Dear Editor,
Molecular techniques are the gold standard method for the diagnosis of AML with mutated nucleophosmin gene (NPM1mut). However, their worldwide availability is limited and they provide limited insight into disease heterogeneity. Hence, surrogate markers of NPM1mut are used for fast diagnostic screening of the disease [1], including, among others, immunohistochemical detection of cytoplasmic NPM1 (NPM1c) [2], cup-like nuclear morphology [3], normal karyotype, and/or recurrent flow cytometry profiles -e.g., CD34 negativity, and/or a phenotype resembling acute promyelocytic leukemia (APL)- [4]. Nevertheless, some of these methods are also not widely available, they show limited sensitivity (e.g., low or absent NPM1c expression, particularly among monoblastic/monocytic AML-NPM1mut) [5], frequently lack standardized procedures [1], and they might also bring limited information about disease heterogeneity.
AML-NPM1mut leukemia cells present with heterogeneous cytomorphology and immunophenotypic patterns of lineage commitment and antigen expression, including phenotypes associated with FLT3-internal tandem duplication (ITD) and a poor outcome [6]. In parallel, neutrophil-lineage commitment of AML-NPM1mut cells has been linked with underlying TET2 and IDH1/2 mutations, while the presence of monocytic lineage-committed and immature NPM1mut cells has been related to DNMT3A mutations; indeed, these three patient subgroups show increasingly worse outcomes [4]. Strikingly, NPM1mutFLT3-ITD− patients displaying immature immunophenotypes show strong similarities with NPM1mutFLT3-ITD+ cases regarding leukemia cell transcriptomic profiles, response to therapy and poorer outcomes [7]. Thus, baseline flow cytometric characterization of AML-NPM1mut leukemia cell heterogeneity might contribute to guiding treatment decisions in these patients. Despite the above associations, specific immunophenotypic patterns of AML-NPM1mut remain to be fully defined.
Herewith, we performed a detailed flow cytometric characterization of different subsets of BM leukemia cells from 377 AML patients, including 201 AML-NPM1mut, 144 AML-NPM1wt and 32 APL patients, based on the EuroFlow 8-color acute leukemia orientation tube (ALOT) and the AML/MDS antibody panel (Supplementary methods and Supplementary Table 1). FLT3-ITD was detected in 33% of AML-NPM1mut cases, 19% of AML-NPM1wt and 34% of APL patients. Our aim was to identify reliable phenotypic profiles for fast screening of NPM1mut and/or FLT3-ITD to guide subsequent molecular diagnostic approaches that can be applied worldwide and provide a better understanding of disease heterogeneity.
Our data confirm that AML-NPM1mut patients usually present with high BM leukemia cell percentages at similar levels to APL (Supplementary Table 2). However, NPM1mut cells displayed highly heterogeneous immunophenotypes, consisting of three main BM cell populations: (1) immature leukemia cells showing stem cell-like features (i.e., CD117+HLADR+, 46% of cases; (2) neutrophil lineage-committed CD117+/het HLA-DR− (45%), and/or; (3) monocytic-lineage AML cells expressing CD64+/hi HLA-DR+ and variable CD117 levels (54% of cases) (Supplementary Fig. 1). The differential immunophenotypes observed for these AML cell populations in AML-NPM1mut vs. AML-NPM1wt are detailed in Supplementary Results and Supplementary Table 3. Noteworthy, the relative distribution of AML cell populations defined seven distinct immunophenotypic patterns: (1) a predominant expansion of one (of the above three) leukemia cell population (≥80% of total BM leukemia cells; n = 3 profiles), and; (2) mixed expansions of >1 leukemia cell population (each representing ≥20% of all BM AML cells; n = 4 patterns) (Supplementary Fig. 1). The AML-NPM1mut patients from the former group more frequently showed predominant expansions of neutrophil- (28% of cases), followed by monocytic-lineage (19%) and immature leukemia cells (13%). Conversely, mixed leukemia cell expansions included mixed (1) immature and monocytic (23%), (2) monocytic and neutrophil (7%), (3) immature and neutrophil (5%) and (4) immature plus neutrophil- and monocytic-lineage AML cells (5% of cases).
