Abstract
Background & Aims
Finite duration of treatment associated with HBsAg loss is the current goal for improved therapeutic approaches against chronic HBV infection, as it indicates elimination or durable inactivation of intrahepatic covalently closed circular DNA (cccDNA). To assist drug development, the definition of early predictive markers of HBsAg loss by assessing their value in reflecting intrahepatic cccDNA levels and transcriptional activity is essential. Fine needle aspirates (FNAs) have recently emerged as a less invasive alternative to core liver biopsy (CLB) and showed to be useful for investigating intrahepatic immune responses. The aim of this study was to optimise and validate the use of FNA vs. CLB to evaluate the intrahepatic viral reservoir.
Methods
Paired FNA/CLB samples were obtained from patients with HBeAg+ chronic hepatitis (n = 4), HBeAg− chronic hepatitis (n = 4), and HBeAg− chronic infection (n = 1). One HBeAg+ patient was undergoing tenofovir treatment. HBV 3.5-kb RNA and cccDNA were quantified by droplet digital PCR.
Results
cccDNA was quantifiable in all but one FNA/CLB pair, showing the highest levels in untreated HBeAg+ patients, except for the tenofovir-treated patient. Similarly, 3.5-kb RNA was detectable in all but one FNA sample and showed higher levels in HBeAg+ patients. When comparing cccDNA and 3.5-kb RNA quantification in FNA vs. CLB samples, no statistically significant differences were identified.
Conclusions
These results demonstrate the possibility to quantify cccDNA and assess its transcriptional activity in patients with chronic hepatitis B by combining FNA and droplet digital PCR. This supports the use of FNA in clinical trials to evaluate the intrahepatic viral reservoir during the development of new antivirals and immunomodulatory agents.
Impact and implications
Chronic hepatitis B infection is characterised by a complex interplay between immune responses and viral replication in the liver, which determines the long-term outcome of the disease. In this study, we show that fine needle aspiration of the liver, a less-invasive alternative to core biopsies, allows the assessment of the hepatic viral reservoir.
Keywords: HBV, FNA, Liver biopsy, cccDNA
Graphical abstract
Highlights
-
•
Our study provides the first comparison regarding cccDNA levels and transcriptional activity between CLB and FNA samples.
-
•
No significant differences between FNA and CLB samples were seen in the quantification of HBV cccDNA and 3.5-kb RNA.
-
•
These data support the use of FNAs in clinical trials to evaluate the intrahepatic viral reservoir during drug development.
Introduction
HBV infection represents a major public health burden, as close to 300 million individuals are chronically infected worldwide with a high risk of developing cirrhosis and hepatocellular carcinoma.1 At the virological level, the mechanism of chronicity is based on persistence of the viral genome as an episomal structure referred to as covalently closed circular DNA (cccDNA), which remains in the hepatocyte nuclei as a viral reservoir.2 As a consequence, available therapies effectively limit viral replication but rarely lead to HBV elimination. Thus, there has been a renewed interest in the development of novel antivirals and immunomodulatory compounds aimed at achieving a cure for HBV infection.3
Chronic HBV infection is a highly heterogeneous disease characterised by variable immune responses and viral loads at different phases. Thus, assessment of the intrahepatic immune compartment in combination with quantification of cccDNA levels and transcriptional activity will be essential in longitudinal studies aiming to evaluate new curative strategies and to define practical treatment endpoints.4 Core liver biopsy (CLB) has remained a mainstay for the analysis of liver histology and staging of chronic hepatitis B (CHB), and thus for the assessment of host and viral parameters associated with HBV infection. Management of chronic liver diseases is evolving towards the use of less invasive methods. In this context, fine needle aspirates (FNAs) represent a promising alternative that could allow sequential liver sampling during the natural history of the disease or antiviral therapy while minimising risk and discomfort to patients.5 As FNAs offer a limited amount of material, it is crucial to use sensitive methods for their analysis. Recently, characterisation of FNAs by single-cell technologies has provided a great opportunity to dissect intrahepatic immune responses.6 However, quantification of intrahepatic viral parameters using FNAs remains to be explored.
