software SPSS 16.0 was used for data analysis. One-way ANOVA and SNK analysis were used to analyze data between multiple groups and two groups, respectively. All data were exhibited as mean ± SD; p<0.05 was considered statistically significant.
Results
OPN is associated with the malignant progression of NSCLC
To clarify the relationship between the expression level of OPN and NSCLC, the protein expression of OPN in NSCLC cell lines A549, H1355, H1299 and H460 and the control 16HBE was detected. As shown in Figure 1A, OPN was highly expressed in the four types of NSCLC cell lines, especially in A549 and H460, while it was low expressed in 16HBE cells. In addition, the expression of OPN, VEGF and E-cadherin in tissue samples was evaluated by immunohistochemistry. The level of VEGF and E-cadherin was increased in the samples following the incremental expression of OPN (Figure 1B). Moreover, the follow-up data of clinical samples showed that the level of OPN was negatively correlated with the disease-free survival (DFS) and the overall survival (OS) of patients (Figure 1 C,D). These results suggest that OPN is relative to the malignant progression of NSCLC.
Deletion of OPN inhibits the proliferation of NSCLC cells
Considering that the effect of OPN level on the proliferation of NSCLC cells was unclear, the expression of OPN was suppressed in A549 and H460 cells by shRNA technology and then the proliferative potential of cells was evaluated. The knockdown efficiency of OPN in these two cell lines was about 40% (Figure 2A). The results of CCK-8 assay showed that low-expression of OPN significantly decreased the cell viability (Figure 2B), which indicated that the deletion of OPN could inhibit the multiplication of NSCLC cells.
Figure 1.
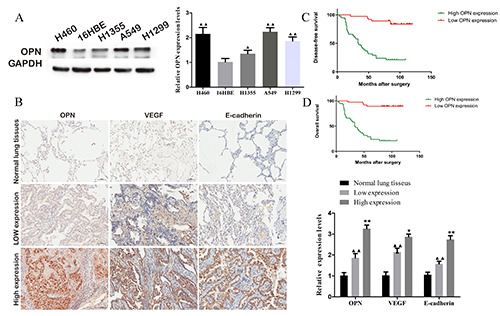
High-expressed osteopontin (OPN) was associated with the metastasis and the survival of NSCLC. A) OPN was high expressed in NSCLC cell lines. B) The expression of VEGF and E-cadherin in NSCLC tissues was positively correlated with the expression of OPN; scale bar: 100 μm. The disease-free survival (C) and overall survival (D) of NSCLC patients were negatively correlated with the expression of OPN.
Table 1.
Primer sequences used in RT-qPCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| miR-34c | AGTTACTAGGCAGTGTAGTTAGCTG | TCTTTTTACCTGGCCGTGTG |
| miR-466 | ATGGTTCGTGGGATACACATACACGCA | GCAGGGTCCGAGGTATTC |
| miR-7-5p | TGTTGTTTTGTGAT | GTGCAGGGTCCGAGGT |
| U6 | ATA CAGAGAAA GTTAGCACGG | GGAATGCTTCAAAGAGTTGTG |
| Notch1 | CCAGCATCACCTGCCTGTTA | CCAAGTCTGACGTCCCTCAC |
| Notch2 | CCAGGAGAGGTGTGCTTGTT | AATGCCCTGGATGGAAAATGG |
| Notch3 | CCTTTGGAGTCTGCCGTGAT | GTTCAGGCATGGGTCTTGC |
| Notch4 | CGAGGAAGATACGGAGTGGC | CTGCTCTGGTGGGCATACAT |
| P-actin | CTCGCCTTTGCCGATCC | TCTCCATGTCGTCCCAGTTG |
Deletion of OPN inhibits the differentiation of OCs
Next, we intended to ascertain the effect of OPN level in NSCLC cells on the differentiation of OCs. To this end, the OC precursor RAW264.7 cells were incubated with the media of A549 cells transfected with sh-NC (hereinafter referred to as sh-NC CM group) and sh-OPN (hereinafter referred to as sh-OPN CM group), respectively. The proportion of RANK-positive RAW264.7 cells in sh-NC CM group was the highest (43%), while the proportion of RANKpositive RAW264.7 cells treated with blank medium and sh-OPN A549 cell medium was 10.3% and 19.3%, respectively (Figure 3A). In addition, the protein expression of Cathepsin K and CTR, two markers of OC, was detected. The results revealed that the media of sh-OPN A549 cells could significantly down-regulate the expression of Cathepsin K and CTR in RAW264.7 (Figure 3B). Furthermore, the formation rate of F-actin ring in sh-NC CM group was the highest, and sh-OPN could inhibit the formation of F-actin ring in RAW264.7 (Figure 3C). The above results confirmed that the deletion of OPN could inhibit the differentiation and generation of OCs.
