Abstract
Background:
Accumulating evidence has indicated a possible connection between post-stroke cognitive impairment (PSCI) and gut microbiota imbalance. To further investigate this association, the present work was designed to systematically assess the dissimilarity of gut microbiota between PSCI and healthy individuals or stroke patients.
Methods:
A meta-analysis and systematic review was conducted by searching various databases including PubMed, Web of Science, Embase, VIP, CNKI, and Wangfang for relevant studies. The pooled outcomes were used to estimate the combined dissimilarity of gut microbiota composition between PSCI and healthy individuals or patients with stroke.
Results:
Nine eligible studies were included in this meta-analysis. The results showed that there were no significant changes in observed richness indexes (Chao1 and ACE) and Shannon index. Notably, a significant decrease in Simpson index was observed in PSCI patients in comparison to the healthy individuals (–0.31, 95% CI: –0.62 to –0.01, P = 0.04). Moreover, the microbiota composition at the phylum level (increased abundance of Proteobacteria), family level (increased abundance of Bacteroidaceae, Lachnospiraceae, and Veillonellaceae; decreased abundance of Enterobacteriaceae), and genus level (increased abundance of Bacteroides, Clostridium XIVa, and Parabacteroides; decreased abundance of Prevotella and Ruminococcus) was found to be significantly different between PSCI and controls.
Conclusion:
This meta-analysis suggests a significant shift of observed species and microbiota composition in PSCI compared to healthy individuals or patients with stroke.
Keywords: diversity, gut microbiota, meta-analysis, post-stroke cognitive impairment
1. Introduction
Stroke is one of the leading causes of death and disability worldwide[1] with Post-stroke cognitive impairment (PSCI) encompassing any type of cognitive impairment ranging from mild to severe, which may occur following a stroke event.[2] Recent studies suggest that PSCI is far more prevalent than previously thought; for example, 10-year stroke survivors have an incidence rate of up to 61%.[3] Besides, recurrent ischemic stroke in high-risk patients under adequate pharmacotherapy, such as antiplatelet therapy, could be associated with PSCI. The lesion location and related data are essential for predicting cognitive recovery, indicating the affected neural substrates.[4–6] Furthermore, common risk factors like hypertension and diabetes may increase the chances of PSCI ensuing from stroke.[7] However, due to the lack of scale application, early symptoms are often overlooked, caused by the absence of diagnosis and treatment. Therefore, it is of great importance to identify potential biomarkers for early detection and diagnosis of PSCI, as it usually takes 3 months or more before onset.[8]
Recent research has revealed that the gut microbiota (GM) structure of patients with neuropsychiatric disorders can be compromised.[9,10] Studies have demonstrated that the fecal microbial diversity and composition of Alzheimer Disease patients are significantly different from those of healthy controls.[11] Jiang et al discovered that the GM structure of patients with active severe depression was altered, with a notable elevation of Bacteroidetes, Proteobacteria, Bacteroidetes, and Enterobacteriaceae, and a marked decrease in Firmicutes and Faecalibacterium.[12] Moreover, following stroke, the signature of GM ecological disruption has been identified as an increased abundance of opportunistic pathogenic bacteria and decreased levels of butyrate-producing bacteria.[13] Accumulating evidence suggests that GM plays an important role in cognitive impairment in various disorders,[14–16] and GM has been confirmed as a noninvasive biomarker for disease diagnosis.
Recently, researchers have observed a reduction in the ɑ diversity and abundance of certain microbial populations in patients with PSCI when compared to healthy control subjects or stroke patients.[17–20] However, due to conflicting reports, more investigation is necessary to explore the relationship between the GM and PSCI. To further understand the possible contribution of GM to the development of PSCI, a meta-analysis was conducted to analyze the changes of microbial populations at different levels between PSCI patients and healthy controls or stroke patients. The aim of this study was to evaluate the degree of agreement of the changes in GM and to identify those microbial populations that may serve as potential biomarkers.
2. Materials and Methods
2.1. Search strategy
A rigorous search was conducted until February 2023 in PubMed, Web of Science, Embase, Cochrane databases, Wangfang, VIP and CNKI without language restrictions, using the search terms “stroke,” “cognitive impairment,” and either “gut” or “intestinal,” to identify studies eligible for inclusion in a meta-analysis. The Preferred Reporting Items for Systematic Reviews and Meta-analyses criteria was used as the guideline for this meta-analysis.
2.2. Inclusion and exclusion criteria
Studies relevant to PSCI and GM were identified using the following criteria: comparison of gut microbial composition between PSC and healthy control subjects or stroke patients; availability of adequate data for consolidated comparison; and access to the full-text. Reviews, animal studies, and conference summaries that did not meet these criteria were excluded.
2.3. Data extraction and quality assessment
The authors, date of publication, country, sample size, and detection methods of all studies included in the analysis are presented herein. The data from 16S rRNA-sequencing and the relative abundances of GM were abstracted independently by 2 reviewers, and any disagreements were resolved by consensus. The quality assessment of the studies included was conducted according to the criteria recommended by the Cochrane Non-Randomized Studies Methods Group, with special consideration given to selection, comparability, and outcome details.[21]
2.4. Statistical analysis
All statistical analyses were conducted using STATA SE 15 software, with data extraction executed by WebPlotDigitizer tool. The standardized mean differences (SMD) were applied to compare the richness and diversity indexes, as well as GM between the PSCI and control groups. Heterogeneity was assessed by I2 statistic, and a fixed-effect model was employed when I2 > 50%, otherwise, the random-effect model was used.
