Abstract
BACKGROUND AND OBJECTIVES:
Histopathological features and molecular biomarkers have been studied as potential prognostic factors. This study aimed to investigate the clinical features, molecular phenotypes, and survival prognosis of isocitrate dehydrogenase (IDH)-mutant (IDHmt) gliomas with histone H3 alterations (H3-alterations).
METHODS:
A total of 236 and 657 patients with whole-exome sequencing data were separately collected from the Chinese Glioma Genome Atlas and The Cancer Genome Atlas databases. Survival analysis of patients with glioma was performed using Kaplan–Meier survival curves stratified by histone H3 status. Univariate and multivariate analyses were used to identify the associations between histone H3 status and other clinicopathological factors with survival in patients with IDH-mutant gliomas.
RESULTS:
Diffuse gliomas with H3 alterations are more likely to be high grade in 2 cohorts (P = .025 and P = .021, respectively). IDHmt glioma patients with H3-alteration had significantly less life expectancy than histone H3 wild-type (P = .041 and P = .008, respectively). In the Chinese Glioma Genome Atlas cohort, Karnofsky performance scores ≤ 80 (HR 2.394, 95% CI 1.257-4.559, P = .008), extent of resection (HR 0.971, 95% CI 0.957-0.986, P < .001), high WHO grade (HR 6.938, 95% CI 2.787-17.269, P < .001), H3-alteration (HR 2.482, 95% CI 1.183-4.981, P = .016), and 1p/19q codeletion (HR 0.169, 95% CI 0.073-0.390, P < .001) were independently associated with IDHmt gliomas. In the The Cancer Genome Atlas cohort, age (HR 1.034, 95% CI 1.008-1.061, P = .010), high WHO grade (HR 2.365, 95% CI 1.263-4.427, P = .007), and H3-alteration (HR 2.501, 95% CI 1.312-4.766, P = .005) were independently associated with IDHmt gliomas.
CONCLUSION:
Identification and assessment of histone H3 status in clinical practice might help improve prognostic prediction and develop therapeutic strategies for these patient subgroups.
KEY WORDS: Diffuse gliomas, IDH mutation, Histone H3, Prognosis, Survival analysis
ABBREVIATIONS:
- CGGA
Chinese Glioma Genome Atlas
- EOR
extent of resection
- H3-alteration
histone H3 alteration
- H3-WT
histone H3 wild-type
- HGG
high-grade glioma
- IDH
isocitrate dehydrogenase
- HR
hazard ratio
- IDHmt
IDH-mutant
- IDH-WT
IDH wild-type
- KPS
Karnofsky performance scores
- LGG
low-grade glioma
- OS
overall survival
- TCGA
The Cancer Genome Atlas
- TERT
telomerase reverse transcriptase promoter
- TP53
tumor protein p53
- WES
whole-exome sequencing
- WHO
World Health Organization.
According to the 2021 WHO Classification of Tumors of the Central Nervous System1 (2021 WHO CNS), diffuse gliomas are classified based on the genetic variations (eg, isocitrate dehydrogenase [IDH] and histone H3 status) that allow for a comprehensive neuropathological diagnosis. A growing number of specific molecular markers can provide valuable prognostic information.2,3 Therefore, molecular characteristics have been added to the biomarkers for estimating prognosis and developing targeted drugs for various tumors.4 The prognosis of diffuse gliomas can be better mapped out using molecular pathology. IDH1/2 status, chromosome 1p/19q codeletion, O6-methylguanine-DNA methyltransferase (MGMT) promoter, and other molecular parameters have become important genes/molecular profiles for diffuse gliomas.5-7 Two newly defined entities of pediatric-type diffuse gliomas are termed “diffuse midline glioma, H3 K27-altered” and “diffuse hemispheric glioma, H3 G34-mutant,” which were associated with histone H3 alterations (H3-alterations) and poor prognosis presented in the 2021 WHO CNS.1,8,9 Histone H3 is 1 of the 5 major histone families in eukaryotic chromatin, and its sequence variation and modifications play a crucial role in genome maintenance and gene regulation.10 Nonetheless, these data are based on particular histone H3 subtypes (H3.1 and H3.3) and come from a single center or database. It is essential to note that H3-alterations may have different clinical implications and may not necessarily have only 2 special molecular features.
Our primary goal of this study was to assess whether there is a potential link between clinical characteristics and the status of histone H3, and we collected whole-exome sequencing (WES) data to investigate the genomic landscape of H3-alterations from the CGGA database. For this reason, we are particularly interested in exploring the relationship between clinicopathological characteristics and clinical prognosis from CGGA and TCGA data sets. The results provided evidence for the interrogation of H3-alterations in current molecular diagnostics and prognostication of diffuse gliomas.
METHODS
Whole-Exome Sequencing (WES) Data
We retrospectively evaluated all the patients with diffuse glioma who underwent surgical resection between March 2008 and May 2018 with WES data in the CGGA.11 The last follow-up data were updated in June 2022. All genomic DNA samples were obtained from patientsʼ tumor/blood and performed using next-generation sequencing technology.11-13 Detailed methods can be found in the study by Zhao et al.11 In the CGGA database (http://www.cgga.org.cn/download.jsp), we investigated all the genomic alteration types of histone H3 genes, while performing patientsʼ subclassification according to different genes and alteration modes. In addition, we obtained relevant WES data with H3-alterations from the TCGA database (https://portal.gdc.cancer.gov/) to further validate our findings.
