Abstract
INTRODUCTION:
Colorectal cancer (CRC) is a potentially life-threatening complication of long-standing ulcerative colitis (UC). MicroRNAs (miRNA) are epigenetic regulators that have been involved in the development of UC-associated CRC. However, their role as potential mucosal biomarkers of neoplastic progression has not been adequately studied.
METHODS:
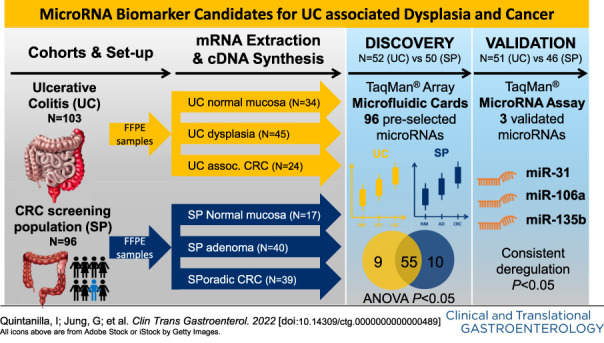
In this study, we analyzed the expression of 96 preselected miRNAs in human formalin-fixed and paraffin-embedded tissue of 52 case biopsies (20 normal mucosa, 20 dysplasia, and 12 UC-associated CRCs) and 50 control biopsies (10 normal mucosa, 21 sporadic adenomas, and 19 sporadic CRCs) by using Custom TaqMan Array Cards. For validation of deregulated miRNAs, we performed individual quantitative real-time polymerase chain reaction in an independent cohort of 50 cases (13 normal mucosa, 25 dysplasia, and 12 UC-associated CRCs) and 46 controls (7 normal mucosa, 19 sporadic adenomas, and 20 sporadic CRCs).
RESULTS:
Sixty-four miRNAs were found to be differentially deregulated in the UC-associated CRC sequence. Eight of these miRNAs were chosen for further validation. We confirmed miR-31, -106a, and -135b to be significantly deregulated between normal mucosa and dysplasia, as well as across the UC-associated CRC sequence (all P < 0.01). Notably, these miRNAs also confirmed to have a significant differential expression compared with sporadic CRC (all P < 0.05).
DISCUSSION:
UC-associated and sporadic CRCs have distinct miRNA expression patterns, and some miRNAs indicate early neoplastic progression.
INTRODUCTION
Inflammatory bowel disease (IBD) has been associated to a higher risk of developing colorectal cancer (CRC) (1,2), and a recent large population-based cohort study concluded that the risk of diagnosis of CRC in patients with IBD has not declined significantly in the past 35 years (3).
Current surveillance strategies aim to identify dysplasia, which is considered a premalignant lesion and thus associated with a high risk to develop CRC. Although there is only evidence from case series and cohort studies, most of the societies recommend 1- to 5-yearly colonoscopy for surveillance starting from 8 to 10 years of disease onset, unless specific risk factors for dysplasia, such as primary sclerosing cholangitis, are also present (4,5).
However, there are several shortcomings when it comes to detect and diagnose dysplasia accurately. First, their low detection rates during surveillance colonoscopy (6,7). In fact, interval cancers are significantly more frequent in IBD-associated cases compared with sporadic cases and are most likely due to not detected or not completely resected dysplastic lesions (8). Second, the poor interobserver agreement among both endoscopists and pathologists to distinguish dysplastic from inflammation-associated morphologic alterations. Third, this poor interobserver agreement is also applicable when differentiating low-grade dysplasia from high-grade dysplasia (9). And fourth, the difficulty in reliably distinguishing colitis-associated dysplasia from sporadic adenomas by histopathologists (10,11).
The difficulties to accurately detect and diagnose dysplasia as the premalignant lesion of colitis-associated cancer have led to the search for more robust, objective, and minimally or noninvasive biomarkers (12). MicroRNAs (miRNAs) are ideal candidates for biomarkers because of their small size, stability in biological samples, availability in blood samples (circulating miRNAs), ability to regulate hundreds of mRNAs, and their relatively small total number compared with mRNAs. Moreover, miRNAs have been linked to pathogenic processes such as inflammation signaling, endothelial-mesenchymal transition, cancer stem cells, and metastatization (13–15).
In UC-associated CRC (UC-CRC), the evidence about the role of miRNAs in inflammation and carcinogenesis is less abundant compared with sporadic CRC (Sp-CRC), but several studies have found promising preliminary results (16–18). Although most of the previous studies had focused on disease-specific expression patterns in both ulcerative colitis (UC) and Crohn's disease, some recent studies aimed to identify differentially expressed miRNAs in UC-CRC (19–22). However, there is only a handful of studies that analyzed single miRNAs associated to UC-CRC, but their sample sizes were rather low, they were not validated with independent cohorts, dysplasia was not considered, or specificity for UC-associated CRC over sporadic CRC was not addressed.
Despite the burgeoning knowledge of miRNAs as regulators in both maintained inflammation and inflammation-associated carcinogenesis, these processes are not fully understood, and more research is needed to find biomarkers for clinical use. Our study aimed to find miRNAs that are differentially and specifically expressed in the colorectal carcinogenesis associated with UC and could serve as biomarkers to improve the detection of UC-associated dysplasia (UC-Dys).
MATERIAL AND METHODS
Patients and sample cohorts
The study group included 103 tissue specimens from 94 patients with UC who were referred for dysplasia screening or diagnosed with a CRC arising on a colitic area and fulfilling all the following eligibility criteria: (i) endoscopically and pathologically confirmed UC proximal to the rectum, (ii) disease duration of at least 8 years, and (iii) absence of clinical activity (i.e., noninvasive 6-point partial Mayo Score (23) <3), to avoid bias introduced by inflammatory processes. To be considered as a UC-associated lesion, dysplasias and CRCs had to be arisen in areas previously affected by chronic inflammation. The control group included 96 tissue specimens from 96 different individuals from the FIT-based organized Barcelona-Eixample-Esquerra population CRC Screening Program, in which all individuals aged 50–69 years are invited to participate, and a FIT cutoff of ≥20 μg of hemoglobin/g of feces is used to indicate a colonoscopy. Personal history of CRC, adenoma, or inflammatory bowel disease, a family history of CRC (defined as those individuals with 2 first-degree relatives with CRC or 1 diagnosed before the age of 60), known hereditary CRC syndromes, severe coexisting illness, colonoscopy performed within the past 5 years, previous colectomy, and contraindication for colonoscopy are considered definitive or temporary exclusion criteria for screening.