The distribution of leukemia cell subsets was consistent with a lower maturation arrest of AML-NPM1mut vs. NPM1wt cells, associated with a lower frequency and size of immature leukemia cell expansions (p < 0.001), while depicting a higher prevalence of more differentiated AML cells committed to the neutrophil (p < 0.001) and/or the monocytic lineage (p = 0.02) (Supplementary Table 2 and Supplementary Fig. 1). These findings might contribute to explain the overall higher sensitivity to chemotherapy of AML-NPM1mut [7].
Despite AML-NPM1mut may originate from CD34+ hematopoietic progenitor cells (HPC) [8], most frequently they lack CD34 (7% vs. 94% NPM1wt CD34+ cells). These cells may expand in BM due to consistent expression of HOX genes [9], which has been directly associated with NPM1 cytoplasmic dislocation [10]. However, CD34lo expression is not specific to AML-NPM1mut, and it has been found to be independent of NPM1 dislocation [9]. In line with these observations, we also found CD34lo expression among NPM1wt immature, neutrophil and monocytic lineage-committed AML cells, and thereby, this phenotype is of limited specificity for AML-NPM1mut.
Beyond CD34lo expression, other immunophenotypic features of AML-NPM1mut cells supported a less pronounced maturation blockade vs. other AML patients. Hence, AML-NPM1mut immature cells retained a higher capability for neutrophil lineage maturation with higher expression of CyMPO (p = 0.04), CD15 and CD33 (p < 0.001), associated with downregulation of the early monocytic markers CD64 (p = 0.05) and HLA-DR (p = 0.004), in line with the higher frequency of neutrophil (vs. monocytic) lineage-commitment observed for AML-NPM1mut cells. Furthermore, AML-NPM1mut cases more frequently showed immature AML cells with aberrant CD7 positivity (60% vs. 32% cases), but they rarely expressed CD56 (1% vs. 15%) and NuTdT (3% vs. 20%, respectively) (p < 0.001) (Supplementary Fig. 2 and Supplementary Table 3). Multivariate logistic regression analysis revealed that decreased CD34 and HLA-DR, together with upregulation of CD15 and CD7 (but not NuTdT), was the best combination of markers expressed on immature leukemia cells to predict for NPM1mut in AML (Table 1).
Table 1.
Univariate and multivariate logistic regression analysis of those immunophenotypic features of BM leukemia cells associated with NPM1 mutation (A) and FLT3-ITD (B) from AML patients (n = 377).
| AML-NPM1mut vs. AML-NPM1wt and APL | ||||||
|---|---|---|---|---|---|---|
| (A) Variables and leukemia cell subsets | Univariate analysis | Multivariate analysis | ||||
| OR | 95% CI | p-value | OR | 95% CI | p-value | |
| CD34+ and/or CD117+HLADR+ leukemia cells | ||||||