Patient Enrollment and Clinicopathological Information
This study obtained clinicopathological information on IDH status, MGMT promoter methylation, 1p/19q status, pathology, clinical features, MRI information, and follow-up data in patients with diffuse gliomas from the CGGA database. We defined midline violation as tumor border encroaching basal ganglia, brainstem, corpus callosum, third ventricle, thalamus, and other midline structures.14 Our inclusion criteria were (1) preoperative and postoperative MRI (T2-weighted and/or fluid-attenuated inversion recovery); (2) characteristics information including sex, age at diagnosis, pathological diagnosis, WHO grade, and preoperative Karnofsky performance scores (KPS); and (3) IDH status and follow-up data. We excluded patients with a survival of fewer than 30 days because the death of these individuals might be due to perioperative complications. Because of incomplete information on early data in retrospective studies, telomerase reverse transcriptase (TERT) promoter, tumor protein p53 (TP53), and other molecular data were unavailable in some samples and were not included in our analyses. The extent of resection (EOR) was evaluated according to the volume of the removed tumor measured on postoperative MRI by 2 experienced neuroradiologists.
Similarly, the clinical and follow-up data of diffuse gliomas obtained from the TCGA website15,16 were used as a validation cohort. These data were supplemented by other published literature.16 Our data were extracted from a publicly available database, and some variables (preoperative KPS, EOR, and midline violation) were missing. We abide to the principles of the Declaration of Helsinki, and our study was a retrospective study for which all data were kept anonymous and done after agreement from the local ethics committee and with the patients' informed consent.
Statistical Analyses
In our study, we classified 2 subgroups with different WHO grades as low-grade glioma (LGG, WHO grade 2) and high-grade glioma (HGG, WHO grade 3 and 4). Overall survival (OS) was measured from the surgery date to documented death or the last follow-up.
We tested numerical variables using the unpaired, two-tailed t-test and categorical variables using Fisherʼs exact test or χ2, depending on the situation. The Fisher's exact test was performed to analyze categorical data on account of the χ2 test violation assumptions. If some statistical variables were not eligible for normal distribution, the Mann–Whitney U-test was used instead of the t-test. Survival data were analyzed using the Kaplan–Meier method, and between-group differences in survival were tested using the log-rank (Mantel–Cox) test. In the univariate analysis, variables with a P-value <.05 were entered into the multivariate analysis. A Cox proportional hazards model was used to analyze the multivariate survival analysis of specific variables, and the proportional hazards assumption was tested. In this study, statistical analyses were performed with SPSS software (version 22.0, SPSS Inc) and a P-value <.05 was considered statistically significant.
RESULTS
Patient Characteristics
We scrutinized all 286 patients with glioma with WES data from the CGGA database, and 50 patients were excluded (Figure 1A). These excluded samples were due to a lack of follow-up and necessary clinical data (preoperative KPS, adjuvant therapy after surgery, etc.). Similarly, a total of 657 samples from the TCGA database were retained to validate our findings (Figure 1B). Patient demographics and baseline characteristics from CGGA and TCGA databases were separately shown in Table 1 and Supplementary Table 1, http://links.lww.com/NEU/D776. As shown in Table 1, there was no statistically significant difference in sex, age, presenting symptoms, and KPS score between these 2 subgroups of patients with H3-alterations and histone H3 wild-type (H3-WT). The proportion of lesions on the left side in H3-alteration tumors was significantly higher than in H3-WT ones (61.1% vs 39.6%, χ2 test, P = .019). The ratio of HGG was significantly higher in patients with H3-alterations (81.5% vs 65.4%, χ2 test, P = .025).
FIGURE 1.

Flowcharts of the patients with glioma in the CGGA and TCGA databases. A, In the CGGA database, 236 patients with glioma were classified into 2 subgroups based on histone H3 status. B, In the CGGA database, 657 patients with glioma were classified into 2 subgroups based on histone H3 status. CGGA, Chinese Glioma Genome Atlas; IDH, isocitrate dehydrogenase; OS, overall survival; TCGA, The Cancer Genome Atlas; WES, whole-exome sequencing.
TABLE 1.
