Abstract
The placement of a nonhyperthermophilic order Mesoaciditogales as the earliest branching clade within the Thermotogota phylum challenges the prevailing hypothesis that the last common ancestor of Thermotogota was a hyperthermophile. Yet, given the long branch leading to the only two Mesoaciditogales described to date, the phylogenetic position of the order may be due to the long branch attraction artifact. By testing various models and applying data recoding in phylogenetic reconstructions, we observed that early branching of Mesoaciditogales within Thermotogota is strongly supported by the conserved marker genes assumed to be vertically inherited. However, based on the taxonomic content of 1,181 gene families and a phylogenetic analysis of 721 gene family trees, we also found that a substantial number of Mesoaciditogales genes are more closely related to species from the order Petrotogales. These genes contribute to coenzyme transport and metabolism, fatty acid biosynthesis, genes known to respond to heat and cold stressors, and include many genes of unknown functions. The Petrotogales comprise moderately thermophilic and mesophilic species with similar temperature tolerances to that of Mesoaciditogales. Our findings hint at extensive horizontal gene transfer (HGT) between, or parallel independent gene gains by, the two ecologically similar lineages and suggest that the exchanged genes may be important for adaptation to comparable temperature niches.
Keywords: Mesoaciditoga, Athalassotoga, Petrotogales, Thermotogota, horizontal gene transfer, temperature adaptation
Significance.
The high-temperature phenotype is often referenced when conjecturing about characteristics of the last common ancestor of all present-day organisms. Such inferences rely on accuracy of phylogenetic trees, especially with respect to lineages that branch closest to the last common ancestor. Here, we examined evolutionary history of Mesoaciditogales, an early-branching lineage within Thermotogota phylum, which is one of the early-diverging groups of bacteria. Thermotogota is composed of thermophiles, hyperthermophiles, and mesophiles, which collectively can grow between 20 and 90 °C, making it challenging to infer the growth temperature of their common ancestor. Our analysis revealed a complex evolutionary history of Mesoaciditogales’ genome content impacted by horizontal gene transfer (HGT), highlighting the challenges of ancestral phenotype inferences using present-day genomes.
Introduction
The Thermotogota is a bacterial phylum encompassing thermophiles, hyperthermophiles, and mesophiles, with their optimum growth temperatures (OGTs) ranging from 37 to 80 °C. Since the first isolated members of the phylum were hyperthermophiles (Patel et al. 1985; Huber et al. 1986; Antoine et al. 1997), the extant Thermotogota species have long been assumed to have descended from a hyperthermophilic last common ancestor (LCA) (Zhaxybayeva et al. 2009; Butzin et al. 2013; Green et al. 2013). Later discoveries of thermophilic and mesophilic members of the phylum were inferred to be secondary adaptations of some Thermotogota members to lower OGTs (Pollo et al. 2015), notably in species from the families Kosmotogaceae and Petrotogaceae of the order Petrotogales.
In 2013, a new Thermotogota member was identified—the moderate thermophile Mesoaciditoga lauensis—and the 16S rRNA phylogeny placed this novel organism as an early-diverging taxon of the phylum (Reysenbach et al. 2013). Shortly after, a sister taxon of M. lauensis, Athalassotoga saccharophila, was described (Itoh et al. 2016). Currently, these taxa are the only two characterized species of the Mesoaciditogales order. In most 16S rRNA phylogenetic trees, Mesoaciditogales branch off before all other described Thermotogota diversify (Itoh et al. 2016; L’Haridon et al. 2019). The OGTs of M. lauensis and A. saccharophila are 57–60 and 55 °C (Itoh et al. 2016), respectively, which are far below the threshold of 80 °C for hyperthermophily. As a result, Mesoaciditogales’ placement challenges inferences about the hyperthermophilic phenotype of the phylum's LCA and existing hypotheses about how Thermotogota taxa have adapted to different growth temperatures. It also affects inferences about phenotype of the LCA of the Bacteria domain (Stetter 1996; Galtier et al. 1999; Brochier and Philippe 2002; Barion et al. 2007; Boussau et al. 2008; Catchpole and Forterre 2019).
Despite deeply branching in most 16S rRNA phylogenies, Mesoaciditogales share some common features with a subset of later-diversified Thermotogota. First, M. lauensis and A. saccharophila can grow at temperatures as low as 45 and 30 °C, respectively (Reysenbach et al. 2013; Itoh et al. 2016), making their temperature tolerances similar to multiple species in the Petrotogales order, whose OGTs range from 37 to 70 °C (Davey et al. 1993; Wery et al. 2001; L’Haridon et al. 2021). Second, outside of Mesoaciditogales, M. lauensis’ 16S rRNA gene has the closest nucleotide identity to that of the Kosmotoga spp. (Reysenbach et al. 2013). Finally, in two recent 16S rRNA gene phylogenies, Mesoaciditogales branch within Petrotogales (Steinsbu et al. 2016; Mori et al. 2020).
Observation of conflicting relationships between Mesoaciditogales and the rest of Thermotogota in different analyses raises the possibility that the position of Mesoaciditogales is an artifact. There are two reasons to question either of the two inferred Mesoaciditogales’ positions within Thermotogota. First, Mesoaciditogales’ 16S rRNA gene sequences are very divergent from the rest of Thermotogota, and therefore, the early-branching position of Mesoaciditogales could be due to long branch attraction (Felsenstein 1978; Bergsten 2005). Second, OGT affects GC-content in rRNA stems (Galtier and Lobry 1997; Green et al. 2013) and amino acid composition of proteins (Zeldovich et al. 2007; Sauer and Wang 2019). As a result, standard phylogenetic models may not adequately account for heterogeneity in sequence composition of Thermotogota, which are adapted to a wide range of OGTs.
In this study, we investigate phylogenetic placement of Mesoaciditogales in relationship to other Thermotogota by analyzing 1) 16S rRNA gene sequences from Thermotogota genomes and direct environmental amplifications and 2) protein-coding genes from the genomes of M. lauensis and A. saccharophila, representative Thermotogota isolates and metagenome-assembled genomes (MAGs) of other Thermotogota. Our data set is designed to break long branches leading to M. lauensis and A. saccharophila, and we use substitution models and data recoding tailored to deal with heterogeneous data sets. Our analyses support the early-branching position of Mesoaciditogales within Thermotogota but also reveal that some parts of Mesoaciditogales genomes share evolutionary history with genomes of bacteria from Petrotogales order.
Results
16S rRNA Gene Tree Supports the Early-Branching Placement of Mesoaciditogales within Thermotogota but Produces an Unexpected Position of Kosmotogaceae
To shorten the lengths of the Mesoaciditogales branches in comparison to the rest of the phylum members, we augmented the data set of the 16S rRNA genes from 55 described Thermotogota species with the environmental 16S rRNA gene sequences most closely related to Mesoaciditogales. There is a substantial variation in GC-content of Thermotogota 16S rRNA genes due to correlation of the GC-content of stems in the secondary structure with OGT (Green et al. 2013). Indeed, among 277 sequences in our data set, 23 out of 63 16S rRNA genes from the described species and 16 of the 203 environmental sequences failed the χ2 test of compositional homogeneity. The data set as a whole also failed the Bowker test (Dutheil and Boussau 2008) under a homogenous model (Global Bowker's test, P = 0.0019). These results suggest that the data set is compositionally heterogeneous, raising the possibility of an incorrect phylogenetic inference under a homogeneous model. To address this issue, we reconstructed trees under both homogeneous and nonhomogeneous models.
Phylogenies under both types of models have identical relations among the Thermotogota genera, and Mesoaciditogales clade branches off before the rest of the described Thermotogota species diversify (supplementary fig. S1, Supplementary Material online). The addition of environmental sequences shortens the lengths of the Mesoaciditogales branches in comparison to the rest of the phylum members, reducing the possibility that the early-branching position of Mesoaciditogales is due to the long branch attraction artifact (Felsenstein 1978; Bergsten 2005). However, both trees exhibit a nonconventional relationship among Thermotogota families: the Kosmotogaceae family groups closest to Mesoaciditogales order instead of being a sister clade to the Petrotogaceae family. Notably, this relationship is different from the Mesoaciditogales relationships with Petrotogales observed in some 16S rRNA phylogenies (Steinsbu et al. 2016; Mori et al. 2020).