3. Results
3.1. Characteristics of included studies
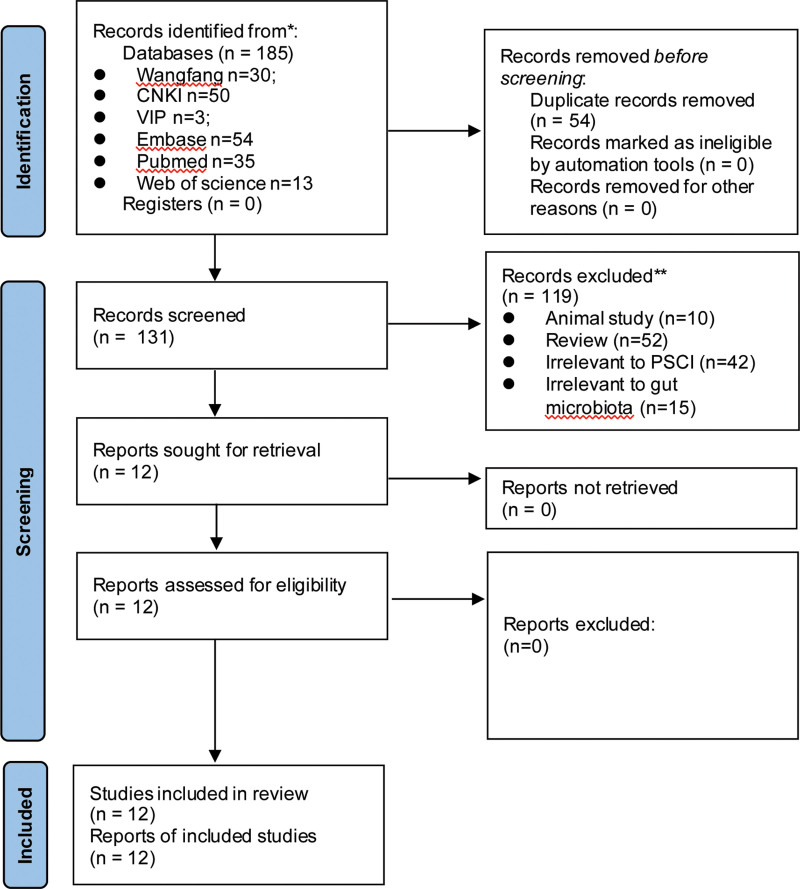
As presented in Figure 1, a total of 185 relevant articles were identified in 6 databases, with 54 of them being duplicates. After a rigorous assessment of titles and abstracts, 119 articles were eliminated due to not meeting the criteria, resulting in 12 eligible studies[17–20,22–29] included in the meta-analysis. Consequently, the analysis included 800 subjects, 456 of whom were suffering from PSCI and 344 in the control group. The pertinent characteristics of the eligible articles are displayed in Table 1. All 12 selected researches were conducted in 6 provinces in China, with 6 of them being published in Chinese. The microbiota of the eligible studies was evaluated with high-throughput sequencing of the V4 region, V3 to V4 region, or the entire length of the 16S rRNA gene. The assessment of the methodological quality revealed that eleven studies had excellent quality[17–20,22–24,26–29] and one had decent quality.[25]
Figure 1.
Flow chart of the search strategy and study selection progress.
Table 1.
Details of the eligible studies in this meta-analysis.
| First author | Yr | Province | Exp. | Ctr. | Total number | cExp. | cCtr. | Male | Female | Smoke | Alcohol | Diabetes | Hypertension | Age | BMI | MoCA | Country | 16S region | Seq. Tech. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yinting Huang | 2021 | Guangdong | PSCI | nPSCI | 56 | 29 | 27 | 34 | 22 | NA | NA | 31.03 | 58.62 | 62.76 | NA | 5.38 | China | V3–V4 | MiSeq Benchtop Sequencer |
| Yongqiang Liu | 2020 | Shanghai | PSCI | nPSCI | 65 | 30 | 35 | 49 | 16 | NA | NA | NA | NA | 64.90 | 23.41 | 17.46 | China | V3–V4 | Illumina MiSeq PE250 |
| Huidi Wang | 2022 | Guangdong | PSCI | nPSCI | 83 | 34 | 49 | 68 | 15 | 44.10 | 32.40 | 35.30 | 70.60 | 61.50 | 23.70 | NA | China | V4 | Illumina iSeq 100 |
| Feng Rongjian | 2021 | Sichuan | PSCI | HC | 47 | 24 | 23 | 26 | 21 | 54.17 | 45.83 | NA | NA | 63.17 | 24.73 | 13.25 | China | Whole 16S region | PacBio Sequel |
| Li Yamei | 2022 | Sichuan | PSCI | nPSCI(HC) | 36 | 12 | 12 (12) | 20 | 16 | NA | NA | NA | NA | 60.75 | 25.20 | 19.92 | China | Whole 16S region | PacBio Sequel |
| Li Yonghua | 2022 | Shandong | PSCI | nPSCI(HC) | 67 | 29 | 18 (20) | 34 | 33 | 27.60 | 17.20 | 37.90 | 44.80 | 68.50 | NA | 13.30 | China | V3–V4 | MiSeq |
| Liu Zhirong | 2021 | Sichuan | PSCI | HC | 47 | 24 | 23 | 26 | 21 | 50.00 | 45.80 | NA | NA | 63.01 | 25.34 | 13.25 | China | Whole 16S region | PacBio Sequel |
| Song Xinna | 2021 | Guangdong | PSCI | nPSCI | 141 | 120 | 21 | NA | NA | NA | NA | NA | NA | NA | NA | NA | China | NA | MiSeq |
| Yi Ling | 2020a | Zhejiang | PSCI | nPSCI | 93 | 53 | 40 | 62 | 31 | 17.00 | 41.50 | 24.50 | 77.40 | 72.20 | 24.70 | 13.70 | China | V3–V4 | MiSeq Benchtop Sequencer |
| Yi Ling | 2020b | Zhejiang | PSCCID | nPSCCID | 66 | 41 | 25 | 31 | 35 | 51.20 | NA | 29.30 | 58.50 | 69.63 | 25.14 | 13.17 | China | V3–V4 | MiSeq Benchtop Sequencer |
| Feng Dan | 2022 | Sichuan | PSCI | HC | 39 | 20 | 19 | 22 | 17 | NA | NA | NA | NA | 61.25 | 24.96 | 12.15 | China | Whole 16S region | PacBio |
| Du Jun | 2022 | Jiangsu | VCIND | HC | 60 | 40 | 20 | 29 | 31 | 35.00 | 42.50 | NA | NA | 61.73 | NA | 22.08 | China | V3–V4 | NA |
BMI = body mass index, cCtr = cases in control group, Ctr = control group, Exp = experimental group, HC = healthy control, MoCA = montreal cognitive assessment, NA = not available, nExp = cases in experiment group, nPSCI = non-PSCI, PSCI = post-stroke cognitive impairment, Seq. Tech = sequencing technique.