Herein, we optimised a method for the quantification of HBV RNA and cccDNA in FNAs using droplet digital PCR (ddPCR). Using this procedure, we compared HBV levels in paired FNA/CLB samples and showed similar profiles between both sample types. These results highlight the potential application of FNAs in combination with ddPCR as a research tool for the assessment of the intrahepatic HBV reservoir.
Patients and methods
Patient characteristics
Paired FNA/CLB samples were obtained from patients with HBeAg+ chronic hepatitis (n = 4), HBeAg− chronic hepatitis (n = 4), and HBeAg− chronic infection (n = 1). One patient with HBeAg+ chronic hepatitis was on treatment with tenofovir disoproxil fumarate (TDF) at the time of sampling. A short description of patients’ clinical parameters is found in Table 1. Patients with CHB who were included in this study were undergoing percutaneous liver biopsy for diagnostic purposes of disease assessment; tissue surplus to diagnostic requirements was used in tandem with an FNA performed simultaneously, where patients had agreed to undergoing an FNA for research purposes. Written informed consent was obtained from all patients, and the study was approved by the local research ethics committee (Brent Research Ethics Committee, reference: 16/LO/1699) and complied with the Declaration of Helsinki.
Table 1.
Clinical characteristics of patients.
| ID | Age (years) | Sex | Ethnic origin | Genotype | HBsAg (log IU/ml) | HBeAg | HBeAb | HBV DNA (log IU/ml) | HBcrAg (log U/ml) | HBV RNA (log IU/ml) | ALT | Ishak fibrosis stage (/6) | EASL category | Additional notes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 259 | 30 | Male | Caucasian – European | D | 4.88 | Neg | Pos | 2.19 | 3.7 | N.D. | 55 | 2 | HBeAg− chronic hepatitis | Nil |
| 262 | 30 | Female | Asian – Bangladeshi | n.d. | 4.34 | Pos | Neg | 9.88 | 9.1 | 8.8 | 100 | 3 | HBeAg+ chronic hepatitis | Nil |
| 265 | 33 | Male | Asian – Bangladeshi | D | 3.77 | Neg | Pos | 3.98 | 3 | N.D. | 34 | 1 | HBeAg− chronic hepatitis | Nil |
| 267 | 59 | Male | Asian – Bangladeshi | n.d. | 2.81 | Neg | Pos | 3.15 | <2 | d.n.q. | 13 | 0 | HBeAg− chronic infection | Nil |
| 279 | 35 | Male | Asian – Bangladeshi | C | 1.41 | Pos | Neg | 5.16 | 6.6 | 3.5 | 65 | 3 | HBeAg+ chronic hepatitis | Nil |
| 280 | 35 | Male | Asian – Pakistani | n.d. | 3.89 | Pos | Neg | 1.30 | 5.3 | 2.8 | 54 | 2 | HBeAg+ chronic hepatitis | On treatment (TDF) |
| 281 | 42 | Male | Afro Caribbean | n.d. | 2.26 | Neg | Pos | 1.79 | 3 | N.D. | 35 | 1 | HBeAg− chronic hepatitis | Nil |
| 283 | 22 | Male | Afro Caribbean | E | 4.16 | Pos | Neg | 5.67 | 5.5 | 2.6 | 25 | 2 | HBeAg+ chronic hepatitis | Nil |
| 284 | 33 | Male | Asian – Bangladeshi | A | 4.42 | Neg | Pos | 4.64 | 4 | 1.8 | 19 | 1 | HBeAg− chronic hepatitis | Nil |
| 285 | 54 | Female | Caucasian | — | — | — | — | — | — | — | 69 | n.d. | — | AIH; portal-based inflammation |
| 52 | 67 | Female | Caucasian | — | — | — | — | — | — | — | 47 | 1 | — | Cytolysis |
| 59 | 52 | Female | Caucasian | — | — | — | — | — | — | — | 67 | 1 | — | Cytolysis, steatosis 5% |
AIH, autoimmune hepatitis; ALT, alanine aminotransferase; d.n.q., detected, not quantified; HBcrAg, hepatitis B core-related antigen; HBeAb, hepatitis B e antibody; n.d., not determined; N.D., not detected; TDF, tenofovir disoproxil fumarate.