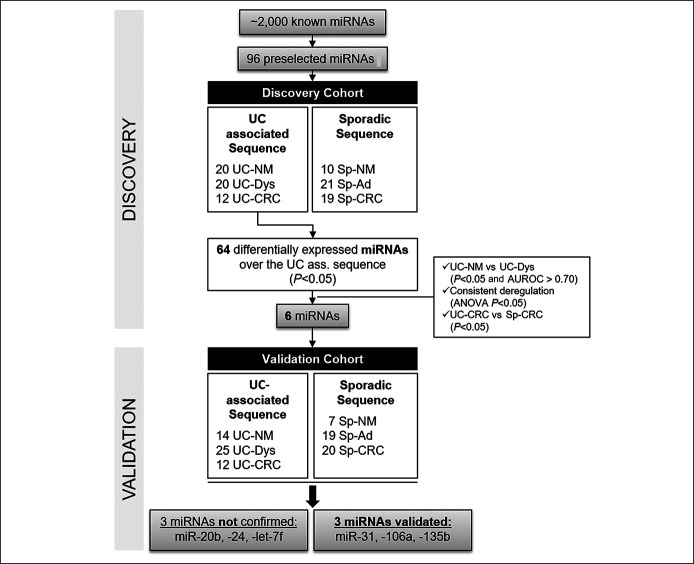
The samples from the study group and the control group were each divided to form 2 independent cohorts: (i) a discovery cohort with 52 samples from the study group (20 UC normal mucosa biopsies [UC-NM], 20 UC-associated dysplastic lesions [UC-Dys], and 12 UC-associated colorectal cancers [UC-CRC]) and 50 samples from the control group (10 normal mucosa biopsies [Sp-NM], 21 sporadic adenomas [Sp-Ad], and 19 sporadic CRC [Sp-CRC]) (Figure 1). All samples derived from different unique patients except for 3 patients of the study group who provided 1 sample of normal mucosa and 1 sample of dysplasia and 2 patients who provided 2 samples of dysplasia. (ii) The validation cohort was composed of 51 samples from the study group: 14 UC-NM, 25 UC-Dys, and 12 UC-CRC and 46 samples from the control group: 7 Sp-NM, 19 Sp-Ad, and 20 Sp-CRC (Figure 1). All samples derived from different patients except for 4 patients who provided 2 samples of dysplasia. Sporadic adenomas in patients with UC were disregarded in this study.
Figure 1.
Study flowchart. AUROC, area under the receiver operating curve; Sp-Ad, sporadic adenoma; Sp-CRC, sporadic colorectal cancer; Sp-NM, sporadic normal mucosa; UC, ulcerative colitis; UC-CRC, ulcerative colitis–associated colorectal cancer; UC-Dys, ulcerative colitis–associated dysplasia; UC-NM, UC-associated normal mucosa.
Tissue samples of normal mucosa, UC-associated dysplasia, and sporadic adenoma were obtained by colonoscopy, whereas CRC specimens derived from surgical blocks. All samples were fixed by immersion in buffered formalin immediately after the colonoscopy or surgery and then processed and embedded in paraffin (FFPE). All specimens were evaluated by pathologists at each participating institution, according to the Vienna classification and the seventh edition of the American Joint Committee on Cancer tumor, node, metastasis grading system. Before RNA extraction, microdissection of relevant tissue was performed by using a biopsy punch device.
RNA extraction from FFPE specimens
RecoverAll Total Nucleic Acid Isolation Kit for FFPE (ref. AM1975, Invitrogen; Thermo Fisher Scientific, Waltham, MA) was used to extract total RNA from tissue cores, according to the manufacturer's protocol. RNA concentration was determined with a NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE).
Quantitative reverse-transcription PCR assays
For the discovery phase, 96 miRNAs were selected after a comprehensive literature and database research based on their previously described functions in cell cycle control, signal transduction, cell-cell interaction, inflammation, and tumorigenesis, as well as their potential biomarker role in colorectal cancer or colitis-associated cancer. We used 96-well Custom TaqMan Array Microfluidic Cards (ref. 4342261; Applied Biosystems, Thermo Fisher Scientific, Waltham, MA) for the evaluation of miRNA expression levels in patient samples. All RNA samples were first reverse-transcribed to cDNA by using the TaqMan MicroRNA Reverse-Transcription Kit (ref. 4366596; Applied Biosystems) according to the manufacturer's instructions. Briefly, 100 ng of RNA was reverse-transcribed in a final volume of 15 μL per reaction under the following conditions: 30 minutes at 16°C, 30 minutes at 42°C, and 5 minutes at 85°C. Then, the cDNA was preamplified for 14 cycles using the TaqMan PreAmp Master Mix (ref. 4488593; Applied Biosystems) combined with the corresponding Custom TaqMan PreAmp primer pool. The preamplified cDNA was either directly processed or stored at −20°C but for no longer than 64 hours. For quantitative real-time polymerase chain reaction (qRT-PCR), each of the 4 microfluidic card's ports were filled with 100 μL of reaction volume from a master mix consisting of 225 μL TaqMan Universal Master Mix II No UNG (ref. 4440048; Applied Biosystems), 4.5 μL of preamplified and 1:4 diluted cDNA (at a concentration of 0.142 ng/μL), and nuclease-free water up to 450 μL. The qRT-PCR was run on a 7900HT Fast Real-Time PCR instrument (ref. 4351405; Applied Biosystems) using the following conditions: hold for 10 minutes at 95°C, then 15 seconds at 95°C, and 1 minute at 60°C for 40 cycles. All qRT-PCR reactions were performed in duplicate for each sample and miRNA assay. Ct values were calculated from automatic threshold. RNU66, RNU48, RNU44, and RNU6b were initially included as endogenous controls for normalization, but data analysis showed that miR-30a-5p, miR-30e, and miR-28 were more stably expressed across all samples and were therefore used as better controls for our study.