3.2. Differences in richness and microbial diversity between PSCI and controls
A meta-analysis of 10 studies was carried out for quantifying the pooled disparities in the general characteristics of high-throughput sequencing between PSCI and control groups. The indices of richness (Chao1, ACE) and alpha diversity (Shannon and Simpson) were then assessed. The summary of the meta-analysis results can be seen in Table 2. A total of 294 individuals from 7 studies were examined to compare the ACE index of 16S rRNA sequencing results between PSCI patients and control groups. A non-significant difference was observed between PSCI and the control group (SMD, 0.10, 95% CI: −0.55–0.74, P = .770) as per the random-effect model, due to high heterogeneity (I2 = 85.8%). When compared to healthy individuals (SMD, −0.3, 95% CI: −1.22 to 0.43, P = .350) or stroke patients (SMD, 0.75, 95% CI: −0.11 to 1.60, P = .08), no considerable variation was found in ACE index of PSCI patients. Data from 8 studies on Chao1 indicated an insignificant difference between PSCI and controls (SMD, −0.31, 95%CI: −0.77 to 0.15, P = .191). Compared to the healthy group, PSCI patients showed lower Chao1 (SMD, −0.57, 95%CI: −0.93 to −0.21, P = .002), though no important discrepancy was noticed when PSCI patients were compared to stroke patients (SMD, 0.04, 95%CI: −0.84 to 0.92, P = .927). Data from ten studies with 485 participants on Shannon demonstrated no considerable variation between PSCI and the control group (SMD, 0.08, 95%CI: −0.31 to 0.47, P = .680), while no remarkable difference was observed either between PSCI patients and healthy individuals (SMD, −0.12, 95%CI: −0.60 to 0.37, P = .639) or stroke patients (SMD, 0.29, 95%CI: −0.35 to 0.94, P = .375). Nine studies provided Simpson index data and the pooled effect was calculated using a fixed-effect model due to the low heterogeneity. The results revealed a significant discrepancy between PSCI and controls (SMD, −0.21, 95%CI: −0.40 to −0.02, P = .034). Compared to the healthy group, PSCI patients had significantly lower Simpson index (SMD, −0.31, 95%CI: −0.62 to −0.01, P = .042), while no considerable variation was seen between PSCI and stroke patients (SMD, −0.12, 95%CI: −0.38 to 0.11, P = .279).
Table 2.
Meta-analysis of diversity of gut microbiota of patients with PSCI.
| Terns | Number of studies | Participants | I2 | P | Effect [95%CI] | z | P |
|---|---|---|---|---|---|---|---|
| ACE | 7 | 294 | 85.8 | <.001 | 0.10 [−0.55, 0.74] | 0.293 | .770 |
| HC | 4 | 167 | 84.9 | <.001 | −0.39 [−1.22, 0.43] | −0.934 | .350 |
| nPSCI | 3 | 127 | 79.5 | <.001 | 0.75 [−0.11, 1.60] | 1.718 | .08 |
| Chao 1 | 8 | 464 | 77.9 | <.001 | −0.31 [−0.77, 0.15] | −1.307 | .191 |
| HC | 4 | 178 | 25.3 | .260 | −0.57 [−0.93, −0.21] | −3.128 | .002 |
| nPSCI | 4 | 286 | 88.3 | <.001 | 0.04 [−0.84, 0.92] | 0.091 | .927 |
| Shannon | 10 | 485 | 76.9 | <.001 | 0.08 [−0.31, 0.47] | 0.412 | .680 |
| HC | 5 | 258 | 68.1 | .014 | −0.12 [−0.60, 0.37] | −0.469 | .639 |
| nPSCI | 5 | 227 | 83.9 | <.001 | 0.29 [−0.35, 0.94] | 0.888 | .375 |
| Simpion | 9 | 530 | 0 | .626 | −0.21 [−0.40, −0.02] | −2.117 | .034 |
| HC | 4 | 178 | 0 | .542 | −0.31 [−0.62, −0.01] | −2.030 | .042 |
| nPSCI | 5 | 352 | 0 | .520 | −0.12 [−0.38, 0.11] | −1.082 | .279 |
HC = healthy control, nPSCI = non-PSCI, PSCI = post-stroke cognitive impairment.