To further evaluate if there are specific taxa responsible for placement of Kosmotogaceae near Mesoaciditogales, we performed likelihood mapping analysis (Schmidt and von Haeseler 2007) on an alignment that contained only described Thermotogota species and an outgroup. Surprisingly, only 37.6% of quartets strongly support a relationship consistent with an early-branching position of Mesoaciditogales. The plurality of quartets (45.6%) strongly support grouping of Mesoaciditogales with Petrotogales (supplementary fig. S2A, Supplementary Material online). In an additional likelihood mapping analysis, aimed at evaluating if relationships between Mesoaciditogales and Petrotogales orders are due to specific relationship of Mesoaciditogales with either Kosmotogaceae or Petrotogaceae families, the plurality of quartets (45.7%) strongly support Mesoaciditogales grouping with Petrotogaceae, while only 14.8% of the quartets cluster Mesoaciditogales with Kosmotogaceae (supplementary fig. S2B, Supplementary Material online). These surprising relationships between Mesoaciditogales and Petrotogales are consistent with the observation that the 16S rRNA genes of M. lauensis and A. saccharophila are generally most similar to those of the described Kosmotogaceae (median 76.8% nt identity) and Petrotogaceae (76.1%) species than to those of Thermotogales (74.8%), although there is an overlap of pairwise identities to specific members of these taxonomic groups (supplementary tables S1 and S2, Supplementary Material online).
The Mesoaciditogales and Petrotogales relationships could be an artifact of nucleotide composition biases. Specifically, due to similarities in OGT between described species of Mesoaciditogales and Petrotogales, the GC-content of their 16S rRNA genes (either for the full-length sequence or for stem regions only) is more similar to each other than to that of Thermotogales (supplementary tables S3 and S4, Supplementary Material online). Additionally, 16S rRNA phylogenies could be discordant with the evolutionary histories of other genes in a genome due to recombination or horizontal gene transfer (HGT) (Hassler et al. 2022). If the relationship between Mesoaciditogales and Petrotogales 16S rRNA genes is not an artifact, we should observe it in phylogenies of other genes. Therefore, we expanded our analyses to genes encoding ribosomal proteins and other broadly conserved proteins, which are widely used as universal markers to infer organismal relationships (Wolf et al. 2001; Gevers et al. 2005; Yutin et al. 2012).
Trees Reconstructed from Ribosomal Proteins and Single-Copy Core Proteins Also Support the Early-Branching Placement of Mesoaciditogales
The tree based on 50 concatenated ribosomal proteins from the described Thermotogota species supports the deeply branching placement of Mesoaciditogales (supplementary fig. S3, Supplementary Material online). However, unlike 16S rRNA gene phylogenies, the topology shows the conventional placement of Kosmotogaceae as a sister group to Petrotogaceae (Nesbø et al. 2021). Since this tree can also suffer from the effects of long branch attraction artifacts, we added sequences from MAGs that broke multiple long branches, including the branch leading to Mesoaciditogales. No change to the topology was observed (supplementary fig. S4, Supplementary Material online). Individually, the majority of ribosomal proteins (33 out of 50) strongly support the early-branching position of Mesoaciditogales, and only 6 of 50 ribosomal proteins place Mesoaciditogales taxa with Petrotogales (supplementary fig. S5, Supplementary Material online).
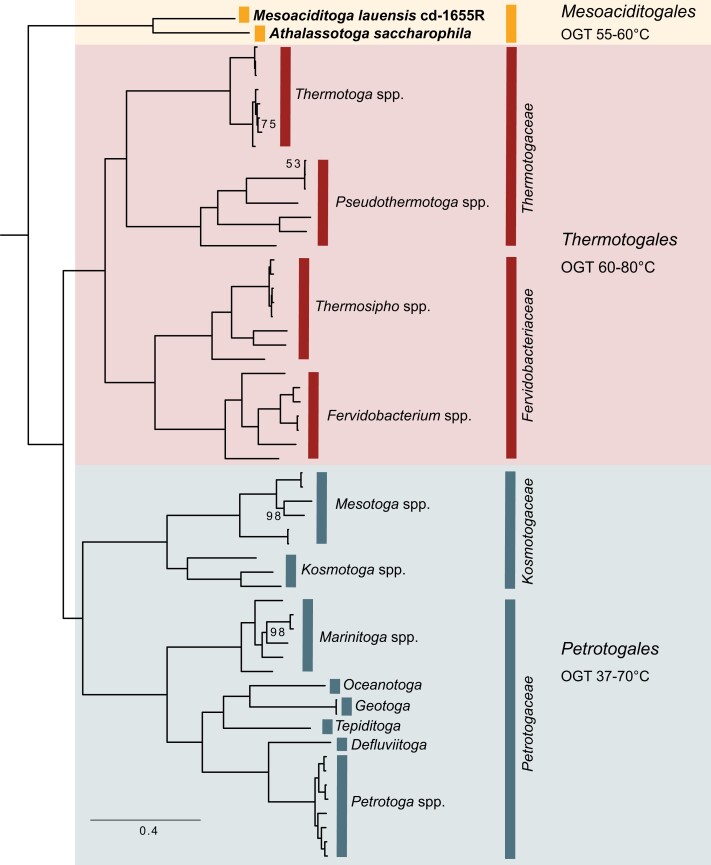
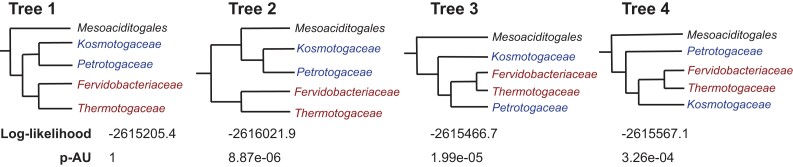
Expanding the ribosomal protein families to 232 protein families encoded by single-copy core (SCC) genes, defined as genes present in at least 75% of described Thermotogota species, did not change the phylogeny (fig. 1). We further tested the strength of this phylogenetic signal by comparing the likelihoods of several alternative relationships between Mesoaciditogales and other Thermotogota families. All alternative topologies were rejected (an approximately unbiased [AU] test, P < 0.001; fig. 2).
Fig. 1.
—The ML phylogenetic tree of the described Thermotogota species. The tree was reconstructed from amino acid sequences of 232 SCC genes. Bootstrap support values below 100% are shown as values at the branches; all unlabeled branches have 100% bootstrap support. Scale bar, substitutions per site. The tree with taxa labels of individual branches is available in the figshare repository (see Materials and Methods).
Fig. 2.
—Support of the four different relationships among Thermotogota families by the concatenated SCC data set. Tree 1 represents the relationships shown in figure 1 and is the null hypothesis. The tested alternative relationships are shown as Tree 2, Tree 3, and Tree 4. “Log-likelihood” is logarithm of likelihood of the SCC alignment given the tree. “p-AU” is the P value of the AU test.
Accounting for the Amino Acid Composition Bias of Thermotogota Proteins Does Not Affect the Early-Diverging Position of Mesoaciditogales
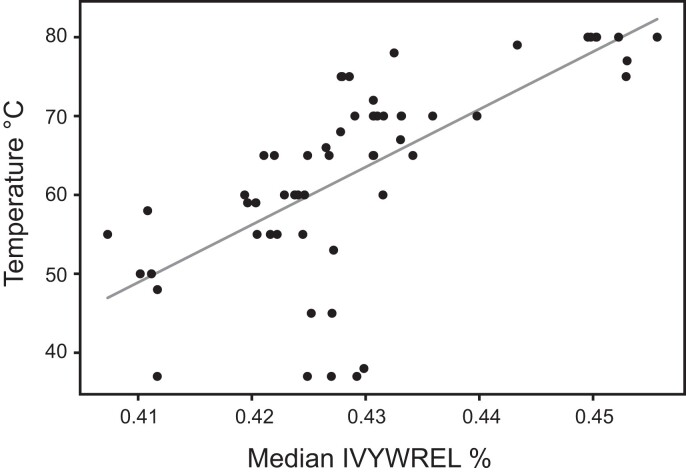
Optimal growth temperature results not only in GC bias in 16S rRNA genes but also in overrepresentation of certain amino acids in proteins, such as IVYWREL (Zeldovich et al. 2007), and these biases have been observed in Thermotogota (fig. 3; Zhaxybayeva et al. 2009; Nesbø et al. 2012; Pollo et al. 2015).
Fig. 3.
—Correlation of OGT in the described Thermotogota species with the median IVYWREL amino acid composition bias in proteins encoded in their genomes (R2 = 0.46, mean squared error = 78.06, y = 731.0x − 250.8). Each point represents a genome of a species. The median IVYWREL bias was calculated from IVYWREL biases of all cytosolic proteins encoded in a genome (see Materials and Methods for details).
