Abstract
Siglecs are important lectins found in different types of immune cells and function as regulatory molecules by recognizing self-associated glycans and converting extracellular interactions into signals for inhibiting immune cell functions. Although many Siglecs have been found to show broad specificities and recognize different types of sulfated oligosaccharides, Siglec-8 and Siglec-9 displayed a high degree of specificity for sialyl N-acetyllactosamine (sLacNAc) with sulfations at O6-positions of the galactose (6’-sulfation) and N-acetylglucosamine (6-sulfation), respectively. Siglec-3 was recently discovered to bind sLacNAc both sulfations. In addition to a conserved arginine residue for binding to sialic acid residue, the sequence variety in the CC’ loop may provide binding specificities to sulfated oligosaccharides in Siglecs. Thus, the present study employed molecular models to study the impact of different residues in the CC’ loops of Siglec-8/9/3 to the recognitions of 6-sulfations in Gal and/or GlcNAc of sLacNAc. The negatively charged residues in the CC’ loop of Siglec-9 formed unfavorable electrostatic repulsions with the 6-sulfate in Gal and resulted no recognitions, in contrast to the favorable interactions formed between the positively charged residues in the CC’ loop of Siglec-8 and the 6-sulfate in Gal resulting strong specificity. A two-state binding model was proposed for Siglec-3 recognizing 6-sulfations in Gal and GlcNAc of sLacNAc, as the neutral residues in the CC’ loop of Siglec-3 could not form strong favorable interactions to lock the 6-sulfate in Gal within a single binding pose or strong unfavorable interactions to repel the 6-sulfate in Gal. The oligosaccharide adopted two distinctive binding poses and oriented the sulfate groups to form interactions with residues in the CC’ loop and G-strand. The present study provided a structural mechanism for the sequence variety in the CC’ loop of Siglec-8/9/3 determining the recognitions to the sulfated oligosaccharides and offered insights into the binding specificities for Siglecs.
Keywords: Siglecs, Sialic acid, Molecular model, Two-state binding, GLYCAM, Binding pose, Energy prediction
Graphical Abstract
1. Introduction
Siglecs, sialic acid binding immunoglobulin-like lectins, is a major subfamily of I-type lectins [1]. Most of the Siglecs in human have been found in different types of cells in immune system, where they recognize self-associated glycans and convert these extracellular interactions into signals for inhibiting immune cell functions [2], [3], [4], [5], [6], [7]. Their functions as regulatory molecules have been discovered in various diseases, such as Alzheimer disease, asthma, and cancers [5], [6], [8], [9], [10], [11]. Each Siglec has an N-terminal V-set Ig domain containing a sialoglycan binding site with a conserved arginine residue that forms strong interactions with the carboxylate group in sialic acid for ligation [7], [12]. The V-set domain is composed of 10 different β-strands (A, A’, B, B’, C, C’, D, E, F, and G) and 9 loops, in which the conserved arginine residue locates in the F-strand [9].
Different endogenous Siglecs recognize distinctive sialoglycan ligands, often with sufficient specificities and affinities, to initiate response signals important to the function of the cells on which they are expressed [8], [13], [14]. Relatively rapid evolution for Siglecs permits its members to develop specificity for different types of sialoglycans with α2–3 [15], [16], [17], [18], [19], [20], α2–6 [15], [16], [19], [20], and/or α2–8 [20], [21], [22], [23], [24] linkages, as well as underlying glycans with various compositions and/or modifications [2], [18], [25], [26]. Sulfation is a ubiquitous and important post-translational modification of many biological molecules including carbohydrates [27], [28]. At least 8 of the 15 known Siglecs (−2/3/5/7/8/9/14/15) have shown enhanced binding affinities with the presence of the additional sulfate group(s) [10]. While many of them have broad specificities, Siglec-8 and Siglec-9 display exceptionally narrow specificity for two types of sulfated sialyl N-acetyllactosamine (sLacNAc: Neu5Acα2–3Galβ1–4GlcNAcβ1-R) motifs: 6’-sulfo-sLacNAc (6-sulfation in Gal) and 6-sulfo-sLacNAc (6-sulfation in GlcNAc), respectively [29], [30].