Patient Demographics and Baseline Characteristics From CGGA Database (N = 236)
| Characteristic | H3-alteration | H3-WT | P valuea | Total |
|---|---|---|---|---|
| Total (%) | 54 (22.9) | 182 (77.1) | 236 (100) | |
| Sex (%) | .949 | |||
| Male | 30 (55.6) | 102 (56.0) | 132 (55.9) | |
| Female | 24 (44.4) | 80 (44.0) | 104 (44.1) | |
| Age at diagnosis, y, median (range) | 40 (15-66) | 41 (10-76) | .590 | 41 (10-76) |
| Presenting symptom (%) | .273 | |||
| Headache | 19 (35.2) | 41 (22.5) | 60 (25.4) | |
| Epilepsy | 17 (31.5) | 63 (34.6) | 80 (33.9) | |
| Neurological dysfunction | 20 (37.0) | 66 (36.3) | 86 (36.4) | |
| Incidental | 6 (11.1) | 33 (18.1) | 39 (16.5) | |
| Preoperative KPS (%) | .648 | |||
| Median (IQR) | 90 (80-90) | 90 (80-90) | 90 (80-90) | |
| >80 | 35 (64.8) | 124 (68.1) | 159 (67.4) | |
| ≤80 | 19 (35.2) | 58 (31.9) | 77 (32.6) | |
| Side of the lesion (%) | .019 | |||
| Left | 33 (61.1) | 72 (39.6) | 105 (44.5) | |
| Right | 17 (31.5) | 92 (50.5) | 109 (46.2) | |
| Bilateral | 4 (7.4) | 18 (9.9) | 22 (9.3) | |
| Tumor location (%) | .599 | |||
| Frontal | 36 (66.7) | 115 (63.2) | 151 (64.0) | |
| Temporal | 22 (40.7) | 68 (37.4) | 90 (38.1) | |
| Parietal | 15(27.8) | 35 (19.2) | 50 (27.5) | |
| Occipital | 3 (5.6) | 6 (3.3) | 9 (3.8) | |
| Insular | 13 (24.1) | 21 (11.5) | 34 (14.4) | |
| Otherb | 12 (22.2) | 32 (17.6) | 44 (18.6) | |
| EOR (IQR) | 100.0% (79.2%-100%) | 100.0% (86.9%-100.0%) | .112 | 100.0% (85.4%-100.0%) |
| Treatment after surgery (%) | .480 | |||
| Chemoradio-therapy | 40 (74.1) | 116 (63.7) | 156 (66.1) | |
| Radiotherapy | 7 (13.0) | 39 (21.4) | 46 (19.5) | |
| Chemotherapy | 4 (7.4) | 13 (7.1) | 17 (7.2) | |
| None | 3 (5.5) | 14 (7.7) | 17 (7.2) | |
| Midline violation (%) | .442 | |||
| Nonmidline glioma | 42 (77.8) | 150 (82.4) | 192 (81.4) | |
| Midline glioma | 12 (22.2) | 32 (17.6) | 44 (18.6) | |
| Grade (%) | .025 | |||
| LGG | 10 (18.5) | 63 (34.6) | 73 (30.9) | |
| HGG | 44 (81.5) | 119 (65.4) | 163 (69.1) | |
| IDH status (%) | .093 | |||
| IDHmt | 26 (48.1) | 111 (61.0) | 137 (58.1) | |
| IDH-WT | 28 (51.9) | 71 (39.0) | 99 (41.9) | |
| 1p/19q status (%) | .665 | |||
| Noncodel | 34 (63.0) | 115 (63.2) | 149 (63.1) | |
| Codel | 15 (27.8) | 59 (32.4) | 74 (31.4) | |
| NA | 5 (9.3) | 8 (4.4) | 13 (5.5) | |
| MGMT promoter status | .898 | |||
| Unmethylated | 13 (24.1) | 42 (23.1) | 55 (23.3) | |
| Methylated | 27 (50.0) | 83 (45.6) | 110 (46.6) | |
| NA | 14 (25.9) | 57 (31.3) | 71 (30.1) |
CGGA, Chinese Glioma Genome Atlas; EOR, extent of resection; H3-alteration, histone H3 alteration; H3-WT, histone H3 wild-type; HGG, high-grade glioma; IDH, isocitrate dehydrogenase; IDHmt, IDH-mutant; IDH-WT, IDH wild-type; IQR, interquartile range; KPS, Karnofsky performance score; LGG, low-grade glioma; MGMT, O6-methylguanine-DNA methyltransferase; NA, not applicable.
Results in bold represent P < 0.05.
Comparison between H3-alteration gliomas and H3-WT gliomas.
Basal ganglia, brainstem, corpus callosum, third ventricle, thalamus, and other midline structures.
We found that the ratio of diffuse gliomas with H3-alterations in HGGs was significantly higher than in LGGs in both cohorts (Figure 2A, χ2 test, P = .025 and P = .021, respectively). However, there was no statistical difference between the proportion of H3-alterations in IDH-mutant and IDH wide-type gliomas in both cohorts (Figure 2B, χ2 test, P = .093 and P = .096, respectively).
FIGURE 2.
Associations between H3-alterations and WHO grade or IDH status in diffuse gliomas from CGGA and TCGA databases. A, Among all sequenced tumors, H3-alteration was significantly increased in high-grade gliomas in the CGGA and TCGA cohorts (χ2 test, P = .025 and P = .021, respectively). B, The proportion of IDH-WT gliomas with H3-alteration was higher than IDHmt gliomas, but statistically significant differences were not found in the CGGA and TCGA cohorts (χ2 test, P = .093 and P = .096, respectively). CGGA, Chinese Glioma Genome Atlas; H3-alteration, histone H3 alteration; HGG, high-grade glioma; IDH, isocitrate dehydrogenase; IDHmt, IDH-mutant; IDH-WT, IDH wild-type; LGG, low-grade glioma; TCGA, The Cancer Genome Atlas.
