In the Annals of Translational Medicine, the paper titled “Oncolytic adenovirus-mediated dual knockdown of survivin and OCT4 improves therapeutic efficacy in esophageal cancer” (1) describes a gene therapy approach for treating esophageal cancer using an oncolytic adenovirus to deliver silencing short hairpin RNA (shRNA) targeting survivin and OCT4 genes. The study found that this approach led to a significant reduction in the expression of both genes and resulted in enhanced cancer cell death in vitro and tumor growth inhibition in vivo with direct intra-tumoral injection.
The authors used a dual-promoter adenovirus vector which allowed for the efficient delivery of two different shRNAs simultaneously to the cancer cells. The study provides a promising therapeutic strategy for esophageal cancer and highlights the potential of using oncolytic adenoviruses to deliver dual gene knockdown as a cancer treatment.
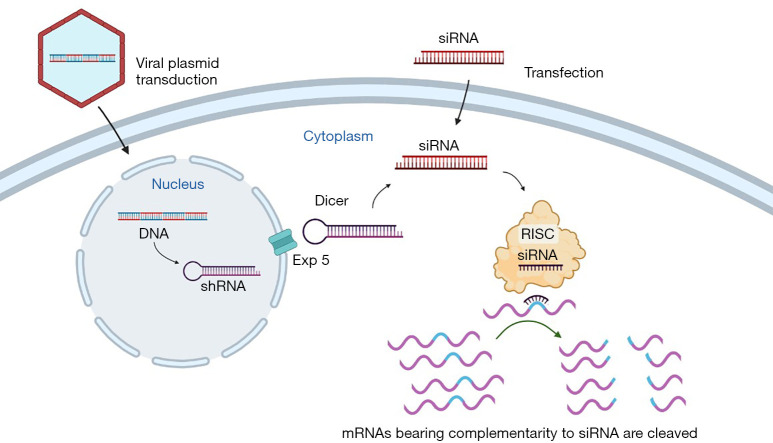
There are two main strategies for use in gene silencing, vector-based shRNAs and double-stranded small interfering RNAs (siRNAs). They have different mechanisms of action for protein knockdown. The pathway for shRNA is more complex. The adenovirus-DNA plasmids enter the nucleus. After that, shRNAs are produced and then exported to cytoplasm via exportin-5 after it is processed by Drosha. Dicer removes the loop sequence of shRNA in the cytoplasm. Following this, the next steps are the same as for siRNAs. The siRNA associates with RNA-induced silencing complex (RISC) uses the siRNA as a template for recognizing complementary mRNA. It binds to the mRNA and cleaves it to silence its expression (Figure 1) (2,3).
Figure 1.
The mechanisms of silencing for siRNA and shRNA. The figure is prepared by https://www.biorender.com under license. siRNA, small interfering RNA; shRNA, short hairpin RNA; RISC, RNA-induced silencing complex.
The double-stranded siRNAs and vector-based shRNAs are two fundamental methods to RNA interfering. While both shRNAs and siRNAs can be used to inhibit protein expression, their methods of action differ. For shRNA, the pathway is more intricate. Plasmids with Adenovirus DNA reach the nucleus. After being expressed in the nucleus, shRNAs are processed by Drosha and transported to the cytoplasm by Exportin-5, where they interact with Dicer and the loop sequence is removed. They continue after this in a manner similar to that of siRNAs (introduced into cells as short duplexes and identified by Dicer). A multiprotein complex is known as the RISC. RISC recognizes complementary mRNA by using siRNA or miRNA as a template. Following the removal of one of the RNA strands by RISC, they target mRNAs with a complementary sequence, resulting in their degradation (Figure 1) (2,3).
Oncolytic adenoviruses are viruses that have been modified to selectively infect and kill cancer cells. They are designed to replicate within cancer cells, leading to their destruction. The safety of oncolytic adenoviruses is a major concern because of the potential for the virus to cause harm to normal cells. While the virus is generally safe and well-tolerated, there have been reports of serious side effects, including immune responses, and cytotoxicity, as well as cancer (4).
An alternative strategy to silence these genes is nanoparticle delivery. Nanoparticles are small particles (usually less than 100 nm in diameter) that can be engineered to deliver drugs or other therapeutic agents directly to cancer cells. They are generally considered safe because they are biocompatible, biodegradable, and can be eliminated from the body relatively quickly. However, there have been some concerns about the potential toxicity of certain types of nanoparticles, particularly if they accumulate in certain organs or tissues (5-7).
