Abstract
Subnival glasshouse plants provide a text-book example of high-altitude adaptation with reproductive organs enclosed in specialized semi-translucent bracts, monocarpic reproduction and continuous survival under stress. Here, we present genomic, transcriptomic and metabolomic analyses for one such plant, the Noble rhubarb (Rheum nobile). Comparative genomic analyses show that an expanded number of genes and retained genes from two recent whole-genome duplication events are both relevant to subnival adaptation of this species. Most photosynthesis genes are downregulated within bracts compared to within leaves, and indeed bracts exhibit a sharp reduction in photosynthetic pigments, indicating that the bracts no longer perform photosynthesis. Contrastingly, genes related to flavonol synthesis are upregulated, providing enhanced defense against UV irradiation damage. Additionally, anatomically abnormal mesophyll combined with the downregulation of genes related to mesophyll differentiation in bracts illustrates the innovation and specification of the glass-like bracts. We further detect substantial accumulation of antifreeze proteins (e.g. AFPs, LEAs) and various metabolites (e.g. Proline, Protective sugars, procyanidins) in over-wintering roots. These findings provide new insights into subnival adaptation and the evolution of glasshouse alpine plants.
Subject terms: Evolutionary ecology, Plant evolution, Genome evolution, Molecular ecology, Abiotic
A chromosome-level genome assembly for the glasshouse plant Rheum nobile (noble rhubarb) combined with phylogenetic, transcriptomic, and metabolomic analyses provides insight into how alpine plants have adapted to growth in extreme environments.
Introduction
Subnival ecosystems, existing just below the permanent snowline in mountainous regions, are subject to high levels of irradiation, freezing temperatures and hypoxia throughout the year, is the most inhospitable climate zone at the highest altitude among terrestrial ecosystems, making it nearly lifeless for most higher animals and plants1. Such ecosystems are widely distributed in the high mountains of North America, Europe, and Asia (especially the Qinghai-Tibet Plateau [QTP])2 (Fig. 1a). All resident plant species in this zone face enormous challenges for survival. Thus, under selective pressure from a variety of stressful environmental factors, plants in this area have developed highly complex and elaborate adaptive evolutionary traits to ensure their survival and reproduction in extreme environments. For example, ‘glasshouse-like’, ‘cushion’ and ‘woolly’ phenotypes3. Of which, the “glasshouse structure” is one of textbook example for high-altitude adaptation in subnival belt, in which plants develop cream-colored translucent bract-like leaves during the reproductive phase. These bract-like leaves forming a glasshouse that plays a crucial role in ensuring successful reproduction and completing the plant’s life cycle. Alpine ‘glasshouse’ plants have been recorded in a range of plant families, for example the Ranunculaceae, Caryophyllaceae, Lamiaceae, Asteraceae and Polygonaceae.
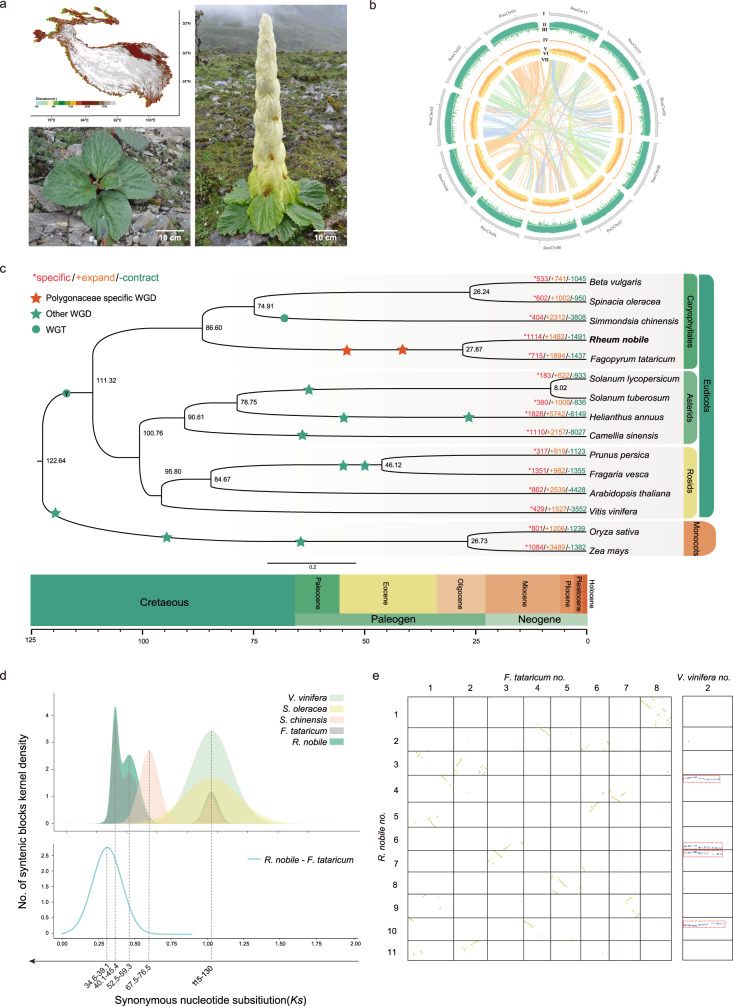
Fig. 1. Typical habitat of noble rhubarb (Rheum nobile) and genome evolution analyses.
a Altitude across the Qinghai-Tibet plateau (top left), a vegetative individual (bottom left) and a reproductive individual (right) of noble rhubarb. b Overview of the two Rheum genomes, the tracks (moving inward) show: (I) chromosomes; (II) gene numbers (0–100); (III) GC content (0.25–0.45); (IV) TE density (0–100%); (V) Gypsy density (0–90%); (VI) Copia density (0–60%); and (VII) identified syntenic blocks, calculated using 500 kb sliding windows. c Phylogenetic tree of 16 species and dating of WGD events. Gains and losses of gene families in sub-branches are highlighted in orange and green, respectively. Red, green and blue stars indicate Polygonaceae-specific WGDs, other reported WGDs, and whole-genome triplication events, respectively. d Distribution of average synonymous substitutions (Ks) between syntenic blocks after evolutionary rate correction. e Dotplots of interspecific syntenic blocks among Vitis vinifera, Fagopyrum tataricum, and R. nobile.
As an iconic of alpine plant, Noble rhubarb (Rheum nobile Hook. F. et Thoms., Polygonaceae) were considered by Hooker and Fitch (1855) as “certainly the most striking of the many fine alpine plants of Sikkim”4. This giant herb occurs sparsely across subnival belts in the QTP at elevations between 4000 and 6000 m5–7 and at flowering possesses stacked layers of large and showy bracts that securely conceal the entire inflorescence, to produce a pagoda-shaped ‘glasshouse’ phenotype (Fig. 1b)7,8. These bracts aids reproduction in multiple ways5,6,9–15, which could increasing flower and fruit temperature within bracts and that higher by up to 10 and 8 °C, respectively15, than when bracts were removed or in the ambient conditions on sunny days, preventing pollen grains from being washed away by rain9,15, and intensity of ultraviolet-B (UV-B) radiation reaching flowers (or fruits) was decreased by 93–98% by bracts5,11,15, but there have been no previous attempts to investigate the genetic bases of subnival adaptation in these plants. Before flowering, plants develop for several years as a rosette during which they are subject to extremely low temperatures and strong winds, particularly during winter periods. Senescence of above ground parts is a response to such conditions16, but tissues below ground continue to survive rasing questions on how this is achieved and the mechanisms involved.
Here, we present a high-quality chromosome-scale genome assembly of the Noble rhubarb and detect the genetic legacy of two recent whole-genome duplication events that might have promoted its subnival adaption. In addition, we examine changes in gene expression and chemical production in its floral bracts and rosette roots based on transcriptomic and metabolomic analyses. Using these data we further investigate possible evolutionary adaptations underlying stress tolerance in this ‘glasshouse’ plant through detailed examination of expressed genes. Finally, subnival adaptation to extreme freezing environments during overwintering was examined using a multiomics approach. The availability of a reference genome sequence of R. nobile, together with information on the molecular basis and genetic mechanisms of adaptation to subnival environments, provide valuable resources for future evolutionary investigations of high alpine plants exposed to extreme ecological stress.
