Abstract
INTRODUCTION:
We previously reported a lower fecal abundance of Ruminococcus spp., Faecalibacterium prausnitzii, and Coprococcus spp. in nonalcoholic fatty liver disease (NAFLD). In this article, we assess the associations between hepatic gene expression, the specific taxa, and bacterial pathways.
METHODS:
The relationships between hepatic genes that were differentially expressed in patients with NAFLD vs healthy controls (HC) and the abundance of these specific taxa were studied. Inferred functional metagenomic analysis using Piphillin was also performed to investigate associations with bacterial pathways.
RESULTS:
Fifteen patients with NAFLD and 6 HC participated. Of 728 hepatic genes examined, 176 correlated with the abundance of Ruminococcus spp., 138 with F. prausnitzii, and 92 with Coprococcus spp. For Ruminococcus spp., genes were enriched in gene ontology (GO) terms related to apoptotic process, response to external and cytokine stimuli, and regulation of signaling. Several genes related to the Kyoto Encyclopedia of Genes and Genomes pathway insulin resistance were correlated with F. prausnitzii. The hepatic genes associated with F. prausnitzii were enriched in GO terms related to cellular response to different stimuli, apoptotic process, and regulation of metabolic pathways. For Coprococcus spp., only the GO term response to external stimulus was enriched. There was a distinct pattern of associations between hepatic genes and bacterial taxa in NAFLD vs HC. For bacterial pathways, 65 and 18 hepatic genes correlated with bacterial metabolic functions in NAFLD and HC, respectively.
DISCUSSION:
Hepatic gene expression related to insulin resistance, inflammation, external stimuli, and apoptosis correlated with bacterial taxa. Patients with NAFLD showed a higher presence of bacterial pathways associated with lipid metabolism.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) includes simple steatosis (SS) and nonalcoholic steatohepatitis (NASH) and can lead to fibrosis, cirrhosis, and, potentially, hepatocellular carcinoma (HCC). NAFLD has a complex pathogenesis that involves several mechanisms, including insulin resistance (IR) (1) and an altered hepatic gene expression pattern (2) where genes related to molecular processes such as cell adhesion, extracellular matrix, cell motion, and integrin signaling are associated with NAFLD and disease severity (2). In addition, our own group identified differences in hepatic expression of cancer-related genes and retinol metabolism among healthy controls (HC), SS, and NASH (3–5).
Intestinal microbiota (IM) may also influence NAFLD pathogenesis (6,7). Several studies reported dysbiosis in patients with NAFLD (8), with some bacteria associated with disease severity. Among those studies, there were consistent findings of reduced fecal Coprococcus and Faecalibacterium in NAFLD but conflicting data regarding Ruminococcus. We found a reduced abundance of Ruminococcus spp., F. prausnitzii, and Coprococcus spp. in patients with biopsy-proven NAFLD, independent of body mass index (BMI) and IR (9). Coprococcus, F. prausnitzii, and some Ruminococcus species are butyrate-producers (10). Because butyrate plays an essential role in colonocyte metabolism and gut health, a lower abundance of these species may be linked to increased intestinal permeability in NAFLD (11). Intestinal permeability can facilitate the translocation of bacteria and bacterial products resulting in chronic inflammation and IR (8,12), potentially affecting hepatic gene expression (13). Anti-inflammatory and insulin-sensitizing properties have also been reported for F. prausnitzii (10) through the expression of microbial anti-inflammatory molecules (14), all of which could affect NAFLD. This is supported by a recent mouse study showing benefits from oral gavage of F. prausnitzii on liver parameters related to NAFLD with increased insulin sensitivity and adiponectin expression in visceral adipose tissue (15). Two human studies (16,17) also suggest a beneficial effect of F. prausnitzii on metabolic parameters related to NAFLD. Ruminococcus, on the other hand, is a heterogeneous genus, which includes beneficial and deleterious bacteria. This could explain conflicting reports where higher Ruminococcus can be associated with fibrosis severity (6), whereas others found a lower abundance in patients with NASH or NAFLD (9,18).
Very few studies assessed the relationship between the IM and hepatic gene expression. A mouse study showed a close correlation between altered IM and hepatic gene expression, particularly for genes related to pathways involved in the immune response and metabolism during liver regeneration (19). One pediatric study in NAFLD reported a relationship between IM and hepatic gene expression related to bile acid metabolism (20). Finally, in nondiabetic obese women with liver steatosis, molecular networks linking IM and host phenome to hepatic steatosis were reported (13). In that study, several hepatic genes were significantly associated with low microbial gene richness, but no specific taxon was studied. Among the overlapping genes coassociated with hepatic steatosis and low microbial gene richness, lipoprotein lipase was among the most strongly correlated with hepatic steatosis. Short/branched-chain acyl-CoA dehydrogenase and insulin receptor were the most strongly anticorrelated. This suggests a molecular basis for the observation that individuals with low microbial gene richness have a reduced capacity to respond to insulin.