The amino acid compositional biases are known to affect phylogenetic inferences that employ standard, homogeneous models (Foster and Hickey 1999; Li et al. 2014). Unfortunately, all sequences in the alignment of the concatenated SCC proteins failed IQ-TREE's χ2 compositional heterogeneity test (P < 0.05; df = 19), which is likely linked to the above-described amino acid bias.
To correct for a skewed composition of amino acids, we explored two sequence recoding strategies on the alignment of the concatenated SCC proteins. In the first approach, we searched for a set of amino acid bins that “minimize the maximum chi-squared statistic” of heterogeneity, as implemented in the “MaxMin-ChiSq” program (Susko and Roger 2007). However, the program did not find any binning of amino acids that reduced heterogeneity in the alignment (homogeneity was rejected at P < 0.002 even for the highest-ranked bin choices), indicating that the alignments are still heterogenous despite the attempted recoding.
In the second approach, we manually selected a recoding that controls for IVYWREL bias in spirit of the Dayhoff 6-state recoding, which is commonly used to reduce heterogeneity while accounting for similarity in physiochemical properties of certain amino acids (Hrdy et al. 2004). The reconstructed phylogeny has the same major topological relationships at the genus and higher taxonomic levels in comparison to the tree obtained using nonrecoded SCC proteins (supplementary fig. S6, Supplementary Material online).
Phylogenetic mixed models can be more effective at accounting for heterogeneity than recoding (Le et al. 2008; Hernandez and Ryan 2021). We used two different mixture models to test for changes in topologies: the posterior mean site frequency (PMSF) model (Wang et al. 2018) and the general heterogeneous evolution on a single topology (GHOST) model (Crotty et al. 2020), which could account for possible heterotachous evolution due to OGT's impact on mutation rates in different lineages and at different sites in a protein sequence (Zeldovich et al. 2007; Crotty et al. 2020). The topologies of trees reconstructed using both models (supplementary figs. S7 and S8, Supplementary Material online) are consistent with the SCC proteins’ tree built under a homogeneous model.
Overall, and despite the challenges of compositional biases, phylogenies of marker genes strongly support the early divergence of Mesoaciditogales from the LCA of Thermotogota.
Evolutionary Histories of Many Mesoaciditogales and Petrotogales Genes Are Intertwined
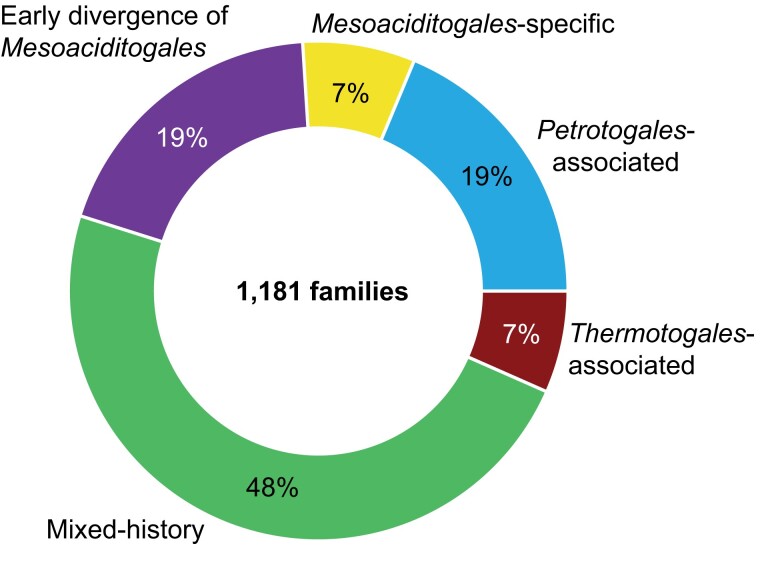
The analyzed 232 single-copy gene families form only a small portion of any Thermotogota genome, which encode between 1,564 and 3,097 protein-coding genes across the described Thermotogota species (M. lauensis and A. saccharophila genomes encode 2,054 and 2,004 protein-coding genes, respectively). The histories of these conserved core genes often do not reflect the complex evolution of prokaryotic genomes, which are substantially impacted by HGT (Philippe and Douady 2003; Gogarten and Townsend 2005; Zhaxybayeva et al. 2009; Doolittle and Brunet 2016; Arnold et al. 2022). Therefore, we expanded our analyses to 1,181 gene families detected in both M. lauensis and A. saccharophila.
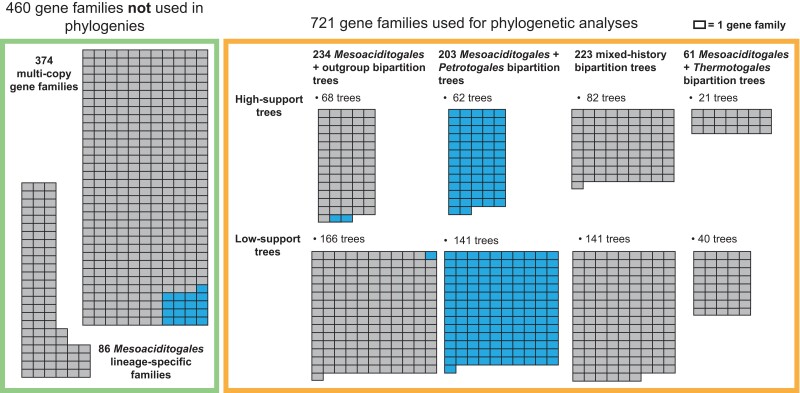
To investigate placement of Mesoaciditogales within Thermotogota on the individual gene family trees, we developed an approach that we dubbed the “minimal bipartition” analysis. In this method, we represent each gene family phylogeny as a set of bipartitions. The expected early-branching position of Mesoaciditogales would produce a bipartition that separates Mesoaciditogales and the outgroup taxa from other Thermotogota. However, this bipartition would not exist in a tree where Mesoaciditogales branched later. Our algorithm first identifies the tree bipartitions that contain M. lauensis and A. saccharophila and the outgroup taxa and then finds the smallest set of other Thermotogota taxa that are required to create each split. This “minimum bipartition” process can quantify the relationship of Mesoaciditogales to other Thermotogota without visual inspection of individual gene trees. Of the 1,181 gene families, 721 (including 190 of 232 SCC families) met the criteria for a minimum bipartition analysis (see Materials and Methods section for details).
In 234 of the 721 gene families’ trees (32%), Mesoaciditogales branch before other taxa in the Thermotogota phylum (“Mesoaciditogales + outgroup” in fig. 4), although three of these gene families are not found in the order Thermotogales. In 203 out of 721 trees (28%), Petrotogales taxa join the minimum bipartition (“Mesoaciditogales + Petrotogales” in fig. 4). Only eight of these 203 families are found solely in Mesoaciditogales and Petrotogales taxa, indicating that many genes that are present across the Thermotogota phylum have evolutionary history similar to the 16S rRNA gene phylogeny discussed above. In 223 of the 721 trees (31%), minimum bipartition includes a mixture of Petrotogales and Thermotogales (“mixed-history” in fig. 4), suggesting that evolutionary histories of these gene families likely involve multiple HGT events, which are common in Thermotogota (Zhaxybayeva et al. 2009; Pollo et al. 2015).
Fig. 4.
—The distribution of phylogenetic relationships in Thermotogota gene families that contain Mesoaciditogales. Each rectangle represents one gene family. Gene families in the green box (on the left) were analyzed only for taxonomy of their members. Gene families in the orange box (on the right) underwent phylogenetic reconstruction. The latter gene families are subdivided into categories based on the taxonomic composition of their “minimum bipartition” and the support of that bipartition (high-support trees have QP-IC score ≥ 0.3 and bootstrap support ≥ 50%). Petrotogales-associated gene families (see text for definition) are shaded in blue.
Many of the 721 trees have low support for the above-described bipartitions, which may be due to short sequence length, limited number of informative sites, or poor alignment quality. However, 233 “high-support” trees (quadripartition internode certainty [QP-IC] score ≥ 0.3 and bootstrap support ≥ 50% for the minimum bipartition) result in similar fractions of trees supporting the above-described scenarios (fig. 4).
Additionally, among the 460 gene families not used for phylogenetic analyses, 17 are found exclusively in Mesoaciditogales and Petrotogales taxa (fig. 4). Combined with the 203 “Mesoaciditogales + Petrotogales” families and the three “Mesoaciditogales + outgroup” gene families found exclusively in Mesoaciditogales and Petrotogales species, a total of 223 of 1,181 gene families found in both Mesoaciditogales genomes (19%) share their recent evolutionary history with their Petrotogales sp. homologs. We refer to this set of 223 families as “Petrotogales-associated” gene families.