Genomic Landscape of Histone H3 Alterations in Diffuse Gliomas
In the CGGA cohort, we divided all the alterations of H3 family genes into 3 categories: 16 (10.3%) mutations, 129 (82.7%) amplifications, and 11 (7.1%) deletions. The landscape of clinicopathological features of 54 sequenced tumors with H3-alterations in the CGGA cohort is shown in Figure 3. Among all 54 sequenced gliomas with H3-alterations, we discovered that 20 family genes of the histone H3 were altered, in which 8 (40%) mutant genes occurred in 10 (18.5%) patients. In the TCGA cohort, mutations were found in only 2 of 20 (10.0%) patients with LGG with H3-alterations. More information and all H3-alterations are provided in Supplementary Data, http://links.lww.com/NEU/D777.
FIGURE 3.
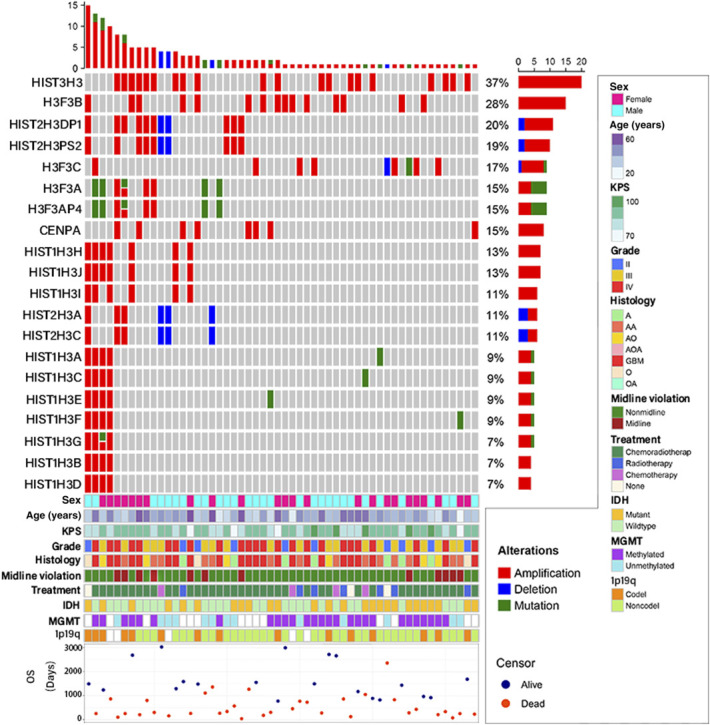
The landscape of clinicopathological features of 54 sequenced tumors with H3-alterations in the CGGA cohort. Twenty genes from 54 patients were observed altered and included 17 (85.0%) protein-coding genes and 3 (15%) pseudogenes. HIST3H3 amplification was the most prevalent actionable alteration. As for the genetic alteration of histone H3, 8 genes were mutated in 10 (18.5%) of 54 patients. All 16 mutations included 5 missense mutations, 1 start-lost mutation, and 10 multiple mutations. Interestingly, all 10 patients with LGG with H3-alterations were not found to have a mutation. CGGA, Chinese Glioma Genome Atlas; H3-alteration, histone H3 alteration; IDH, isocitrate dehydrogenase; KPS, Karnofsky performance score; MGMT, O6-methylguanine-DNA methyltransferase; TCGA, The Cancer Genome Atlas; OS, overall survival.
Predictive Value of Histone H3 Alterations in Patients With Diffuse Gliomas
Based on these 2 databases, Kaplan–Meier curves of OS stratified by histone H3 status were shown in Figure 4. Among all 236 patients from CGGA, diffuse gliomas with H3-alteration had a worse prognosis than those with H3-WT (P = .020). However, we did not find a statistically significant difference between these 2 subgroups in the TCGA cohort (P = .191). We further conducted survival analyses on the different patient subgroups stratified by the status of H3-alterations and IDH mutation in CGGA and TCGA databases. For IDH-mutant gliomas from the CGGA and TCGA cohorts, patients with H3-alterations showed a worse survival prognosis than those with H3-WT (Figure 5A and 5B, P = .041 and P = .008, respectively). However, for IDH wild-type gliomas, no significant difference was found in overall survival between these 2 subgroups (Figure 5C and 5D, P = .620 and P = .990, respectively).
FIGURE 4.
Kaplan–Meier curves of overall survival stratified by histone H3 status. A, Overall survival of patients with glioma with H3-alteration was significantly shorter in the Chinese Glioma Genome Atlas cohort (P = .020). B, In the The Cancer Genome Atlas cohort, a downward trend of overall survival with H3-alteration can be seen, but the effect was not statistically significant (P = .191).
FIGURE 5.
Kaplan–Meier curves of overall survival stratified by histone H3 status and IDH status. A, In the CGGA cohort, OS of patients with glioma with IDHmt was significantly different between H3-WT and H3-alteration (P = .041). B, In the TCGA cohort, OS of patients with glioma with IDHmt was significantly different between H3-WT and H3-alteration (P = .008). C, As a contrast, in the CGGA cohort, a statistically significant correlation was not identified in histone H3 status (P = .620). D, In the TCGA cohort, a statistically significant correlation was not identified in histone H3 status (P = .990). CGGA, Chinese Glioma Genome Atlas; H3-alteration, histone H3 alteration; H3-WT, histone H3 wild-type; IDH, isocitrate dehydrogenase; IDHmt, IDH-mutant; IDH-WT, IDH wild-type; OS, overall survival; TCGA, The Cancer Genome Atlas.