3.3. Differences in the microbial composition
3.3.1. Phylum level.
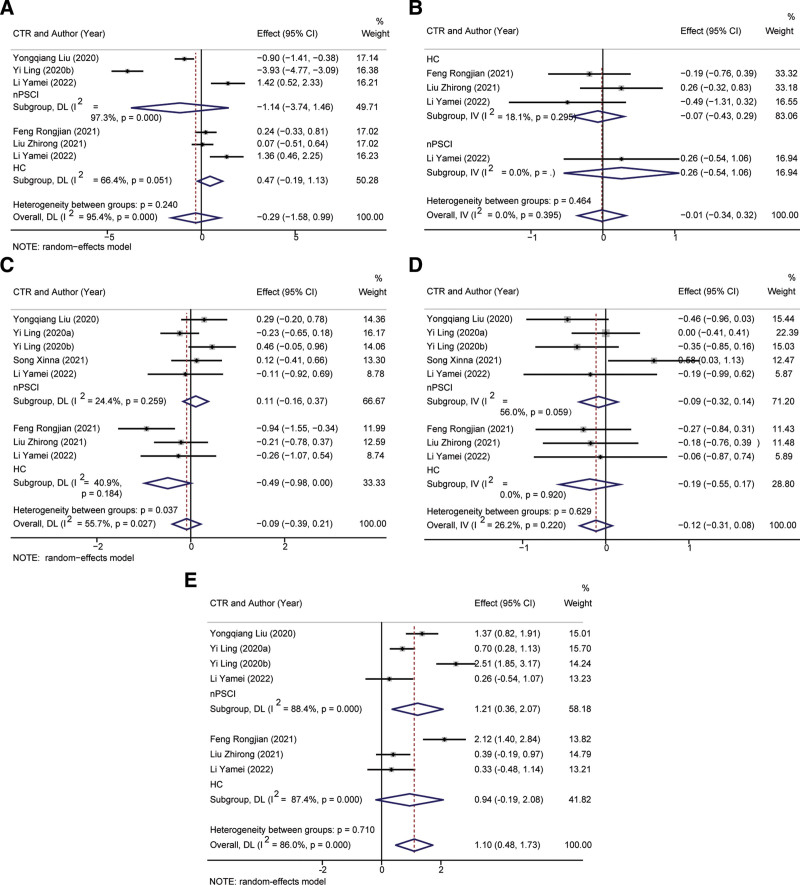
Analysis of Bacteroidetes in 273 individuals revealed no significant difference in abundance between PSCI and control groups (SMD, −0.29, 95%CI: −1.58 to 0.99, P = .653, Fig. 2A). Verrucomicrobiota exhibited significantly increased relative abundance in PSCI compared to control groups (SMD, 0.01, 95%CI: −0.54 to 1.06, P = .934, Fig. 2B). Random-effect remodel was utilized to assess pooled differences in Bacteroidetes between PSCI and controls, with no significant disparity observed (SMD = −0.09, 95%CI: −0.39 to 0.21, P = .566, Fig. 2C). Similarly, no significant difference was determined in relative abundance of Firmicutes between PSCI and controls (Fig. 2D). Seven studies provided data on Proteobacteria abundance, and the meta-analysis indicated a significantly higher relative abundance in PSCI than stroke patients (P = .006) and no significant difference from healthy individuals (Fig. 2E).
Figure 2.
Forest plot of meta-analysis of gut mcrobita in post-stroke cognitive impairment (PSCI) at the phylum levels. (A) Bacteroidetes, (B) Verrucomicrobiota, (C) Bacteroidetes, (D) Firmicutes, (E) Proteobacteria.
3.3.2. Family level.
Table 3 presents the results of a meta-analysis on the levels of family in the GM of PSCI patients. Results indicated that, compared with the control group, there was a significant increase in the abundance of Bacteroidaceae, Lachnospiraceae, and Veillonellaceae in the GM, while the abundance of Enterobacteriaceae was significantly reduced, and the other families showed no significant difference. Further comparisons between the levels of family in the GM of PSCI patients and that of healthy people and stroke patients were made. Results suggested that there was no significant difference between PSCI patients and healthy people in terms of the abundance of the family, while compared with stroke patients, a significant increase in Bacteroidaceae, Lachnospiraceae, and Veillonellaceae, and a significant decrease in Enterobacteriaceae were observed.
Table 3.
Meta-analysis of gut microbiota of PSD at the family levels.
| Gut microbiota | Number of studies | Participants | I2 | P | Effect [95%CI] | z | P |
|---|---|---|---|---|---|---|---|
| Bacteroidaceae | 4 | 289 | 88.0 | <.001 | −1.15 [−1.90, −0.39] | −2.985 | .003 |
| HC | 1 | 71 | 0 | NA | -0.13 [−0.70, 0.44] | −0.452 | .651 |
| nPSCI | 3 | 218 | 75.0 | .018 | −1.47 [−2.06, −0.90] | −4.942 | 0 |
| bifidobacteriaceae | 4 | 289 | 85.0 | <.001 | −0.46 [−1.09, 0.17] | −1.445 | .149 |
| HC | 1 | 71 | 0 | NA | −0.53 [−1.36, 0.30] | −0.885 | .376 |
| nPSCI | 3 | 218 | 89.6 | <.001 | −0.26 [−0.83, 0.31] | −1.245 | .213 |
| Enterobacteriaceae | 4 | 289 | 97.0 | <.001 | 2.91 [0.92, 4.90] | 2.862 | .004 |
| HC | 1 | 71 | 0 | NA | 0.23 [−0.34, 0.80] | 0.788 | .431 |
| nPSCI | 3 | 218 | 91.0 | <.001 | 3.79 [2.34, 5.24] | 5.131 | 0 |
| Lachnospiraceae | 4 | 289 | 38.3 | .182 | −0.57 [−0.81, −0.33] | −4.658 | 0 |
| HC | 1 | 71 | 0 | NA | −0.29 [−0.87, 0.28] | −1.004 | .315 |
| nPSCI | 3 | 218 | 47.3 | .150 | −0.63 [−0.89, −0.36] | −4.664 | 0 |
| lactobacillaceae | 3 | 242 | 97.8 | <.001 | 0.63 [−1.22, 2.48] | 0.672 | .502 |
| HC | 1 | 71 | 0 | NA | 0.04 [−0.53, 0.61] | 0.145 | .884 |
| nPSCI | 2 | 171 | 98.5 | <.001 | 0.84 [−1.79, 3.47] | 0.627 | .53 |
| Prevotellaceae | 4 | 289 | 97.5 | <.001 | 1.47 [−0.47, 3.42] | 1.483 | .138 |
| nPSCI | 4 | 289 | 97.5 | <.001 | 1.47 [−0.47, 3.42] | 1.483 | .138 |
| Ruminococcaceae | 4 | 289 | 68.7 | .023 | 0.11 [−0.32, 0.54] | 0.509 | .611 |
| HC | 1 | 71 | 0 | NA | −0.35 [−0.93, 0.23] | −1.195 | .232 |
| nPSCI | 3 | 218 | 65.7 | .054 | 0.24 [−0.20, 0.69] | 1.074 | .283 |
| streptococcaceae | 3 | 242 | 96.3 | <.001 | 1.35 [−0.08, 2.78] | 1.851 | .064 |
| HC | 1 | 71 | 0 | NA | 0.25 [−0.33, 0.82] | 0.85 | .396 |
| nPSCI | 2 | 171 | 97.1 | <.001 | 1.72 [−0.14, 3.58] | 1.811 | .07 |
| Veillonellaceae | 3 | 242 | 0 | .470 | −0.27 [−0.53, −0.01] | −2.047 | .041 |
| nPSCI | 3 | 242 | 0 | .470 | −0.27 [−0.53, −0.01] | −2.047 | .041 |
CI = confidence interval, HC = healthy control, NA = not available, nPSCI = non-PSCI, PSCI = post-stroke cognitive impairment.