| <26.5% of all leukemia cells | 2.0 | 1.4–2.5 | <0.001 | |||
| CD34 (<35%) | 4.8 | 2.6–8.6 | <0.001 | 7.7 | 3.5–17.0 | <0.001 |
| CD33 (>96%) | 1.4 | 1.0–2.0 | 0.04 | |||
| CD105 (<9.5%) | 0.4 | 0.2–0.6 | 0.001 | |||
| HLA-DR (<97%) | 0.3 | 0.3–0.7 | 0.001 | 0.2 | 0.1–0.5 | <0.001 |
| CD15 (>6.6%) | 0.3 | 0.1–0.6 | <0.001 | 0.2 | 0.1–0.5 | <0.001 |
| CD7 (>3%) | 1.5 | 1.0–2.2 | 0.02 | 2.0 | 1.1–3.8 | 0.02 |
| CD56 negative | 0.3 | 0.1–0.6 | 0.002 | |||
| NuTdT negative | 0.2 | 0.1–0.5 | <0.001 | 0.2 | 0.1–0.5 | 0.002 |
| Neutrophil-committed leukemia cells | ||||||
| >21.5% of all leukemia cells | 1.6 | 1.2–2.2 | 0.008 | - | - | - |
| CD34 (<5%) | 4.0 | 2.4–6.6 | <0.001 | 7.0 | 2.9–17.5 | <0.001 |
| CD71 (<70%) | 2.5 | 1.6–3.9 | <0.001 | 3.0 | 1.1–7.9 | 0.02 |
| CD105 (>3%) | 5.1 | 2.7–9.8 | <0.001 | 4.8 | 1.9–12.4 | 0.001 |
| CD64 (<30%) | 4.3 | 2.5–7.5 | <0.001 | 4.4 | 1.8–10.9 | 0.001 |
| CD13 (<92%) | 3.2 | 2.0–5.1 | <0.001 | - | - | - |
| CD56 (>5%) | 5.6 | 2.1–14.5 | <0.001 | - | - | - |
| Monocytic-committed leukemia cells | ||||||
| Any asynchronous pattern | 6.5 | 3.8–11.1 | <0.001 | - | - | - |
| Asynchronous CD300e+CD14- profile | 85.0 | 12.0–610 | <0.001 | 49.0 | 3.7–641 | 0.004 |
| Asynchronous CD35+CD14- profile | 11.4 | 5.5–23 | <0.001 | - | - | - |
| CD34+ (<3.8%) | 3.7 | 2.4–5.7 | <0.001 | - | - | - |
| Any asynchronous pattern plus CD34+ (<3.8%) | 34.3 | 11.0–108 | <0.001 | 223.9 | 3.7–641.5 | <0.001 |
| CD117 (<5.9%) | 3.7 | 2.1–6.5 | 0.001 | - | - | - |
| CD13 (<77%) | 4.3 | 2.7–6.8 | <0.001 | 0.2 | 0.04–1.0 | 0.05 |
| CD123 (>83%) | 2.9 | 1.8–4.5 | <0.001 | 0.3 | 0.1–0.8 | 0.02 |
| CD15+ (>77%) | 3.4 | 2.2–5.4 | <0.001 | - | - | - |
| CD36 (>87%) | 3.2 | 2.0–5.1 | <0.001 | - | - | - |
| FLT3-ITD+ vs. FLT3-ITD- | ||||||
| (B) Variables and leukemia cell subsets | ||||||
| AML-NPM1mut patients | ||||||
| CD34+ and/or CD117+HLADR+ leukemia cells | ||||||
| CD34+ (>3%) | 5.3 | 1.9–14.8 | 0.001 | - | - | - |
| CD38 (<95%) | 5.6 | 2.2–14.1 | <0.001 | 0.1 | 0.01–0.8 | 0.03 |
| CD7 (>55%) | 5.4 | 2.2–13.9 | <0.001 | 7.2 | 1.0–48.5 | 0.04 |
| CD25 (>25%) | 7.1 | 1.3–37.5 | 0.02 | - | - | - |
| Neutrophil-committed leukemia cells | ||||||
| CD117 (<69%) | 5.7 | 2.1–15.5 | 0.001 | 9.4 | 2.7–32.4 | <0.001 |
| CD123 (>84%) | 4.6 | 1.7–12.6 | 0.003 | 7.6 | 2.2–26.0 | 0.001 |
| CD13 (>56%) | 2.6 | 1.0–6.8 | 0.05 | - | - | - |
| AML-NPM1wt patients | ||||||
| % total BM blasts (>40%) | 3.7 | 1.2–11.6 | 0.02 | - | - | - |
| CD34+ and/or CD117+HLADR+ leukemia cells | ||||||
| CD34+ (<57%) | 4.3 | 1.6–11.5 | 0.004 | 3.8 | 1.0–15.3 | 0.05 |
| CD25 (>10%) | 6.9 | 1.4–33.8 | 0.01 | 7.9 | 1.5–40.3 | 0.01 |
OR odds ratio, CI confidence interval.