Deletion of OPN promotes the differentiation and mineralization of OBs
Further, we aimed to find out the effect of OPN level in NSCLC cells on the differentiation of OBs. The SaOS-2 cells were divided into sh-NC CM and sh-OPN group and treated with corresponding culture medium. Flow cytometry results showed that the osteocalcin-positive rate of SaOS-2 cells in control group was 46.6%, and 22.3% in sh-NC CM group, while the osteocalcin-positive rate of SaOS-2 cells in sh-OPN CM group was increased to 33.9% (Figure 4A). The same results were exhibited in the IF staining of cells (Figure 4B). It was observed that the number of calcium nodules in SaOS-2 cells decreased and the area of calcium nodules decreased significantly in sh-NC CM group, while the number of calcium nodules increased notably in sh-OPN CM group (Figure 4C). Moreover, by comparison of the test groups and control group, the level of RANKL in sh-NC CM group was remarkably increased, and the level of M-CSF and OPG was decreased, while the expression of RANKL, M-CSF and OPG was dramatically reversed in SaOS-2 cells treated with the medium of sh-OPN A549 cells (Figure 4D). These outcomes suggested that the deletion of OPN could promote the differentiation and mineralization of OBs.
OPN regulates miR-34c/ Notch1 pathway
Here, we found that after OPN was knocked down, the expression level of miR-34c in A549 and H460 cells markedly reduced, while the expression of miR-466 and miR-7-5p did not change significantly (Figure 5A). Next, we detected the levels of miR-34c in the medium of the transfected A549 and H460 cells, and RAW264.7 and SaOS-2 cells incubated with the culture medium. The content of miR-34c was decreased both in the medium and in the cells (Figure 5B), which indicated that OPN could repress the expression of miR-34c. Notch is known to be the putative target of miR-34c, and there are four kinds of Notch receptors. We found that only the mRNA level of Notch1 was markedly decreased in miR-34c-overexpressed A549 and H460 cells (Figure 5C). Therefore, we hypothesized that Notch1 might be the direct target of miR-34c. This hypothesis was confirmed by dual-luciferase reporter assay. When miR-34c was up-regulated, the luciferase activity of Notch1-WT 3’-UTR-transfected cells was dramatically decreased. However, the activity of Notch1-MUT 3’-UTR-transfected cells did not change obviously (Figure 5D). In addition, after incubating RAW264.7 and SaOS-2 cells with the medium of sh-OPN A549 cells, the level of Notch1 protein in these two kinds of cells was increased, while this elevation was abolished by the transfection of miR-34c mimics (Figure 5E), which also demonstrated the regulation of miR-34c on the expression of Notch1. These results confirmed that OPN could regulate miR-34c/ Notch1 pathway in RAW264.7 and SaOS-2 cells.
Figure 2.