Genes That Share Recent Evolutionary Histories with Petrotogales Have Functions Important for Responding to Environmental Conditions
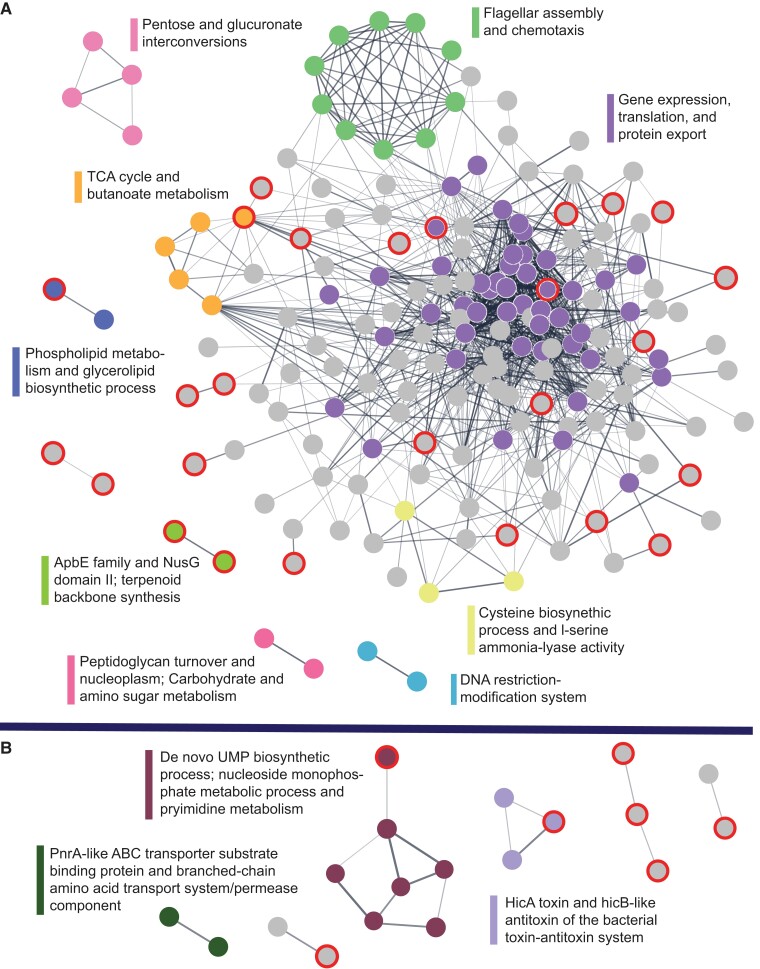
The 223 Petrotogales-associated gene families contain 243 M. lauensis genes (supplementary file 1, Supplementary Material online), which can be further subdivided in two sets: 195 genes (from 195 gene families) that are present widely in the Thermotogota and 48 genes (from 28 gene families) found solely in Mesoaciditogales and Petrotogales taxa. Many of these 243 genes (as highlighted below) are known to be involved in response to environmental changes. We investigated functional connections among the 243 genes by reconstructing STRING association networks (Szklarczyk et al. 2015) and examining their Clusters of Orthologous Groups (COG) categories (Galperin et al. 2021).
In the set of 195 widely distributed genes, products of 182 genes are predicted by STRING to interact with at least one other protein from the set (fig. 5A). The largest of these networks is enriched in genes involved in gene expression, translation, and protein export and contains a highly connected subnetwork of genes relating to bacterial chemotaxis. In the set of 48 genes, 16 genes form a few small networks (fig. 5B), including a network of genes involved in de novo Uridine MonoPhosphate (UMP) biosynthesis and nucleoside monophosphate and pyrimidine metabolism, several networks of genes involved in transport of amino acids and carbohydrates, a network of lipid metabolism genes, and a network related to toxin–antitoxin systems. Thus, many of the 243 Petrotogales-associated genes are likely functionally connected or even belong to the same biochemical pathway or molecular complex.
Fig. 5.
—The STRING protein–protein interaction networks of M. lauensis genes from the 223 Petrotogales-associated gene families. (A) Networks of genes that are present widely in Thermotogota (182 connected proteins). (B) Networks of genes found solely in Mesoaciditogales and Petrotogales (22 connected proteins). Singleton proteins are not shown. The edges are weighted by the confidence of the interactions, with a minimum association threshold of 0.4. Subnetworks enriched for certain functions have nodes highlighted in color and their “STRING Cluster” annotations listed. Gray nodes are not part of the enriched subnetworks. Outlined nodes (in red) are from proteins of unknown function.
The aforementioned genes for de novo UMP biosynthesis are assigned to the “coenzyme transport and metabolism” (H) COG category. Another gene in the H COG category, glyceraldehyde 3-phosphate dehydrogenase, is known to be differentially regulated in Kosmotoga olearia (order Petrotogales) depending on temperature (Pollo et al. 2017). More broadly, genes from the H COG category are overrepresented among the 243 genes (Fisher's exact test, P = 0.005). Within the STRING's “gene expression, translation, and protein export” subnetwork and among 13 genes with posttranslational modification, protein turnover, and chaperone functions (COG category O) are two genes that encode GroEL and a serine protease DO chaperone. These two proteins are implicated in the response to heat stress in K. olearia (Pollo et al. 2017). Among genes assigned to “Translation, ribosomal structure and biogenesis” (J) COG category are genes encoding ribosomal protein L12P and ribosome-binding factor A (rbfA), which, along with other ribosomal proteins, are known to exhibit changes in expression linked to cold response (Jones and Inouye 1996; Barria et al. 2013; Pollo et al. 2017). Two genes that form the small “peptidoglycan turnover and nucleoplasm” subnetwork (fig. 5A) are part of fatty acid synthesis and membrane envelope biogenesis (COG category M), along with six additional genes. One poorly characterized gene (COG category R), putatively annotated to encode enoyl-ACP reductase II, is connected to two lipid transport and metabolism genes (COG category I) within the large STRING network of Petrotogales-associated genes. In the Mesoaciditogales genomes, this gene is located in a neighborhood of genes involved in fatty acid biosynthesis and is therefore likely involved in that biochemical pathway. Based on a gene neighborhood analysis using DOE IMG/M website (IMG Gene ID 2582540014; accessed June 14 2023) (Chen et al. 2017), the gene is likely acquired from a Firmicutes (Thermincola).
There are 55 additional genes of yet unknown function or with only general function assigned in silico (COG category R or S) (supplementary file 1, Supplementary Material online). Eleven of these genes have temperature-responsive homologs in K. olearia (Pollo et al. 2017), and 26 are Mesoaciditogales–Petrotogales specific. Products of some of these uncharacterized genes are predicted to interact with other Petrotogales-associated genes (fig. 5). Hence, there are likely undiscovered genes important for specific environmental adaptations.
Discussion
Our phylogenetic analyses of commonly used phylogenetic markers (16S rRNA and SCC genes) support the early-branching position of the order Mesoaciditogales within the Thermotogota phylum (fig. 1; supplementary fig. S1, Supplementary Material online). This inference does not change when taxa are added to break long branches, sequences are recoded to adjust for compositional biases, and nonhomogeneous models are used to correct for compositional heterogeneity across Thermotogota phylum.
However, expanding the analyses beyond these commonly used marker genes reveals that many gene families in Mesoaciditogales have alternative evolutionary histories (figs. 4 and 6). It is not surprising that genomic content of Mesoaciditogales has been impacted by extensive HGT, as it is common in other Thermotogota (Zhaxybayeva et al. 2009; Nesbø et al. 2015; Haverkamp et al. 2021) as well as in bacteria in general (Philippe and Douady 2003; Gogarten and Townsend 2005; Doolittle and Brunet 2016; Arnold et al. 2022).
Fig. 6.
—Summary of phylogenetic relationships of 1,181 gene families found in both Mesoaciditogales with respect to other Thermotogota.
Notably, nearly a fifth of Mesoaciditogales gene families are more closely related to taxa from Petrotogales order (fig. 6), many of which grow optimally at lower temperatures than members of Thermotogales order (fig. 1). Some Petrotogales species have been found in similar environments as Mesoaciditogales and Mesoaciditogales-like MAGs (L’Haridon et al. 2019; Mori et al. 2020; Nesbø et al. 2021; L’Haridon et al. 2023), which could provide an opportunity for HGT between members of these two taxonomic groups. Alternatively, the Mesoaciditogales and Petrotogales lineages may have independently acquired these homologs via other taxa, such as Firmicutes.