Our goal was to examine potential associations between global hepatic gene expression and specific taxa (Ruminococcus spp., F. prausnitzii, and Coprococcus spp.) that we previously found to be significantly less abundant in patients with NAFLD, independent of BMI and IR. In addition, inferred functional metagenomic analysis and correlation with the hepatic genes and the 3 specific taxa were analyzed.
METHODS
Study design, participants, and data collection
This is a secondary analysis of data from 2 previous cross-sectional studies that compared clinical parameters and IM or hepatic gene expression between adults with NAFLD (SS and NASH) and healthy living liver donors as controls (3,9). All participants who had both IM and hepatic gene expression assessed (7 SS, 8 NASH, and 6 HC) by 16S rRNA gene sequencing and quantitative polymerase chain reaction (Ruminococcus spp., F prausnitzii, and Coprococcus spp.) (9) and microarray profiling (3), respectively, were included. The 16S rRNA gene sequencing data were used here to infer bacterial metabolic functions (see Methods, Supplementary Digital Content, http://links.lww.com/CTG/A767). The 728 genes used in the present analysis are the ones that were differentially expressed among HC, SS, and NASH, defined as at least a 2-fold upregulation or downregulation between at least 2 of these groups (3). These data, in conjunction with the quantitative polymerase chain reaction and gene expression data, were used to identify relationships between the hepatic gene expression and the fecal microbiota. Participants, sample collection, and methods were previously described (3,9) (see Materials/Patients and Methods, Supplementary Material, http://links.lww.com/CTG/A767).
Statistical analysis
Statistical analyses were performed with IBM SPSS Statistics Version 24 (IBM, Armonk, NY) and R software (21). P < 0.05 was considered significant except for correlations where a P < 0.001 was considered significant.
Data availability
Hepatic gene expression data are available from the NCBI gene expression omnibus, Accession No: GSE89632. IM data are not publicly available because of privacy restrictions, but some blinded data may be made available from the corresponding author on request.
RESULTS
Clinical and biochemical characteristics of patients with NAFLD and HC
Clinical and biochemical characteristics of patients with NAFLD and HC are shown in Table S1 (see Supplementary Digital Content, http://links.lww.com/CTG/A767). The results are consistent with those reported previously in the larger study populations (3,9).
Associations between hepatic gene expression and fecal concentration of Ruminococcus spp., F. prausnitzii, and Coprococcus spp.
In total, 176 genes correlated significantly with Ruminococcus spp. (70 negatively and 106 positively), 138 genes with F. prausnitzii (75 negatively and 63 positively), and 92 genes with Coprococcus spp. (41 negatively and 51 positively). See Tables S2–S4, Supplementary Digital Content, http://links.lww.com/CTG/A767 for a full list of correlated genes for each taxon.
A Venn diagram comparing the significantly correlated genes among the 3 bacterial taxa (Figure 1) shows 6 shared genes: RNF43, IFIT2, FOSB, SIPA1L2, SOCS2, and MYADM. Some can be relevant to NAFLD. RNF43 is a negative regulator of the Wnt pathway, which is related to inflammation and cancer (22) and was negatively correlated with all 3 bacterial taxa. SOCS2, which serves as a cytokine‐inducible negative regulator of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway (23), was positively correlated with all 3 bacterial taxa. IFIT2 and FOSB show potential relationship to cell proliferation and cancer growth, whereas SIPA1L2 and MYADM have unknown relevance to NAFLD. Table S5 (see Supplementary Digital Content, http://links.lww.com/CTG/A767) includes the full list of 324 genes included in the Venn diagram.
Figure 1.
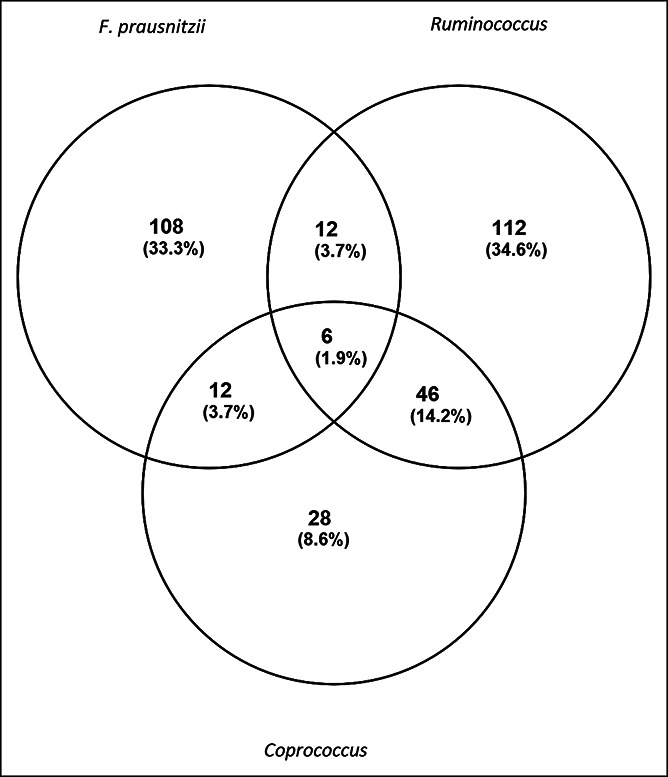
Venn diagram of hepatic genes that were differentially expressed in nonalcoholic fatty liver disease vs healthy controls and were significantly correlated with Ruminococcus spp., F. prausnitzii, and Coprococcus spp.