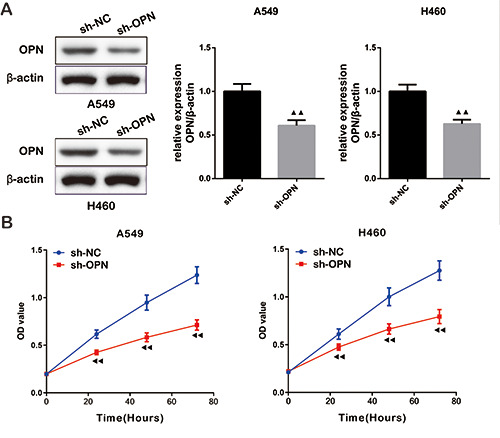
Deletion of osteopontin (OPN) inhibited the proliferation of NSCLC cells. A) The knockdown efficiency of sh-OPN in A549 and H460 cells was verified by Western blot. B) CCK-8 assay confirmed that OPN deletion inhibited the viability of NSCLC cells.
OPN affects bone metabolism through Notch pathway
To investigate whether OPN affects bone metastasis of NSCLC by inhibiting miR-34c and regulating Notch pathway, RAW264.7 cells and SaOS-2 cells were treated with Notch pathway inhibitor LY3039478. The RANK-positive rate of RAW264.7 cells in the sh-NC + LY3039478 group was increased, and in the sh-OPN group it was decreased, by comparison with the sh-NC group; The RANK-positive rate of RAW264.7 cells in sh-OPN + LY3039478 group was noticeably increased compared with that in sh-OPN group (Figure 6A). In contrast, the osteocalcin-positive rate of SaOS-2 cells in sh-NC + LY3039478 group was lower than that in sh-NC group, and in the sh-OPN group it was higher than that in the sh-NC group; The osteocalcin-positive rate in the sh- OPN + LY3039478 group was reversed in comparison with sh- OPN group (Figure 6B). Alizarin red staining also confirmed that LY3039478 could inhibit the mineralization of OBs. The amount of calcium nodules in OBs remarkably increased when OPN was knocked down, while knocking down OPN and inhibiting Notch pathway decreased the mineralization level of OBs (Figure 6C). These results indicated that OPN was involved in the regulation of bone metabolism by regulating Notch pathway.
Figure 3.
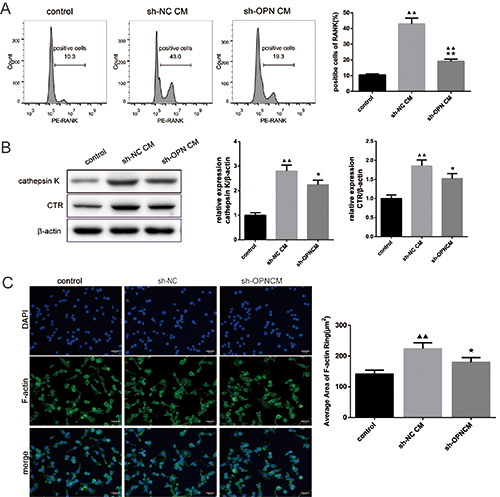
Deletion of osteopontin (OPN) inhibited the differentiation of osteoclasts. A) RANK-positive rate of RAW264.7 cells was detected by flow cytometry. B) The protein expressions of OCs markers Cathepsin K and CTR were detected by Western blot. C) The area of F-actin ring was evaluated by immunofluorescence staining.
Figure 4.