COX Regression Analyses of Survival in Patients With IDH-Mutant Gliomas
To further investigate the influence that H3-alterations and other clinical factors may exert on the prognosis of IDH-mutant gliomas, we performed univariate and multivariate COX regression analyses of survival, respectively, in 137 patients from the CGGA database and 337 patients from the TCGA database. 1p/19q codeletion status was missing for 2 of 137 IDH-mutant gliomas in the CGGA cohort. Of the 337 IDH-mutant gliomas in TCGA, 1p/19q codeletion status was missing for only 1 H3 wild-type. MGMT status was missing for 47 of 137 IDH-mutant gliomas in the CGGA cohort. All there unknown MGMT statuses were H3 wild-type in the TCGA cohort.
Of the CGGA cohort, in univariate analysis, preoperative KPS (>80 vs ≤80, HR 2.039, 95% CI 1.142-3.624, P = .016), EOR (HR 0.958, 95% CI 0.945-0.972, P < .001), treatment after surgery (P = .022), midline violation (nonmidline vs midline, HR 3.029, 95% CI 1.598-4.677, P = .001), grade (LGG vs HGG, HR 5.795, 95% CI 2.793-12.023, P < .001), histone H3 status (H3-WT [histone H3 wild-type] vs H3-alteration [histone H3 alteration], HR 1.921, 95% CI 1.017-3.629, P = .044), and 1p/19q codeletion status (noncodel vs codel, HR 0.154, 95% CI 0.069-0.342, P < .001) were all associated with OS in IDH-mutant gliomas (Table 2). In multivariate analysis, preoperative KPS (>80 vs ≤80, HR 2.394, 95% CI 1.257-4.599, P = .008), EOR (HR 0.971, 95% CI 0.957-0.986, P < .001), grade (LGG vs HGG, HR 6.938, 95% CI 2.787-17.269, P < .001), histone H3 status (H3-WT vs H3-alteration, HR 2.482, 95% CI 1.183-4.981, P = .016), and 1p/19q codeletion status (HR 0.169, 95% CI 0.073-0.390, P < .001) were significant prognostic factors. In the TCGA cohort, some parameters (preoperative KPS, EOR, and midline violation) were not included in these analyses because these data were not measured or not disclosed. In univariate analysis, age (HR 1.034, 95% CI 1.009-1.060, P = .008), grade (LGG vs HGG, HR 2.513, 95% CI 1.360-4.647, P = .003), and histone H3 status (H3-WT vs H3-alteration, HR 2.279, 95% CI 1.213-4.280, P = .010) were associated with survival in IDH-mutant gliomas (Table 3). In multivariate analysis, age (HR = 1.034, 95% CI 1.008-1.061, P = .010), grade (low-grade vs high-grade, HR 2.365, 95% CI 1.263-4.427, P = .007), and histone H3 status (wild-type vs alteration, HR 2.501, 95% CI 1.312-4.766, P = .005) were significant prognostic factors for OS. To further test our hypothesis, we selected H3-alterations and other clinical factors for COX regression analyses of survival in patients with IDH wild-type gliomas. We found histone H3 was no longer an independent prognostic factor in 2 cohorts (Supplementary Tables 2 [http://links.lww.com/NEU/D778] and 3 [http://links.lww.com/NEU/D779]). In summary, H3-alteration was particularly proved to be an independent prognostic risk factor for IDH-mutant gliomas from both CGGA and TCGA databases.
TABLE 2.
Univariate and Multivariate Analyses of Survival of IDH-Mutation Glioma From CGGA Database
| Covariates | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age at diagnosis | 0.976 | 0.945-1.008 | .145 | |||
| Preoperative KPS | ||||||
| KPS >80 | 1 | Reference | — | 1 | Reference | — |
| KPS ≤80 | 2.039 | 1.142-3.642 | .016 | 2.394 | 1.257-4.559 | .008 |
| EOR (%) | 0.958 | 0.945-0.972 | <.001 | 0.971 | 0.957-0.986 | <.001 |
| Treatment after surgery | .022 | .132 | ||||
| Chemoradiotherapy | 1 | Reference | — | 1 | Reference | — |
| Radiotherapy | 0.382 | 0.187-0.781 | .008 | 0.745 | 0.342-1.622 | .459 |
| Chemotherapy | 0.431 | 0.075-1.291 | .108 | 0.245 | 0.055-1.049 | .066 |
| None | 0.310 | 0.132-1.403 | .162 | 2.420 | 0.594-9.857 | .217 |
| Midline violation | ||||||
| Nonmidline glioma | 1 | Reference | — | 1 | Reference | — |
| Midline glioma | 3.029 | 1.598-4.677 | .001 | 1.390 | 0.674-2.868 | .373 |
| Grade | ||||||
| Low-grade | 1 | Reference | — | 1 | Reference | — |
| High-grade | 5.795 | 2.793-12.023 | <.001 | 6.938 | 2.787-17.269 | <.001 |
| Histone H3 status | ||||||
| H3-WT | 1 | Reference | — | 1 | Reference | — |
| H3-alteration | 1.921 | 1.017-3.629 | .044 | 2.482 | 1.183-4.981 | .016 |
| 1p/19q codeletion status | ||||||
| Noncodel | 1 | Reference | — | 1 | Reference | — |
| Codel | 0.154 | 0.069-0.342 | <.001 | 0.169 | 0.073-0.390 | <.001 |
| MGMT promoter status | ||||||
| Unmethylated | 1 | Reference | — | |||
| Methylated | 1.406 | 0.636-3.108 | .399 | |||
CGGA, Chinese Glioma Genome Atlas; EOR, extent of resection; H3-alteration, histone H3 alteration; H3-WT, histone H3 wild-type; IDH, isocitrate dehydrogenase; KPS, Karnofsky performance score; MGMT, O6-methylguanine-DNA methyltransferase.