A heatmap (Figure 2) visualizes the significant positive and negative correlations between the 324 significant genes illustrated in Figure 1 and the 3 bacterial taxa, categorized by disease type (HC, SS, or NASH). It reveals a distinct pattern of associations between genes and bacterial taxa in patients with NAFLD vs HC.
Figure 2.
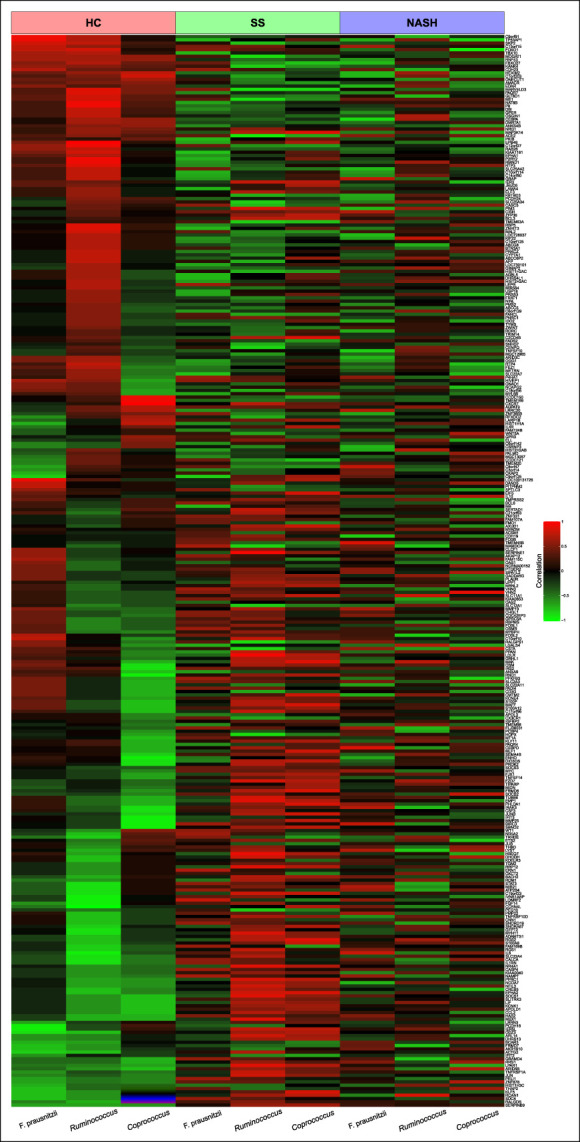
Heatmap illustrating the correlation between gene expression and the 3 bacterial taxa of interest, categorized by disease type. The disease categories (top row: red, HC; green, SS; blue, NASH), the genes (rows), and bacterial taxa (columns). Green highlighted genes are negatively correlated, and red highlighted genes are positively correlated. The intensity of the color indicates the strength of the correlation evaluated by coefficient of correlations. HC, healthy controls; NASH, nonalcoholic steatohepatitis; SS, simple steatosis.
We then looked at the 20 genes showing the strongest positive or negative correlations with each of the 3 bacterial taxa. Ruminococcus spp. was negatively correlated with the expression of genes from 4 terms related to negative regulation of phosphate metabolic process and phosphorylation (4.6- to 5.5-fold enrichment) and cellular response to cytokine stimulus (3.9-fold, P = 0.0022). Based on annotation clustering, the genes most frequently associated with F. prausnitzii were enriched in genes related to cellular response to different stimuli, the apoptotic process, and regulation of metabolic processes.
Genes that correlated significantly with Ruminococcus spp. were enriched in 44 functional annotations, mainly related to the terms “response to lipopolysaccharide” (4.7-fold, P = 0.0370), “positive regulation of transcription from RNA polymerase II promoter” (3.3-fold, P < 0.0001), and “response to cytokine” (3.3-fold, P = 0.0017). Based on annotation clustering, the genes most frequently associated with Ruminococcus spp. were related to the apoptotic process, response to external and cytokine stimuli, and regulation of signaling.
For Coprococcus spp., only the functional annotation “response to external stimulus” was significantly enriched (2.5-fold, P = 0.0170).
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed no enrichment for the genes correlated to any of the 3 bacterial taxa.