Clinical laboratory tests
Baseline CHB characterisation, including HBeAg and hepatitis B e antibody (HBeAb) status, HBV DNA, HBsAg, hepatitis B core-related antigen (HBcrAg), HBV RNA, alanine aminotransferase (ALT) levels, HBV genotype, and histological staging, was performed as previously described.4,7,8
FNA and liver biopsy collection
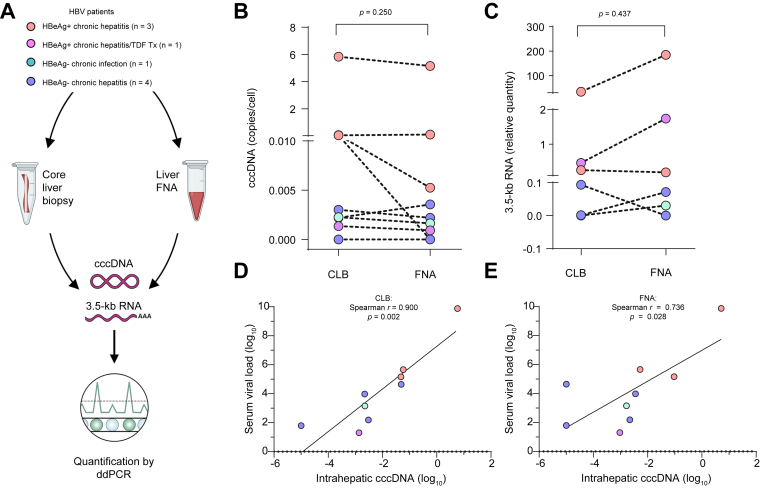
The FNA material was collected in 50-ml falcon tubes containing 40 ml of RPMI and centrifuged at 1,500 rpm for 10 min. After discarding the supernatant, samples were treated with 2 ml of red blood cell lysis buffer (BioLegend, San Diego, CA, USA) for 5 min on ice. The reaction was stopped with 40 ml of PBS and centrifuged at 1,500 rpm for 10 min. Supernatants were discarded, pellets were resuspended in 1 ml of PBS, and cells were counted. Between 105 and 106 cells were recovered from each FNA. Finally, samples were centrifuged at 8,000 rpm for 3 min, the remaining media was aspirated, and pellets were resuspended in freeze media (90% FBS, 10% DMSO) to be stored at -80 °C. For FNA samples in which very low starting material was available, DNA extraction was favoured over RNA (e.g. Fig. 1B and C).
Fig. 1.
FNAs allow quantification of intrahepatic HBV markers by ddPCR.
(A) Experimental approach for the quantification of HBV parameters in FNA/CLB samples by ddPCR. (B and C) Quantification of cccDNA (B) and 3.5-kb RNA species (C) by ddPCR comparing FNAs with CLBs (Wilcoxon matched-pairs signed-rank test). Values represent cccDNA/HBB and 3.5-kb RNA/GUSB ratios. (D and E) Intrahepatic cccDNA levels quantified in CLB (D) and FNA samples (E) in correlation with serum HBV DNA levels (Spearman’s rank correlation coefficient). cccDNA, covalently closed circular DNA; CLB, core liver biopsy; ddPCR, droplet digital PCR; FNA, fine needle aspirate; GUSB, glucuronidase beta; HBB, haemoglobin subunit beta; TDF, tenofovir disoproxil fumarate.
Liver biopsy samples of approximately 5 mm in length, surplus to diagnostic requirements, were immersed directly into freeze media and stored at -80 °C for further experimentation as below.
HBV DNA/RNA extraction
Liver samples were processed for the extraction of viral nucleic acids using the Quick-DNA/RNA Microprep Plus kit (Zymo Research, Irvine, CA, USA). Total DNA was digested for cccDNA quantification using Plasmid-Safe ATP-Dependent DNase (Lucigen, Middleton, WI, USA), followed by purification with the DNA Clean & Concentrator kit (Zymo Research). RNA samples were prepared using TURBO DNase (Thermo Fisher Scientific, Waltham, MA, USA), followed by overnight precipitation. Reverse transcription was performed on equivalent volumes of each FNA/CLB sample and using SuperScript IV VILO (Thermo Fisher Scientific).