3.3.3. Genus level.
Table 4 presents the results of the meta-analysis at the genus level. Compared to the control group, significantly increased abundances of Bacteroides, Clostridium XIVa, and Parabacteroides were observed in PSCI patients, while significantly decreased abundances of Blautia, Prevotella, and Ruminococcus were detected. In contrast, compared to the healthy group, no significant differences in the abundances of all genera were found in PSCI patients. Additionally, compared to the stroke patients, significantly decreased abundances of Bacteroides, Clostridium XIVa, and Parabacteroides, as well as significantly increased abundances of Prevotella and Ruminococcus were observed in PSCI patients.
Table 4.
Meta-analysis of gut microbiota of PSD at the genus levels.
| Gut microbiota | Number of studies | Participants | I2 | P | Effect [95%CI] | z | P |
|---|---|---|---|---|---|---|---|
| Akkermansia | 4 | 152 | 93.2 | <.001 | −0.28 [−1.66, 1.09] | −0.404 | .686 |
| HC | 2 | 63 | 91.5 | .001 | 0.00 [−1.84, 1.85] | 0.003 | .997 |
| nPSCI | 2 | 89 | 96.3 | <.001 | −0.56 [−3.23, 2.12] | −0.408 | .683 |
| Bacteroides | 9 | 463 | 96.3 | <.001 | 1.43 [0.25, 2.61] | 2.369 | .018 |
| HC | 3 | 112 | 95.9 | <.001 | 0.48 [−1.64, 2.60] | 0.444 | .657 |
| nPSCI | 6 | 351 | 96.8 | <.001 | 1.91 [0.38, 3.43] | 2.453 | .014 |
| Bifidobacterium | 8 | 407 | 96.6 | <.001 | −0.88 [−2.24, 0.48] | −1.266 | .206 |
| HC | 3 | 112 | 93.4 | <.001 | −1.16 [−2.85, 0.53] | −1.345 | .179 |
| nPSCI | 5 | 295 | 97.7 | <.001 | −0.70 [−2.80, 1.39] | −0.658 | .511 |
| Blautia | 5 | 183 | 94.1 | <.001 | −1.98 [−3.54, −0.42] | −2.482 | .013 |
| HC | 3 | 112 | 94.6 | <.001 | −1.75 [−3.81, 0.31] | −1.665 | .096 |
| nPSCI | 2 | 71 | 96.4 | <.001 | −2.33 [−5.80, 1.14] | −1.317 | .188 |
| Clostridium XIVa | 3 | 224 | 92.9 | <.001 | 1.12 [0.01, 2.24] | 1.973 | .048 |
| nPSCI | 3 | 224 | 92.9 | <.001 | 1.12 [0.01, 2.24] | 1.973 | .048 |
| Enterococcus | 3 | 135 | 98.8 | <.001 | −4.20 [−10.80, 2.40] | −1.247 | .212 |
| HC | 2 | 88 | 99.0 | <.001 | −2.31 [−10.09, 5.47] | −0.582 | .561 |
| nPSCI | 1 | 47 | 0 | NA | −8.03 [−9.78, −6.27] | −8.941 | 0 |
| Escherichia/Shigella | 8 | 416 | 94.9 | <.001 | −0.34 [−1.33, 0.66] | −0.666 | .505 |
| HC | 3 | 112 | 97.7 | <.001 | −2.36 [−5.83, 1.12] | −1.329 | .184 |
| nPSCI | 5 | 304 | 90.5 | <.001 | 0.58 [−0.23, 1.38] | 1.406 | .16 |
| Eubacterium | 5 | 183 | 96.9 | <.001 | 1.77 [−0.43, 3.98] | 1.573 | .116 |
| HC | 3 | 112 | 97.0 | <.001 | 0.80 [−1.87, 3.48] | 0.588 | .556 |
| nPSCI | 2 | 71 | 97.8 | <.001 | 3.30 [−2.19, 8.79] | 1.177 | .239 |
| Faecalibacterium | 8 | 424 | 97.8 | <.001 | 1.27 [−0.59, 3.12] | 1.34 | .18 |
| HC | 2 | 73 | 97.6 | <.001 | 1.27 [−2.61, 5.15] | 0.641 | .521 |
| nPSCI | 6 | 351 | 98.2 | <.001 | 1.27 [−1.07, 3.61] | 1.063 | .288 |
| Klebsiella | 5 | 320 | 98.3 | <.001 | −1.56 [−4.11, 0.99] | −1.2 | .23 |
| HC | 1 | 49 | 0 | NA | −6.94 [−8.45, −5.42] | −8.982 | 0 |
| nPSCI | 4 | 271 | 97.5 | <.001 | −6.94 [−8.45, −5.42] | −0.173 | .863 |
| Lachnoclostridium | 4 | 225 | 94.4 | <.001 | −0.28 [−1.42, 0.86] | −0.483 | .629 |
| HC | 1 | 49 | 0 | NA | −0.93 [−1.53, −0.33] | −3.029 | .002 |
| nPSCI | 3 | 176 | 95.0 | <.001 | −0.07 [−1.56, 1.43] | −0.087 | .93 |
| Parabacteroides | 3 | 187 | 4.9 | .350 | 2.30 [1.92, 2.69] | 11.71 | 0 |
| nPSCI | 3 | 187 | 4.9 | .350 | 2.30 [1.92, 2.69] | 11.71 | 0 |
| Prevotella | 4 | 280 | 98.4 | <.001 | −2.60 [−5.19, −0.01] | −1.965 | .049 |
| nPSCI | 4 | 280 | 98.4 | <.001 | −2.60 [−5.19, −0.01] | −1.965 | .049 |
| Roseburia | 3 | 224 | 93.4 | <.001 | 0.63 [−0.47, 1.73] | 1.117 | .264 |
| nPSCI | 3 | 224 | 93.4 | <.001 | 0.63 [−0.47, 1.73] | 1.117 | .264 |
| Ruminococcus | 6 | 359 | 98.2 | <.001 | −2.52 [−4.95, −0.09] | −2.034 | .042 |
| HC | 2 | 73 | 99.2 | <.001 | −1.07 [−11.61, 9.47] | −0.199 | .842 |
| nPSCI | 4 | 286 | 97.7 | <.001 | −3.21 [−5.54, −0.87] | −2.692 | .007 |
| Streptococcus | 7 | 342 | 96.5 | <.001 | −0.51 [−1.88, 0.85] | −0.737 | .461 |
| HC | 3 | 112 | 97.4 | <.001 | −0.66 [−3.65, 2.34] | −0.431 | .666 |
| nPSCI | 4 | 230 | 96.8 | <.001 | −0.42 [−2.11, 1.27] | −0.487 | .626 |
| Subdoligranulum | 5 | 183 | 96.6 | <.001 | 0.62 [−1.44, 2.68] | 0.589 | .556 |
| HC | 3 | 112 | 96.3 | <.001 | −0.81 [−3.21, 1.60] | 1.239 | .215 |
| nPSCI | 2 | 71 | 97.3 | <.001 | 2.83 [−1.65, 7.30] | −0.658 | .511 |
CI = confidence interval, HC = healthy control, NA = not available, nPSCI = non-PSCI, PSCI = post-stroke cognitive impairment.