Results
Assembly and annotation of the high-quality genome
A high-quality chromosome-level genome sequence of Rheum nobile was produced using multiple technologies. In total, 217.64 Gb of Illumina reads (~135× depth), 93.26 Gb PacBio HiFi reads (~29× depth), and 189.62 Gb of high-throughput chromosome conformation capture (Hi-C) reads (~119× depth) were generated (Supplementary Table 1; depths based on estimated genome size, Supplementary Fig. 1a). Based on Hi-C integrated assembly of hifiasm, a primary assembly with long stretches of phased blocks and two phased contig graph were obtained, and the primary assembly were used in subsequent analysis. The sequence (1.57 Gb, N50 = 6.72 Mb) was very close to the estimated genome size (~1.59 Gb) based on k-mer distribution analysis (and Supplementary Table 2). After preliminary assembly, both purge_haplotigs and purge_dup were used to generate the final contig-level assembly to retain only one copy of each of the contigs from heterozygous regions. While the distribution of reads depth indicated there is no peak of heterozygous regions in purge_haplotype (Supplementary Fig. 1b), and purge_dup identified ~2 Mb junk region but not duplicate haplotig, these results also suggest high accuracy of the assembled genome. Then, the Hi-C dataset was further used to cluster and order contigs to generate a chromosome-level genome assembly. In total, 99.33% of assembled sequence could be anchored into 11 chromosomes (Fig. 1b, Supplementary Fig. 1c and Supplementary Table 3). To comprehensively assess the accuracy, continuity and completeness of our R.nobile genome, four analyses were used to evaluate the assembly quality. In total, the raw Illumina paired-end reads were mapped to the assembled genome with mapping rates and coverage of 99.92% and 99.67%, respectively (Supplementary Table 4). And the accuracy and integrenity of the assembly were evaluated by consensus quality value (QV score), k-mer completeness and long terminal repeat (LTR) Assembly Index (LAI) score. Of which, QV score and k-mer completeness was evaluated at 51.96 and 99.62% using Merqury, respectively (Supplementary Table 5) and LAI score of 25.08 were estimated (Supplementary Table 5), which suggested R. nobile assembly exceeds the minimum reference standard of 6.7.Q40 (e.g., >1.0 Mb contig) and 7.C.Q50 (e.g., chromosome-level, QV score>50, k-mer completeness > 95%, functional completeness>90%) set for eukaryotic species by the Earth BioGenome Project. Together, these two indices indicate high base accuracies and continuityfor our R. nobile genome. Moreover, Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis indicated that 97.3% of the conserved single-copy eukaryotic genes were completely captured in the R. tanguticum genome assembly (Supplementary Table 6).
The majority (83.69%) of the genome consisted of transposable elements (TEs) distributed across all chromosomes (Fig. 1b and Supplementary Table 6). Long-terminal repeat elements (LTRs) dominated, accounting for 71.75% of all TE sequences (Supplementary Table 6). Copia and Gypsy were the two major families of LTRs, comprising 0.22 Gb and 0.60 Gb of the genome, respectively. Both LTR families showed a sudden increase in frequency in the genome ~3 million years ago (Mya), with Copia subsequently undergoing a rapid decrease while Gypsy families remained relatively stable in frequency (Supplementary Fig. 1d). We predicted a total of 37,770 protein-coding genes in the genome with complete BUSCO values of 95.6% (Supplementary Tables 7 and 8). Average protein-coding gene length was 4314 bp and mean exon number ~5 (Supplementary Table 7). Approximately 90.7% of genes were functionally annotated through Blast searches of five functional databases (Supplementary Table 9). In addition, 2341 transcription factors and 10,947 non-coding RNAs were identified (Supplementary Table 10, 11).
Genome evolution
Gene sequences from 15 species (R. nobile and four other, four asterids, four rosids and two monocots [rice and maize]) were clustered and assigned to 46,684 gene families. Of these, 1098 single-copy gene families were determined and used in phylogenetic analysis. Both coalescent-based and concatenation-based phylogenies with high support (posterior probability values of 1 and bootstrap values of 100) showed consistent topology with those previously report (Fig. 1c and Supplementary Fig. 2)17–20. Noble rhubarb was estimated to have diverged from Tartary buckwheat (Fagopyrum tataricum, Polygonaceae) ~ 27.87 million years ago (Mya) (Fig. 1c). Dating further showed that Polygonaceae species diverged from Amaranthaceae (including beet [Beta vulgaris] and spinach [Spinacia oleracea]) and Simmondsiaceae (including jojoba [Simmondsia chinensis]) ~86.60 Mya, and that Caryophyllales diverged from asterids and rosids ~111.32 Mya (Fig. 1c).
We identified 1482 expanded gene families in R. nobile (Fig. 1c) with functions mainly associated with DNA damage repair, possibly of adaptive significance in subnival areas subject to high UV radiation (Supplementary Data 1 available on Figshare; 10.6084/m9.figshare.19662933). Gene functions associated with leaf morphogenesis and cotyledon morphogenesis were also enriched (Supplementary Data 1 available on Figshare; 10.6084/m9.figshare.19662933) and may combine in the formation of the ‘glass-house’-like plant organ. In addition, 1,114 unique gene families with functional enriched categories were mainly implicated in plant defense (Supplementary Data 1 available on Figshare; 10.6084/m9.figshare.19662933).
The distribution of average synonymous substitution rates (Ks) of intra-genomic collinear gene blocks indicated that two polyploidization events occurred in the evolutionary histories of both R. nobile and Tartary buckwheat (Fig. 1d and Supplementary Data 2 available on Figshare; 10.6084/m9.figshare.19662933) in addition to the γ event (whole-genome triplication) shared by all core eudicots21,22. Analyses of synteny depths supported this (Fig. 1e and Supplementary Fig. 3a). Previously, it was thought that only one additional WGD event had occurred in Tartary buckwheat17. Because beet and spinach genomes have undergone complex chromosome rearrangement events during their evolution23, we selected the genome of Vitis vinifera as a reference for further analyses. This genome underwent only the γ event with fewer subsequent chromosomal rearrangements. We obtained synteny depth ratios of 1:4 between V. vinifera and both Polygonaceae species (Fig. 1e), indicating that the two recent shared polyploidization events in these species were both whole-genome duplications (WGDs). The two recent WGDs among Polygonaceae species received further support from phylogenetic analyses of all identified collinear genes (Supplementary Fig. 3b), and after correction for evolutionary rates24 were estimated to have occurred 52.35-59.32 and 40.19-45.43 Mya, respectively, that is before divergence of R. nobile from Tartary buckwheat (Fig. 1c, d). Notably, compared to other four Caryophyllales species that used in this study, including the Tartary buckwheat which shared the same polyploid history, the rhubarb genome exhibited a higher retention of genes originating from WGD events (Supplementary Fig. 4). And these genes were enriched (inter alia) in the GO terms that related to freezing stress response (GO:0050826), and some GO terms related to osmotic stress response (e.g.: GO:0047484, GO:1901002, GO:1901000, GO:1902074), which may be related to its cold stress response, as freezing stress often leads to cellular dehydration and subsequent osmotic stress. And some GO terms that related to hypoxia response (e.g.: GO:0036293, GO:0001666, GO:0070482) were only enriched in the R. nobile genome. Moreover, numerous WGD originated genes were also enriched in GO terms that related flavonoid biosynthetic process (e.g.: GO:0009963, GO:0009699, GO:0009813, GO:0051555, GO:0009962), which also is an important class of secondary metabolites play a role in plant defense to multiple stressors, indicating that R. nobile may have partially acquired the ability to adapt to harsh environments through specific retention of WGD originated genes (Supplementary Data 3 available on Figshare; 10.6084/m9.figshare.19662933).