Another important consideration is the incorporation of the therapeutic agent into the patient’s genome. While RNA delivered via nanoparticles does not integrate into the genome, DNA plasmids for shRNA as used in this paper can, and this can be a potential safety issue. If the virus integrates into the patient’s DNA, it can function for long periods and could potentially cause mutations and lead to cancer or other diseases (8,9).
There are many challenges regarding using shRNA or siRNA. One disadvantage of siRNA delivery is that the siRNA concentration becomes diluted as cells divide. Also due to high concentration of siRNA in cytoplasm, there is risk of off-target effects. There are many drawbacks besides the advantages of shRNA. Using shRNA may generate stable knockdown cell lines which is a danger. Stable shRNA production is time-consuming, since it may take months to prepare (10).
In addition, shRNA therapies are based on DNA plasmids. The shRNA is produced by DNA. But unlike RNA, DNA can be incorporated into the genome and function for a long time. RNA is quickly degraded, but not DNA. This raises safety concerns for the DNA approach (11-13).
ShRNA is often expressed abundantly. Cells cannot process it accurately. Overexpression can occur in loss of target-specificity. ShRNA treatment has the additional drawback of being unsuitable for certain cell types (10).
Overall, both nanoparticle and oncolytic adenovirus therapies have potential benefits and drawbacks, and safety is a critical consideration for both. Nanoparticles are generally considered safe and have a short duration of expression, while oncolytic adenoviruses have a longer duration of expression and also carry potential safety concerns, such as the risk of integration into the genome.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: The study was supported by Johns Hopkins Research Gift Fund (JWH).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Translational Medicine. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-23-1646/coif). The authors have no conflicts of interest to declare.
References
- 1.Li C, Zhu M, Lu Q, et al. Oncolytic adenovirus-mediated dual knockdown of survivin and OCT4 improves therapeutic efficacy in esophageal cancer. Ann Transl Med 2023;11:193. 10.21037/atm-22-4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Keefe EP. siRNAs and shRNAs: Tools for protein knockdown by gene silencing. Mater Methods 2013;3,197. 10.13070/mm.cn.3.197 [DOI] [Google Scholar]
- 3.Lingor P. Regulation of Cell Death and Survival by RNA Interference – The Roles of miRNA and siRNA. In: Cecconi F, D'Amelio M. editors. Apoptosome. Dordrecht: Springer; 2010:95-117. [Google Scholar]
- 4.Keshavarz M, Mohammad Miri S, Behboudi E, et al. Oncolytic virus delivery modulated immune responses toward cancer therapy: Challenges and perspectives. Int Immunopharmacol 2022;108:108882. 10.1016/j.intimp.2022.108882 [DOI] [PubMed] [Google Scholar]
- 5.Jin S, Ye K. Nanoparticle-mediated drug delivery and gene therapy. Biotechnol Prog 2007;23:32-41. 10.1021/bp060348j [DOI] [PubMed] [Google Scholar]
- 6.Vago R, Collico V, Zuppone S, et al. Nanoparticle-mediated delivery of suicide genes in cancer therapy. Pharmacol Res 2016;111:619-41. 10.1016/j.phrs.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 7.Hashida M. Role of pharmacokinetic consideration for the development of drug delivery systems: A historical overview. Adv Drug Deliv Rev 2020;157:71-82. 10.1016/j.addr.2020.06.015 [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Troilo PJ, Griffiths TG, 2nd, et al. Characterization of integration frequency and insertion sites of adenovirus DNA into mouse liver genomic DNA following intravenous injection. Gene Ther 2022;29:322-32. 10.1038/s41434-021-00278-2 [DOI] [PubMed] [Google Scholar]
- 9.Ghosh S, Brown AM, Jenkins C, et al. Viral Vector Systems for Gene Therapy: A Comprehensive Literature Review of Progress and Biosafety Challenges. Appl Biosaf 2020;25:7-18. 10.1177/1535676019899502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore CB, Guthrie EH, Huang MT, et al. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol 2010;629:141-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naso MF, Tomkowicz B, Perry WL, 3rd, et al. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017;31:317-34. 10.1007/s40259-017-0234-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bijlani S, Pang KM, Sivanandam V, et al. The Role of Recombinant AAV in Precise Genome Editing. Front Genome Ed 2021;3:799722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanlon KS, Kleinstiver BP, Garcia SP, et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 2019;10:4439. 10.1038/s41467-019-12449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as