TaqMan MicroRNA Assays (ref. 4449142; Applied Biosystems) were used to validate differential miRNA expression in 97 samples by qRT-PCR. RNA was diluted to 4 ng/μL, and 10 ng was used as a template for each reverse-transcription reaction. All qRT-PCR reactions were performed on a ViiA 7 Real-Time PCR instrument (ref. 4453534; Applied Biosystems) in triplicate for each sample and miRNA. Thermal conditions were as follows: hold 10 minutes at 95°C, then 15 seconds at 95°C, and 1 minute at 60°C for up to 50 cycles to detect any expression possible. CT values were calculated from automatic threshold, and only those below 40 were regarded for final analysis. As in the discovery phase, miR-30a-5p, miR-30e, and miR-28 were used as endogenous controls.
Statistical analysis
Statistics of clinical and pathological characteristics were calculated with GraphPad Prism (version 9.1.2). The unpaired Student t test and Fisher exact test were used where applicable for differences between study groups. All statistics of experimental data including all plots were calculated and generated with R software. We did not perform an interplate normalization because the principal component analysis did not show a batch effect. Quantitative RT-PCR results were analyzed by standard 2-class unpaired Welch t test and ANOVA where applicable. Finally, for each selected miRNA, box and whisker plots and receiver operating characteristic (ROC) curves were generated, and the area under the curve (AUC) was analyzed to test the performance of the different models.
RESULTS
Characteristics of the study cohorts
Patients included in the discovery and validation cohorts presented similar distribution of sex and age at study inclusion and diagnosis between all cases and controls. However, for the discovery phase, patients of the study group were slightly younger (52 vs 57 years, P = 0.02), and advanced-stage tumors (III/IV) were twice as frequent in the UC-associated CRC group compared with the sporadic CRC group (41.6 vs 21%, respectively), but which was not statistically significant (Table 1). For a few patients in both study groups, more than 1 sample was available. Precisely, for 3 patients, 1 sample of normal mucosa and 1 sample of dysplasia were available; for 6 patients, 2 samples of dysplasia were available. By contrast, all samples of both control groups came from different patients, resulting in a total of 102 samples from 97 patients in the discovery cohort and 97 samples from 93 patients in the validation cohort. Regarding the group of UC-associated dysplasia, in the discovery cohort, 18 and 2 of 20 were low-grade and high-grade dysplasia, respectively, whereas in the validation cohort, 100% were low-grade dysplasia. All sporadic adenomas were tubular adenomas with low-grade dysplasia except for a single one in the discovery cohort that was a tubulovillous adenoma with high-grade dysplasia (Table 1 and Figure 1).
Table 1.
Clinical characteristics of the patients within the different groups
| Discovery (N = 97) | Validation (N = 93) | |||||
| Study group | Control group | P | Study group | Control group | P | |
| Total patients, n | 47 | 50 | 47 | 46 | ||
| Sex | ||||||
| Female, n (%) | 16 (34) | 23 (46) | 0.30 | 17 (36%) | 23 (50) | 0.21 |
| Age | ||||||
| Mean age at inclusion, yr (SD) | 52 (13.8) | 57 (3.1) | 0.02 | 56 (12.2) | 57 (3.5) | 0.64 |
| Mean age at diagnosis UC, y (SD) | 36 (18.4) | NA | NA | 38 (14.1) | NA | NA |
| UC-associated dysplasia, n | 20 | 25 | ||||
| Low-grade/high-grade, n (%) | 18/2 (90/10) | 25/0 (100/0) | ||||
| Sporadic adenoma, n | 21 | 0.61 | 19 | 0.99 | ||
| Low-grade/high-grade, n (%) | 20/1 (95/5) | 19/0 (100/0) | ||||
| Sex | ||||||
| Female, n (%) | 7 (35) | 9 (43) | 0.75 | 10 (40) | 10 (53) | 0.54 |
| Age | ||||||
| Mean age at inclusion, yr (SD) | 53 (13.0) | 58 (0.8) | 0.08 | 56 (12.9) | 58 (1.2) | 0.51 |
| Mean age at diagnosis UC, yr (SD) | 36 (14.6) | NA | NA | 38 (14.7) | NA | NA |
| Carcinomas, n | 12 | 19 | 12 | 20 | ||
| Sex | ||||||
| Female, n (%) | 3 (25) | 9 (47) | 0.27 | 5 (42) | 9 (45) | 0.99 |
| Age | ||||||
| Mean age at inclusion, yr (SD) | 56.9 (16.4) | 54.4 (2.9) | 0.52 | 60 (12.8) | 55 (2.7) | 0.09 |
| Mean age at diagnosis CRC, yr (SD) | 57.8 (17.5) | 54.4 (2.9) | 0.42 | 58 (12.5) | 55 (2.7) | 0.32 |
| Δt between diagnosis UC and CRC, yr (SD) | 13.5 (11.3) | NA | NA | 14.9 (9.3) | NA | NA |
| TNM, n (%) | ||||||
| In situ | 0 (0) | 6 (31.6) | 0 (0) | 0 (0) | ||
| I | 2 (16.7) | 9 (47.4) | 4 (33.3) | 9 (45) | ||
| II | 4 (33.3) | 0 (0) | 5 (41.7) | 2 (10) | ||
| 0.22 | 0.21 | |||||
| III | 1 (8.3) | 2 (10.5) | 3 (25) | 6 (30) | ||
| IV | 4 (33.3) | 2 (10.5) | 0 (0) | 1 (5) | ||
| Unknown | 1 (8.3) | 0 (0) | 0 (0) | 2 (10) | ||
| Locationa | ||||||
| Proximal/distal, n (%) | 3/8 (25/67) | 2/17 (11/89) | 0.33 | 3/6 (25/50) | 5/15 (25/75) | 0.67 |
| Unknown | 1 (8) | 0 (0) | 3 (25) | 0 (0) | ||
| Adjuvant treatment, n (%) | ||||||
| Yes/no | 5/5 (42/42) | 4/15 (21/79) | 0.20 | 9/3 (75/25) | 11/9 (55/45) | 0.45 |
| Unknown | 2 (16.7) | 0 (0) | 0 (0) | 0 (0) | ||
CRC, colorectal cancer; TNM, tumor, node, metastasis; UC, ulcerative colitis; NA, not applicable. P-values <0.05 are highlighted in bold.