HBV cccDNA and HBV 3.5-kb RNA quantification by ddPCR
The quantification of absolute copy numbers of intrahepatic cccDNA and 3.5-kb RNA species was performed following the International Coalition to Eliminate HBV (ICE-HBV) panel recommendations.9,10 Primers and probes are listed in the Supplementary CTAT Table. In brief, the 22-μl ddPCR reaction included 2 × ddPCR Supermix for probes (Bio-Rad, Hercules, CA, USA), 900 nM primers, 250 nM probe, and 5 μl of DNA/RNA sample. The ddPCR reaction for the quantification of sodium taurocholate cotransporting polypeptide (NTCP) RNA was performed using QX200™ ddPCR™ EvaGreen Supermix (Bio-Rad). Samples were partitioned into the Automated Droplet Generator with specific oil for probes (Bio-Rad), amplified in the C1000 Touch thermal cycler (Bio-Rad), and analysed with the QX100 Droplet Reader using QuantaSoft software (v1.7.4, Bio-Rad, Hercules, CA, USA). Haemoglobin subunit beta (HBB) and glucuronidase beta (GUSB) served as internal reference for HBV DNA and RNA quantification, respectively. Negative controls (distilled water) and a positive control were included in each run. The analysis of HBV-negative FNA/CLB samples (n = 3) confirmed the specificity of the technique and served as reference for the setting of thresholds discriminating positive and negative droplets (Fig. S1 and Table 1). Expression of the hepatocyte marker NTCP was performed as a means to estimate the proportion of this cell population in FNA/CLB samples. Absolute copy numbers of cccDNA and 3.5-kb RNA species were normalised with the NTCP/GUSB ratio to account for variations in hepatocyte proportion.
Statistical analyses
Comparison between paired FNA/CLB samples was performed using a Wilcoxon matched-pairs signed-rank test. Correlations between serum HBV DNA and intrahepatic cccDNA were done with a Spearman’s rank correlation coefficient, using the raw data as input. Plots presented in Fig. 1D, E show log-transformed data, following addition of 10-5 as a constant to deal with zero values. All analyses were performed using GraphPad Prism (v9.4, GraphPad Software, San Diego, CA, USA).
Results
To evaluate the potential relevance of FNAs as an alternative to CLBs in the assessment of intrahepatic HBV parameters, we took advantage of paired samples obtained by each method from a group of patients representing different HBV phases and treatment regimens (Fig. 1A and Table 1). Quantification of intrahepatic HBV cccDNA levels (cccDNA/HBB ratio) by ddPCR revealed no significant difference when comparing the results obtained from paired FNA/CLB samples (p = 0.250, Wilcoxon matched-pairs signed-rank test) (Fig. 1B). Similarly, quantification of intrahepatic 3.5-kb RNA species resulted in comparable levels using both sample types, following normalisation to GUSB expression (p = 0.437) (Fig. 1C).
To consider potential differences in the proportion of hepatocytes between FNAs and CLBs, we quantified the expression of NTCP mRNA in both sample types. As expected, FNAs presented significantly lower NTCP levels (p = 0.031, mean = 0.48) compared with CLBs (mean = 3.53) (Fig. S2A). However, normalisation of each viral parameter according to the NTCP/GUSB ratio still resulted in comparable cccDNA (p = 0.312) and 3.5-kb RNA levels (p = 0.218) in CLB vs. FNA (Fig. S2B and C). Moreover, and in agreement with previous reports,7 a positive correlation between serum HBV DNA and intrahepatic cccDNA from CLBs was observed (r = 0.900, p = 0.002, Spearman’s rank correlation coefficient) (Fig. 1D). This significant correlation was equally observed in the case of cccDNA levels obtained from FNA samples (r = 0.736, p = 0.028) (Fig. 1E). Finally, a similar trend of association was found between serum HBcrAg and cccDNA levels from both CLB (r = 0.742, p = 0.041) and FNA samples (r = 0.716, p = 0.052) (Fig. S3A and B).