Gene expressions and metabolites accumulations of the innovative ‘glasshouse’-like bracts
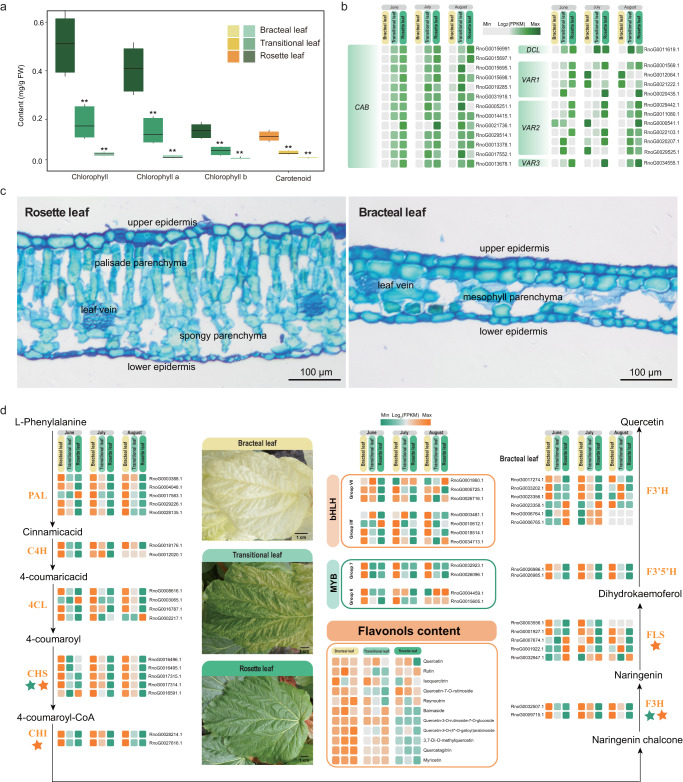
‘Glasshouse’ plants are characterized by large and showy semi-translucent bracts concealing a compound raceme. To explore the genetic basis of bract formation, we initially compared the regulatory mechanisms of gene expression underlying structural differences between bracts and leaves. Fewer chloroplasts with severe developmental defects are also evident in bracts6. For transcriptome analyses we collected 27 samples of three tissues (bracts, transitional leaves and rosette leaves) at three different times throughout the growing season (June, July and August) (Fig. 2, Supplementary Fig. 5 and Supplementary Table 12). We detected 5142 genes that were expressed significantly less strongly in bracts than in transitional leaves or rosette leaves (Supplementary Fig. 5). The functions of these genes were mainly associated with photosynthesis, such as ‘photosynthetic, light harvesting’, ‘photosynthetic electron transport chain’, ‘photosynthesis’, ‘chlorophyll biosynthetic process’ and ‘carotenoid biosynthesis’ (Supplementary Data 4 available on Figshare; 10.6084/m9.figshare.19662933) with genes related to both chlorophyll and carotenoid biosynthesis also downregulated in bracts (Supplementary Figs. 6 and 7). Bracts were found to have 87.27 and 95.63% lower chlorophyll a/b contents than transitional and rosette leaves, and exhibited similar reductions for carotenoid contents (ca. 88.78 and 93.52%, respectively) (Fig. 2a). Key genes involved in chloroplast and mesophyll development in leaves were weakly expressed in bracts (Fig. 2b, Supplementary Fig. 8), for example, multiple copies of CHLOROPHYLL A/B BINDING PROTEIN (CAB)25,26. Also, most VARIEGATED (VAR) genes were expressed significantly less strongly in bracts (Fig. 2b). VAR127,28 and VAR227,29 play important roles in photoprotection and development of thylakoid membranes, while absence of VAR330, which is required for chloroplast and palisade cell development, results in lower levels of chlorophylls and carotenoids. It was further evident that DEFECTIVE CHLOROPLASTS AND LEAVES (DCL)31 genes involved in palisade morphogenesis were expressed extremely weakly in bracts (Fig. 2b). This was of interest as cross-sections of rosette leaf (left) and bracteal leaf (right) mesophyll parenchyma showed that bracts comprised only 3–4 layers of cells and lack a distinct palisade and spongy parenchyma present in rosette leaves (Fig. 2c).
Fig. 2. Innovation of glasshouse-like bracts.
a Chlorophyll (green) and carotenoid (orange) contents in three types of leaves (bract, transitional leaf and rosette) of R. nobile. Three independent biological replicates were used to estimate means (n = 3 biologically independent samples) and standard deviations (SD) in each case. For each photosynthetic pigment, t tests were applied to test the significance of differences in contents between bracts and leaves (rosette and transitional). Double asterisks indicate p < 0.01. b Heatmap of expression of genes involved in chloroplast and mesophyll development in leaves. Gene expression profiles (in fragments per kilobase of exon per million mapped fragments) in the three types of leaves at three time points in the growth season (here June, July and August) are shown from left to right. Colors from gray to green indicate gene expression levels from low to high. Gene names are shown beside each heatmap. CAB, CHLOROPHYLL A/B BINDING PROTEIN; DCL, DEFECTIVE CHLOROPLASTS AND LEAVES; VAR1, VARIEGATED 1; VAR2, VARIEGATED 2; VAR3, VARIEGATED 3. c Cross-sections of rosette leaf (leaf) and bracteal leaf (right) of R. nobile that stained with 0.1% toluidine blue. Bar = 100 μm. d Expression patterns of genes associated with flavonoid biosynthesis and accumulation in indicated tissues, obtained using same tissue panel as in (b), with low to high expression/content indicated by colors from green to orange. PAL phenylalanine ammonia-lyase, C4H cinnamate-4-hydroxylase, 4CL 4-coumarate CoA ligase 4, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone 3-hydroxylase, FLS flavonol synthase, F3′5′H flavonoid 3′,5′-hydroxylase; F3′H flavonoid 3′-hydroxylase, MYB myeloblastosis transcription factor, bHLH basic helix–loop–helix transcription factor.
The distinctive structure of bracts enables high light transmittance, thus warming the reproductive organs inside the ‘glasshouse’. In addition, the high content of UV-absorbing compounds (e.g., flavonols and related phenylpropanoid derivatives) protect reproductive organs from the damaging effects of UV irradiation11. We quantified flavonoids based on our metabolomic data, and detected substantially higher levels of flavonols, especially quercetin and myricetin derivatives (e.g., quercetin-3-O-rutinoside-7-O-glucoside, 3,7-di-O-methylquercetin, myricetin and myricetin-3-O-rutinoside) in bracts than in leaves (Fig. 2d Supplementary Data 5 available on Figshare; 10.6084/m9.figshare.19662933). Transcriptome data further indicated that 4863 genes associated with ‘Flavonoid biosynthesis’ and ‘Phenylpropanoid biosynthetic process’ (Supplementary Data 6 available on Figshare; 10.6084/m9.figshare.19662933) were more highly expressed in bracts than transitional or rosette leaves. Moreover, all genes encoding the nine enzymes involved in the flavonoid biosynthetic pathway were more strongly expressed in bracts than in leaves at each time of analysis (Fig. 2d). This was also the case for transcription factors (including both bHLHs (Groups VII and IIIf) and MYBs (Groups 6 and 7)) related to regulation of flavonoid biosynthesis (Fig. 2d)32–36. And 5 mature miRNAs were also found showed high similarity (>85%) with miRNAs involved in regulating flavonoid biosynthesis in Arabidopsis (e.g.: miR166, miR177 and miR396) may also participated in the flavonoid accumulation in R. nobile (Supplementary Table 13). Taken together, these results suggest continuously strong biosynthesis and concentration of flavonoids in bracts.
Approximately 26.5% (1288 genes) of upregulated genes in bracts are remnants of whole-genome duplications and linked to ecological functions of bracts. These genes were found not only enriched in auxin efflux, plant epidermis and stomatal complex development, flower development and flavonoid biosynthesis, but also enriched in response to blue light, glutathione metabolic process and shoot system morphogenesis (Supplementary Data 7 available on Figshare; 10.6084/m9.figshare.19662933). These results indicate that WGD contributed to bract development in R. nobile.
Overwintering of the perennial root
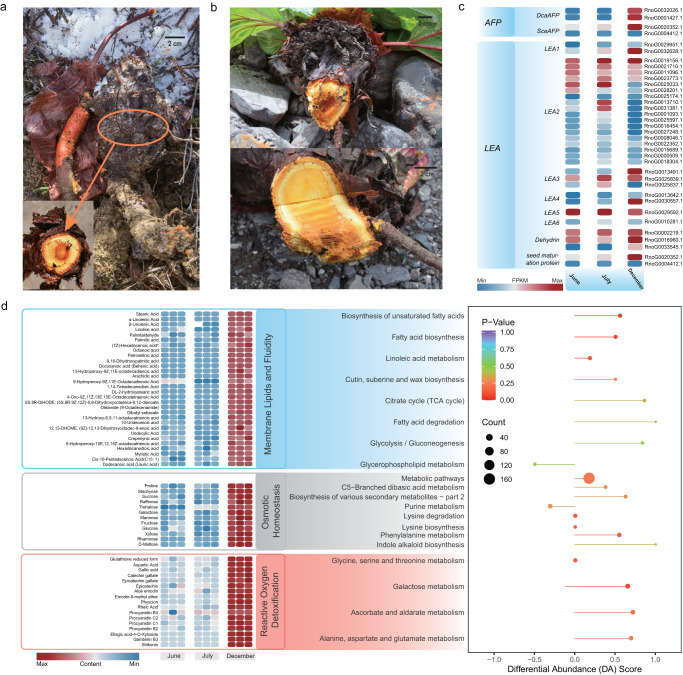
Extremely low temperatures during wintertime in subnival regions pose severe challenges for perennial plants. Overwintering without injury may require multiple adaptations, involving physiological and biochemical modifications, alterations of gene expression, and changes in concentration of specific proteins and metabolites37–40. To assess such changes in roots of R. nobile, root samples collected during the growing season (June and July) and wintertime (December) were subjected to transcriptome and metabolic analyses (Fig. 3a, b, Supplementary Fig. 9 and Supplementary Table 14). Transcription was downregulated during winter of genes involved in regulation of auxin-mediated signaling pathways, plant hormone signal transduction and several biogenesis and developmental processes (Supplementary Data 8 available on Figshare; 10.6084/m9.figshare.19662933). Ribosomal function and biogenesis, however, were maintained in a state of transcriptional readiness during this period to initiate protein synthesis, with transcription of genes involved in these pathways upregulated (Supplementary Data 9 available on Figshare; 10.6084/m9.figshare.19662933). Thus, ribosome biogenesis and recycling of cytosolic ribosomes41,42 continued even at very low temperatures. In addition, the expression of two gene families encoding antifreeze proteins (AFP)43–47, and 11 Late Embryogenesis Abundant (LEA) genes48 involved in cold tolerance of plants, were upregulated in roots during the winter. These changes are likely to increase low temperature tolerance by reducing freezing points of all active systems (Fig. 3c).