Neutrophil and monocytic lineage-committed AML-NPM1mut cells were typically characterized by prominent asynchronous maturation profiles. Thus, despite their CD34lo phenotype, neutrophil lineage AML-NPM1mut cells showed (vs. their NPM1wt counterpart) more immature features, including downregulation of the neutrophil lineage markers CD15, CD71, CD13 and CD64 (p ≤ 0.05), associated with higher levels of the immature antigens CD123 and CD105 (p ≤ 0.01). In contrast to NPM1mut immature leukemia cells, NPM1mut neutrophil-lineage cells barely expressed CD7 but more frequently showed aberrant positivity for CD56 (24% vs. 7%, respectively; p = 0.03) and to a lesser extent also for CD9 and CD4 (p ≤ 0.02). Furthermore, compared with neutrophil-lineage APL cells, AML-NPM1mut neutrophil-lineage cells downregulated CD34, CD13, CD64 and CD71, while they upregulated CD105 (p ≤ 0.001), and aberrant CD56 expression, but showed lower rates of CD203c and CD7 (p ≤ 0.02) (Supplementary Fig. 2 and Supplementary Table 3). Multivariate analysis revealed that the unique CD34loCD71loCD64loCD105+ profile had the highest predictive value for NPM1mut among neutrophil lineage AML cells (Table 1).
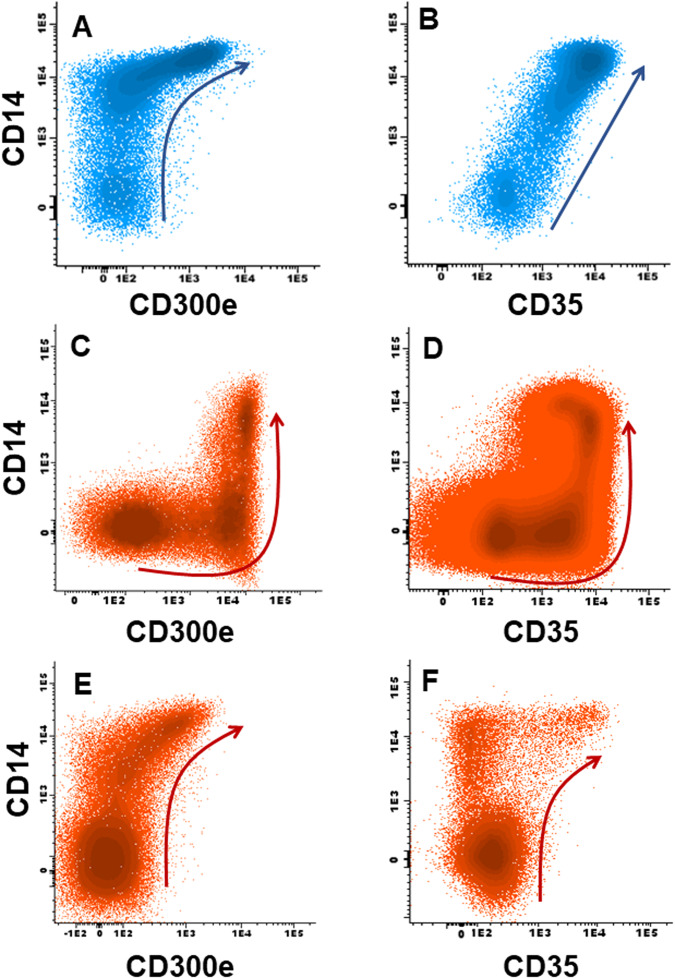
Finally, NPM1mut monocytic-committed leukemia cells showed (vs. AML-NPM1wt) decreased expression of immature markers (i.e., CD34, CD117; p < 0.001), while upregulated the monocytic-associated antigens CD4 (p = 0.04), CD11b (p = 0.03), CD15, CD36, and CD300e in addition to CD123 (p ≤ 0.006). However, these more markedly mature monocytic features coexisted with asynchronous downregulation of other monocytic-associated markers (i.e., CD13, CD71, CD14 and CyMPO; p ≤ 0.002) and a higher frequency of AML-NPM1mut cases showing aberrant CD56 (p = 0.03). Altogether, these phenotypes defined three unique asynchronous monocytic maturation profiles present in most AML-NPM1mut cases (90%) vs. a minority of NPM1wt patients (24%; p < 0.001): [11] (1) abnormal (early) upregulation of CD300e prior to CD14 (CD300e+CD14−: 74% vs. 3% NPM1wt cases, p < 0.001); and/or, either (2) early expression of CD35 prior CD14 (CD35+CD14−: 72% vs. 9%, p < 0.001), or; (3) early upregulation of CD14 prior CD35 (CD14+CD35−: 6% vs. 13%, respectively; p = 0.02) (Fig. 1 and Supplementary Table 4). Noteworthy, the presence of CD300e+CD14− and/or CD35+CD14− leukemia cells showing CD34lo expression emerged as the most specific phenotypes for AML-NPM1mut (odds ratio: 223.9; p < 0.001) (Table 1).