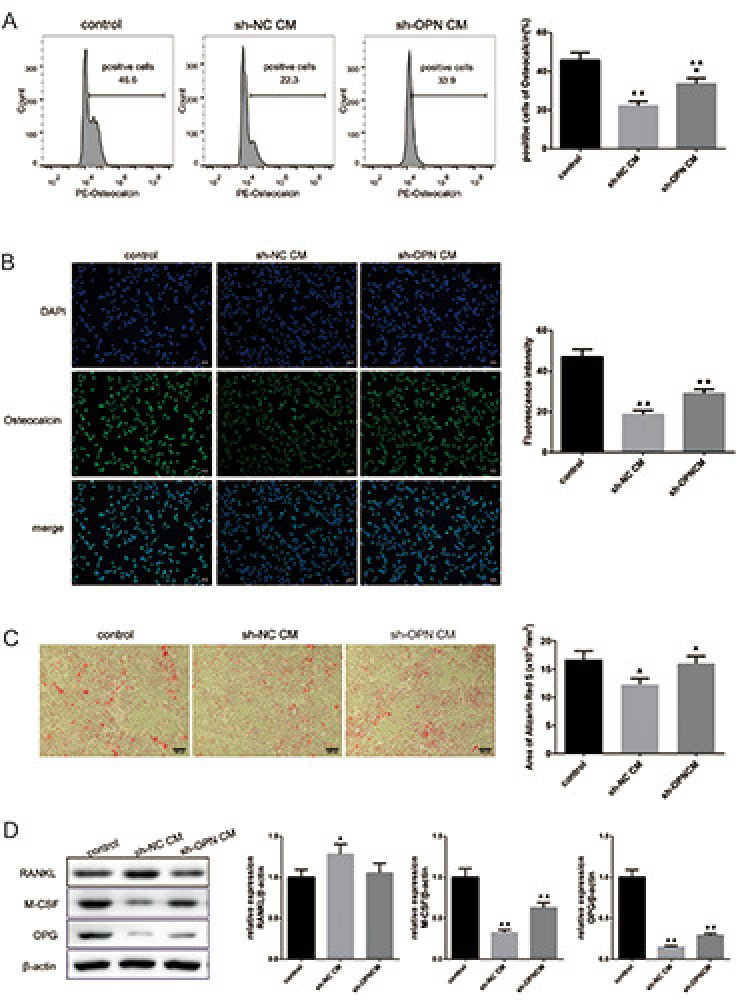
Deletion of osteopontin (OPN) promoted the differentiation and mineralization of osteoblasts. A) Osteocalcin-positive rate of SaOS-2 cells was detected by flow cytometry. B) The expression of osteocalcin in SaOS-2 cells was observed by immunofluorescence staining, and the fluorescence intensity was exhibited on the right. C) Alizarin Red S staining was performed on SaOS-2 cells to evaluate the mineralization. D) The protein expression of RANKL, M-CSF and OPG was detected by Western blot.
Discussion
Tumor bone metastasis and bone destruction is a continuous and complex process. In normal bone metabolism, OC-mediated bone resorption and OB-mediated bone regeneration are in a dynamic balance, so as to maintain the stability of bone mass.20 Previous studies have verified that many substances are involved in the regulation of bone metabolic balance. Li and colleagues found that ubiquitin-specific proteinase 26 (USP26) is a regulator of bone homeostasis, USP26 knockdown inhibited the osteogenic activity of mesenchymal cells by inhibiting β-catenin, and promoted the OC differentiation of bone marrow monocytes by inhibiting NF-κBα (IκBα).21 Liang et al. reported that ELMO1 can activate the small GTPase Rac1, a key regulator of OC differentiation, which is important for OC differentiation and bone resorption.22 OPN plays an essential role in bone matrix mineralization, resorption and bone remodeling. 23 The increased expression of OPN was found in three NSCLC cell lines. Moreover, in clinical samples, the level of VEGF and Ecadherin, which are closely related to tumor metastasis, was increased in lung tissues with high expression of OPN, suggesting that high OPN level may be closely related to the distal metastasis of NSCLC. Besides, we found that the survival rate of NSCLC patients with high expression of OPN was significantly reduced, which may be associated with tumor metastasis. The multiplication ability of the NSCLC cells decreased with the knockdown of OPN, indicating that the level of OPN affected the progression of NSCLC. Studies have demonstrated that tumor cells, OCs, OBs and bone stromal cells interact to promote the formation and development of osteolytic bone metastases.24 Here, we found that the expression of OC markers RANK, Cathepsin K, and CTR was decreased and the F-actin ring was also decreased in OC progenitors after co-cultured with sh- OPN A549 cell medium. This suggests that the deletion of OPN in human NSCLC cells inhibits the differentiation of precursor cells into OCs. Ishii et al. also confirmed that OPN is a promoter of OC formation in arthritis.25 However, the process of bone metabolism is not completed in the isolated microenvironment where OCs exist alone. The effect of OPN on OBs may also affect the differentiation of OCs. This research confirmed that the loss of OPN was conducive to osteogenic induction and mineralization. In terms of the direct regulation of OBs and OCs differentiation, RANKL, M-CSF and OPG expressed by OBs and RANK expressed on the surface of OCs have been proved to be the most critical and direct regulators of bone remodeling process.26 RANKL, M-CSF and OPG secreted by OBs play important roles in the activation, differentiation and maturation of OCs.27 In this study, we found that the protein expression of RANKL decreased, while M-CSF and OPG increased in SaOS-2 cells after co-cultured with sh-OPN A549 cell medium. This implies that the deletion of OPN in NSCLC may favor OB-mediated bone formation and mineralization, while inhibiting osteolytic activity of OCs, thereby reducing the risk of osteolytic bone destruction in NSCLC.