Fig. 3. Adaptive genetic basis of perennial root of R. nobile to overwinter.
a, b Images illustrating the morphology of R. nobile in its natural habitat during winter and summer, respectively, with zoomed lateral section of thick taproots sampled for sequencing. c Expression profiles (in fragments per kb of exon per million mapped fragments) of two genes encoding antifreeze proteins in roots collected at three time points (June, July and December). Low to high expression is indicated by colors from blue to red. d Metabolomic regulation of responses to the extremely low temperatures during wintertime in the subnival belt. Levels of three classes of metabolites in roots in indicated months are presented as heatmaps (left). Differential abundance scores (DA Score) calculated for corresponding KEGG pathways are presented in the lollipop chart (right). Horizontal coordinates of DA Scores reflect overall changes in levels of all metabolites in a given metabolic pathway. Scores of 1 and −1 indicate trends of up- and down-regulation of all identified metabolites in the pathway, respectively. The length of the line segment indicates the absolute value of DA Score, and the size of the dots at the end points of each line segment indicates the number of differential metabolites in the pathway. Low to high expression is indicated by colors from blue to red.
Metabolomic analyses provided further evidence for root overwintering ability in extremely cold subnival habitats (Fig. 3d and Supplementary Fig. 9). We identified 1,091 metabolites in root tissues relevant to survival in freezing conditions (Fig. 3d and Supplementary Data 10 available on Figshare; 10.6084/m9.figshare.19662933) with multiple KEGG categories related to biosynthesis of unsaturated fatty acid enriched in overwintering roots (e.g., ‘Biosynthesis of unsaturated fatty acids’, ‘Linolenic acid metabolism’ and ‘Fatty acid biosynthesis’) (Fig. 3d). Similarly, diverse compatible osmolytes were significantly more abundant in winter roots, e.g., proline, which is required to maintain cytosolic acidity and membrane integrity37, and various soluble sugars (sucrose, raffinose, fructose, trehalose, etc.) that can lower a cell’s osmotic potential and stabilize cellular and protein structures (Fig. 3d)37. These modifications likely offset the adverse effects of low temperatures on cell membrane properties and functions caused by reduced membrane fluidity37,49,50. Freezing temperatures can also induce severe oxidative stress through generation of reactive oxygen species (ROS). Excessive accumulation of ROS leads to cellular injury, and ultimately death of plants by damaging the photosystem II reaction center and membrane lipids37. Multiple antioxidant metabolites (e.g., reduced glutathione, procyanidins, gallic acid and epicatechin gallate) were significantly more abundant in winter roots, as were several categories of metabolites related to the Ascorbate-Glutathione (AsA-GSH) cycle (‘Ascorbate and aldarate metabolism’, ‘Alanine, aspartate and glutamate metabolism’, ‘Glycine, serine and threonine metabolism’, etc.), which play key roles in ROS scavenging (Fig. 3d)51,52. These results suggest, therefore, that improved membrane stability, osmolarity, and resistance to ROS during periods of very low temperatures have developed in R. nobile through adjustment of physiological pathways and modification of metabolic profiles.
Notwithstanding the above, comparisons show a major distinction exists between transcriptional and metabolic responses of R. nobile to freezing stress, as overwintering roots had massive levels of associated metabolites but correlated pathways were retained in relatively quiescent transcriptional states. This may arise from a time lag between transcript and metabolite accumulation. It is essential for plants to accumulate sufficient relevant metabolites to survive in sub-zero temperatures during winter, while correlated transcripts may be largely generated during autumn37,53,54.
Discussion
Driven by extreme selection pressures, alpine subnival plants have developed diverse specialized adaptive traits and evolutionary strategies to survive and reproduce in high stress environments3,55. To explore the underlying genetic basis of some of these adaptations, we generated a chromosome-level genome assembly of the ‘glasshouse’ R. nobile, an iconic subnival species that occurs at elevations between 4000 m and 6000 m in the QTP. The R. nobile genome was ~1.57 Gb in size, and 99.33% of the sequences were assigned to 11 pseudochromosomes based on Hi-C auxiliary assembly. The higher scaffolding rate of contigs and all four indices (mapping rate, BUSCO, QV and LAI) highlighted the higher quality of our R. nobile genome assembly than recently published R. nobile assembly56 (Supplementary Table 15). To obtain highly reliable gene models, initial gene set were further filtered and a gene set containing 37,770 genes was finally obtained which number close to its close relative buckwheat, and with high functional completeness (>90%). Thus, a more accurate picture of gene order and genome structure and a valuable genetic resource of alpine plant were provided for future study related to adaptive evolution of plants.
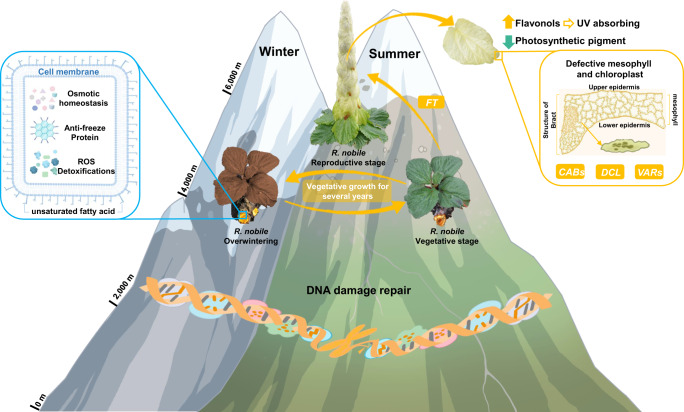
In contrast to previous indications of only one recent WGD event occurring Polygonaceae species (Fagopyrum tataricum) or failure distinguishing for WGD or WGT in R. nobile assembly which published recently, we revealed two recent WGD events shared by Polygonaceae species, dated to ~40.2-45.4 and ~52.5-59.3 Mya, respectively. It was because when multiple round WGD events occurred in a very short time, the Ks distribution or Ks peak fitting may induce errors as only give one peak (corresponding one WGD event). This is a common phenomenon and the errors about the WGDs in Simmondsia chinensis, Carthamus tinctorius and Olea europaea had been reported57. In our study, we identified two adjacent Ks peaks in a very short time (Ks range: 0.3–0.5) (Fig. 1d), suggesting two WGD events occurred in R. nobile and F. tataricum. Our synteny gene dotplot analyses between the two Polygonaceae species and V. vinifera (Fig. 1e and Supplementary Fig. 3a) showed clear evidences of synteny depth ratio of 1:4 between V. vinifera and the two Polygonaceae species, which means every chromosome of V. vinifera could be find four copies in the two Polygonaceae species and also could be another evidence for two rounds of WGDs. Also, the synteny genes between the species were extracted to construct the phylogenetic tree, and most phylogenetic topology of gene trees supporting the shared WGD events between the two species(Supplementary Fig. 3b). Moreover, WGD is considered to contribute greatly to adaptive evolution because it provides additional genetic materials for divergent selection to act on as a consequence of neofunctionalization and increased gene product dosage58,59. Notably in R. nobile, a high retention was evident of duplicated genes that were annotated likely to promote subnival adaptation. Thus, responses to hypoxia and osmotic stress, and cellular responses to lipid were significantly enriched, which is highly suggestive of the important role of WGD events in adaption to extremely freezing temperatures. In addition, expanded gene families were detected that were mainly related to DNA damage repair and leaf development (Fig. 4), possibly enabling adaptation to high UV radiation stress at subnival elevations and formation of the ‘glasshouse’ reproductive structure.