We hypothesize that evolutionary histories of these genes reflect HGT-facilitated adaptation of Petrotogales and Mesoaciditogales to similar ecological niches. For instance, M. lauensis and Kosmotogaceae species share genes needed to grow on acetate that are not found in other Thermotogota species (Nesbø et al. 2019). In another example, de novo UMP biosynthesis is a highly conserved process that requires specific adaptations in hyperthermophilic environments (Thia-Toong et al. 2002)—which may explain why homologs of Mesoaciditogales UMP-related genes are found in Petrotogales, but not in the hyperthermophilic Thermotogales. Other central carbon metabolism genes, such as glyceraldehyde 3-phosphate dehydrogenase and the small networks of metabolism-related genes, may be specifically adapted to “ideal” environmental conditions, since metabolism is maximally upregulated at optimal temperatures and growth conditions (Pollo et al. 2017). The similarity of ribosome-associated genes (encoding ribosomal proteins and rbfA) suggests that the ribosomes in Mesoaciditogales and Petrotogales might be fine-tuned to their preferred growth temperature range, consistent with earlier studies of temperature adaptations of K. olearia and other bacteria (Barria et al. 2013; Pollo et al. 2017).
Several genes that are known to have temperature-dependent expression in K. olearia were among the Petrotogales-associated genes. Glyceraldehyde 3-phosphate dehydrogenase is upregulated in K. olearia (order Petrotogales) at its OGT of 65 °C (Pollo et al. 2017)—a temperature close to Mesoaciditoga's OGT. The two genes that are known to be part of K. olearia's heat stress response, GroEL and a serine protease DO chaperone, have homologs across Thermotogota, but the similarity between these Petrotogales and Mesoaciditogales homologs may be due to specialized adaptations to specific ranges of temperature stress, or they may have shared their evolutionary history through HGT. In K. olearia, enoyl-ACP reductase II contributes to changes in the cellular membrane composition in response to growth temperature, and its gene is also predicted to be horizontally acquired (Pollo et al. 2017). This and other Mesoaciditogales’ and Petrotogales-associated genes related to fatty acid synthesis, membrane biogenesis, and lipid metabolism are likely important for membrane remodeling in response to shifting temperatures (Suutari and Laakso 1994; Pollo et al. 2017). While these genes represent only a fraction of the 243 Petrotogales-associated genes, many of the remaining genes are predicted to have functional associations with the highlighted genes. This raises a possibility that their involvement in environmental adaptations may have not yet been identified.
Since HGT likely had a strong role in shaping Mesoaciditogales’ genome content, the question about the OGT of the LCA of Thermotogota remains open. It is possible that within Thermotogota, adaptations to lower temperature happened multiple times, independently, and that the LCA of the phylum was a hyperthermophile as previously hypothesized (Zhaxybayeva et al. 2009; Green et al. 2013). To resolve this question, further analyses of gene gain, loss, and HGT across the phylum are needed.
Materials and Methods
Retrieval and Analyses of 16S rRNA Genes
Thermotogota 16S rRNA gene sequences from 55 described Thermotogota species, whose strains are available in the DSMZ collection [https://www.dsmz.de/], were downloaded from GenBank (Benson et al. 2018) on November 8, 2020. Eight sequences were added as an outgroup: Hydrogenivirga caldilitoris IBSK3 (NR_024824), Aquifex aeolicus VF5 (NR_114796), Hydrogenobacter thermophilus TK-6 (NR_074870), Thermocrinis ruber OC 1/4 (NR_121741), Persephonella marina EX-H1 (NR_102828), Dictyoglomus thermophilum H-6-12 (NR_029235), Staphylococcus aureus ATCC 12600 (NR_118997), and Coprothermobacter platensis (Y08935). The sequences were aligned using SILVA's Incremental Aligner (SINA) v1.2.11 (Pruesse et al. 2012). The multiple sequence alignment was trimmed using trimAl v1.4.rev22 with the –gappy-out setting (Capella-Gutiérrez et al. 2009), resulting in the 1,548 nt alignment. Pairwise nucleotide identity among the Thermotogota 16S rRNA genes was calculated from the trimmed alignment using in-house scripts.
The data set was expanded by adding environmental sequences, which were retrieved using M. lauensis cd-1655RT 16S rRNA gene (NR_125611) as a query in a web-based BLASTN search (Altschul et al. 1990) of the nt database (Benson et al. 2018) with low complexity filter on, E value threshold of 0.0001, a maximum of 5,000 target sequences, and all other parameters as default (the search was performed in February 2021). All matches with >80% sequence identity to the query were added to the data set of 16S rRNA genes from 55 described Thermotogota species, and the whole data set was realigned with SINA v1.2.11 (Pruesse et al. 2012). This resulted in an alignment of 277 sequences that contain 2,168 sites.
The maximum likelihood (ML) tree under a homogeneous model was constructed in IQ-TREE v1.6.7 (Nguyen et al. 2015) under the GTR+F+R4 model chosen by ModelFinder (Kalyaanamoorthy et al. 2017). Accuracy of the topology was assessed via bootstrap analysis of 100 pseudo-samples.
The heterogeneity of sequence composition was assessed using two methodologies. First, χ2 test was performed on individual sequences, as implemented in IQ-TREE v1.6.7 (Nguyen et al. 2015). Second, Bowker's test for nonstationarity was performed on whole data set using TestNH v.1.3.0 from the Bio++ suite (Dutheil and Boussau 2008). The ML tree and model (GTR+G4 substitution model, alpha = 0.6) reconstructed in IQ-TREE (see above) was used as the starting tree, and a homogenous model was fit using the ML method implemented in the Bio++ bppml program. Then, the Bowker test was performed on the Bio++ homogenous tree with a P value threshold of 0.05 using 1,000 parametric bootstraps.
A ML tree reconstruction under a nonhomogenous model was performed using nhPhyML v0.1 (Boussau and Gouy 2006). Transition/transversion ratio was estimated from the data set, and site rate variation was modeled under the G4 model. The rooted tree reconstructed under the homogenous model in IQ-TREE was provided as the starting tree.
Stem regions of the 16S rRNA genes were predicted using Infernal v.1.1.4 (Nawrocki and Eddy 2013). GC-content of full-length 16S rRNA genes and of their stem regions were calculated using in-house scripts.
Retrieval of Genomes and Assignment of Taxonomy
Thermotogota genomes and MAGs (412 in total) were obtained either from NCBI's “Assembly” database (Kitts et al. 2016) or from the IMG database (Chen et al. 2017) in December 2020. Duplicated genomes found in both databases were removed. Additionally, 42 MAGs were shared with us privately by Eric Boyd (Montana State University), Håkon Dahle (University of Bergen), and Brett Baker (University of Texas in Austin). The genome list was checked against the list of strains of Thermotogota available in the DSMZ collection (https://www.dsmz.de/, last accessed December 2020) to ensure the inclusion of all described species with available genomes, and against the NCBI's Taxonomy database (Schoch et al. 2020) to verify that MAGs were classified as Thermotogota (NCBI taxid 200918). Genomes were assessed for completeness and contamination using CheckM (Parks et al. 2015). Among MAGs, only genomes that were estimated to be at least 50% complete and less than 10% contaminated were retained (i.e., we retained MAGs of at least medium quality per definition in Bowers et al. 2017). Fifteen genomes from bacteria belonging to other phyla were used as an outgroup. Since Thermotogota are strictly anaerobic bacteria, the aerobic taxa were included in the outgroup to reduce the potential impact of HGT on gene phylogenies.
This procedure resulted in 172 genomes (154 genomes and MAGs of Thermotogota, 15 genomes of outgroup species, and 3 unassigned MAGs that grouped with the outgroup), which were used in the subsequent analyses. GenBank and IMG accession numbers, as well as genome completeness information, are listed in supplementary file 2, Supplementary Material online.
Genomes were assigned taxonomy based on the Genome Taxonomy Database (GTDB) (Chaumeil et al. 2020). While NCBI's Taxonomy database (Schoch et al. 2020) classifies Kosmotogaceae as a family within the order Kosmotogales, the GTDB nomenclature places both the Kosmotogaceae and Petrotogaceae families into the order Petrotogales. We use the GTDB nomenclature throughout this manuscript. Sixty of the 172 genomes belong to strains of the described Thermotogota species.
Phylogenetic Reconstruction from Genes Encoding Ribosomal Proteins
Amino acid sequences of genes encoding ribosomal proteins were identified by using 50 Thermotoga maritima ribosomal proteins as queries in BLASTP v. 2.6.0 searches (Altschul et al. 1990) against the data set of the above-described 172 genomes. Results were filtered for coverage and quality (E < 0.0001, and the match length is between 60% and 140% of the query length) and then verified to be annotated as ribosomal proteins.