Inferred functional metagenomic analysis and its association with hepatic gene expression
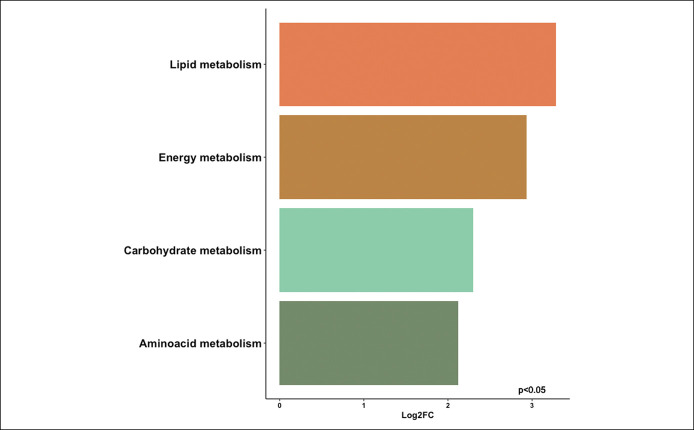
We then performed inferred functional metagenomic analysis using Piphillin, and we identified 4 major bacterial KEGG orthologs related to bacterial lipid, carbohydrate, energy, and amino acid metabolism that were highly abundant in patients with NAFLD compared with HC (Figure 3). The pathways were grouped at KEGG level II classification, next to metabolism (level I classification). The list of pathways is shown in Table S9 (see Supplementary Digital Content, http://links.lww.com/CTG/A767).
Figure 3.
Inferred functional metagenomics by Piphillin demonstrating the Log2fold change (Log2FC) of differentially abundant bacterial metabolic processes (lipid, carbohydrate, energy, and amino acid metabolism) in the microbiome of patients with nonalcoholic fatty liver disease relative to healthy controls (P < 0.05).
Next, we investigated the association between bacterial metabolic functions and differentially expressed hepatic genes. In total, 65 and 18 genes correlated with bacterial metabolic pathways in patients with NAFLD and HC, respectively (Figure 4; see Tables S10 and S11, Supplementary Digital Content, http://links.lww.com/CTG/A767). For example, STAT3 had a positive correlation with bacterial lipid metabolism (rho = 0.698, P value = 0.0103) in NAFLD, and TNFSF10 showed a negative correlation with bacterial carbohydrate metabolism in HC (rho = −0.762, P value = 0.0368). Considering our previous study showing no significant differences in IM between SS and NASH (9) and having here no clear difference in the patterns of associations between hepatic genes and bacterial taxa in SS vs NASH (Figure 2), we did not compare the bacterial metabolic functions of these 2 subgroups.
Figure 4.
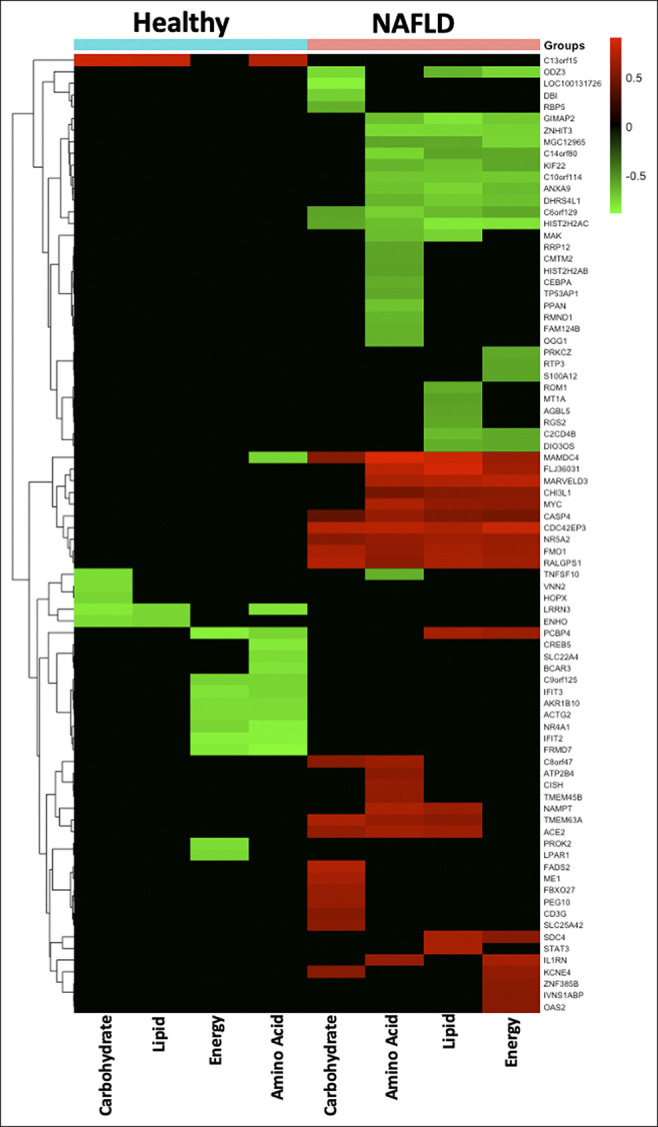
Heatmap illustrating the correlation between gene expression and bacterial functional content, categorized by disease type. The disease categories (top row: cyan, HC; coral, NAFLD), the genes (rows), and bacterial function (columns) are clustered hierarchically. Green highlighted genes are negatively correlated, red highlighted genes are positively correlated, and black color indicates no correlation. HC, healthy controls; NAFLD, nonalcoholic fatty liver disease.