Results in bold represent P < 0.05.
TABLE 3.
Univariate and Multivariate Analyses of Survival of IDH-Mutation Glioma From TCGA Database
| Covariates | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age at diagnosis | 1.034 | 1.009-1.060 | .008 | 1.034 | 1.008-1.061 | .010 |
| Grade | ||||||
| LGG | 1 | Reference | — | 1 | Reference | — |
| HGG | 2.513 | 1.360-4.647 | .003 | 2.365 | 1.263-4.427 | .007 |
| Histone H3 status | ||||||
| H3-WT | 1 | Reference | — | 1 | Reference | — |
| H3-alteration | 2.279 | 1.213-4.280 | .010 | 2.501 | 1.312-4.766 | .005 |
| Treatment after surgery | .272 | |||||
| Chemoradiotherapy | 1 | Reference | — | |||
| Radiotherapy | 0.710 | 0.311-1.619 | .416 | |||
| Chemotherapy | 0.482 | 0.170-1.373 | .172 | |||
| None | 0.126 | 0.184-1.231 | .476 | |||
| 1p/19q status | ||||||
| Noncodel | 1 | Reference | — | |||
| Codel | 0.566 | 0.297-1.079 | .084 | |||
| MGMT promoter status | ||||||
| Unmethylated | 1 | Reference | — | |||
| Methylated | 0.989 | 0.304-3.212 | .985 | |||
H3-alteration, histone H3 alteration; H3-WT, histone H3 wild-type; HGG, high-grade glioma; IDH, isocitrate dehydrogenase; LGG, low-grade glioma; MGMT, O6-methylguanine-DNA methyltransferase; TCGA, The Cancer Genome Atlas.
Results in bold represent P < 0.05.
DISCUSSION
In this research, we discussed the observation of diffuse IDH-mutant glioma with H3-alterations, which had distinctive clinical features, molecular patterns, and survival prognosis. In 2012, genome sequencing studies brought the remarkable finding that cancer-related mutations can occur in the histones themselves.17,18 More studies have expanded the range of malignancies known to harbor mutations in histone H3 to include leukemia, giant cell tumors of the bone, chondroblastoma, pediatric soft tissue sarcoma, and chondrosarcoma.19 Consequently, many studies have reported the genomic landscape of specific H3-alterations in gliomas and their relationship with clinical characteristics and prognosis.20-22 They have mainly focused on the H3.1 and H3.2 replicative histones and H3.3 and CENPA histone variants, while we tried to extend the scope to the whole histone H3 family as much as possible. We proposed that H3-alterations may cause poor outcomes and have particular clinical characteristics in patients with glioma. In the CGGA cohort, amplifications of histone H3 family genes were identified as the most common alterations. It may reveal strong genetic heterogeneity among the histone H3 family genes associated with diffuse glioma in their clinical behavior. Therefore, further investigation of various H3-alterations in genomics and epigenetics is needed in the future.
The findings in our study revealed that patients with genetic alterations in the histone H3 family showed shorter survival and tended to occur in HGGs. The previous studies also reported that H3-alterations were associated with WHO grade and molecular patterns relevant to the biology of gliomas.20 Patients with glioma with IDH mutation have a relatively good prognosis.5,7 However, the IDH-mutant gliomas were often also molecularly heterogeneous at presentation, and in oligodendrogliomas, combined 1p and 19q loss were significantly associated with longer survival. Thus, we assumed that it could be further refined according to the different statuses of histone H3 in patients with glioma. In the study, IDH status was not correlated with H3-alterations in the 2 cohorts. Then, we further stratified patients with diffuse glioma into subgroups according to the status of IDH mutation and H3-alterations. This finding indicated that IDH-mutant gliomas with H3-alteration were more malignant than IDH-mutant gliomas with H3-WT. It suggested that histone H3 status testing could be valuable for developing a future grading approach for IDH-mutant gliomas.
We performed univariate and multivariate analyses to identify clinical characteristics and prognostic variables associated with survival outcomes in patients with IDH-mutant gliomas. In the COX regression analyses of the CGGA cohort, preoperative KPS, EOR, grade, histone H3 status, and 1p/19q codeletion status were independent prognostic factors, as indicated by other studies.23-29 EOR has emerged as one of the most important prognostic factors for gliomas. This also confirms that surgery is the preferred treatment for gliomas. However, we could not validate it using the TCGA cohort because of insufficient data of EOR. Future validation is needed in cohort with more samples. However, H3-alteration was proved to be an independent prognostic factor for patients with IDH-mutant glioma in the CGGA and TCGA cohorts. We also performed univariate and multivariate analyses to further explore the influence of H3-alterations on IDH wild-type gliomas, and we found histone H3 was no longer an independent prognostic factor in 2 cohorts (Supplementary Tables 2 [http://links.lww.com/NEU/D778] and 3 [http://links.lww.com/NEU/D779). The underlying mechanisms, H3-mutation on gliomas with different IDH status and their implications, should be further evaluated in basic research studies.