For the 60 strains of described Thermotogota species and all 15 outgroup species, the retrieved sequences for each of the 50 ribosomal proteins were aligned in MAFFT v7.305b using -linsi setting (Katoh and Standley 2013). The alignments were concatenated with in-house scripts into one alignment; in cases where a genome did not have a detected ribosomal protein, gaps were inserted into the alignment. The phylogenetic tree was reconstructed using the trimmed concatenated alignment in IQ-TREE v1.6.7 (Nguyen et al. 2015) using the LG+F+R5 substitution model selected by built-in ModelFinder (Kalyaanamoorthy et al. 2017). Accuracy of the topology was assessed via bootstrap analysis of 100 pseudo-samples.
The alignment, concatenation, and tree reconstruction were repeated using the 50 ribosomal proteins from all 172 genomes to assess how the inclusion of MAGs would affect the topology. For this expanded phylogenetic analysis, the LG+F+R5 model was again selected by ModelFinder (Kalyaanamoorthy et al. 2017).
Likelihood Mapping
Likelihood mapping analyses for 16S rRNA gene and ribosomal protein alignments were carried out in TREE-PUZZLE 5.3.rc16 (Schmidt and von Haeseler 2007).
For 16S rRNA sequence analyses, the trimmed alignment of sequences from the described species was used (1,548 nucleotide sites for 63 rRNA sequences, including outgroup). All 11,232 possible quartets were evaluated using approximate parameter estimation for computational efficiency. GTR model was used for the substitution model with nucleotide frequencies estimated from the alignment, and rate heterogeneity was modeled using gamma distribution with four rate categories. Two tests were performed: In the first test, all sequences were assigned into one of the four taxonomic groups, Mesoaciditogales, Thermotogales, Petrotogales, or the outgroup species; in the second test, sequences from Thermotogota were divided into Mesoaciditogales, Thermotogales, Petrotogaceae, or Kosmotogaceae, while the outgroup species were excluded.
For the ribosomal protein analyses, all 50 protein alignments were analyzed individually. For each protein, all possible quartets were analyzed using approximate parameter estimation for computational efficiency. The LG substitution matrix was used for the substitution model with amino acid frequencies estimated from the alignment, and rate heterogeneity was modeled using gamma distribution with four rate categories. All sequences were assigned to one of four taxonomic groups: Mesoaciditogales, Thermotogales, Petrotogales, or outgroup.
Calculation of Bias towards IVYWREL Amino Acids
For each protein encoded in 60 genomes from the described Thermotogota species, the proportion of IVYWREL amino acids was calculated using in-house scripts (available in the figshare repository). Transmembrane proteins were predicted using Phobius v.1.01 (Käll et al. 2004). The linear regression analysis of median IVYWREL of the nontransmembrane proteins and OGT of the 60 species was performed using scikit-learn v.0.23.2 (Pedregosa et al. 2011). Optimal growth temperatures were retrieved from each species’ defining publication and cross-checked against strain and species data from BacDive (Reimer et al. 2019) when available.
Identification of Gene Families
Gene families (orthogroups) in the 172 genome data set were identified in OrthoFinder v.2.5.1 (Emms and Kelly 2019). Within OrthoFinder, BLASTP was used for the sequence search and MAFFT v.7.305b for sequence alignment. FASTA files for gene families are available in the figshare repository.
Analyses of Single-Copy Gene Families
Single-copy gene families conserved across the Thermotogota phylum (“SCC” gene families) were defined as orthogroups that contained genes from at least 45 of the 60 genomes from the described Thermotogota species. This threshold was chosen to maximize the number of gene families, while retaining genes that are found across the majority (>50%) of taxa in each of the three Thermotogota orders (fig. 1). The selection resulted in 232 gene families, which include all 50 ribosomal proteins described above. The sequence alignments were retrieved from OrthoFinder and were concatenated using in-house scripts. The concatenated alignment was trimmed with TrimAl v1.4.rev22 using the -gappyout setting (Capella-Gutiérrez et al. 2009). This trimmed concatenated alignment was used throughout the phylogenetic analyses described in this section.
Compositional heterogeneity of sequences was assessed using the χ2 testing performed in IQ-TREE v1.6.12 (Nguyen et al. 2015). The phylogenetic tree was reconstructed in IQ-TREE v1.6.12 using the LG+F+R8 model selected by ModelFinder (Kalyaanamoorthy et al. 2017). Accuracy of the topology was assessed via bootstrap analysis of 100 pseudo-samples.
Alternative topology testing was carried out in IQ-TREE v1.6.12 (Nguyen et al. 2015). The SCC protein alignment was used to reconstruct ML phylogenetic trees that followed a set of topological constraints, which were imposed to define various relationships among Thermotogota families (fig. 2). All trees were reconstructed under the LG+F+R8 model, which was selected as the best fitting model for the SCC alignment during the unconstrained tree reconstruction. The likelihoods of the constrained trees were compared to the likelihood of the unconstrained SCC protein phylogeny using the AU test (Shimodaira 2002).
The alignment recoding was carried out using two approaches. In the first approach, recoding states that minimize sequence heterogeneity were searched using MinMax-ChiSq v1.1 (Susko and Roger 2007), which assessed all bin sizes between 2 and 20 amino acids and considered 5,000 random choices of starting bins for each bin size. In the second approach, recoding was used to adjust for the proportion of IVYWREL amino acids. Specifically, amino acids were recoded into ten states (A, G, P, S, T, C, DENQ, HKR, ILMV, and FWY). This ten-state recoding is modified from the Dayhoff recodings (Embley 2003; Hrdy et al. 2004), which groups amino acids into states based on physiochemical characteristics. Per recommendation by Hernandez and Ryan (2021), the model was tailored to our specific case as follows. Four states that involve IVYWREL amino acids as defined by Dayhoff 6-state recoding (DENQ, HKR, ILMV, and FWY) were retained, since IVYWREL bias is correlated with OGT in Thermotogota. The remaining six amino acids were allowed to be their own states since frequencies of these amino acids in proteins are presumed to be less affected by OGT. The recoding was carried out using in-house scripts (available in the figshare repository). The phylogeny was built with RAxML v8.2.11 using the GTR+G model for multistate inference.
Phylogenetic trees were also reconstructed under two nonhomogenous phylogenetic models as implemented in IQ-TREE v1.6.12 (Nguyen et al. 2015). First, the PMSF model was used to account for site-specific compositional frequencies. The SCC phylogeny was provided as a guide tree to compute the PMSF amino acid profiles for each alignment site. A phylogeny was built using the obtained profiles under the LG+C20+F+G model. Second, the heterogeneous, edge-unlinked GHOST model (Crotty et al. 2020) was used to correct for possible heterotachy. Specifically, a phylogeny was built under the LG+F0*H4 model. The LG substitution model was selected due to it being the best fit substitution matrix under the homogeneous model; F0 was used to assign separate base frequencies to each class, and *H4 was used to estimate model parameters separately for each of the four mixture classes. Accuracy of the reconstructions under both models was assessed using 1,000 ultrafast bootstrapping replicates.
Identification of Minimum Bipartitions
For these analyses, 721 gene families that contain a single gene from M. lauensis genome, a single gene from A. saccharophila genome, sequences from more than five additional Thermotogota genomes, and at least three outgroup sequences were selected. For each gene family, the ML tree was reconstructed in IQ-TREE v. v1.6.12 using the MAFFT sequence alignment calculated in OrthoFinder and the best fitting substitution model selected by ModelFinder (Kalyaanamoorthy et al. 2017). Accuracy of the tree reconstruction was determined by analysis of 100 bootstrap replicates. Additionally, a quartet-based internode certainty score was used to complement bootstrap support by providing a measure of branch incongruence alongside the measure of topological accuracy (Zhou et al. 2020). This “QP-IC” was calculated using the ML tree and the trees from bootstrap replicates.
Phylogenetic trees were converted to a set of bipartitions using bitstring representation in DendroPy v.4.5.2 (Sukumaran and Holder 2010). First, for each gene family tree, the largest bipartition that contained only outgroup taxa was labeled as the “outgroup bipartition.” Gene families with outgroup bipartition that contained less than three taxa were excluded from further analyses. Then, all bipartitions that included M. lauensis, A. saccharophila, and the taxa in the outgroup bipartitions were identified. Among these bipartitions, the bipartition that contained the smallest number of other taxa was labeled as the “minimum bipartition.” Trees of 20 gene families, in which M. lauensis and A. saccharophila did not group together, were excluded from further analyses. For each minimum bipartition, we identified the lowest taxonomic rank of the set of Thermotogota taxa that joined the Mesoaciditogales. Taxonomic rank of the set was determined by using the GTDB taxonomy classification of the 60 strains from the described species.