4. Discussion
Recently, reports have indicated changes in the composition of the GM and disruptions to the intestinal metabolic process in those suffering from PSCI, potentially influencing brain activity through the microbiota-gut-brain axis.[30] Nonetheless, the results of various studies have been discordant. In order to include as much information as possible, we determined the abundance of microbial communities instead of the raw datasets.[31] In this systematic review, we used 12 case-control studies to compare the gut microbial communities of PSCI and healthy individuals, or of stroke patients. We observed a significantly decreased Simpson index, suggesting reduced diversity in the GM of PSCI patients, although the other indices showed no significant variations. At the phylum level, Proteobacteria was found to be significantly more abundant in PSCI patients. At the family level, Bacteroidaceae, Lachnospiraceae, and Veillonellaceae were significantly increased, while Enterobacteriaceae was significantly decreased. At the genus level, Bacteroides, Clostridium XIVa, and Parabacteroides were significantly more abundant in PSCI patients, while Blautia, Prevotella, and Ruminococcus were significantly decreased.
Dynamic alterations in the composition and metabolic byproducts of the gut microbiome are influenced by both internal and external factors. Recent studies have found that it may not only have an indirect impact on cerebral infarcts,[32] but may also directly influence their occurrence, progression and prognosis.[33] This gut microbiome-gut-brain axis, the bi-directional interaction mechanism between the gut microbiome and the brain, encompasses the vagus nerve, endocrine, metabolic and immunologic pathways, and facilitates mutual communication.[34] In 2020, initial reports indicated the changes in the gut microbiome of patients with PSCI, demonstrating a notably modified diversity and relative abundance in comparison to those individuals without cognitive deficits after stroke, thus indicating a possible relationship between gut microbiome dysfunction and PSCI.[26] Nonetheless, the characteristics of the gut microbiome disruptions associated with PSCI remain unknown, prompting further investigation into the changes of the gut microbiome in patients exhibiting early cognitive decline after stroke, and their potential as markers for early recognition, intervention and prognosis of PSCI.
Proteobacteria are present in different parts of the human body, such as the oral cavity, skin, gastrointestinal tract and vagina, with their diverse shapes and physiological functions. Its unique oxygen requirement keeps it in the gastrointestinal environment, and the imbalance of intestinal microecology can lead to an increase in the number of Proteobacteria, which can be observed in post-neonatal gastroenteric diversion, metabolic disorders or obese patients. In addition, Proteobacteria have also been shown to induce insulin resistance and obesity in mouse models, and may constitute a risk factor for cognitive impairment after stroke, and an increase in Proteobacteria can be observed in the intestinal environment of elderly and autistic children. However, different results were obtained in the studies of Alzheimer mouse models regarding the changes of Proteobacteria.
Our study of GM demonstrated a marked transition from a high prevalence of Enterobacteriaceae to a cohort of organisms that is enriched with Bacteroidaceae, which is typically associated with a healthy microbiome.[35] Gram-negative members of the Enterobacteriaceae family are characterized by the presence of lipopolysaccharide in the outer membrane, which can trigger an immune response through Toll-like receptor transduction pathways.[36,37] Inflammatory conditions of the gut can be beneficial for the proliferation of Enterobacteriaceae pathobionts, as they can use host-derived factors for anaerobic respiration and outcompete strictly anaerobic microbes that inhabit the gut.[38] Increase abundance of Enterobacteriaceae can further fuel a dysbiosis, resulting in an inflammatory status of the gut epithelium.[39] Lachnospiraceae family are known for producing beneficial butyrate salts, which can be beneficial for the integrity of intestinal epithelial cells.[40] Deficiencies of Lachnospiraceae may lead to exacerbated intestinal inflammation, increased production of toxins, and impaired intestinal epithelial barrier.[41] It has also been reported that a decrease in Lachnospiraceae abundance is associated with longer duration of Parkinson disease and decreased cognitive ability.[41] On the other hand, a higher abundance of Veillonellaceae has been linked with greater severity of schizophrenia[42] as well as a worse prognosis of cancer immunotherapy.[43]
By analyzing the abundance at the genus level, we can achieve a better understanding of the composition differences of the GM in patients with PSCI, as these genera and species have been widely accepted as having a close correlation with human health. Among them, Bacteroides plays a critical role in the health of the host, with impairment of the normal microecological balance of the host potentially triggering endogenous infections or colitis.[44] Previously, it had been hypothesized that Clostridium XIVa could produce butyrate,[45] which may help suppress systemic inflammatory responses. Parabacteroides merdae has been found to protect against cardiovascular damage through an increase in branched-chain amino acid catabolism.[46] Moreover, Parabacteroides distasonis has been observed to alleviate obesity and metabolic dysfunctions via the production of succinate and secondary bile acids.[47] The lipopolysaccharide derived from Parabacteroides goldsteinii has anti-inflammatory properties and has been reported to significantly ameliorate chronic obstructive pulmonary disease through acting as an antagonist of toll-like receptor 4 signaling pathway.[48] The oral ingestion of Blautia wexlerae has been observed to effect metabolic alterations and anti-inflammatory activities in mice, which has resulted in a reduction of both high-fat diet-induced obesity and diabetes.[49] Additionally, studies have indicated that a lack of gut Ruminococcus can lead to the development of antibiotic-associated diarrhea.[50] Not withstanding, the abundance and functions of different genera or species may vary across different diseases, and although these species’ functions have been partially elucidated, their roles in the development of PSCI remain to be further explored.