Figure 5.
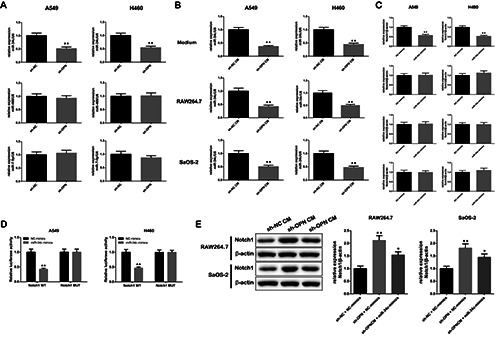
OPN regulated miR-34c/ Notch1 pathway. A) The relative expressions of miR-34c, miR-466 and miR-7-5p after knocking down OPN were analyzed by RT-qPCR. B) The expression of miR-34c was reduced both in the medium of sh-OPN A549 and H460 cells and in the RAW264.7 and SaOS-2 cells treated by the medium of sh-OPN A549 and H460 cells. C) The relative expression of Notch 1 was decreased when transfected with miR-34c, while the level of Notch 2, 3 and 4 was basically unchanged. D) Dual-luciferase reporter assay was performed to verify the direct targeting relationship between miR-34c and Notch 1. E) The changes of Notch 1 protein in RAW264.7 and SaOS-2 cells treated with sh-NC + NC-mimics, sh-OPN + NC-mimics and sh-OPN CM + miR-34c mimics.
MiRNAs are important regulators involved in the progression of a variety of tumors.28 Studies have indicated that miR-34c is closely related to bone metastasis of cancer.29 Here, we found that the expression of miR-34c was inhibited after knockdown of OPN. However, the pathway through which the low expression of OPN mediates the suppression of miR-34c still needs to be further explored. Duan et al. found that OPN inhibited miR-29a expression in human retinal capillary endothelial cells, which may be related to OPN’s ability to regulate the excitation of NF-κB, since NF-κB has been proved to regulate the transcription of miR-29.30 Whether OPN regulates miR-34c expression through the same pathway in NSCLC remains to be studied. Bae et al. demonstrated that miR-34c directly targets and regulates multiple members including Notch1, Notch2 and Jag1 of Notch signaling pathway in OBs to affect bone development.18 Here, we confirmed that miR-34c can directly target Notch1 in NSCLC cells A549 and H460. In addition, OPN can affect the expression of Notch1 in RAW264.7 and SaOS-2 cells by regulating miR-34c, and Notch pathway inhibitor LY3039478 can promote OC differentiation and inhibit OB mineralization. Previous works also showed that Notch pathway could inhibit the differentiation of OC precursors.31 Bai et al. found that inactivation of Notch1 in OCs inhibited the expression of OPG, thereby promoting the differentiation and bone resorption capacity of OCs.32 Therefore, the activation of Notch1 expression in the Notch pathway is beneficial to inhibit the differentiation of OCs and reduce the occurrence of osteolytic response.