Bipartitions were considered “highly supported” if their QP-IC score ≥ 0.3 and bootstrap support ≥ 50%, and otherwise were considered “low-support” bipartitions. The analyses were carried out using in-house scripts (available in the figshare repository).
Identification of Taxonomic Rank for Gene Family Composition
We assigned a lowest taxonomic rank to the 1,181 gene families containing Mesoaciditogales, including gene families not used in the phylogenetic analyses. Taxonomic rank was determined by excluding all Mesoaciditogales genomes and then using GTDB taxonomic classification in conjunction with ETE 3 (Huerta-Cepas et al. 2016) to summarize which Thermotogota genera outside the Mesoaciditogales contributed genes to each family.
COG Assignment and COG Categories’ Enrichment Calculations
Protein-coding genes from M. lauensis and A. saccharophila genomes were used as queries in the BLASTP v.2.6.0+ search of the NCBI COG database (2020 release; downloaded in March 2021) (Galperin et al. 2021). Forty-one percent of genes in these two genomes received a COG category assignment.
The distribution of COG categories in 1,181 gene families that contained both Mesoaciditogales species was compared to the distribution of COG categories in a subset of 223 “Petrotogales-associated” gene families (see Results for definition). Families were assigned a COG category by using the COG category of the Mesoaciditogales gene as a proxy. The significance of the difference was assessed using a one-sided Fisher's exact test, with a P value threshold of ≤0.05.
Reconstruction of Protein–Protein Association Networks
Protein–protein association networks were inferred using the STRING database and its web-based tools (Szklarczyk et al. 2015) (accessed on July 21, 2022). At the time of the analyses, the STRING database included the genome of M. lauensis cd-1655R but did not contain the genome of A. saccharophila. Therefore, the networks were reconstructed using only M. lauensis protein-coding genes. Specifically, functional associations were searched within the set of the 243 M. lauensis genes encompassed in the 223 “Petrotogales-associated” gene families (supplementary file 1, Supplementary Material online).
The 243 genes were separated into two sets: genes from families that also included genes from Thermotogales (195 genes) and genes from families that were present only in Mesoaciditogales and Petrotogales (48 genes). For each set, the separate association networks were inferred. For both networks, the default association threshold was used (0.4). Gene interaction tables, functional annotation files, and images of the original STRING networks are available in the figshare repository. The two STRING association networks were imported into Cytoscape v.3.8 (Su et al. 2014), and the Cytoscape tool “StringApp” (Doncheva et al. 2019) was used to retrieve functional annotations of selected clusters within each network. The functions for all genes in Mesoaciditoga lauensis’ genome were used as the background when calculating enrichment values.
Phylogenetic Tree Visualization
All trees were visualized with iTOL's web platform version 6 (Letunic and Bork 2021).
Supplementary Material
Acknowledgments
We thank Eric Boyd (Montana State University), Håkon Dahle (University of Bergen), and Brett Baker (University of Texas in Austin) for sharing MAGs, from which we extracted Mesoaciditogales’ homologs used in this study. The work was supported by Dartmouth Fellowship and Cramer funds to A.A.F. and Dartmouth Dean of Faculty funds to O.Z.
Contributor Information
Anne A Farrell, Department of Biological Sciences, Dartmouth College, Hanover, New Hampshire, USA.
Camilla L Nesbø, Department of Biological Sciences, University of Alberta, Edmonton, Alberta, Canada; Department of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, Ontario, Canada.
Olga Zhaxybayeva, Department of Biological Sciences, Dartmouth College, Hanover, New Hampshire, USA; Department of Computer Science, Dartmouth College, Hanover, New Hampshire, USA.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
The genomes and 16S rRNA sequences used for this research are publicly available via NCBI Nucleotide (https://www.ncbi.nlm.nih.gov/nucleotide/), NCBI Assembly (https://www.ncbi.nlm.nih.gov/assembly), or the Joint Genome Institute Integrated Microbial Genomes (https://img.jgi.doe.gov/). Out of the 172 nonredundant Thermotogota genomes and MAGs used in this study, 141 are publicly available, and their accession numbers are listed in supplementary file 2, Supplementary Material online. For the remaining 31 MAGs shared with us but not yet publicly available, the genes used in our analyses are available as part of the orthogroup data, described below. Amino acid sequences of proteins in orthogroups and 16S rRNA gene sequences in FASTA format, alignments, phylogenetic trees in Newick format, likelihood mapping results, STRING network analysis files, program log files, and in-house scripts are deposited in a figshare repository available at https://doi.org/10.6084/m9.figshare.23303717.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Antoine E, et al. 1997. Thermosipho melanesiensis sp. nov., a new thermophilic anaerobic bacterium belonging to the order Thermotogales, isolated from deep-sea hydrothermal vents in the Southwestern Pacific Ocean. Int J Syst Bacteriol. 47:1118–1123. [DOI] [PubMed] [Google Scholar]
- Arnold BJ, Huang I-T, Hanage WP. 2022. Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Microbiol. 20:206–218. [DOI] [PubMed] [Google Scholar]
- Barion S, Franchi M, Gallori E, Di Giulio M. 2007. The first lines of divergence in the Bacteria domain were the hyperthermophilic organisms, the Thermotogales and the Aquificales, and not the mesophilic Planctomycetales. Biosystems 87:13–19. [DOI] [PubMed] [Google Scholar]
- Barria C, Malecki M, Arraiano CM. 2013. Bacterial adaptation to cold. Microbiology(Reading) 159:2437–2443. [DOI] [PubMed] [Google Scholar]
- Benson DA, et al. 2018. Genbank. Nucleic Acids Res. 46:D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten J. 2005. A review of long-branch attraction. Cladistics 21:163–193. [DOI] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M. 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456:942–945. [DOI] [PubMed] [Google Scholar]
- Boussau B, Gouy M. 2006. Efficient likelihood computations with nonreversible models of evolution. Syst Biol. 55:756–768. [DOI] [PubMed] [Google Scholar]
- Bowers RM, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 35:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Philippe H. 2002. A non-hyperthermophilic ancestor for Bacteria. Nature 417:244–244. [DOI] [PubMed] [Google Scholar]
- Butzin NC, et al. 2013. Reconstructed ancestral myo-inositol-3-phosphate synthases indicate that ancestors of the Thermococcales and Thermotoga species were more thermophilic than their descendants. PLoS One 8:e84300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole RJ, Forterre P. 2019. The evolution of reverse gyrase suggests a nonhyperthermophilic last universal common ancestor. Mol Biol Evol. 36:2737–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2020. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IA, et al. 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45:D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty SM, et al. 2020. GHOST: recovering historical signal from heterotachously evolved sequence alignments. Syst Biol. 69:249–264. [DOI] [PubMed] [Google Scholar]
- Davey ME, Wood WA, Key R, Nakamura K, Stahl DA. 1993. Isolation of three species of Geotoga and Petrotoga: two new genera, representing a new lineage in the bacterial line of descent distantly related to the “Thermotogales”. System Appl. Microbiol. 16:191–200. [Google Scholar]
- Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. 2019. Cytoscape StringApp: network analysis and visualization of proteomics data. J. Proteome Res. 18:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Brunet TD. 2016. What is the tree of life? PLoS Genet. 12:e1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil J, Boussau B. 2008. Non-homogeneous models of sequence evolution in the Bio++ suite of libraries and programs. BMC Evol Biol. 8:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM. 2003. Mitochondria and hydrogenosomes are two forms of the same fundamental organelle. Philos Trans R Soc Lond B Biol Sci. 358:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst Biol. 27:401–410. [Google Scholar]
- Foster PG, Hickey DA. 1999. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol. 48:284–290. [DOI] [PubMed] [Google Scholar]
- Galperin MY, et al. 2021. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49:D274–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Lobry JR. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in Prokaryotes. J Mol Evol. 44:632–636. [DOI] [PubMed] [Google Scholar]
- Galtier N, Tourasse N, Gouy M. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220–221. [DOI] [PubMed] [Google Scholar]
- Gevers D, et al. 2005. Re-evaluating prokaryotic species. Nat Rev Microbiol. 3:733–739. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. 2005. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 3:679–687. [DOI] [PubMed] [Google Scholar]