Fig. 4. A scheme of subnival adaptation in R. nobile.
Indicated are: expanded gene families and WGD-retained genes for reducing DNA damage caused by strong UV radiation; accumulation of anti-freeze proteins and metabolites facilitating overwintering.
The innovative development of translucent cream-colored bracts which combine to conceal the inflorescence and produce a pagoda-shaped ‘glasshouse’ phenotype4,7,8, likely insures successful flowering and seed development of this giant monocarpic perennial5,6,9–12,14,15,60. Tissue-specific genetic modifications affecting multiple regulatory pathways are likely associated with the adaptability of this trait. In support, we detected significant downregulation in the bracts of key genes involved in chloroplast and mesophyll development (CABs), palisade morphogenesis (DCL), and photoreception and thylakoid membrane development (VAR1, VAR2 and VAR3). Anatomical abnormality of mesophyll and dramatic decrease of content for both of two photosynthetic pigments (chlorophylls and carotenoids) in bracts further verified the results of transcriptomics, and all above completely reveal specialization of adaptive function of bracts. On the other hand, avoidance of high UV irradiation is likely to result from the significantly enhanced activities detected for enzymes involved in the flavonoid biosynthesis, and instead of only a handful of genes in previous studies, we found whole pathway of flavonoid biosynthesis, even related transcription factors (R2R3-MYB and bHLH) which were confrimed regulate the accumulation flavonoid all exhibit tissue-specific expression. Moreover, retention of a high proportion of duplicated genes up-regulated in bracts and enrichment of genes related to flavonoid biosynthesis suggest that WGD substantially contributed to the innovative function of bracts. In combination, these changes are likely to have increased the translucence of bracts while maintaining a robust tolerance to strong UV exposure.
Overwintering vegetative growth is also critical for this monocarpic perennial to survive subnival extremes over multiple years. However, research on R. nobile is mostly focused on its ‘glasshouse’ reproductive structure, and there is very little research related to the freezing tolerance mechanisms. In addition to the genetic changes involved in the production of the ‘glasshouse’ bract, several changes in gene expression and metabolic accumulation were detected firstly in the roots of R. nobile that are likely to improve their survival during freezing winters. These included alterations to expression of cold tolerance genes leading to accumulation of several protective proteins including antifreeze proteins (AFPs) and late embryogenesis abundance (LEAs), plus changes in concentrations of proteins and metabolites (unsaturated fatty acids, soluble sugars, proline, and antioxidants) enormously enhance accomplishment of basic requirements of carbon source, energy, membrane stability, osmolarity, and resistance against ROS produced during freezing stress (Fig. 4).
In combination, our findings have deepened considerably our current understanding of the likely genetic changes that have occurred to enable ‘glasshouse-like’ plants to survive and reproduce under some of the most hostile conditions that plants grow in.
Materials and methods
Genome sample collection and sequencing
Fresh leaf tissue was sampled from a wild individual of R. nobile growing on Mount Segrila, Tibet, China (29°37′5.87″ N, 94°38′59.84″E, 4,628 m) and immediately stored in liquid nitrogen before sending to Grandomics (Wuhan, China) for genomic sequencing. High molecular weight genomic DNA was prepared by the CTAB method then purified with a QIAGEN® Genomic kit (Cat. No. 13343, QIAGEN). To obtain Illumina short reads, DNA libraries with 500 bp inserts were constructed and sequenced using an Illumina HiSeq 4000 platform. In addition, high-molecular-weight DNA was prepared and used to construct PacBio SMRTbell libraries using a SMRTbell Express Template Prep Kit 2.0, following the manufacturers’ recommended protocols. The SMRTbell libraries were sequenced using a PacBio Sequel II system and consensus (HiFi) reads were generated using ccs software (https://github.com/pacificbiosciences/unanimity). Hi-C (high-throughput chromosome conformation capture) sequencing was performed as follows: sampled DNA was cross-linked with 1% formaldehyde to capture interacting DNA segments, chromatin was digested with the Dpn II restriction enzyme, and libraries were constructed and sequenced using the Illumina HiSeq 4000 platform.
Genome size estimation and assembly
Before estimating genome sizes, short Illumina reads were filtered using fastp (v.0.20.0)61 with default parameters. Clean reads were then used to generate K-mer (21 bp) frequencies by Jellyfish (v.2.2.10)62, and the resulting histogram was exported into GenomeScope (v.1.0.0)63. Hifiasm (0.15.3-r339)64 was used for de novo assembly of R. nobile’s genome with default parameters with HiFi sequencing data. A subprogram of Purge Haplotigs (v.1.1.2)65 and purge_dup (v.1.2.5)66 was used to identify uncollapsed duplications. The quality of the assembly was comprehensively sessed by using four methods: (i) Mapping the Illumina paired-end reads to our final assembly shows high completeness of the genome when high mapping rates are obtained; (ii) BUSCO (Benchmarking Universal Single-Copy Orthologs) (v.5.2.1)67 was used with the embryophyta_odb10 database and a high percent of complete BUSCOs also indicates high completeness of the genome; (iii) the consensus quality value (QV score) evaluated using Merqury (v.1.3)68 indicates high base accuracies of the genome with a high QV score; (iv) the long terminal repeat (LTR) Assembly Index (LAI) evaluated using LTR_retriver (v2.8.5)69 serves as the gold standard for genome benchmarking when LAI > 20. Clean Hi-C data were mapped to contig sequences by BWA-MEM (0.7.10-r789)70 and valid interaction pairs were extracted. Based on those chromatin interactions, 3D-DNA (v.180922)71 was employed to cluster, order, and orient the contigs into pseudo-chromosomes. Juicebox72 was used to visualize the chromatin interactions among the assembled pseudo-chromosomes, and then we manually corrected and validated the obvious Hi-C assembly errors to generate the final chromosome assembly.
Repeat element identification and gene prediction
RepeatMasker (v.4.1.0)73 and RepeatProteinMasker (v.4.1.0)73 were used to identify repetitive elements in noble rhubarb genome based on homology alignments between our assembly sequences and Repbase (v.16.10). We then applied the de novo approach to improve the sensitivity of our repeat identification. Briefly, RepeatModeler (v.2.0)74 and LTR_Finder (v.1.06)75 were used to construct a repeat library, then RepeatMasker73 was employed to generate de novo predictions.
A combination of transcriptome-based, homology-based, and de novo approaches was used to accurately predict high-quality protein-coding genes. To predict genes ab initio, Augustus (v.3.2.3)76, GenScan (v.2.0)77, and GlimmerHMM (v.3.0.4)78 were employed with the Arabidopsis thaliana training set. GeMoMa (v.1.7.1)79 was used for homology-based prediction, together with protein sequences from A. thaliana80, Beta vulgaris19, Fagopyrum tataricum17, Prunus persica81, and Spinacia oleracea82 (Supplementary Table 16). For transcriptome-based prediction, the fresh leaf tissues that from the genome sequencing individual were collected for the annotation, then the de novo transcriptome assemblies of the leaf tissues were aligned to the genomes to resolve gene structures using PASA. EVidenceModeler (EVM, v.1.1.1)83 was used to generate consensus sets of gene models obtained using the three approaches (transcriptome-based, homology-based, and de novo approaches). To obtain highly reliable gene models, we fitted genes that were only supported by transcriptome-based prediction and with just one exon, and also those only supported by the ab initio process with less than three exons. Protein-coding genes were functionally annotated by BLASTP (v.2.7.1+)84 (E-value < 1 × 10−5) searches against SwissProt and TrEMBL databases. InterProScan (v.5.28)85 was used to annotate protein domains by searching the InterPro databases. Gene Ontology (GO) terms for each gene were obtained from the corresponding InterProScan results. Pathways in which each gene might be involved were assigned by BLAST searches against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database86. Transcription factors in noble rhebarb were detected using iTAK87. Non-coding RNAs (ncRNAs) were annotated using cmscan from INFERNAL (v1.1.2) (http://eddylab.org/infernal). The mature miRNAs were further identified using miRPara (v6.0)88 based on the results of cmscan subprogram, and the target genes of the mature miRNAs were predicted by psRobot (v1.2)89 with default parameters.