Proximal and distal to the splenic flexure.
MiRNAs are differentially expressed across the colitis-associated cancer sequence
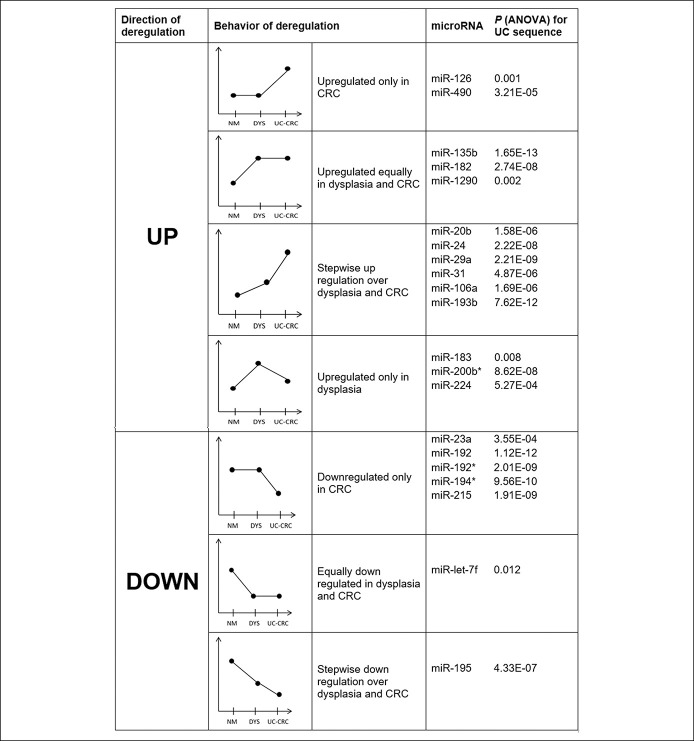
We found 64 miRNAs that were differentially deregulated across the UC-associated CRC sequence, whereas 65 miRNAs were altered in the sporadic CRC sequence (see Supplementary Table S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A803). We compared the 2 sequences of differentially expressed miRNAs (ANOVA P < 0.05) and found that most of the miRNAs were shared between the 2 sequences. However, we also found 9 miRNAs that were only differentially regulated in the UC-associated sequence, whereas 10 were only significant for the sporadic sequence (see Supplementary Table S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A803). We next filtered the significantly deregulated miRNAs by those who showed the largest difference in relative expression levels between normal mucosa (UC-NM) and UC-associated CRC (UC-CRC). For a difference greater than ±1.0, we retrieved 21 miRNAs that were upregulated and 11 that were downregulated (see Supplementary Table S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A803). Interestingly, within each group (i.e., upregulated and downregulated), they showed different types of change in expression level. For instance, within the upregulated group, although some were only overexpressed in the cancer or in the dysplasia with respect to normal mucosa, others showed a consistent upregulation across the UC-associated sequence (Figure 2). We also compared the miRNA expression profiles between UC-CRCs and Sp-CRCs and retrieved 47 miRNAs that showed statistically significant differences between both entities. Thirty of these miRNAs showed a more than 2-fold difference, 9 of which were higher expressed in the UC-associated CRCs, whereas 21 were higher expressed in the sporadic CRCs (Figure 3a; see Supplementary Table S2, Supplementary Digital Content 2, http://links.lww.com/CTG/A804).
Figure 2.
Behavior of selected differentially deregulated miRNAs across the UC-associated CRC sequence in the discovery phase. CRC, colorectal cancer; miRNA, microRNA; UC, ulcerative colitis.
Figure 3.
(a) Heatmap of selected microRNAs differentiating the expression patterns of UC-associated CRC (UC-CRC) in light blue and sporadic CRC (Sp-CRC) in pink. (b) Heatmap of selected microRNAs differentiating the expression patterns of UC-associated dysplasia (UC-DYS) in light blue and normal colonic mucosa of patients with UC (UC-NM) in pink.
miRNAs are potential mucosal biomarkers for dysplasia in patients with ulcerative colitis
When compared with normal colonic mucosa of patients with UC, dysplastic lesions showed a distinct miRNA expression profile (Figure 3b; see Supplementary Table S3, Supplementary Digital Content 3, http://links.lww.com/CTG/A805). In accordance with our main objective of discovering tissue biomarkers for identification of dysplasia associated to UC and which at the same time were highly specific for neoplastic progression associated to UC rather than the sporadic adenoma-carcinoma sequence, we used the following criteria for selecting the miRNAs for the validation phase: (i) miRNAs that showed a significant deregulation between normal mucosa, dysplasia, and cancer, (ii) miRNAs that showed a consistent deregulation across the UC-associated sequence meaning that those miRNAs dysregulated in the dysplasia cases that did not show a dysregulation of higher magnitude in cancer were not regarded as meaningful, (iii) miRNAs with a significantly different expression level in the colitis-associated cancers compared with the sporadic cancers (Figure 3A; see Supplementary Table S2, Supplementary Digital Content 2, http://links.lww.com/CTG/A804), and (iv) miRNAs that showed a differential expression between normal mucosa of patients with UC and UC-associated dysplasia (Figure 3B; see Supplementary Table S3, Supplementary Digital Content 3, http://links.lww.com/CTG/A805). Accordingly, we only considered those miRNAs for which the levels of specificity according to their ROC curves were at least 0.7 or higher when discriminating dysplasia from corresponding normal mucosa.
We identified 6 miRNAs from the discovery phase that fulfilled each of these criteria: miR-20b, -24, -31, -106a, and -135b, all of which were upregulated across the sequence, and miR-let-7f, which was downregulated (Table 2).