- Green AG, Swithers KS, Gogarten JF, Gogarten JP. 2013. Reconstruction of ancestral 16S rRNA reveals mutation bias in the evolution of optimal growth temperature in the Thermotogae phylum. Mol Biol Evol. 30:2463–2474. [DOI] [PubMed] [Google Scholar]
- Hassler HB, et al. 2022. Phylogenies of the 16S rRNA gene and its hypervariable regions lack concordance with core genome phylogenies. Microbiome 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp THA, et al. 2021. Newly identified proviruses in Thermotogota suggest that viruses are the vehicles on the highways of interphylum gene sharing. Environ Microbiol. 23:7105–7120. [DOI] [PubMed] [Google Scholar]
- Hernandez AM, Ryan JF. 2021. Six-state amino acid recoding is not an effective strategy to offset compositional heterogeneity and saturation in phylogenetic analyses. Syst Biol. 70:1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdy I, et al. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432:618–622. [DOI] [PubMed] [Google Scholar]
- Huber R, et al. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 °C. Arch Microbiol. 144:324–333. [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, et al. 2016. Athalassotoga saccharophila gen. nov., sp. nov., isolated from an acidic terrestrial hot spring, and proposal of Mesoaciditogales ord. nov. and Mesoaciditogaceae fam. nov. in the phylum Thermotogae. Int J Syst Evol Microbiol. 66:1045–1051. [DOI] [PubMed] [Google Scholar]
- Jones PG, Inouye M. 1996. Rbfa, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 21:1207–1218. [DOI] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 338:1027–1036. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts PA, et al. 2016. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 44:D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O. 2008. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 363:3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Haridon S, Gouhier L, John ES, Reysenbach A-L. 2019. Marinitoga lauensis sp. nov., a novel deep-sea hydrothermal vent thermophilic anaerobic heterotroph with a prophage. Syst Appl Microbiol. 42:343–347. [DOI] [PubMed] [Google Scholar]
- L’Haridon S, Nesbø C, Farrell A, Zhaxybayeva O. 2021. Kosmotoga. In: Trujillo ME, Dedysh S, DeVos P, Hedlund B, Kämpfer P, Rainey FA, Whitman WB, editors. Bergey's manual of systematics of Archaea and Bacteria: John Wiley & Sons. Ltd. 10.1002/9781118960608.gbm01863 [DOI] [Google Scholar]
- L’Haridon S, Nesbo C, Farrell A, Zhaxybayeva O. 2023. Mesoaciditoga. In: Trujillo ME, Dedysh S, DeVos P, Hedlund B, Kämpfer P, Rainey FA, Whitman WB, editors. Bergey's manual of systematics of Archaea and Bacteria: John Wiley & Sons. Ltd. 10.1002/9781118960608.gbm01867 [DOI] [Google Scholar]
- Li B, Lopes JS, Foster PG, Embley TM, Cox CJ. 2014. Compositional biases among synonymous substitutions cause conflict between gene and protein trees for plastid origins. Mol Biol Evol. 31:1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sakurai K, Hosoyama A, Kakegawa T, Hanada S. 2020. Vestiges of adaptation to the mesophilic environment in the genome of Tepiditoga spiralis gen. nov., sp. nov. Microbes Environ. 35:ME20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbø CL, et al. 2012. Mesotoga prima gen. nov., sp. nov., the first described mesophilic species of the Thermotogales. Extremophiles 16:387–393. [DOI] [PubMed] [Google Scholar]
- Nesbø CL, et al. 2015. Evidence for extensive gene flow and Thermotoga subpopulations in subsurface and marine environments. ISME J. 9:1532–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbø CL, et al. 2019. Genomic analysis of the mesophilic Thermotogae genus Mesotoga reveals phylogeographic structure and genomic determinants of its distinct metabolism. Environ Microbiol. 21:456–470. [DOI] [PubMed] [Google Scholar]
- Nesbø CL, L’Haridon S, Zhaxybayeva O, Farrell AA. 2021. Kosmotogaceae. In: Trujillo ME, Dedysh S, DeVos P, Hedlund B, Kämpfer P, Rainey FA, Whitman WB, editors. Bergey's manual of systematics of Archaea and Bacteria: John Wiley & Sons. Ltd. 10.1002/9781118960608.fbm00361 [DOI] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. Checkm: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BKC, Morgan HW, Daniel RM. 1985. Fervidobacterium nodosum gen. nov. and spec. nov., a new chemoorganotrophic, caldoactive, anaerobic bacterium. Arch Microbiol. 141:63–69. [Google Scholar]
- Pedregosa F, et al. 2011. Scikit-learn: machine learning in python. J Mach Learn Res. 12:2825–2830. [Google Scholar]
- Philippe H, Douady CJ. 2003. Horizontal gene transfer and phylogenetics. Curr Opin Microbiol. 6:498–505. [DOI] [PubMed] [Google Scholar]
- Pollo SMJ, et al. 2017. Genomic insights into temperature-dependent transcriptional responses of Kosmotoga olearia, a deep-biosphere bacterium that can grow from 20 to 79 °C. Extremophiles 21:963–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollo SMJ, Zhaxybayeva O, Nesbø CL. 2015. Insights into thermoadaptation and the evolution of mesophily from the bacterial phylum Thermotogae. Can J Microbiol 61:655–670. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer LC, et al. 2019. Bacdive in 2019: bacterial phenotypic data for high-throughput biodiversity analysis. Nucleic Acids Res. 47:D631–D636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A-L, et al. 2013. Mesoaciditoga lauensis gen. nov., sp. nov., a moderately thermoacidophilic member of the order Thermotogales from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 63:4724–4729. [DOI] [PubMed] [Google Scholar]
- Sauer DB, Wang D-N. 2019. Predicting the optimal growth temperatures of prokaryotes using only genome derived features. Bioinformatics 35:3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, von Haeseler A. 2007. Maximum-likelihood analysis using TREE-PUZZLE. Curr Protoc Bioinformatics. Chapter 6:Unit 6.6. [DOI] [PubMed] [Google Scholar]
- Schoch CL, et al. 2020. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 51:492–508. [DOI] [PubMed] [Google Scholar]
- Steinsbu BO, Røyseth V, Thorseth IH, Steen IH. 2016. Marinitoga arctica sp. nov., a thermophilic, anaerobic heterotroph isolated from a Mid-Ocean Ridge vent field. Int J Syst Evol Microbiol. 66:5070–5076. [DOI] [PubMed] [Google Scholar]
- Stetter KO. 1996. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 18:149–158. [Google Scholar]
- Su G, Morris JH, Demchak B, Bader GD. 2014. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 47:8.13.1–8.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. 2010. Dendropy: a python library for phylogenetic computing. Bioinformatics 26:1569–1571. [DOI] [PubMed] [Google Scholar]
- Susko E, Roger AJ. 2007. On reduced amino acid alphabets for phylogenetic inference. Mol Biol Evol. 24:2139–2150. [DOI] [PubMed] [Google Scholar]
- Suutari M, Laakso S. 1994. Microbial fatty acids and thermal adaptation. Crit Rev Microbiol. 20:285–328. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, et al. 2015. STRING V10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thia-Toong T-L, et al. 2002. Genes of de novo pyrimidine biosynthesis from the hyperthermoacidophilic crenarchaeote Sulfolobus acidocaldarius: novel organization in a bipolar operon. J Bacteriol. 184:4430–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-C, Minh BQ, Susko E, Roger AJ. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst Biol. 67:216–235. [DOI] [PubMed] [Google Scholar]
- Wery N, et al. 2001. Marinitoga camini gen. nov., sp. nov., a rod-shaped bacterium belonging to the order Thermotogales, isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 51:495–504. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Puigbò P, Koonin EV, Wolf YI. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7:e36972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. 2007. Protein and DNA sequence determinants of thermophilic adaptation. PLoS Comput Biol. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, et al. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci U S A. 106:5865–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, et al. 2020. Quartet-based computations of internode certainty provide robust measures of phylogenetic incongruence. Syst Biol. 69:308–324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomes and 16S rRNA sequences used for this research are publicly available via NCBI Nucleotide (https://www.ncbi.nlm.nih.gov/nucleotide/), NCBI Assembly (https://www.ncbi.nlm.nih.gov/assembly), or the Joint Genome Institute Integrated Microbial Genomes (https://img.jgi.doe.gov/). Out of the 172 nonredundant Thermotogota genomes and MAGs used in this study, 141 are publicly available, and their accession numbers are listed in supplementary file 2, Supplementary Material online. For the remaining 31 MAGs shared with us but not yet publicly available, the genes used in our analyses are available as part of the orthogroup data, described below. Amino acid sequences of proteins in orthogroups and 16S rRNA gene sequences in FASTA format, alignments, phylogenetic trees in Newick format, likelihood mapping results, STRING network analysis files, program log files, and in-house scripts are deposited in a figshare repository available at https://doi.org/10.6084/m9.figshare.23303717.