In this research, we demonstrated for the first time that deletion of OPN in NSCLC affects bone metabolism by regulating miR- 34c/Notch1 axis, which may provide a new target and therapeutic strategy for inhibiting bone metastasis in NSCLC. Our research is meaningful and promising, but there are still some limitations, the research was only performed on cultured cells and failed to conduct in vivo studies, which makes it difficult for us to make clear whether inhibiting OPN can inhibit bone metastasis of NSCLC.
In summary, this manuscript confirmed the high expression of OPN in NSCLC is closely related to the malignant progression of NSCLC and the worse survival of patients. The deletion of OPN is beneficial to inhibit the differentiation of OCs and promote the mineralization of OBs. In addition, the deletion of OPN can regulate bone metabolism by regulating miR-34c/Notch1 pathway, thereby inhibiting the occurrence of osteolytic bone metastasis in NSCLC patients.
Figure 6.
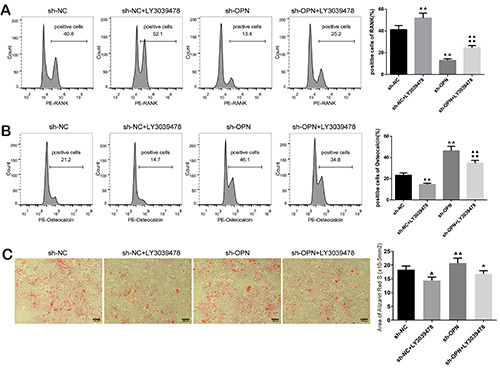
Osteopontin affected bone metabolism through Notch pathway. A) RANK-positive rates of RAW264.7 cells in sh-NC, sh-NC + LY3039478, sh-OPN and sh-OPN + LY3039478 groups were detected by flow cytometry. B) Osteocalcin-positive rates of SaOS-2 cells in the four groups. C) Alizarin Red S staining was performed on the four groups of SaOS-2 cells to evaluate the mineralization.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209-49. [DOI] [PubMed] [Google Scholar]
- 2.Friedlaender A, Addeo A, Russo A, Gregorc V, Cortinovis D, Rolfo CD. Targeted therapies in early stage NSCLC: hype or hope? Int J Mol Sci 2020;21:6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Shi X, Huang D, Li B, Lin N, Pan W, et al. Mutational characteristics of bone metastasis of lung cancer. Ann Palliat Med 2021;10:8818-26. [DOI] [PubMed] [Google Scholar]
- 4.Popper HH. Progression and metastasis of lung cancer. Cancer Metastasis Rev 2016;35:75-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahara Y, Nakase K, Nojiri M, Kato R, Shinomiya S, Oikawa T, et al. Relationship between clinical features and gene mutations in non-small cell lung cancer with osteoblastic bone metastasis. Cancer Treat Res Commun 2021;28:100440. [DOI] [PubMed] [Google Scholar]
- 6.Lamort AS, Giopanou I, Psallidas I, Stathopoulos GT. Osteopontin as a link between inflammation and cancer: the thorax in the spotlight. Cells 2019;8:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berge G, Pettersen S, Grotterod I, Bettum IJ, Boye K, M.landsmo GM. Osteopontin--an important downstream effector of S100A4-mediated invasion and metastasis. Int J Cancer 2011;129:780-90. [DOI] [PubMed] [Google Scholar]
- 8.Bruha R, Vitek L, Smid V. Osteopontin - A potential biomarker of advanced liver disease. Ann Hepatol 2020;19:344-52. [DOI] [PubMed] [Google Scholar]
- 9.Shi L, Hou J, Wang L, Fu H, Zhang Y, Song Y, et al. Regulatory roles of osteopontin in human lung cancer cell epithelial-to-mesenchymal transitions and responses. Clin Transl Med 2021;11:e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu K, Tian X, Oh SY, Movassaghi M, Naber SP, Kuperwasser C, et al. The fibroblast Tiam1-osteopontin pathway modulates breast cancer invasion and metastasis. Breast Cancer Res 2016;18:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin X, Yan M, Wang X, Xu Q, Wang X, Zhu X, et al. Cancerassociated fibroblast-derived IL-6 promotes head and neck cancer progression via the osteopontin-NF-kappa B signaling pathway. Theranostics 2018;8:921-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues LR, Teixeira JA, Schmitt FL, Paulsson M, Lindmark-Mansson H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol Biomarkers Prev 2007;16:1087-97. [DOI] [PubMed] [Google Scholar]