Table 2.
Results for discovery phase and validation phase for 8 validated microRNAs (bold = positively validated)
| miRNA | Discovery | Validation | ||||||
| UC-NM vs UC-Dys, Student P | UC-NM vs UC-Dys, AUROC | UC-CRC sequence, ANOVA P | UC-CRC vs Sp-CRC, Student P | UC-NM vs UC-Dys, Student P | UC-NM vs UC-Dys, AUROC | UC-CRC sequence, ANOVA P | UC-CRC vs Sp-CRC, Student P | |
| let-7f | 0.003 | 0.720 | 0.012 | 5.20E-04 | 0.707 | 0.554 | 0.006 | 0.384 |
| miR-20b | 2.92E-04 | 0.820 | 1.58E-06 | 0.005 | 0.016 | 0.734 | 3.10E-06 | 0.110 |
| miR-24 | 1.60E-06 | 0.900 | 2.22E-08 | 0.045 | 0.199 | 0.620 | 0.140 | 7.26E-04 |
| miR-29a | 3.90E-07 | 0.920 | 2.21E-09 | 0.062 | 0.645 | 0.517 | 0.003 | 0.021 |
| miR-31 | 0.002 | 0.760 | 4.87E-06 | 0.020 | 2.64E-04 | 0.774 | 1.90E-06 | 0.015 |
| miR-106a | 6.16E-06 | 0.900 | 1.69E-06 | 0.001 | 0.002 | 0.777 | 0.003 | 0.012 |
| miR-135b | 4.57E-09 | 0.960 | 1.65E-13 | 0.010 | 5.12E-08 | 0.923 | 4.40E-05 | 0.040 |
| miR-195 | 8.42E-04 | 0.800 | 4.33E-07 | 0.611 | 1.45E-04 | 0.831 | 1.40E-04 | 2.44E-04 |
AUROC, area under the receiver operating curve; Sp-CRC, sporadic colorectal cancer; UC-CRC, ulcerative colitis–associated colorectal cancer; UC-Dys, ulcerative colitis–associated dysplasia; UC-NM, ulcerative colitis–associated normal mucosa.
Successful validation of the miRNA biomarkers in the validation cohort
The 6 miRNAs that fulfilled the above conditions were chosen for further validation. Moreover, we decided to include also miR-195 and miR-29a into the analysis because they also correlated well with carcinogenesis and were accurate discriminators for dysplasia, although failed to show statistically significant differences between sporadic and UC-associated cancer. As shown in Table 2 (and Supplementary Table S4, Supplementary Digital Content 4, http://links.lww.com/CTG/A806), we positively validated the expression of 3 of 8 miRNAs: miR-31, -106a, and -135b, all of which were upregulated. Box plots and ROC curves of these 3 miRNAs are shown in Figure 4.
Figure 4.
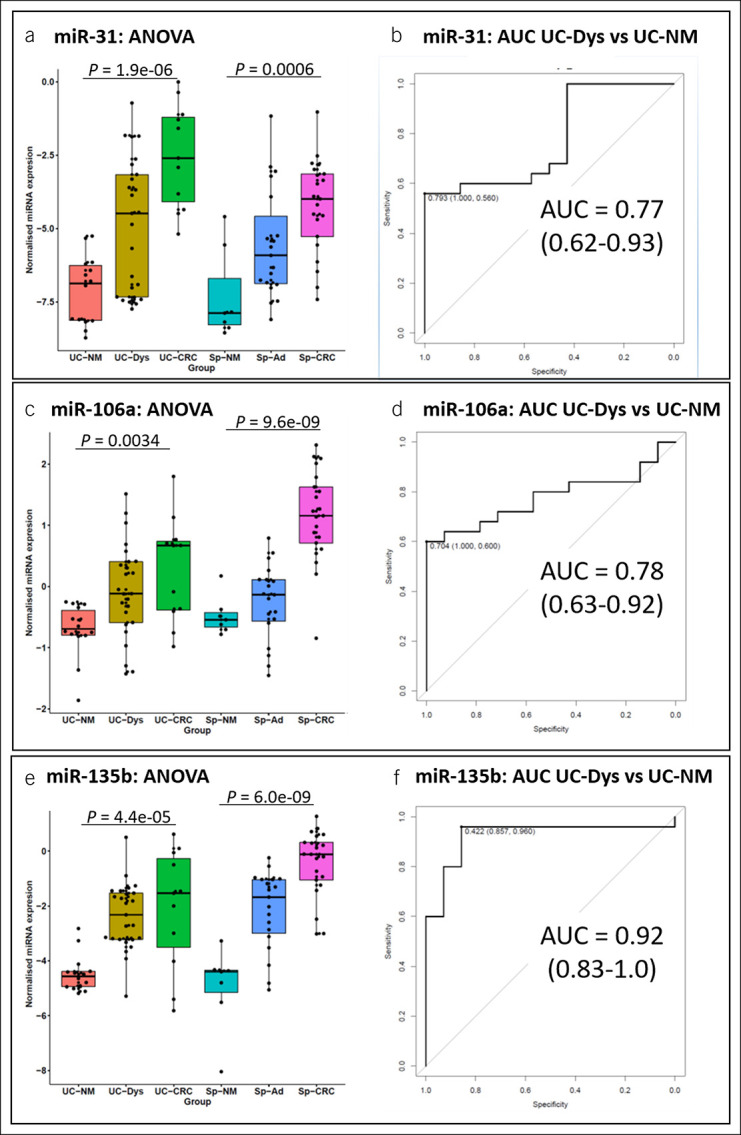
Left: Box and whisker plots for relative expression of validated microRNAs comparing the UC-associated (left) and the sporadic sequence (right): miRNA-31 (a, b), miR-106a (c, d), and miR-135b (e, f). Right: Sensitivities, specificities, and receiver operating curve for discriminating between normal mucosa and dysplasia of the ulcerative colitis cases. Sp-Ad, sporadic adenoma; Sp-CRC, sporadic CRC; Sp-NM, sporadic normal mucosa; UC-CRC, UC-associated colorectal cancer; UC-Dys, UC-associated dysplasia; UC-NM, ulcerative colitis–associated normal mucosa.