Phylogenetic analysis and expansion/contraction of gene families
To investigate the evolutionary trajectories of R. nobile, we selected 14 other species for phylogenetic analysis (Supplementary Table 16): Arabidopsis thaliana80, Beta vulgaris19, Camellia sinensis90, Fragaria vesca91, Fagopyrum tataricum17, Helianthus annuus92, Oryza sativa93, Prunus persica81, Simmondsia chinensis20, Solanum lycopersicum94, Solanum oleracea82, Solanum tuberosum95, Vitis vinifera23, and Zea mays96. An all-vs-all BLASTP84 search (E-value cutoff: 1 × 10−5) was first employed to generate similarity information for all genes. Next, we identified high-quality single-copy genes by applying OrthoMCL (v. 2.0.9–4)97, and constructed a concatenation tree and clusters of gene trees using IQ-TREE (v. 2.0.3-h176a8bc_0, with ‘-m MFP –bb 1000’ settings)98 and a coalescent tree by ASTRAL (v.5.7.8)99. We further estimated divergence times between species with MCMCtree (v.4.8)100 of the PAML package (v.4.8)101. Divergence times between A. thaliana and V. vinifera (115-130 Mya) and B. vulgaris and S. oleracea (22-30 Mya) acquired from TimeTree (http://www.timetree.org/)102 were used as calibration points. Gene family expansions and contractions were further estimated by CAFÉ (v.4.2)103 using the gene cluster information and estimated time tree. The parameter λ was estimated along each branch with the random model, and gene families were classified into four types: expanded, contracted, unique, or unchanged.
Detection of WGD events
Two Polygonaceae species (Rheum nobile and Fagopyrum tataricum), together with Simmondsia chinensis and B. vulgaris of the Caryophyllales, were used for WGD analyses. Synteny blocks and collinear genes were identified by WGDI (v.0.5.3)104 with ‘-icl’ within each species and between Polygonaceae species. Numbers of synonymous substitutions per synonymous site (Ks) between collinear genes were also estimated by ‘-ks’ in WGDI, and a median Ks value was selected to represent each syntenic block, with Ks peak fitting also performed by WGDI with ‘-pf’. Dot plots of collinear genes and synteny blocks were used to obtain syntenic ratios between the species to confirm the polyploidy level of each species. We further used the collinear genes for phylogenomic analyses to check if detected WGDs occurred independently in the histories of R. nobile and F. tataricum. Collinear genes between pairs of these two Polygonaceae species were extracted by WGDI with ‘-at’, and IQ-TREE (v. 2.0.3-h176a8bc_0)98 was used to construct gene trees. For each gene tree, we randomly rooted a collinear gene from one species then checked whether the retained collinear genes from that species could be clustered as a monophyletic clade to support the hypothesis that independent WGDs occurred in these two species’ histories. Finally, we calculated the frequency of gene trees that supported independent WGDs for each species.
Estimation of TE insertion times
The 5ʹ-LTR is usually identical to the 3′-LTR when a retrotransposon is inserted. Thus, in this analysis only LTR sequences identified with complete 5ʹ-LTR and 3ʹ-LTR were used. Each of the 5ʹ-LTR flanking sequences and 3′-flanking sequences was aligned by MUSCLE (v.3.8.31)105 with default parameters and evolutionary distances of aligned sequences were calculated by disMat (EMBOSS: v.6.6.0.0, with parameters -nucmethod 2)106. The mutation rates (per base per year) were claculted by r8s107 (1.6 × 10−9 for R. nobile), then the insertion times were further calculated using the formula: T = K/2r (divergence between LTRs/substitution per site per year).
Chlorophyll and carotenoid concentrations
Total chlorophyll and carotenoids were extracted from bracts, transitional leaves and rosette leaves using the dimethylsulfoxide (DMSO) method108. The absorbance of each resulting extract (with three technical replicates of each of three biological replicates) was spectrophotometrically measured at 647, 663, 652 and 440 nm wavelengths. Total chlorophyll, chlorophyll a/b and carotenoid contents were calculated109.
Cross-sectioning and histological staining
The rosette leavea and bracteal leaves were cut into 0.5–1 cm2 fragments, fixed in FAA solution, and embedded in paraffin. A rotary microtome (RM2235, Leica) was used to section the embedded fragments to a thickness of 10 μm. The sections were stained with 0.1% toluidine blue and observed using an optical microscope (Zeiss, Germany).
Transcriptome sequencing and analysis
For transcriptome analysis, bract, transitional leaf, rosette leaf, and root samples of R. nobile were collected, with three replicates per tissue, on 19 August 2020 and 5 June, 6 June, 6 July, and 4 December 2021. Information on sample collection procedure for each tissue type is given in Supplementary Tables 12 and 14. Total RNA extraction, library construction and sequencing were performed by BGI-Shenzhen Company (Wuhan, China) using a MGI2000 platform and 2 × 150 bp pair-end model. After filtering low quality reads by fastp, clean reads were mapped to the R. nobile genome assembly using HISAT2 (v.2.2.1)110. StringTie (v.2.1.2)111 was used to predict new transcripts, which were combined with gene annotations to obtain a final transcriptome set. DEseq2 (v.1.22.2)112 was used to identify differentially expressed genes (DEGs), defined as those with |log2(fold change) |>1 and FDR significance score (Padj) < 0.05. DEGs were subjected to KEGG and GO enrichment analysis using clusterProfiler113.
Metabolite profiling and analysis
For metabolomic analysis, bract, transitional leaf and rosette leaf, and root samples of R. nobile were collected, with three replicates per tissue (leaves and roots on 6 July 2021, and overwintering on 4 December 2021). Information on sample collection procedure for each tissue type is given in Supplementary Tables 12 and 14. Total metabolite extraction and analysis were performed by Mateware Company (Wuhan, China) using a SHIMADZU Nexera X2 ultra-high pressure liquid chromatography system (www.shimadzu.com.cn) coupled to an Applied Biosystems 4500 Q TRAP electrospray ionization-tandem mass spectrometry system (www.appliedbiosystems.com.cn/). Metabolite data were log2-transformed for statistical analysis to improve normalization. Metabolites from 18 samples were used for Hierarchical Clustering Analysis (HCA), Principal Component Analysis (PCA), and Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) using R software to study accession-specific metabolite accumulation, with significant p and fold change thresholds set to 0.05 and 2.0, respectively. Venn diagrams were used to illustrate the distribution of metabolites, and the KEGG database was used to identify metabolites that were differentially expressed (with a significance threshold of p < 0.01) in (i) bracts compared to transitional leaf and rosette leaves, and (ii) between roots in summer and winter.
Analysis of photosynthetic pigment biosynthesis genes
Chlorophyll biosynthesis involves 15 catalytic steps from L-glutamyl-tRNA to chlorophyll a and chlorophyll b114. Genes encoding these 15 enzymes reportedly play key roles in changes in chlorophyll content in A. thaliana (https://www.arabidopsis.org/). Similarly, carotenoid synthesis involves 15 steps from geranylgeranyl pyrophosphate (GGPP) and 13 enzymatic reactions (KEGG: map00906), with corresponding genes being key determinants of changes in carotene content in A. thaliana. Thus, proteins encoded by these Arabidopsis genes were used to search for homologs in the predicted proteome of R. nobile using BLASTP84 with an e-value of 1e-5, >40% identity value and >40% coverage. In addition, domains of these A. thaliana genes were identified with hmmscan115, then applied in further searches against the R. nobile proteome using hmmsearch115.
Genes related to chloroplast and mesophyll development
Genes reportedly involved in regulation of chloroplast development and mesophyll differentiation were downloaded. Nine (ARC, CAB, CUE1, HB1, PHTO2, SCA3, VAR1, VAR2, and VAR3) were retrieved from the A. thaliana genome and one (DCL) from the tomato genome. These genes were used as queries for BLASTP searches with an e-value of 1e−5, >40% identity value and >40% coverage. Domains of these genes were identified with hmmscan115 and further used to search against the proteome using hmmsearch115.
Genes related to flavonoid biosynthesis
Genes related to flavonoid biosynthesis were retrieved from A. thaliana (KEGG: map00941) and homologs in the R. nobile genome were identified based on the constructed gene families. Homologs of several bHLH and MYB transcription factors that can activate multiple genes’ co-expression and play important roles in flavonoid biosynthesis were identified using iTAK software. Phylogenetic trees of bHLH and MYB proteins were constructed using iqtree.
Antifreeze protein analysis
Antifreeze protein (AFP) improves an organisms’ ability to tolerate low temperatures, but has not been reported in the model plant A. thaliana. Thus, we identified homologs of AFPs from Secale cereale (Gene bank ID: AY590122, AY843521 and AY843522) and Daucus carota (Gene bank ID: A91926.1). These proteins were then used to search for homologs in the predicted proteome of R. nobile using BLASTP84 with an e-value of 1e−5, >40% identity value and >40% coverage. Moreover, HMM profiles of LEAs (LEA1, PF03760; LEA2, PF03168; LEA3, PF03242; LEA4, PF02987; LEA5, PF00477; LEA6, PF10714; Dehydrin, PF0025; SMP, PF04927) were downloaded from the Pfam database (http://pfam.sanger.ac.uk/) and HMMER115 was used to search the encoded protein sequences with default parameters and a filter threshold of 1e−5.