Fig. 1. Monocytic maturation pathways in normal and AML bone marrow.
Maturation pathways of monocytic cells in normal bone marrow (A, B, blue dots), and asynchronous AML-NPM1mut patterns of expression of CD300e+CD14− (C), CD35+CD14− (D). E and F depict normal patterns of acquisition of CD14 vs. CD300e in a patient with AML-NPM1wt while showing an asynchronous CD14+ CD35− phenotype (E, F) among monocytic cells (red dots). Arrows represent the normal (blue) and leukemia (red) maturation pathways of monocytic lineage-committed (gated) CD64hi cells.
Hundreds of neutrophil and monocytic differentiation-associated genes are repressed in AML-NPM1mut, which might be related to NPM1 haploinsufficiency and/or the cytoplasmic relocation and functional blockade of myeloid transcription factors interacting with NPM1c [12, 13]. For instance, the functional reduction of PU.1 represses the PU.1/CEBPA/RUNX1 myeloid transcriptional hub regulating terminal monocytic and neutrophil differentiation [13]. Conversely, other nuclear transcriptional regulators inhibited by NPM1 under physiological conditions are not translocated to the cytoplasm, leading to abnormally high activation of their target genes [14]. Therefore, asynchronous neutrophil and/or monocytic differentiation profiles of AML-NPM1mut cells are consistent with abnormal activation vs. repression of distinct sets of myeloid gene promoters regulated by NPM1.
Expectedly, (non-APL) AML cases with FLT3-ITD showed greater BM leukemia cell infiltration and increased proportions of immature CD34+ leukemia cells, frequently in association with monocytic AML cells, independently of NPM1 comutation (Supplementary Fig. 1I and Supplementary Table 2). Such specific expansion of immature AML cells might be related to the physiological restriction of FLT3 gene expression to BM hematopoietic CD34+ HPC [15].
Noteworthy, FLT3-ITD promoted distinct immunophenotypic profiles in NPM1mut and NPM1wt AML (Supplementary results and Supplementary Fig. 3). Although CD34 and/or CD25 expression has been associated with FLT3-ITD [6], we show that both markers are more frequent among immature NPM1mutFLT3-ITD+ cells. Hence, CD25 positivity and heterogeneous CD34 expression on immature AML cells emerged as the best combination of predictors for FLT3-ITD among AML-NPM1wt cases. Conversely, in AML-NPM1mut cases, FLT3-ITD was strongly associated with a CD7hiCD38lo profile on immature leukemia cells and/or a CD117hetCD123hi phenotype among neutrophil lineage leukemia cells (Table 1).
In summary, the mutational status of NPM1 and FLT3 is associated with unique BM leukemia cell distribution and immunophenotypic profiles, even when only cases with a normal karyotype were considered (data not shown), which might contribute to a fast diagnostic screening of NPM1mut and/or FLT3-ITD in AML, and an improved classification of AML-NPM1mut patients. Further prospective studies are needed to confirm these findings.
Supplementary information
Acknowledgements
This study has been funded by Instituto de Salud Carlos III (ISCIII) through the project PI21/01115 and co-funded by the European Union and the grant of CIBERONC of the Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, Madrid, Spain, and FONDOS FEDER (no. CB16/12/00400); MR was supported by the Ministry of Health of the Czech Republic, grant number NU20J-07-00028.
Author contributions
SM and AO were responsible for study design, extracting and analyzing data and manuscript writing; PL extracted and analyzed data; AY-B, VvdV, AEB, JISG, QL, RA-B, CT, IC, JF-M, AA and MBV were responsible for quality assessment and screening potentially eligible studies; MG-G performed fluorescence in situ hybridization studies; MCC, TG, RG-S, MIPC performed and compiled cytogenetic and molecular studies; NV, LM, EC, PF, ES, JP, MR, JCCB, FJD-G, FR, JDV, RMS, JST, SA, AF and CQC screened potentially eligible studies and clinical data; XC, LGA, LA, JJMvD provided feedback on the report.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sergio Matarraz, Alberto Orfao.