Contrarily, miR-195 showed a statistically significant downregulation in dysplasia (P = 0.0002), but in the UC-CRC group, it was expressed at the same level as in normal mucosa (P = 0.65), results not consistent with those of the discovery phase (see Supplementary Figures S5, Supplementary Digital Content 5, http://links.lww.com/CTG/A807). On the other hand, miR-20b showed a consistent and statistically significant deregulation across the UC-associated and sporadic CRC sequences in both the discovery and validation cohort, but in the latter, it failed to show significant differences between the UC-CRC and Sp-CRC cases (P = 0.11) (Table 2). Finally, miR-24 did not show any differences across the UC-associated CRC sequence, and miR-29a and miR-let-7f did not show differences between normal mucosa and dysplasia (see Supplementary Table S4, Supplementary Digital Content 4, http://links.lww.com/CTG/A806 and Supplementary Figures S5, Supplementary Digital Content 5, http://links.lww.com/CTG/A807).
DISCUSSION
CRC is still a severe and life-threatening complication of long-standing UC; thus, screening and early treatment of premalignant lesions are decisive to decrease morbidity and mortality. Current surveillance strategies aim to identify dysplasia by periodic colonoscopies; however, there are important difficulties to detect and diagnose dysplasia accurately, and colonoscopy is still an invasive procedure, costly, and uncomfortable for patients. Therefore, there is a substantial need to find new biomarkers that can predict dysplasia or cancer noninvasively.
miRNAs represent a crucial part in the complex network of epigenetic regulation and have become potential biomarkers for UC-CRC. However, there is still a need to improve the diagnostic performance based on the detection of miRNAs species and validate the findings. Indeed, there have been previous attempts to distinguish IBD related to sporadic colonic neoplasia with some success, but observations are not consistent. Histological differentiation of colitis-associated CRC from sporadic CRC is not possible. Commonly, studies have considered those CRC arising in an area with known previous inflammation as colitis-associated CRC (24). Also, colitis-associated CRC is often labeled at biobanks as CRC without including any specification of their relationship with colitis, which makes their identification and differentiation from sporadic CRC difficult. We addressed this limitation in our study by including as UC-CRC only those cases that occurred in patients with known longstanding UC and that raised in areas with previous colitis (i.e., a proximal CRC in a patient with distal longstanding UC would not have been included in the study). Moreover, to ensure that our results would not be biased toward inflammation, we only included dysplasias and CRCs from patients with UC in clinical remission. We cannot exclude, however, that there was some degree of microscopic inflammation in some of the samples, but we believe that the impact was minimal. On the other hand, we recruited as sporadic CRCs only those cases diagnosed through a population-based CRC screening program in asymptomatic individuals where patients with IBD are specifically excluded.
One major strength of our study is that we mirror the pathophysiologic cascade of carcinogenesis of both the UC-CRC and Sp-CRC sequence by analyzing samples of the intermediate steps, i.e., dysplasia for UC-CRC and sporadic adenoma for Sp-CRC. By doing so, we found sixty-4 differentially deregulated miRNAs for the UC-CRC sequence, which is comparable with previous studies (20–22,25). We observed that most miRNAs were equally deregulated across the UC-CRC and Sp-CRC sequence, whereas some miRNA species were exclusively deregulated across the UC-CRC sequence, for instance, downregulated miR-192 and upregulated miR-126, indicating that although both processes share a wide range of common features, they have indeed distinct miRNA expression patterns. We also found miRNAs that were associated with both UC-CRC and Sp-CRC development but on statistically significant different levels (e.g., miR-31 being higher expressed in the UC-CRC sequence), which might reflect a different relevance for the carcinogenesis such as faster progression or higher grade of invasion or because of the underlying inflammation in UC-CRC. Hence, another strength of our study was that we considered these significantly different levels as one of the relevant criteria to select miRNAs for further validation.
The results of our study also suggest that miRNAs are linked to different stages of carcinogenesis. For example, miR-126 and -490 were only upregulated in cancer and not in dysplasia, suggesting they are late events, whereas others (e.g., miR-20b, -31, and -106a) seemed to be early events because they were already upregulated in the dysplasia yet on a lower level than in the cancer. We hypothesize that those miRNAs altered early during carcinogenesis and that maintain or further enhance this deregulation in cancer are of most interest as biomarkers because they could be an early predictor for a higher risk to develop cancer. In this study, more than 90% of UC-associated dysplasia were classified as low grade, whereas 95% of adenomas were tubular adenomas with low-grade dysplasia. Hence, the number of samples of high-grade dysplasia was too low to perform a subgroup analysis.
Our main goal was to find miRNAs to be as specific as possible for colitis-associated dysplasia that have a high potential for moving on to cancer. Therefore, we applied very restrictive criteria to identify candidate mucosal miRNA biomarkers that would not only accurately discriminate dysplasia from normal mucosa but also indicate a higher risk of CRC and are differentially expressed in UC-CRC compared with Sp-CRC. Despite this narrow definition, we achieved to identify 6 candidates, 3 of which were positively validated in an independent cohort. All 3 validated miRNAs (i.e., miR-31, -106a, and -135b) have been previously described for various cancer types including CRC and have been associated with different cancer-related pathways such as RAS signaling for miR-31 (26) and Wnt/β-catenin, PI3K/AKT, TGFBR2, and PTEN for miR-135 (27–30). Most intriguingly, miR-31 has been linked to IBD-associated CRC (21,31). On the other hand, miR-106a, whose role in cancer cell proliferation, migration, and invasion has been widely described for CRC (32), has also been proven to distinguish between CD and UC as well as to classify indeterminate IBD, but our findings also suggest a novel role of this miRNA in IBD-associated carcinogenesis (33). MiR-135, which has been proposed as a potential noninvasive biomarker in stool for sporadic CRC and advanced adenoma, has not shown any diagnostic value in patients with IBD so far, and therefore, its association with UC-CRC is a truly novel finding of our study (34).