Statistics and reproducibility
The functional enrichment analysis was performed using the ClusterProfile. The statistical significance of GO terms was evaluated using Fisher’s exact test in combination with FDR correction for multiple testing (P < 0.05). All experiments were carried out at least three times, independently, with similar results. All values are presented as means ± SD. Statistical significance was based on t tests. For RNA-seq and metabolite profiling, three biological repeats were used. For and chlorophyll and carotenoid concentrations, three biological replicates, each represented by three technical replicates from the same individual were used. And the software version and parameters applications in this study were listed in Supplementary Tables 17.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Financial support was equally provided by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31000000 to J.L. and Y.Y.), the Key Science & Technology Project of Gansu Province (22ZD6NA007 to Y.Y.), the Science Fund for Creative Research Groups of Gansu Province (21JR7RA533 to Y.Y.), the Young Talent Development Project of State Key Laboratory of Herbage Improvement and Grassland Agro-ecosystems (No. 2021 + 02 to Y.Y.), andInternational Collaboration 111 Programme (BP0719040). All the computation works were supported by Supercomputing Center of Lanzhou University and Big Data Computing Platform for Western Ecological Environment and Regional Development.
Author contributions
Y.Z.Y. and J.L. led and designed this project. Y.L., Z.N., M.L., C.D., Y.W., D.D.W., Z.L. and Y.Z.Y. performed sample collection. Z.N. and Y.L. performed all the field work and experiments. Y.L., R.X and J.Z. carried out the genome assembly and annotation. M.Z., Z.W. and D.D.W. performed the whole-genome duplication analyses. Y.L. performed the genome and gene family evolution analyses. Z.Z. and H.H. performed the population analyses. Y.B.Y. and J.Y. carried out the typical habitat figure of Noble rhubarb. J.L., Y.Z.Y. and J.L. wrote the manuscript and R.A. and D.S.W. polished the English writing. All of the authors read and approved the final manuscript.
Peer review
Peer review information
Communications Biology thanks Yang Jae Kang, Aureliano Bombarely and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: David Favero.
Data availability
The genome assembly file and genome annotation files (contig level and chromosome level) are available at figshare (10.6084/m9.figshare.19662933). All genomic data (short-reads sequencing data, long-reads sequencing data and HiC sequencing data) and transcriptome data have been deposited at NCBI under the BioProject accession numbers of PRJNA830994 and PRJNA831329, respectively. The source data behind the graphs in Figs. 1d, 2a and 3d are available at Figshare (10.6084/m9.figshare.19662933) as Supplementary Data 2, 5 and 10, respectively. All other Supplementary Datas are also available at Figshare (10.6084/m9.figshare.19662933). All other data are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ying Li, Zhimin Niu.
Contributor Information
Jianquan Liu, Email: liujq@nwipb.ac.cn.
Yongzhi Yang, Email: yangyongzhi2008@gmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05271-6.
References
- 1.Yang Y, et al. Advances in the studies of plant diversity and ecological adaptation in the subnival ecosystem of the Qinghai-Tibet plateau. Chin. Sci. Bull. 2019;64:2856–2864. [Google Scholar]
- 2.Zhang Y, Li B, Zheng D. Datasets of the boundary and area of the Tibetan plateau. Acta Geographica Sin. 2014;69:65–68. [Google Scholar]
- 3.Sun H, et al. Survival and reproduction of plant species in the Qinghai–Tibet plateau. J. Syst. Evolut. 2014;52:378–396. [Google Scholar]
- 4.Hooker, J. & Fitch, W. Illustrations of Himalayan plants. (Lovell Reeve, 1855).
- 5.Song B, et al. Ultraviolet screening increases with elevation in translucent bracts of Rheum nobile (Polygonaceae), an alpine ‘glasshouse’ plant from the high Himalayas. Botanical J. Linn. Soc. 2020;193:276–286. [Google Scholar]
- 6.Zhang D, et al. Ecological functions and differentially expressed transcripts of translucent bracts in an alpine ‘glasshouse’ plant Rheum nobile (Polygonaceae) Planta. 2010;231:1505–1511. doi: 10.1007/s00425-010-1133-x. [DOI] [PubMed] [Google Scholar]
- 7.Ohba, H. The alpine flora of the Nepal Himalayas: An introductory note. The Himalayan plants (University of Tokyo Press, 1988).
- 8.Lancaster, R. Plant hunting in Nepal. (Croom Helm, 1981).
- 9.Omori Y, Ohba H. Pollen development of Rheum nobile Hook.f. & Thomson (Polygonaceae), with reference to its sterility induced by bract removal. Botanical J. Linn. Soc. 1996;122:269–278. [Google Scholar]
- 10.Wang L, et al. Genome-Scale Transcriptome Analysis of the Alpine “Glasshouse” Plant Rheum nobile (Polygonaceae) with Special Translucent Bracts. PLoS ONE. 2014;9:e110712. doi: 10.1371/journal.pone.0110712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwashina T, et al. Flavonoids in translucent bracts of the Himalayan Rheum nobile (Polygonaceae) as ultraviolet shields. J. Plant Res. 2004;117:101–107. doi: 10.1007/s10265-003-0134-2. [DOI] [PubMed] [Google Scholar]
- 12.Omori Y. Selective light transmittance of translucent bracts in the Himalayan giant glasshouse plant Rheum nobile Hook.f. & Thomson (Polygonaceae) Botanical J. Linn. Soc. 2000;132:19–27. [Google Scholar]
- 13.Song, B., Stoll, P., Peng, D., Sun, H. & Stöcklin, J. Demography of the giant monocarpic herb Rheum nobile in the Himalayas and the effect of disturbances by grazing. Ann. Botany mcz178 10.1093/aob/mcz178 (2019). [DOI] [PMC free article] [PubMed]
- 14.Song B, et al. A new pollinating seed‐consuming mutualism between R heum nobile and a fly fungus gnat, B radysia sp., involving pollinator attraction by a specific floral compound. N. Phytol. 2014;203:1109–1118. doi: 10.1111/nph.12856. [DOI] [PubMed] [Google Scholar]
- 15.Song B, et al. Multifunctional bracts enhance plant fitness during flowering and seed development in Rheum nobile (Polygonaceae), a giant herb endemic to the high Himalayas. Oecologia. 2013;172:359–370. doi: 10.1007/s00442-012-2518-2. [DOI] [PubMed] [Google Scholar]
- 16.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506:89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, et al. The Tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol. Plant. 2017;10:1224–1237. doi: 10.1016/j.molp.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, L., Chen, C., Mao, X. & Liu, J. Phylogenomics and evolutionary diversification of the subfamily Polygonoideae. J. Sytem. Evolut. jse.12913 10.1111/jse.12913 (2022).
- 19.Dohm JC, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris) Nature. 2014;505:546–549. doi: 10.1038/nature12817. [DOI] [PubMed] [Google Scholar]
- 20.Sturtevant D, et al. The genome of jojoba (Simmondsia chinensis): A taxonomically isolated species that directs wax ester accumulation in its seeds. Sci. Adv. 2020;6:eaay3240. doi: 10.1126/sciadv.aay3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao Y, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vekemans D, et al. Gamma paleohexaploidy in the stem lineage of core eudicots: Significance for MADS-Box gene and species diversification. Mol. Biol. Evolut. 2012;29:3793–3806. doi: 10.1093/molbev/mss183. [DOI] [PubMed] [Google Scholar]
- 23.The French–Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, et al. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinforma. 2006;7:447. doi: 10.1186/1471-2105-7-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Culligan K, Dixon RA, Chory J. CUE1: A mesophyll cell-specific positive regulator of light-controlled gene expression in arabidopsis. Plant Cell. 1995;7:1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozuka T, Kong S-G, Doi M, Shimazaki K, Nagatani A. Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell. 2011;23:3684–3695. doi: 10.1105/tpc.111.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto W. Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 2003;78:1–9. doi: 10.1266/ggs.78.1. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto W, Tamura T, Hanba-Tomita Y, Sodmergen, Murata M. The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles: VAR1 encodes a chloroplastic FtsH. Genes Cells. 2002;7:769–780. doi: 10.1046/j.1365-2443.2002.00558.x. [DOI] [PubMed] [Google Scholar]
- 29.Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated Regulation and Complex Formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, Chloroplastic FtsH Metalloproteases Involved in the Repair Cycle of Photosystem II in Arabidopsis Thylakoid Membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Næsted H, et al. Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. J. Cell Sci. 2004;117:4807–4818. doi: 10.1242/jcs.01360. [DOI] [PubMed] [Google Scholar]
- 31.Keddie JS, Carroll B, Jones JD, Gruissem W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 1996;15:4208–4217. [PMC free article] [PubMed] [Google Scholar]
- 32.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 33.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 34.Fraser LG, et al. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genomics. 2013;14:28. doi: 10.1186/1471-2164-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hichri I, et al. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 36.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family[W] Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha, S. et al. The Omics of Cold Stress Responses in Plants. in Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives, Volume 2 (ed. Pandey, G. K.) 143–194 (Springer New York, 2015). 10.1007/978-1-4939-2540-7_6.