Discussion
With the development of novel classes of direct-acting antivirals and immune interventions, it has become essential to understand their effect on both the hepatic viral reservoir and the immune microenvironment.5,11 FNAs provide an opportunity to access and analyse immune and parenchymal cells of the infected liver compartment in a less invasive manner than the conventional CLB. Although the study of the liver immune compartment has been reported in patients with chronic HBV6 or HCV infection,12,13 analysis of the viral cccDNA has not been possible owing to sensitivity issues of the available assays. The development of sensitive ddPCR assays has opened the door for the quantification of cccDNA and viral transcripts in patient samples containing limited amounts of material.[14], [15], [16] PCR-based techniques for cccDNA quantification still suffer from difficulties associated with specificity of cccDNA detection and lack of standardisation. However, recent efforts to provide evidence-based guidelines and standardise nucleic acid extraction, digestion, and storage of HBV-infected samples for reliable and reproducible cccDNA analysis represent important milestones that could bring closer the translation of these methods into the clinic.10
Although our study has limitations with respect to the small number of patients, it provides the first direct comparison regarding the analysis of cccDNA levels and transcriptional activity between CLB and FNA samples. Further characterisation of FNA samples will be required to define the proportion of HBV-infected hepatocytes and their relationship to immune infiltrates. Another important question was to normalise results of HBV markers on the number of hepatocytes. Here, we showed that normalisation with hNTCP, HBB, and GUSB demonstrated good concordance between CLB and FNA for both cccDNA and 3.5-kb RNAs, and a correlation between serum HBV DNA and cccDNA for both liver sample types (Fig. 1). To consider the heterogeneity of HBV-induced liver disease, it will be interesting to investigate the viral markers and the immune cells in the liver FNAs from patients across the different phases of the infection.11,17
In conclusion, FNAs are thus amenable to perform not only immunological but also virological analysis of the liver compartment and open new opportunities to study the pathogenesis of CHB and the changes in the liver reservoir induced by novel therapies. Approaches integrating investigations of the liver viral reservoir and immune microenvironment will be essential in the path towards the development of curative therapies.
Financial support
This work is supported by the French National Research Agency ‘Investissements d’Avenir’ program (CirB-RNA project – ANR-17-RHUS-0003), the European Union's Horizon 2020 research and innovation programme under grant agreement no. 847939 (IP-cure-B project), and the ‘Agence Nationale pour la Recherche sur le SIDA et les hepatites virales et les maladies infectieuses emergentes’ (ANRS-MIE) to BT, ML, and FZ. Moreover, financial support was awarded to USG from The Academy of Medical Sciences Starter Grant (SGL021/1030), Seedcorn funding Rosetrees/Stoneygate Trust (A2903), and The Medical Research Foundation (MRF-044-0004-F-GILL-C0823) along with a Barts Charity Project Grant (MGU/0406) awarded to PTFK.
Authors’ contributions
Conceptualisation: BT, UG, PK, ML, FZ. Formal analysis: BT, AARS, MLP. Funding acquisition: BT, ML, UG, PK, FZ. Investigation: BT, AARS, AB, MLP. Methodology: BT, AARS, AB, MLP, UG. Resources: MH, TF, FV, YC, ML, UG, PK, FZ. Supervision: BT, UG, PK, FZ. Visualisation: BT, AARS. Writing – original draft: BT, AARS. Writing – review and editing: all authors.
Data availability statement
De-identified individual participant data that underlie the reported results will be made available upon request 3 months after publication for a period of 5 years after the publication date.
Conflicts of interest
FZ received grants from Assembly, Beam Therapeutics, and Evotec; FZ had consulting activities with Assembly, Blue Jay, Gilead, and GSK.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We apologise to the various colleagues whose original contributions could not be included in this manuscript and are not cited because of space constraints.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100841.
Supplementary data
The following are the supplementary data to this article.