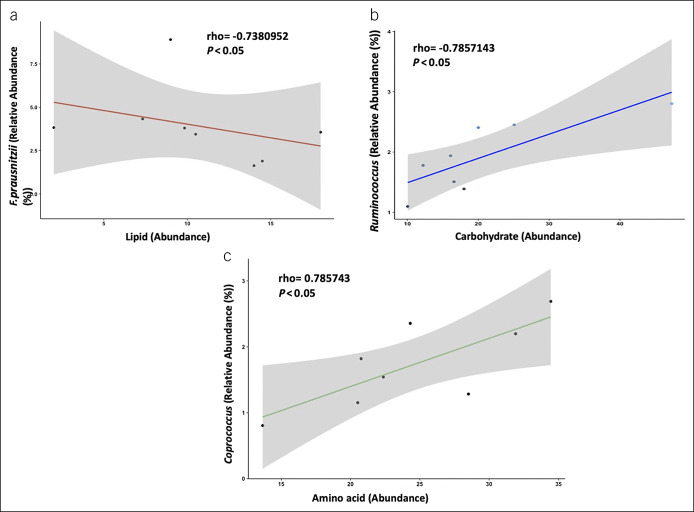
We then looked at correlations between the 3 bacterial taxa and the bacterial functional content. From the correlation analyses, we found, in HC, a notable association between the specific taxa (F. prausnitzii, Ruminococcus, and Coprococcus) and the bacterial KEGG orthologs related to metabolism. F. prausnitzii negatively correlated with lipid metabolism (rho = −0.7380952, P = 0.04583); Ruminococcus and Coprococcus positively correlated with carbohydrate and amino acid metabolism (rho = 0.7857143, P = 0.02793), respectively (Figure 5). However, in NAFLD, no significant correlation was identified between the 3 bacterial taxa of interest and the bacterial function content.
Figure 5.
Significant correlations between the 3 specific bacterial taxa and bacterial pathways involved in metabolic processes for lipid, carbohydrate, and amino acid metabolism in healthy controls. (a) Negative correlation between F. prausnitzii and bacterial lipid metabolism. (b) Positive correlation between Ruminococcus and bacterial carbohydrate metabolism. (c) Positive correlation between Coprococcus and amino acid metabolism. P < 0.05.
DISCUSSION
This study describes associations between hepatic gene expression and 3 fecal bacterial taxa previously found to be associated with NAFLD, independent of IR and BMI (9). Furthermore, we also analyzed the relationship between the hepatic genes, 3 specific taxa, and its predicted functional content. Several genes related to NAFLD that include IR/glucose homeostasis and inflammation at the level of signal transduction correlated with F. prausnitzii abundance. In addition, Ruminococcus spp., F. prausnitzii, and Coprococcus spp. were associated with hepatic genes involved in apoptosis and response to stimuli-like cytokines. A heatmap showed correlation patterns for HC, but there was no clear pattern distinguishing SS from NASH, consistent with our previous observation that the microbiome characteristics in these patients were similar (9). We found that patients with NAFLD had a significantly greater presence of bacterial genes associated with lipid metabolism compared with HC when using inferred functional metagenomics. In addition, the bacterial taxon F. prausnitzii negatively correlated with bacterial gene functions involved in lipid metabolism.
F. prausnitzii is one of the taxa that has the most substantiated relation to NAFLD/NASH (9,18,24). We showed lower abundance of F. prausnitzii in patients with NAFLD compared with HC, which was independent of IR or BMI (9). In this study, hepatic genes that correlated with F. prausnitzii are involved in NAFLD through various mechanisms, including insulin signaling (insulin receptor substrate-1/2), inflammation (IL-6, SOCS3), and their effect on the adipose tissue (leptin receptor) and adipogenesis (sterol regulatory element binding protein-α). This relationship is supported by studies showing that an increase in F. prausnitzii is associated with improvements in IR and other features of the metabolic syndrome (16,17).