- 13.Zhang N, Li F, Gao J, Zhang S, Wang Q. Osteopontin accelerates the development and metastasis of bladder cancer via activating JAK1/STAT1 pathway. Genes Genomics 2020;42:467-75. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colden M, Dar AA, Saini S, Dahiya PV, Shahryari V, Yamamura S, et al. MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2. Cell Death Dis 2017;8:e2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Xu T, Li Y, Fei W, Yang G, Hong Y. Inhibition of CRY2 by STAT3/miRNA-7-5p promotes osteoblast differentiation through upregulation of CLOCK/BMAL1/P300 expression. Mol Ther Nucleic Acids 2020;19:865-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M, Jin H, Xu CX, Bi WZ;Wang Y. MiR-34c inhibits osteosarcoma metastasis and chemoresistance. Med Oncol 2014;31:972. [DOI] [PubMed] [Google Scholar]
- 18.Bae Y, Yang T, Zeng HC, Campeau PM, Chen Y, Bertin T, et al. miRNA-34c regulates Notch signaling during bone development. Hum Mol Genet 2012;21:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae Y, Zeng HC, Chen YT, Ketkar S, Munivez E, Yu Z, et al. miRNA-34c suppresses osteosarcoma progression in vivo by targeting Notch and E2F. JBMR Plus 2022;6:e10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X. Interactions between cancer cells and bone microenvironment promote bone metastasis in prostate cancer. Cancer Commun (Lond) 2019;39:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Qiu M, Chang L, Qi J, Zhang L, Ryffel B, et al. The osteoprotective role of USP26 in coordinating bone formation and resorption. Cell Death Differ 2022;29:1123-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X, Hou Y, Han L, Yu S, Zhang Y, Cao X, et al. ELMO1 regulates RANKL-stimulated differentiation and bone resorption of osteoclasts. Front Cell Dev Biol 2021;9:702916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey S, Karsenty G, Gundberg C, Vashishth D. Osteocalcin and osteopontin influence bone morphology and mechanical properties. Ann NY Acad Sci 2017;1409:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brook N, Brook E, Dharmarajan A, Dass CR, Chan A. Breast cancer bone metastases: pathogenesis and therapeutic targets. Int J Biochem Cell Biol 2018;96:63-78. [DOI] [PubMed] [Google Scholar]
- 25.Ishii T, Ohshima S, Ishida T, Mima T, Tabunoki Y, Kobayashi H, et al. Osteopontin as a positive regulator in the osteoclastogenesis of arthritis. Biochem Biophys Res Commun 2004;316: 809-15. [DOI] [PubMed] [Google Scholar]
- 26.Eriksen EF. Cellular mechanisms of bone remodeling. Rev Endocr Metab Disord 2010;11:219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JM, Lin C, Stavre Z, Greenblatt MB, Shim JH. Osteoblast-osteoclast communication and bone homeostasis. Cells 2020;9:2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech 2021;14:dmm047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Wang L;Zhang R. Corylin suppresses metastasis of breast cancer cells by modulating miR-34c/LINC00963 target. Libyan J Med 2021;16:1883224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan P, Chen S, Zeng Y, Xu H;Liu Y. Osteopontin upregulates Col IV expression by repressing miR-29a in human retinal capillary endothelial cells. Mol Ther Nucleic Acids 2020;20: 242-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Canalis E. Notch and the regulation of osteoclast differentiation and function. Bone 2020;138:115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, et al. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 2008;283:6509-18. [DOI] [PubMed] [Google Scholar]