Despite our promising results, we also acknowledge some limitations of our study. First, the number of samples is still limited; nevertheless, we were able to identify numerous highly relevant biomarker candidates and validated some of them. Second, the design of the study was retrospective, and the samples in the UC-CRC sequence were mostly derived from different patients, which does not allow inferring causality. Third, we did not correlate the results to clinical data, such as grade of inflammation, tumor stage, or disease-free survival, because these data were either not available or the retrospective design did not allow its usage.
In our opinion, the ideal future perspective for miRNAs as biomarkers is their utility as a nonminimally or minimally invasive screening method in blood, stool, or rectal biopsy to identify patients with a high chance of having premalignant or cancerous lesions. If tested positive, in a next step, patients would undergo colonoscopy-driven biopsies or resections to further evaluate and stratify according to individual risk profiles and better rationalize surveillance and treatment strategies.
Our study revealed differentially expressed miRNAs for UC-associated dysplasia and cancer. Most importantly, some of these were found to be specific for colitis-associated compared with sporadic colorectal cancer. They were also further validated in an independent cohort. Based on our and previous results, future research should focus on developing and evaluating noninvasive miRNA panels until they are robust enough as diagnostic tools before entering further validation on a large scale in clinical settings.
DATA AVAILABILITY STATEMENT
The data presented in this study are available in the supplementary tables.
CONFLICTS OF INTEREST
Guarantor of the article: Maria Pellisé, PhD.
Specific author contributions: Conceptualization, M.P., G.J., and I.Q.; methodology, M.P., I.Q., and J.C.; software, J.J.L. and J.S.; validation, G.J., I.Q., and J.J.L.; formal analysis, G.J., I.Q., J.J.L., J.S., and J.C.; investigation, G.J., I.Q., M.C., and J.C.; resources, L.B., M.I.V. E.Q., M.C., M.A., A.C., J.P., and E.R.; data curation, G.J., I.Q., M.J., S.C., and L.M.; writing—original draft preparation, G.J.; writing—review and editing, all authors; visualization, G.J.; supervision, L.M., E.R., F.B., and M.P.; project administration, I.Q. and M.P.; and funding acquisition, M.P. All authors have read and agreed to the published version of the manuscript.
Financial support: This work was funded by grants from the Instituto de Salud Carlos III (PI12/01481; PI19/01050). Project PI19/01050 is funded by Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union. CIBEREHD is funded by the Instituto de Salud Carlos III and Beca Marató de TV3 (201932-30). Parts of this work were also supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d’Oncología de Catalunya (XBTC) and by the Hospital Clínic's Premi Fi de Residència (G.J). None of the funding parties has been involved in collection, analysis, and interpretation of the data.
Potential competing interests: F.B. and M.C. declare that they have received an honorarium for consultancy from Sysmex and Elsevier and speaker's fees from Norgine. M.P. declares that she has received research grants from Fujifilm, consultancy fee from Norgine, and speaker's fee from Olympus, Norgine, Casen Recordati, and Janssen. All other authors have nothing to disclose.
Ethical considerations: The study was conducted according to the guidelines of the Declaration of Helsinki and approved by our Institutional Ethics Committee (CEIC) of Hospital Clínic of Barcelona by May 10 of 2012 (registered 2012/7567). Informed consent for surgical procedures, colonoscopy, biopsy, analysis of specimens, and participation in the study was obtained from all subjects involved in the study.
Study Highlights.
WHAT IS KNOWN
✓ Colorectal cancer (CRC) is a feared complication of ulcerative colitis (UC).
✓ Its prevention is based on endoscopic detection of dysplasia, but this strategy has important limitations.
✓ MicroRNAs (miRNAs) are important regulators of gene expression and are ideal biomarker candidates.
✓ miRNAs have been associated with sporadic and UC-associated CRC.
WHAT IS NEW HERE
✓ Sporadic and UC-associated CRCs have distinct miRNA expression patterns.
✓ miRNAs are potential biomarkers for dysplasia in patients with UC.
✓ Some miRNAs show a gradual increase during neoplastic progression in UC.
✓ MiR-31, -106a, and -135b have been validated as potential mucosal biomarker candidates for neoplastic progression.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A803, http://links.lww.com/CTG/A804, http://links.lww.com/CTG/A805, http://links.lww.com/CTG/A806, and http://links.lww.com/CTG/A807.
Isabel Quintanilla and Gerhard Jung contributed equally to this work.
Contributor Information
Isabel Quintanilla, Email: isabel.quintanilla@nih.gov.
Gerhard Jung, Email: jung@clinic.cat.
Mireya Jimeno, Email: mijira11@hotmail.com.
Juan José Lozano, Email: juanjo.lozano@ciberehd.org.
Julia Sidorova, Email: julia.a.sidorova@gmail.com.
Jordi Camps, Email: JCAMPS@clinic.cat.
Sabela Carballal, Email: CARBALLAL@clinic.cat.
Luis Bujanda, Email: luis.bujanda@osakidetza.eus.
Maria Isabel Vera, Email: maria.vera@uam.es.
Enrique Quintero, Email: equinter@ull.edu.es.
Marta Carrillo-Palau, Email: martacarry@yahoo.es.
Miriam Cuatrecasas, Email: MCUATREC@clinic.cat.
Antoni Castells, Email: CASTELLS@clinic.cat.
Julià Panés, Email: JPANES@clinic.cat.
Elena Ricart, Email: ERICART@clinic.cat.
Leticia Moreira, Email: LMOREIRA@clinic.cat.
Francesc Balaguer, Email: FPRUNES@clinic.cat.
ACKNOWLEDGMENTS
We are indebted to the Tumor Bank of the IDIBAPS Biobank for their support. We thank the patients for their participation. We thank our friend and colleague Montserrat Andreu García, professor at the Pompeu Fabra University (Barcelona) and former founder and leader of the Comprehensive Care Unit for Patients with Inflammatory Bowel Disease at the Department of Gastroenterology, University Hospital del Mar (Barcelona) for providing us with samples and for her invaluable academic advice.