SOCS3 is also of interest because it is induced by various cytokines, including IL-6, IL-10, and interferon-gamma. The protein encoded by this gene can bind to JAK2 kinase and inhibit its activity. Deletion of SOCS3 in the liver of mice increased hepatic insulin sensitivity but paradoxically promoted lipogenesis, leading to the development of NAFLD, inflammation, and obesity (25). SOCS3 deletion in the liver can also result in STAT3 hyperactivation and increase fibrosis (26). Therefore, SOCS3 expression can contribute to NAFLD. Other genes that correlated with F. prausnitzii are involved in IR through their effects on signal transduction (PRKCZ, STAT3, and IRS2), further supporting potential beneficial effects of F. prausnitzii on IR. An important finding was the correlation of F. prausnitzii with genes involved in hepatic lipid storage, glucose homeostasis, and IR (NRG1, NR5A2, and NAMPT). Recombinant NRG1 lowers blood glucose and improves insulin sensitivity (27,28). NR5A2 encodes liver receptor homolog, which can also affect liver histology, because a loss of liver receptor homolog can induce large cytosolic lipid droplets, increased triglycerides, macrovesicular steatosis, liver injury, and glucose intolerance (29). Anti-inflammatory and energy homeostasis properties of F. prausnitzii have been identified mainly related to its capacity to secrete or produce anti-inflammatory molecules that contribute to reduce inflammation (30). In addition, short-chain fatty acids have been shown to act as signaling molecules by binding to G-protein–coupled receptor 43 (Gpr43) reducing IR by promoting glucagon-like peptide 1 secretion in the gut (31,32).
The role of Ruminococcus spp. in NAFLD is not yet clear (6,9,18). In this study, genes correlating with Ruminococcus spp. were found to be involved in apoptosis, response to external and cytokine stimuli, and regulation of signaling, which all could play a role in NAFLD. Of particular interest is the negative correlation with AKR1B10, a marker for disease progression and HCC development (2). AKR1B10 is an enzyme involved in hepatic detoxification process and regulating retinoic acid signaling (33,34). A higher expression of AKR1B10 in NASH (2,3,5) may be due to the presence of lipopolysaccharide and oxidative stress (35,36). In obese women receiving a very-low-calorie diet, fecal Ruminococcus was inversely associated with plasma lipopolysaccharide-binding protein reported (37). This suggests that the higher abundance of Ruminococcus spp. may reduce lipopolysaccharide, which in turn may reduce AKR1B10 expression. In addition, our group reported that the higher expression of AKR1B10 in patients with NASH may reduce retinoic acid levels, favoring disease progression (4). Retinoic acid has been implicated in (i) reducing inflammatory processes and promoting a regulatory/anti-inflammatory environment and (ii) maintaining the gut permeability by reducing the systemic translocation of bacterial lipopolysaccharide (38,39).
Further genes related to this term and positively associated with Ruminococcus spp. were Jun proto-oncogene (JUN) and JunB proto-oncogene (JUNB), which is consistent with a suppressed network of JUN/JUNB in the liver of obese NASH patients, suggesting an hepatoprotective role for JUN/JUNB in the pathogenesis of NASH (40).
It is of interest that apoptosis is related to both F. prausnitzii and Ruminococcus spp. in functional annotation clustering. Previous studies have shown that butyrate, a product of F. prausnitzii, can induce apoptosis through mitochondrial death, reactive oxygen species, and caspases (41). Activation of caspases in the liver promotes DNA fragmentation, cytoskeleton remodeling, and protein degradation in the hepatocyte apoptosis, resulting in progression of NAFLD to NASH (42,43).
We identified 6 genes that were significantly correlated with all 3 bacterial taxa, including RNF43 and SOCS2 with a clear connection to NAFLD. RNF43 is a negative regulator of the Wnt pathway, which is related to inflammation and cancer. SOCS2 serves as a cytokine‐inducible negative regulator of the JAK/STAT pathway identified as a regulator of hepatic homeostasis in a diet-induced hepatic steatosis and IR animal model (23). In addition, SOCS2 has been reported to inhibit proliferation and migration in different cancer types, including HCC (44).
Inferred functional metagenomics revealed the presence of enriched bacterial genes associated with lipid, carbohydrate, energy, and amino acid metabolism. Interestingly, F. prausnitzii showed significant negative correlation with lipid metabolism in HC. Lipid metabolism in bacteria is a long-studied topic, but it is still poorly understood. Several lipids not identified until recently were related to Gram-negative bacteria, particularly Escherichia coli, which play an essential role in the pathogenesis and activation of the innate immune response in the host (45).
To understand the relationship between IM and hepatic gene modulation better, we investigated the correlation between bacterial pathways and hepatic genes. This burgeoning field of IM requires further investigation to prove the causality. The above-mentioned hepatic genes (NAMPT, STAT3, PEG10, and FADS2) which were associated with IR, tumor progression, and biosynthesis of long-chain polyunsaturated fatty acids positively correlated with bacterial carbohydrate and lipid metabolism in patients with NAFLD. Furthermore, the tumor suppressor gene TNFSF10 negatively correlated with F. prausnitzii also showed a negative correlation with bacterial carbohydrate metabolism in HC, thus protecting the host.