Despite the numerous advantages of the present meta-analysis, it has certain limitations. Firstly, there is a marked statistical heterogeneity among the included studies, which is hard to be explained by differences in sample size, geographic area, study methods, etc, due to the small number of studies. Secondly, it is challenging to acquire the primary data from all the included studies, and the application of a digital extraction method could potentially lead to another bias in the findings. Moreover, our study merely addressed the structure and composition of intestinal microbiota, without delving into transcriptome and proteome studies, which could potentially provide a deeper understanding of the functionalities of intestinal microbiota. All these issues should be tackled in future research.
5. Conclusion
In this study, we discovered distinct microbiota distributions in the gut of patients with PSCI compared to healthy controls or stroke patients at the phylum, family and genus levels. The Simpson index was markedly lower in PSCI patients than the control group, without any significant deviation from the norm with regards to richness and diversity indices. Nonetheless, due to the limited sample size and number of studies, as well as considerable disparity among the sampled population, we cannot yet generalize these results to a greater population. To reinforce these findings, further high-quality studies are needed.
Author contributions
Conceptualization: Xiaozhen Hu.
Data curation: Xiaozhen Hu, Yajun Mao, Fang Luo.
Formal analysis: Xiaozhen Hu, Fang Luo.
Funding acquisition: Xiaozhen Hu, Xijun Wang.
Investigation: Xiaozhen Hu.
Methodology: Xiaozhen Hu.
Project administration: Xiaozhen Hu.
Resources: Xiaozhen Hu.
Software: Xiaozhen Hu, Yajun Mao.
Supervision: Xiaozhen Hu, Xijun Wang.
Validation: Xiaozhen Hu, Fang Luo.
Visualization: Xiaozhen Hu, Yajun Mao.
Writing – original draft: Xiaozhen Hu.
Writing – review & editing: Xiaozhen Hu, Xijun Wang.
Abbreviations:
- GM
- gut microbiota
- PSCI
- post-stroke cognitive impairment
- SMD
- standardized mean differences
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The authors have no conflicts of interest to disclose.
This study is funded by Hubei University of Science and Technology Foundation (2016-18X031), and the Chinese Medicine Rehabilitation Service Capacity Improvement Project from the State Administration of Traditional Chinese Medicine 2022.
How to cite this article: Hu X, Mao Y, Luo F, Wang X. Association between post-stroke cognitive impairment and gut microbiota: A PRISMA-compliant systematic review and meta-analysis. Medicine 2023;102:35(e34764).
Contributor Information
Xiaozhen Hu, Email: rehabilitation206@163.com.
Yajun Mao, Email: maoyaj008@136.com.
Fang Luo, Email: 381472047@qq.com.
References
- [1].Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation. 2016;133:e38–360. [DOI] [PubMed] [Google Scholar]
- [2].Verdelho A, Wardlaw J, Pavlovic A, et al. Cognitive impairment in patients with cerebrovascular disease: a white paper from the links between stroke ESO dementia committee. Eur Stroke J. 2021;6:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Delavaran H, Jönsson AC, Lövkvist H, et al. Cognitive function in stroke survivors: a 10-year follow-up study. Acta Neurol Scand. 2017;136:187–94. [DOI] [PubMed] [Google Scholar]
- [4].Munsch F, Sagnier S, Asselineau J, et al. Stroke location is an independent predictor of cognitive outcome. Stroke. 2016;47:66–73. [DOI] [PubMed] [Google Scholar]
- [5].Duering M, Zieren N, Hervé D, et al. Strategic role of frontal white matter tracts in vascular cognitive impairment: a voxel-based lesion-symptom mapping study in CADASIL. Brain. 2011;134(Pt 8):2366–75. [DOI] [PubMed] [Google Scholar]
- [6].Weaver NA, Zhao L, Biesbroek JM, et al. The Meta VCI Map consortium for meta-analyses on strategic lesion locations for vascular cognitive impairment using lesion-symptom mapping: design and multicenter pilot study. Alzheimers Dement (Amst). 2019;11:310–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sun JH, Tan L, Yu JT. Post-stroke cognitive impairment: epidemiology, mechanisms and management. Ann Transl Med. 2014;2:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ballard C, Rowan E, Stephens S, et al. Prospective follow-up study between 3 and 15 months after stroke: improvements and decline in cognitive function among dementia-free stroke survivors >75 years of age. Stroke. 2003;34:2440–4. [DOI] [PubMed] [Google Scholar]
- [9].Bains M, Laney C, Wolfe AE, et al. Vasoactive intestinal peptide deficiency is associated with altered gut microbiota communities in male and female C57BL/6 mice. Front Microbiol. 2689;10:2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nguyen TT, Kosciolek T, Maldonado Y, et al. Differences in gut microbiome composition between persons with chronic schizophrenia and healthy comparison subjects. Schizophr Res. 2019;204:23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhuang ZQ, Shen LL, Li WW, et al. Gut microbiota is altered in patients with alzheimer’s disease. J Alzheimers Dis. 2018;63:1337–46. [DOI] [PubMed] [Google Scholar]
- [12].Jiang H, Ling Z, Zhang Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–94. [DOI] [PubMed] [Google Scholar]
- [13].Yin J, Liao SX, He Y, et al. Dysbiosis of gut microbiota with reduced Trimethylamine-N-Oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc. 2015;4:e002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bajaj JS, Hylemon PB, Ridlon JM, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Carlson AL, Xia K, Azcarate-Peril MA, et al. Infant gut microbiome associated with cognitive development. Biol Psychiatry. 2018;83:148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gao H, Jiang Q, Ji H, et al. Type 1 diabetes induces cognitive dysfunction in rats associated with alterations of the gut microbiome and metabolomes in serum and hippocampus. Biochim Biophys Acta Mol Basis Dis. 2019;1865:165541. [DOI] [PubMed] [Google Scholar]