References
- 1.Polaris Observatory Collaborators Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 2.Martinez M.G., Boyd A., Combe E., Testoni B., Zoulim F. Covalently closed circular DNA: the ultimate therapeutic target for curing HBV infections. J Hepatol. 2021;75:706–717. doi: 10.1016/j.jhep.2021.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Roca Suarez A.A., Testoni B., Zoulim F. HBV 2021: new therapeutic strategies against an old foe. Liver Int. 2021;41(Suppl. 1):15–23. doi: 10.1111/liv.14851. [DOI] [PubMed] [Google Scholar]
- 4.Gill U.S., Pallett L.J., Thomas N., Burton A.R., Patel A.A., Yona S., et al. Fine needle aspirates comprehensively sample intrahepatic immunity. Gut. 2019;68:1493–1503. doi: 10.1136/gutjnl-2018-317071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill U.S., Pallett L.J., Kennedy P.T.F., Maini M.K. Liver sampling: a vital window into HBV pathogenesis on the path to functional cure. Gut. 2018;67:767–775. doi: 10.1136/gutjnl-2017-314873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nkongolo S., Mahamed D., Kuipery A., Vasquez J.D.S., Kim S.C., Mehrotra A., et al. Longitudinal liver sampling in patients with chronic hepatitis B starting antiviral therapy reveals hepatotoxic CD8+ T cells. J Clin Invest. 2023;133 doi: 10.1172/JCI158903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Testoni B., Lebossé F., Scholtes C., Berby F., Miaglia C., Subic M., et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol. 2019;70:615–625. doi: 10.1016/j.jhep.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Scholtès C., Hamilton A.T., Plissonnier M.-L., Charre C., Scott B., Wang L., et al. Performance of the cobas® HBV RNA automated investigational assay for the detection and quantification of circulating HBV RNA in chronic HBV patients. J Clin Virol. 2022;150–151 doi: 10.1016/j.jcv.2022.105150. [DOI] [PubMed] [Google Scholar]
- 9.Lebossé F., Inchauspé A., Locatelli M., Miaglia C., Diederichs A., Fresquet J., et al. Quantification and epigenetic evaluation of the residual pool of hepatitis B covalently closed circular DNA in long-term nucleoside analogue-treated patients. Sci Rep. 2020;10 doi: 10.1038/s41598-020-78001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allweiss L., Testoni B., Yu M., Lucifora J., Ko C., Qu B., et al. Quantification of the hepatitis B virus cccDNA: evidence-based guidelines for monitoring the key obstacle of HBV cure. Gut. 2023;72:972–983. doi: 10.1136/gutjnl-2022-328380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boettler T., Gill U.S., Allweiss L., Pollicino T., Tavis J.E., Zoulim F. Assessing immunological and virological responses in the liver: implications for the cure of chronic hepatitis B virus infection. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pembroke T., Christian A., Jones E., Hills R.K., Wang E.C.Y., Gallimore A.M., et al. The paradox of NKp46+ natural killer cells: drivers of severe hepatitis C virus-induced pathology but in-vivo resistance to interferon α treatment. Gut. 2014;63:515–524. doi: 10.1136/gutjnl-2013-304472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui A., Li B., Wallace M.S., Gonye A.L.K., Oetheimer C., Patel H., et al. Single-cell atlas of the liver myeloid compartment before and after cure of chronic viral hepatitis. J Hepatol. 2023 doi: 10.1016/j.jhep.2023.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villeret F., Lebossé F., Radenne S., Samuel D., Roche B., Mabrut J.Y., et al. Early intrahepatic recurrence of hepatitis B virus infection in liver transplant recipients despite antiviral prophylaxis. JHEP Rep. 2023;5 doi: 10.1016/j.jhepr.2023.100728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caviglia G.P., Abate M.L., Tandoi F., Ciancio A., Amoroso A., Salizzoni M., et al. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: a new tool to detect occult infection. J Hepatol. 2018;69:301–307. doi: 10.1016/j.jhep.2018.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Balagopal A., Grudda T., Ribeiro R.M., Saad Y.S., Hwang H.S., Quinn J., et al. Single hepatocytes show persistence and transcriptional inactivity of hepatitis B. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suslov A., Meier M.-A., Ketterer S., Wang X., Wieland S., Heim M.H. Transition to HBeAg-negative chronic hepatitis B virus infection is associated with reduced cccDNA transcriptional activity. J Hepatol. 2021;74:794–800. doi: 10.1016/j.jhep.2020.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified individual participant data that underlie the reported results will be made available upon request 3 months after publication for a period of 5 years after the publication date.