- 38.Li J, et al. De Novo Transcriptome Sequencing and the Hypothetical Cold Response Mode of Saussurea involucrata in Extreme Cold Environments. IJMS. 2017;18:1155. doi: 10.3390/ijms18061155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, L. et al. Insights into the plateau adaptation of Salvia castanea by comparative genomic and WGCNA analyses. Journal of Advanced Research S2090123222000480 10.1016/j.jare.2022.02.004 (2022). [DOI] [PMC free article] [PubMed]
- 40.Zhang Z, et al. Adaptation to Extreme Antarctic Environments Revealed by the Genome of a Sea Ice Green Alga. Curr. Biol. 2020;30:3330–3341.e7. doi: 10.1016/j.cub.2020.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Beine-Golovchuk O, et al. Plant Temperature Acclimation and Growth Rely on Cytosolic Ribosome Biogenesis Factor Homologs1[OPEN] Plant Physiol. 2018;176:2251–2276. doi: 10.1104/pp.17.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H, et al. HsfA1d promotes hypocotyl elongation under chilling via enhancing expression of ribosomal protein genes in Arabidopsis. N. Phytol. 2021;231:646–660. doi: 10.1111/nph.17413. [DOI] [PubMed] [Google Scholar]
- 43.Griffith M, Ala P, Yang DSC, Hon W-C, Moffatt BA. Antifreeze Protein Produced Endogenously in Winter Rye Leaves 1. Plant Physiol. 1992;100:593–596. doi: 10.1104/pp.100.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu M, Lu C, Shen X, Yin W. Characterization and function analysis of a cold-induced AmCIP gene encoding a dehydrin-like protein in Ammopiptanthus mongolicus. DNA Seq. 2006;17:342–349. doi: 10.1080/10425170600908484. [DOI] [PubMed] [Google Scholar]
- 45.Smallwood M, et al. Isolation and characterization of a novel antifreeze protein from carrot (Daucus carota) Biochem J. 1999;340:385–391. [PMC free article] [PubMed] [Google Scholar]
- 46.Worrall D, et al. A Carrot Leucine-Rich-Repeat Protein That Inhibits Ice Recrystallization. Science. 1998;282:115–117. doi: 10.1126/science.282.5386.115. [DOI] [PubMed] [Google Scholar]
- 47.Yeh S, et al. Chitinase Genes Responsive to Cold Encode Antifreeze Proteins in Winter Cereals. Plant Physiol. 2000;124:1251–1264. doi: 10.1104/pp.124.3.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bies-Ethève N, et al. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008;67:107–124. doi: 10.1007/s11103-008-9304-x. [DOI] [PubMed] [Google Scholar]
- 49.Ding Y, Shi Y, Yang S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. N. Phytol. 2019;222:1690–1704. doi: 10.1111/nph.15696. [DOI] [PubMed] [Google Scholar]
- 50.Kidokoro, S., Shinozaki, K. & Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. S1360138522000085 10.1016/j.tplants.2022.01.008 (2022). [DOI] [PubMed]
- 51.Foyer CH, Noctor G. Ascorbate and Glutathione: The Heart of the Redox Hub1. Plant Physiol. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noctor G, Foyer CH. ASCORBATE AND GLUTATHIONE: Keeping Active Oxygen Under Control. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 53.Guo X, Liu D, Chong K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018;60:745–756. doi: 10.1111/jipb.12706. [DOI] [PubMed] [Google Scholar]
- 54.Wanner LA, Junttila O. Cold-Induced Freezing Tolerance in Arabidopsis. Plant Physiol. 1999;120:391–400. doi: 10.1104/pp.120.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, X. et al. Enhanced habitat loss of the Himalayan endemic flora driven by warming-forced upslope tree expansion. Nature Ecology & Evolution10.1038/s41559-022-01774-3 (2022). [DOI] [PubMed]
- 56.Feng T, et al. The genome of the glasshouse plant noble rhubarb (Rheum nobile) provides a window into alpine adaptation. Commun. Biol. 2023;6:706. doi: 10.1038/s42003-023-05044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, et al. A high-quality Buxus austro-yunnanensis (Buxales) genome provides new insights into karyotype evolution in early eudicots. BMC Biol. 2022;20:216. doi: 10.1186/s12915-022-01420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otto SP. The Evolutionary Consequences of Polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 59.Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Terashima I, Masuzawa T, Ohba H. Photosynthetic characteristics of a giant alpine plant, Rheum nobile Hook. f. et Thoms. and of some other alpine species measured at 4300 m, in the Eastern Himalaya, Nepal. Oecologia. 1993;95:194–201. doi: 10.1007/BF00323490. [DOI] [PubMed] [Google Scholar]
- 61.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vurture GW, et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinforma. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guan D, et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36:2896–2898. doi: 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 68.Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21:245. doi: 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ou S, Jiang N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dudchenko O, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durand NC, et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cels. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. BioinformaticsChapter 4, Unit 4.10 (2009). [DOI] [PubMed]
- 74.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 78.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 79.Keilwagen J, Hartung F, Grau J. GeMoMa: Homology-Based Gene Prediction Utilizing Intron Position Conservation and RNA-seq Data. Methods Mol. Biol. 2019;1962:161–177. doi: 10.1007/978-1-4939-9173-0_9. [DOI] [PubMed] [Google Scholar]
- 80.Zapata, L. et al. Chromosome-level assembly of Arabidopsis thaliana L er reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. U.S.A. 113, E4052-60 (2016). [DOI] [PMC free article] [PubMed]
- 81.Verde I, et al. The Peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics. 2017;18:225. doi: 10.1186/s12864-017-3606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu C, et al. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 2017;8:15275. doi: 10.1038/ncomms15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng Y, et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant. 2016;9:1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 88.Wu Y, Wei B, Liu H, Li T, Rayner S. MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinforma. 2011;12:107. doi: 10.1186/1471-2105-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu H-J, Ma Y-K, Chen T, Wang M, Wang X-J. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012;40:W22–W28. doi: 10.1093/nar/gks554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xia E-H, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 91.Buti M, et al. The genome sequence and transcriptome of Potentilla micrantha and their comparison to Fragaria vesca (the woodland strawberry) Gigascience. 2017;7:giy010. doi: 10.1093/gigascience/giy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Badouin H, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 93.Goff SA, et al. A Draft Sequence of the Rice Genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 94.The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barchi L, et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019;9:11769. doi: 10.1038/s41598-019-47985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiao Y, et al. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524–527. doi: 10.1038/nature22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mirarab S, et al. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30:i541–i548. doi: 10.1093/bioinformatics/btu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Puttick, M. N. MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics btz554 10.1093/bioinformatics/btz554 (2019). [DOI] [PubMed]
- 101.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evolut. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 102.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 103.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 104.Sun, P. et al. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. bioRxiv 10.1101/2021.04.29.441969 (2021). [DOI] [PubMed]
- 105.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 107.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 108.Hiscox JD, Israelstam GF. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979;57:1332–1334. [Google Scholar]
- 109.Lichtenthaler H. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148C:350–382. [Google Scholar]
- 110.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. https://www.liebertpub.com/doi/epdf/10.1089/omi.2011.0118 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed]
- 114.Von Wettstein D, Gough S, Kannangara CG. Chlorophyll biosynthesis. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinforma. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly file and genome annotation files (contig level and chromosome level) are available at figshare (10.6084/m9.figshare.19662933). All genomic data (short-reads sequencing data, long-reads sequencing data and HiC sequencing data) and transcriptome data have been deposited at NCBI under the BioProject accession numbers of PRJNA830994 and PRJNA831329, respectively. The source data behind the graphs in Figs. 1d, 2a and 3d are available at Figshare (10.6084/m9.figshare.19662933) as Supplementary Data 2, 5 and 10, respectively. All other Supplementary Datas are also available at Figshare (10.6084/m9.figshare.19662933). All other data are available from the corresponding authors on reasonable request.