- [17].Feng RJ, Yu Q, Li Y, et al. Dysbiosis of gut microbiota in patients with post-stroke cognitive impairment. Sichuan Da Xue Xue Bao Yi Xue Ban. 2021;52:966–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huang Y, Shen Z, He W. Identification of gut microbiome signatures in patients with post-stroke cognitive impairment and affective disorder. Front Aging Neurosci. 2021;13:706765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ling Y, Gong T, Zhang J, et al. Gut microbiome signatures are biomarkers for cognitive impairment in patients with ischemic stroke. Front Aging Neurosci. 2020;12:511562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ling Y, Gu Q, Zhang J, et al. Structural change of gut microbiota in patients with post-stroke comorbid cognitive impairment and depression and its correlation with clinical features. J Alzheimers Dis. 2020;77:1595–608. [DOI] [PubMed] [Google Scholar]
- [21].von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61:344–9. [DOI] [PubMed] [Google Scholar]
- [22].Du J. Correlation analysis of gut microbiome with syndrome types and TCM constitutions of vascular cognitive impairment no dementia [dissertation, In Chinese]. Nanjing University Of Chinese Medicine, 2022. [Google Scholar]
- [23].Dan F. Characteristics of gut microbiota and its metabolites short chain fatty acids in patients with post-stroke cognitive impairment and effects of rehabilitation intervention [dissertation, In Chinese]. Southwest Medical University, 2022. [Google Scholar]
- [24].Li Y. Research on brain plasticity and gut microbiota change induced by rTMS in patients with cognitive impairment and depression after stroke [dissertation, In Chinese]. University of Electronic Science and Technology of China, 2022. [Google Scholar]
- [25].Li Y, Lu S. Intestinal flora structure in patients with cognitive dysfunction after stroke. BMU J. 2022;45:5–8. [Google Scholar]
- [26].Liu Y, Kong C, Gong L, et al. The association of post-stroke cognitive impairment and gut microbiota and its corresponding metabolites. J Alzheimers Dis. 2020;73:1455–66. [DOI] [PubMed] [Google Scholar]
- [27].Liu Z. The variation of gut microbiota in cognitive dysfunction after stroke and the influence of rehabilitation intervention [dissertation, In Chinese]. Southwest Medical University, 2021. [Google Scholar]
- [28].Song X. Structural characteristics of gut microbiota and its correlation with metabolism and inflammation in patients with post-stroke cognitive impairment [dissertation, In Chinese]. Southern Medical University, 2021. [Google Scholar]
- [29].Wang H, Zhang M, Li J, et al. Gut microbiota is causally associated with poststroke cognitive impairment through lipopolysaccharide and butyrate. J Neuroinflammation. 2022;19:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhuang X, Xiong L, Li L, et al. Alterations of gut microbiota in patients with irritable bowel syndrome: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2017;32:28–38. [DOI] [PubMed] [Google Scholar]
- [32].Pevsner-Fischer M, Blacher E, Tatirovsky E, et al. The gut microbiome and hypertension. Curr Opin Nephrol Hypertens. 2017;26:1–8. [DOI] [PubMed] [Google Scholar]
- [33].Singh V, Roth S, Llovera G, et al. Microbiota dysbiosis controls the neuroinflammatory response after stroke. J Neurosci. 2016;36:7428–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Carabotti M, Scirocco A, Maselli MA, et al. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. 2015;28:203–9. [PMC free article] [PubMed] [Google Scholar]
- [35].Comstock LE. Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe. 2009;5:522–6. [DOI] [PubMed] [Google Scholar]
- [36].Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11:373–84. [DOI] [PubMed] [Google Scholar]
- [37].Park BS, Song DH, Kim HM, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–5. [DOI] [PubMed] [Google Scholar]
- [38].Winter SE, Winter MG, Xavier MN, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lin A, Zheng W, He Y, et al. Gut microbiota in patients with Parkinson’s disease in southern China. Parkinsonism Relat Disord. 2018;53:82–8. [DOI] [PubMed] [Google Scholar]
- [41].Barichella M, Severgnini M, Cilia R, et al. Unraveling gut microbiota in Parkinson’s disease and atypical parkinsonism. Mov Disord. 2019;34:396–405. [DOI] [PubMed] [Google Scholar]
- [42].Zheng P, Zeng B, Liu M, et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci Adv. 2019;5:eaau8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mao J, Wang D, Long J, et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J Immunother Cancer. 2021;9:e003334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Van den Abbeele P, Belzer C, Goossens M, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Qiao S, Liu C, Sun L, et al. Gut Parabacteroides merdae protects against cardiovascular damage by enhancing branched-chain amino acid catabolism. Nat Metab. 2022;4:1271–86. [DOI] [PubMed] [Google Scholar]
- [47].Wang K, Liao M, Zhou N, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26:222–235.e5. [DOI] [PubMed] [Google Scholar]
- [48].Lai HC, Lin TL, Chen TW, et al. Gut microbiota modulates COPD pathogenesis: role of anti-inflammatory Parabacteroides goldsteinii lipopolysaccharide. Gut. 2022;71:309–21. [DOI] [PubMed] [Google Scholar]
- [49].Hosomi K, Saito M, Park J, et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat Commun. 4477;13:2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gu X, Sim J, Lee WL, et al. Gut Ruminococcaceae levels at baseline correlate with risk of antibiotic-associated diarrhea. iScience. 2022;25:103644. [DOI] [PMC free article] [PubMed] [Google Scholar]