REFERENCES
- 1.Stewenius J, Adnerhill I, Anderson H, et al. Incidence of colorectal cancer and all cause mortality in non-selected patients with ulcerative colitis and indeterminate colitis in Malmo, Sweden. Int J Color Dis 1995;10(2):117–22. [DOI] [PubMed] [Google Scholar]
- 2.Manninen P, Karvonen AL, Huhtala H, et al. The risk of colorectal cancer in patients with inflammatory bowel diseases in Finland: A follow-up of 20 years. J Crohns Colitis 2013;7(11):e551–7. [DOI] [PubMed] [Google Scholar]
- 3.Söderlund S, Brandt L, Lapidus A, et al. Decreasing time-trends of colorectal cancer in a large cohort of patients with inflammatory bowel disease. Gastroenterology 2009;136(5):1561–7; quiz 1818–9. [DOI] [PubMed] [Google Scholar]
- 4.Magro F, Gionchetti P, Eliakim R, et al. Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 1: Definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J Crohns Colitis 2017;11(6):649–70. [DOI] [PubMed] [Google Scholar]
- 5.Rutegard M, Palmqvist R, Stenling R, et al. Efficiency of colorectal cancer surveillance in patients with ulcerative colitis: 38 years' experience in a patient cohort from a defined population area. Scand J Surg 2017;106(2):133–8. [DOI] [PubMed] [Google Scholar]
- 6.Eluri S, Parian AM, Limketkai BN, et al. Nearly a third of high-grade dysplasia and colorectal cancer is undetected in patients with inflammatory bowel disease. Dig Dis Sci 2017;62(12):3586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moussata D, Allez M, Cazals-Hatem D, et al. Are random biopsies still useful for the detection of neoplasia in patients with IBD undergoing surveillance colonoscopy with chromoendoscopy? Gut 2018;67:616–24. [DOI] [PubMed] [Google Scholar]
- 8.Sanduleanu S, Rutter MD. Interval colorectal cancers in inflammatory bowel disease: The grim statistics and true stories. Gastrointest Endosc Clin N Am 2014;24(3):337–48. [DOI] [PubMed] [Google Scholar]
- 9.Melville DM, Jass JR, Morson BC, et al. Observer study of the grading of dysplasia in ulcerative colitis: Comparison with clinical outcome. Hum Pathol 1989;20(10):1008–14. [DOI] [PubMed] [Google Scholar]
- 10.Neumann H, Vieth M, Langner C, et al. Cancer risk in IBD: How to diagnose and how to manage DALM and ALM. World J Gastroenterol 2011;17(27):3184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata I. The present status and problems with diagnosis and management of dysplasia/colitic cancer in ulcerative colitis. Clin J Gastroenterol 2008;1(4):139–44. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, Lai LA, Brentnall TA, et al. Biomarkers for colitis-associated colorectal cancer. World J Gastroenterol 2016;22(35):7882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Bao Y, Yang W. Regulatory miRNAs in colorectal carcinogenesis and metastasis. Int J Mol Sci 2017;18(4):890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahmani F, Avan A, Hashemy SI, et al. Role of Wnt/β-catenin signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J Cell Physiol 2018;233(2):811–7. [DOI] [PubMed] [Google Scholar]
- 15.Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis Model Mech 2017;10(3):197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchison J, Cohen Z, Onyeagucha BC, et al. How microRNAs influence both hereditary and inflammatory-mediated colon cancers. Cancer Genet 2013;206(9-10):309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bocchetti M, Ferraro M, Ricciardiello F, et al. The role of microRNAs in development of colitis-associated colorectal cancer. Int J Mol Sci 2021;22(8):3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James J, Riis L, Malham M, et al. MicroRNA biomarkers in IBD-differential diagnosis and prediction of colitis-associated cancer. Int J Mol Sci 2020;21(21):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: A review. Therap Adv Gastroenterol 2015;8(1):4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanaan Z, Rai SN, Eichenberger MR, et al. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat 2012;33(3):551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olaru AV, Selaru FM, Mori Y, et al. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis 2011;17(1):221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olaru AV, Yamanaka S, Vazquez C, et al. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis 2013;19(3):471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder K, Tremaine W, Ilstrup D. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med 1987;317(26):1625–9. [DOI] [PubMed] [Google Scholar]
- 24.Sebastian S, Hernández V, Myrelid P, et al. Colorectal cancer in inflammatory bowel disease: Results of the 3rd ECCO pathogenesis scientific workshop (I). J Crohns Colitis 2014;8(1):5–18. [DOI] [PubMed] [Google Scholar]
- 25.Tan YG, Zhang YF, Guo CJ, et al. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med 2013;6(12):972–6. [DOI] [PubMed] [Google Scholar]
- 26.Sun D, Yu F, Ma Y, et al. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol Chem 2013;288(13):9508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valeri N, Braconi C, Gasparini P, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014;25(4):469–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B, Liu Y, Zhao L, et al. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog 2017;56(12):2669–80. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Liang H, Bai M, et al. miR-135b promotes cancer progression by targeting transforming growth factor beta receptor II (TGFBR2) in colorectal cancer. PLoS One 2015;10(6):e0130194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang S, Fang J, Wang S, et al. MicroRNA135b regulates the stability of PTEN and promotes glycolysis by targeting USP13 in human colorectal cancers. Oncol Rep 2015;33(3):1342–8. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Bai J, Zhang L, et al. Conditional knockout of microRNA-31 promotes the development of colitis associated cancer. Biochem Biophys Res Commun 2017;490(1):62–8. [DOI] [PubMed] [Google Scholar]
- 32.Feng B, Dong TT, Wang LL, et al. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One 2012;7(8):e43452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Cao Q, Zhang J, et al. MicroRNA expression patterns in indeterminate inflammatory bowel disease. Mod Pathol 2013;26(1):148–54. [DOI] [PubMed] [Google Scholar]
- 34.Wu W, Wang Z, Yang P, et al. MicroRNA-135b regulates metastasis suppressor 1 expression and promotes migration and invasion in colorectal cancer. Mol Cell Biochem 2014;388(1–2):249–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in the supplementary tables.