In summary, our study has examined the relationship between hepatic gene expression and 3 bacterial taxa, Ruminococcus spp., F. prausnitzii, and Coprococcus spp. The associations that were found may contribute to our understanding of potential mechanisms by which the IM contributes to the development and progression of NAFLD. A strength of the study is that all subjects were well-characterized with a liver biopsy and collected clinical parameters. In addition, the inclusion of healthy liver donors as controls with liver biopsies allowed us to study the entire spectrum of liver histology from normal liver through SS and NASH. We also focused on 3 bacterial taxa that we previously showed to be associated with NAFLD, independently of IR and BMI. The limitation is that this is a cross-sectional study, and the analysis is purely correlative, which does not allow us to establish a cause-effect relationship. Nevertheless, the data further support the notion that the IM can affect hepatic gene expression and influence NAFLD pathogenesis through regulation of systemic processes.
In conclusion, in NAFLD, Ruminococcus spp., F. prausnitzii, and Coprococcus spp., previously shown to be associated with NAFLD independent of IR and BMI, correlated with hepatic gene expression related to IR, inflammation, external stimuli, and apoptosis. This study also reveals a higher presence of bacterial pathways associated with lipid metabolism in patients with NAFLD, along with their correlation with hepatic genes involved in IR. Although not causative, these associations suggest a potential link between the IM and hepatic gene expression. Underlying mechanisms and consequences for the NAFLD pathogenesis warrant further investigation.
CONFLICTS OF INTEREST
Guarantor of the article: Johane P. Allard, MD.
Specific author contributions: P.P., B.M.A., and J.P.A. were responsible for the conception and design of the study. All listed authors were involved in the generation, collection, assembly, analysis, and/or interpretation of data and in drafting or revision of the manuscript; all authors approved the final version of the manuscript.
Financial support: This project was funded by the Canadian Institutes of Health Research (NMD-86922, MOP-89705), and the Canadian Liver Foundation. E.M.C. holds the Lawson Family Chair in Microbiome Nutrition Research at the University of Toronto. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential competing interests: None to report.
Clinical trial number: NCT02148471.
Study Highlights.
WHAT IS KNOWN
✓ Altered hepatic gene expression or intestinal dysbiosis was shown to contribute to nonalcoholic fatty liver disease (NAFLD) pathogenesis.
✓ We reported lower fecal abundance of Ruminococcus spp., Faecalibacterium prausnitzii, and Coprococcus spp. in NAFLD.
WHAT IS NEW HERE
✓ We found a distinct pattern of associations between hepatic genes and bacterial taxa in NAFLD vs healthy controls.
✓ Several associations related to insulin resistance and inflammation known to play a role in NAFLD.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A767.
Contributor Information
Paulina Pettinelli, Email: ppettinelli@gmail.com.
Bianca M. Arendt, Email: bianca.arendt@gmail.com.
Katherine J.P. Schwenger, Email: kschweng@uhnresearch.ca.
Saranya Sivaraj, Email: saranya1411@gmail.com.
Elena M. Comelli, Email: mamatha.bhat@uhn.ca.
Wendy Lou, Email: wendy.lou@utoronto.ca.
ACKNOWLEDGMENT
We thank the Princess Margaret Genomics Centre, Toronto, Canada, for the analysis of hepatic gene expression (www.pmgenomics.ca).
REFERENCES
- 1.Khan RS, Bril F, Cusi K, et al. Modulation of insulin resistance in nonalcoholic fatty liver disease. Hepatology 2019;70:711–24. [DOI] [PubMed] [Google Scholar]
- 2.Starmann J, Falth M, Spindelbock W, et al. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One 2012;7:e46584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arendt BM, Comelli EM, Ma DW, et al. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology 2015;61:1565–78. [DOI] [PubMed] [Google Scholar]
- 4.Pettinelli P, Arendt BM, Teterina A, et al. Altered hepatic genes related to retinol metabolism and plasma retinol in patients with non-alcoholic fatty liver disease. PLoS One. 2018;13:e0205747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govaere O, Cockell S, Tiniakos D, et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci Transl Med. 2020;12:eaba4448. [DOI] [PubMed] [Google Scholar]
- 6.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013;58:120–7. [DOI] [PubMed] [Google Scholar]
- 8.Sharpton SR, Ajmera V, Loomba R. Emerging role of the gut microbiome in nonalcoholic fatty liver disease: From composition to function. Clin Gastroenterol Hepatol 2019;17:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Da Silva HE, Teterina A, Comelli EM, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep 2018;8:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014;38:996–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallianou N, Liu J, Dalamaga M. What are the key points in the association between the gut microbiome and nonalcoholic fatty liver disease? Metabol Open 2019:1:9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung C, Rivera L, Furness JB, et al. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol 2016;13:412–25. [DOI] [PubMed] [Google Scholar]