In our COX multivariate analysis, radiotherapy, chemotherapy, and MGMT methylation did not significantly change the OS in the 2 cohorts. These results suggested that standard radiotherapy or chemotherapy might not be viable treatment options for tumors with these genetic characteristics. However, it is worth noting that radiotherapy is over-represented in both databases (66.1% in the CGGA and 65.4% in the TCGA). As shown in Table 1 and Supplementary Table 1, http://links.lww.com/NEU/D776, the ratio of treatment for different histone H3 status did not reach a statistically significant in both databases (χ2 test, P = .480 and P = .129, respectively). For IDH-mutant gliomas, 81 patients (59.1%) and 172 (51.0%) patients received chemoradiotherapy. There was no study specifically for routine chemoradiotherapy of the different statuses of histone H3 in IDH-mutant gliomas. We need more prospective data so that we can reach to a conclusion. Currently, several histone H3-targeting strategies have emerged as potential treatment paradigms. GD2-targeted CAR-T cells were used in H3K27M glioma orthotopic xenograft models with good results, and clinical trials have shown that it has been well tolerated.30 Another study has reported that H3K27M gliomas are vulnerable to transcriptional disruption using bromodomain inhibition or CDK7 blockade.31 Novel targeted drugs are critical for this specific subtype of patients with malignant gliomas in the future.
Previous studies established the importance of molecular biomarkers in diagnosing and monitoring diffuse gliomas.1,3,4,29,32 As molecular biomarker research advances, stratification of gliomas according to their molecular characteristics will become a critical component of future precision therapy.23-26,29,33,34 Identification and assessment of histone H3 status in clinical practice might help improve prognostic prediction and develop therapeutic strategies for these patient subgroups.
Limitations
There are 3 main limitations in our research. First, a bias exists in patient selection, because all the patients undergoing surgery had a craniotomy in the CGGA cohort. Some patients were eliminated because of unresectable tumors or unavailable for surgery. Second, our study was a retrospective analysis with many samples coming from earlier data. Some molecular biomarkers were not available to ensure the cohort had a sufficient number, so we had to exclude some samples without other molecular biomarkers for analyses. And last, a more in-depth investigation of the mechanisms of H3-alterations and glioma is needed.
CONCLUSION
Our study mainly represents a potentially significant discovery that diffuse IDH-mutant gliomas with H3-alterations are distinguished by unique clinical characteristics, molecular expression profile, and survival prognosis. An understanding of the characteristics associated with this type of tumor may provide insight into successful therapeutic options aimed at prolonging survival and establishing accurate treatment in the future.
Acknowledgments
The authors thank Dr Kun Yao, Ms. Hua Huang, and Ms. Shuqing Sun for tissue sample collection and clinical data retrieval.
Supplementary Material
Footnotes
Supplemental digital content is available for this article at neurosurgery-online.com.
Contributor Information
Cheng Cheng, Email: chenny1990@126.com.
Di Wang, Email: doctorwd750@163.com.
Mingchen Yu, Email: yumc@mail.ccmu.edu.cn.
You Zhai, Email: zhaiyou1994@aliyun.com.
Changqing Pan, Email: 3116862492@qq.com.
Bo Liang, Email: 13436829433@163.com.
Jiazheng Zhang, Email: 942483157@qq.com.
Chen Wang, Email: chenneuro@163.com.
Yiyun Yin, Email: yinyiyun@126.com.
Lianwang Li, Email: ttyyllw@163.com.
Fan Wu, Email: wufan0510284@163.com.
Zhongfang Shi, Email: shizhongfangbj@163.com.
Xing Fan, Email: xingkongyaoxiang@163.com.
Xing Liu, Email: 15846591696@126.com.
Zhiliang Wang, Email: simon910@126.com.
Zheng Zhao, Email: zhaozheng0503@ccmu.edu.cn.
Guanzhang Li, Email: liguanzhang122@163.com.
Funding
This research was funded by grants from the Medical and Health Technology Innovation Project of the Chinese Academy of Medical Sciences (2020-I2M-C&T-A-024), the National Natural Science Foundation of China (82072768), Sino-German Center Cooperation and Exchanges Program (M-0020), and the public welfare development and reform pilot project of Beijing Medical Research Institute (JYY 2022-7) awarded to professor Wei Zhang. This research was also supported by a grant from the China Postdoctoral Science Foundation (2022M712218) awarded to professor Guanzhang Li.
Disclosures
The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
SUPPLEMENTAL DIGITAL CONTENT
Supplementary Data. All patients'clinicopathological information from the CGGA and TCGA databases.
Supplementary Table 1. Patient Demographics and Baseline Characteristics from TCGA database (N = 657).
Supplementary Table 2. Univariate and Multivariate Analyses of Survival of IDH wild-type gliomas from CGGA.
Supplementary Table 3. Univariate and Multivariate Analyses of Survival of IDH wild-type gliomas from TCGA.