Contributor Information
Sergio Matarraz, Email: smats@usal.es.
Alberto Orfao, Email: orfao@usal.es.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-023-00909-4.
References
- 1.Falini B, Martelli MP, Pileri SA, Mecucci C. Molecular and alternative methods for diagnosis of acute myeloid leukemia with mutated NPM1: flexibility may help. Haematologica. 2010;95:529–34. doi: 10.3324/haematol.2009.017822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108:1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 3.Park BG, Chi H-S, Jang S, Park C-J, Kim D-Y, Lee J-H, et al. Association of cup-like nuclei in blasts with FLT3 and NPM1 mutations in acute myeloid leukemia. Ann Hematol. 2013;92:451–7. doi: 10.1007/s00277-012-1645-5. [DOI] [PubMed] [Google Scholar]
- 4.Mason EF, Hasserjian RP, Aggarwal N, Seegmiller AC, Pozdnyakova O. Blast phenotype and comutations in acute myeloid leukemia with mutated NPM1 influence disease biology and outcome. Blood Adv. 2019;3:3322–32. doi: 10.1182/bloodadvances.2019000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolthuis CM, Mulder AB, Verkaik-Schakel RN, Rosati S, Diepstra A, van den Berg E, et al. A single center analysis of nucleophosmin in acute myeloid leukemia: value of combining immunohistochemistry with molecular mutation analysis. Haematologica. 2013;98:1532–8. doi: 10.3324/haematol.2012.079806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelini DF, Ottone T, Guerrera G, Lavorgna S, Cittadini M, Buccisano F, et al. A leukemia-associated CD34/CD123/CD25/CD99+ immunophenotype identifies FLT3-mutated clones in acute myeloid leukemia. Clin Cancer Res. 2015;21:3977–85. doi: 10.1158/1078-0432.CCR-14-3186. [DOI] [PubMed] [Google Scholar]
- 7.Mer AS, Heath EM, Madani Tonekaboni SA, Dogan-Artun N, Nair SK, Murison A, et al. Biological and therapeutic implications of a unique subtype of NPM1 mutated AML. Nat Commun. 2021;12:1054. doi: 10.1038/s41467-021-21233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martelli MP, Pettirossi V, Thiede C, Bonifacio E, Mezzasoma F, Cecchini D, et al. CD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised mice. Blood. 2010;116:3907–22. doi: 10.1182/blood-2009-08-238899. [DOI] [PubMed] [Google Scholar]
- 9.Pianigiani G, Rocchio F, Peruzzi S, Andresen V, Bigerna B, Sorcini D, et al. The absent/low expression of CD34 in NPM1-mutated AML is not related to cytoplasmic dislocation of NPM1 mutant protein. Leukemia. 2022;36:1931–4. 10.1038/s41375-022-01593-2. [DOI] [PMC free article] [PubMed]
- 10.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang Y-H, Ramabadran R, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34:499–512.e9. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matarraz S, Almeida J, Flores-Montero J, Lécrevisse Q, Guerri V, López A, et al. Introduction to the diagnosis and classification of monocytic-lineage leukemias by flow. Cytom B Clin Cytom. 2017;92:218–27. doi: 10.1002/cyto.b.21219. [DOI] [PubMed] [Google Scholar]
- 12.Wang AJ, Han Y, Jia N, Chen P, Minden MD. NPM1c impedes CTCF functions through cytoplasmic mislocalization in acute myeloid leukemia. Leukemia. 2020;34:1278–90. doi: 10.1038/s41375-019-0681-8. [DOI] [PubMed] [Google Scholar]
- 13.Gu X, Ebrahem Q, Mahfouz RZ, Hasipek M, Enane F, Radivoyevitch T, et al. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub that regulates granulomonocytic fates. J Clin Invest. 2018; 128:4260–79. [DOI] [PMC free article] [PubMed]
- 14.Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia. 2014;28:311–20. doi: 10.1038/leu.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdellateif MS, Kassem AB, El-Meligui YM. Combined expression of CD34 and FLT3-internal tandem duplication mutation predicts poor response to treatment in acute myeloid leukemia. Int J Gen Med. 2020;13:867–79. doi: 10.2147/IJGM.S276138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.