- 13.Hoyles L, Fernandez-Real JM, Federici M, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med 2018;24:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quevrain E, Maubert MA, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut 2016;65:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munukka E, Rintala A, Toivonen R, et al. Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J 2017;11:1667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guevara-Cruz M, Flores-López AG, Aguilar-López M, et al. Improvement of lipoprotein profile and metabolic endotoxemia by a lifestyle intervention that modifies the gut microbiota in subjects with metabolic syndrome. J Am Heart Assoc 2019;8:e012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haro C, Montes-Borrego M, Rangel-Zúñiga OA, et al. Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab 2016;101:233–42. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013;57:601–9. [DOI] [PubMed] [Google Scholar]
- 19.Liu HX, Rocha CS, Dandekar S, et al. Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J Hepatol 2016;64:641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao N, Baker SS, Chapa-Rodriguez A, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2018;67:1881–91. [DOI] [PubMed] [Google Scholar]
- 21.Wolfs MG, Gruben N, Rensen SS, et al. Determining the association between adipokine expression in multiple tissues and phenotypic features of non-alcoholic fatty liver disease in obesity. Nutr Diabetes 2015;5:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suryawanshi A, Tadagavadi RK, Swafford D, et al. Modulation of inflammatory responses by Wnt/beta-catenin signaling in dendritic cells: A novel immunotherapy target for autoimmunity and cancer. Front Immunol 2016;7:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zadjali F, Santana-Farre R, Vesterlund M, et al. SOCS2 deletion protects against hepatic steatosis but worsens insulin resistance in high-fat-diet-fed mice. FASEB J 2012;26:3282–91. [DOI] [PubMed] [Google Scholar]
- 24.Wong VW, Tse CH, Lam TT, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis–a longitudinal study. PLoS One 2013;8:e62885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachithanandan N, Fam BC, Fynch S, et al. Liver-specific suppressor of cytokine signaling-3 deletion in mice enhances hepatic insulin sensitivity and lipogenesis resulting in fatty liver and obesity. Hepatology 2010;52:1632–42. [DOI] [PubMed] [Google Scholar]
- 26.Ogata H, Chinen T, Yoshida T, et al. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene 2006;25:2520–30. [DOI] [PubMed] [Google Scholar]
- 27.Zhang P, Kuang H, He Y, et al. NRG1-Fc improves metabolic health via dual hepatic and central action. JCI Insight 2018;3:e98522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ennequin G, Caillaud K, Chavanelle V, et al. Neuregulin 1 treatment improves glucose tolerance in diabetic db/db mice, but not in healthy mice. Arch Physiol Biochem 2018;126:320–5. [DOI] [PubMed] [Google Scholar]
- 29.Miranda DA, Krause WC, Cazenave-Gassiot A, et al. LRH-1 regulates hepatic lipid homeostasis and maintains arachidonoyl phospholipid pools critical for phospholipid diversity. JCI Insight 2018;3:e96151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenoir M, Martin R, Torres-Maravilla E, et al. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 2020;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolhurst G, Heffron H, Lam YS, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012;61:364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura I, Inoue D, Hirano K, et al. The SCFA receptor GPR43 and energy metabolism. Front Endocrinol (Lausanne) 2014;5:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penning TM. The aldo-keto reductases (AKRs): Overview. Chem Biol Interact 2015;234:236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallego O, Ruiz FX, Ardevol A, et al. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc Natl Acad Sci USA 2007;104:20764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bessone F, Razori MV, Roma MG. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell Mol Life Sci 2019;76:99–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw N, Yang B, Millward A, et al. AKR1B10 is induced by hyperglycaemia and lipopolysaccharide in patients with diabetic nephropathy. Cell Stress Chaperones 2014;19:281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott B, Skurk T, Hastreiter L, et al. Effect of caloric restriction on gut permeability, inflammation markers, and fecal microbiota in obese women. Sci Rep 2017;7:11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadro L, Gamble MV, Vogel S, et al. Retinol and retinol-binding protein: Gut integrity and circulating immunoglobulins. J Infect Dis 2000;182(Suppl 1):S97–S102. [DOI] [PubMed] [Google Scholar]
- 39.Abdelhamid L, Luo XM. Retinoic acid, leaky gut, and autoimmune diseases. Nutrients 2018;10:1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baranova A, Schlauch K, Elariny H, et al. Gene expression patterns in hepatic tissue and visceral adipose tissue of patients with non-alcoholic fatty liver disease. Obes Surg 2007;17:1111–8. [DOI] [PubMed] [Google Scholar]
- 41.Pant K, Yadav AK, Gupta P, et al. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol 2017;12:340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guicciardi ME, Gores GJ. Apoptosis: A mechanism of acute and chronic liver injury. Gut 2005;54:1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanda T, Matsuoka S, Yamazaki M, et al. Apoptosis and non-alcoholic fatty liver diseases. World J Gastroenterol 2018;24:2661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254–70. [DOI] [PubMed] [Google Scholar]
- 45.Ebbensgaard A, Mordhorst H, Aarestrup FM, et al. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front Microbiol 2018;9:2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Hepatic gene expression data are available from the NCBI gene expression omnibus, Accession No: GSE89632. IM data are not publicly available because of privacy restrictions, but some blinded data may be made available from the corresponding author on request.