REFERENCES
- 1.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R, Smith-Cohn M, Cohen AL, et al. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14(2):284-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molinaro AM, Taylor JW, Wiencke JK, et al. Genetic and molecular epidemiology of adult diffuse glioma. Nat Rev Neurol. 2019;15(7):405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reifenberger G, Wirsching H-G, Knobbe-Thomsen CB, et al. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434-452. [DOI] [PubMed] [Google Scholar]
- 5.Houillier C, Wang X, Kaloshi G, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75(17):1560-1566. [DOI] [PubMed] [Google Scholar]
- 6.Grzendowski M, Wolter M, Riemenschneider MJ, et al. Differential proteome analysis of human gliomas stratified for loss of heterozygosity on chromosomal arms 1p and 19q. Neuro Oncol. 2010;12(3):243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis DN, Giannini C, Capper D, et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639-642. [DOI] [PubMed] [Google Scholar]
- 9.Lucas C-HG, Mueller S, Reddy A, et al. Diffuse hemispheric glioma, H3 G34-mutant: genomic landscape of a new tumor entity and prospects for targeted therapy. Neuro Oncol. 2021;23(11):1974-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armache A, Yang S, Martínez de Paz A, et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature. 2020;583(7818):852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z, Zhang K-N, Wang Q, et al. Chinese Glioma Genome Atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinformatics. 2021;19(1):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wensel CR, Pluznick JL, Salzberg SL, et al. Next-generation sequencing: insights to advance clinical investigations of the microbiome. J Clin Invest. 2022;132(7):e154944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Synhaeve NE, van den Bent MJ, French PJ, et al. Clinical evaluation of a dedicated next generation sequencing panel for routine glioma diagnostics. Acta Neuropathol Commun. 2018;6(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng L, Gong J, Yu T, et al. Diffuse midline gliomas with histone H3 K27M mutation in adults and children: a retrospective series of 164 cases. Am J Surg Pathol. 2022;46(6):863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400-416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226-231. [DOI] [PubMed] [Google Scholar]
- 18.St Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project, Broniscer A McEachron TA et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe BR, Maxham LA, Hamey JJ, et al. Histone H3 mutations: an updated view of their role in chromatin deregulation and cancer. Cancers (Basel). 2019;11(5):660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hervás-Corpión I, Gallardo-Orihuela A, Catalina-Fernández I, et al. Potential diagnostic value of the differential expression of histone H3 variants between low- and high-grade gliomas. Cancers. 2021;13(21):5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Wang Y, Li Y, et al. Role of molecular biomarkers in glioma resection: a systematic review. Chin Neurosurg J. 2020;6(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maze I, Wenderski W, Noh K-M, et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015;87(1):77-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou Z, Zhang K, Liu X, et al. Molecular subtype impacts surgical resection in low-grade gliomas: a Chinese Glioma Genome Atlas database analysis. Cancer Lett. 2021;522:14-21. [DOI] [PubMed] [Google Scholar]
- 24.Rossi M, Gay L, Ambrogi F, et al. Association of supratotal resection with progression-free survival, malignant transformation, and overall survival in lower-grade gliomas. Neuro Oncol. 2021;23(5):812-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garton ALA, Kinslow CJ, Rae AI, et al. Extent of resection, molecular signature, and survival in 1p19q-codeleted gliomas [published correction appears. J Neurosurg. 2021;134(5):1357-1367. [DOI] [PubMed] [Google Scholar]
- 26.Eseonu CI, Eguia F, ReFaey K, et al. Comparative volumetric analysis of the extent of resection of molecularly and histologically distinct low grade gliomas and its role on survival. J Neuro Oncol. 2017;134(1):65-74. [DOI] [PubMed] [Google Scholar]
- 27.Kavouridis VK, Boaro A, Dorr J, et al. Contemporary assessment of extent of resection in molecularly defined categories of diffuse low-grade glioma: a volumetric analysis. J Neurosurg. 2020;133(5):1291-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostrom QT, Cote DJ, Ascha M, et al. Adult glioma incidence and survival by race or ethnicity in the United States from 2000 to 2014. JAMA Oncol. 2018;4(9):1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang T, Nam D-H, Ram Z, et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021;499:60-72. [DOI] [PubMed] [Google Scholar]
- 30.Mount CW, Majzner RG, Sundaresh S, et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat Med. 2018;24(5):572-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagaraja S, Vitanza NA, Woo PJ, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635-652.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803-820. [DOI] [PubMed] [Google Scholar]
- 33.Zachariah MA, Oliveira-Costa JP, Carter BS, et al. Blood-based biomarkers for the diagnosis and monitoring of gliomas. Neuro Oncol. 2018;20(9):1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweha SR, Chung C, Natarajan SK, et al. Epigenetically defined therapeutic targeting in H3.3G34R/V high-grade gliomas. Sci Transl Med. 2021;13(615):eabf7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data. All patients'clinicopathological information from the CGGA and TCGA databases.
Supplementary Table 1. Patient Demographics and Baseline Characteristics from TCGA database (N = 657).
Supplementary Table 2. Univariate and Multivariate Analyses of Survival of IDH wild-type gliomas from CGGA.
Supplementary Table 3. Univariate and Multivariate Analyses of Survival of IDH wild-type gliomas from TCGA.