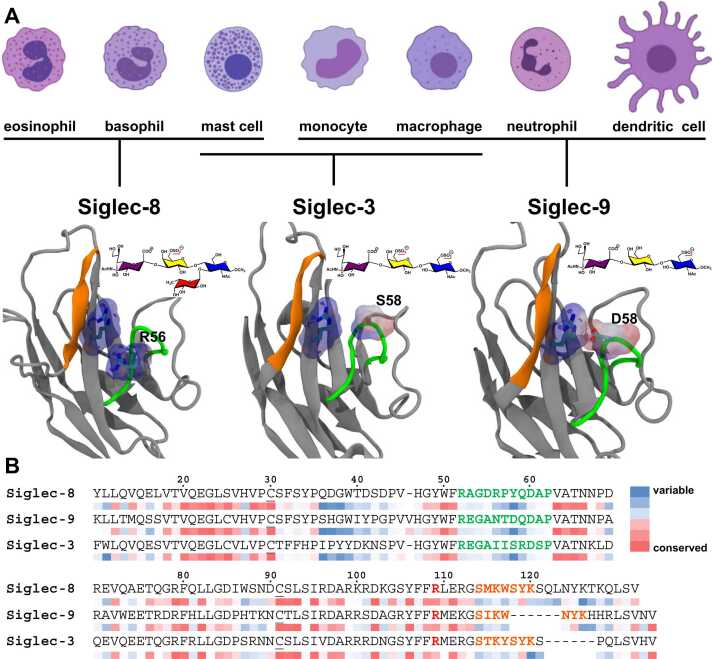
Siglec-8 is an immunoinhibitory lectin expressed on human eosinophils, mast cells, and basophils [31], [32], [33] (Fig. 1). Its engagements with sialoglycan ligands are found to induce eosinophil apoptosis and resolve ongoing allergic inflammation [34], [35]. Glycan microarray results for Siglec-8 suggested a high degree of binding specificity to 6’-sulfo-sLacNAc (6-sulfation in Gal) motif and its fucosylated congener, 6’-sulfo-sialyl-Lewis-X (6’-sulfo-sLex: Neu5Acα2–3 Gal[6 S]β1–4[Fucα1–3]GlcNAcβ1-R) [29], [30]. Structural studies based on the Siglec-8 – 6’-sulfo-sLex complex revealed the molecular basis for this specificity: the carboxylate in sialic acid residue formed strong interactions with the conserved arginine (R109) in the F-strand, and the galactose residue orients its pyranose ring to form favorable CH-π stacking with a tyrosine (Y58) and the 6’-sulfo group at the O6-position of Gal to make favorable interactions to R56 and Q59 on the CC’ loop [36]. It is speculated that the interactions formed by sialic acid residue provide sufficient affinity, while those formed by the sulfate moiety generate the unique specificity among members of the Siglec family. On the other hand, glycan microarray results for Siglec-9 suggested a high degree of specificity for the 6-sulfation in GlcNAc, 6-sulfo-sLacNAc and 6-sulfo-sLex [30], despite that Siglec-9 is also an immunoinhibitory lectin that is broadly expressed on human leukocytes including monocytes, macrophages, neutrophils, dendritic cells, and displays functions related to inflammation and cancer immune evasion [19] (Fig. 1). Although the experimentally measured structure for the Siglec-9 – 6-sulfo-sLacNAc or Siglec-9 – 6-sulfo-sLex complex is currently unavailable, the amino acid sequence of the CC’ loop in Siglec-9 has been found to be markedly different from those in Siglec-8 in terms of their electrostatic properties. The positively charged R56 and the neutral Y58 in Siglec-8 were replaced by a polar asparagine residue and a negatively charged aspartate residue, respectively. Similar to Siglec-8 and Siglec-9, Siglec-3 is also an immunoinhibitory lectin found on monocytes, macrophages, mast cells [37], [38] (Fig. 1). Recent experimental studies suggested that the binding strength for the Siglec-3 – sLacNAc complex could be enhanced by the sulfations at the O6-positions of the Gal and GlcNAc residues in sLacNAc [39]. Siglec-3 also contains a sialoglycan binding site with a conserved arginine residue (R109), but unlike the CC’ loops in the Siglec-8 and Siglec-9 with charged residues, there are no charged residues on the CC’ loop of Siglec-3.
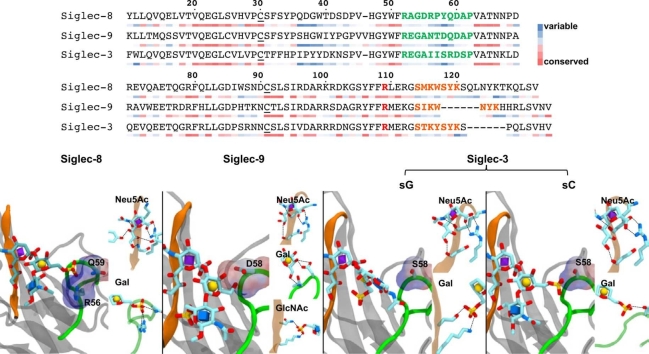
Fig. 1.
A. Siglec-8, 9, and 3 lectin domain structures (grey) and their preferred ligands (schematic structures), as well as the immune cells of their expressions. The backbones for the residues in the CC’ loop and the G-strand were colored in green and orange, respectively. The side chains of the key residues in the CC’ loop and the conserved R109 were shown in licorice representation and molecular surface colored by their electrostatic potentials [40]: red, negative; blue, positive. Monosaccharide rings in the ligands are filled according to the SNFG nomenclature [41], [42]. B. Sequence alignment of Siglec-8/9/3. The sections for the CC’ loop and the G-strand are colored in green and orange, respectively. The cystine residues forming disulfide bond are indicated with underline. The conserved arginine residue is marked bold red. The sequence conservation scores were calculated by ConSurf [43] and shown as colored bars underneath each corresponding amino acid.
The sequence variety in the CC’ loop of Siglecs may have caused different binding specificities to different sulfated oligosaccharides (Fig. 1). Understanding the structural mechanisms for these specificities in relationship with the sequence variety is an important and challenging field. Elucidating the precise terms deciding the specific Siglec – ligand pairs is crucial for mediating the biological functions of Siglecs and dissecting their signaling pathways. Molecular modeling studies have been successfully employed to study the mechanisms for lectin – oligosaccharide interactions [44], [45], [46], [47], including Siglecs – sialoglycans complexes [48], [49], [50], [51], [52], [53], [54], [55], [56], [57]. The present study employed molecular models for the Siglec-9 and Siglec-3 complexes built based on the experimentally measured structures for the Siglec-8 – 6’-sulfo-sLex complex in molecular dynamics (MD) simulations and energy predictions to study the structural mechanisms for their specificities. MD simulations for the Siglec-9 – 6-sulfo-sLacNAc and Siglec-9 – 6’-sulfo-sLacNAc complexes showed that the 6-sulfo-sLacNAc ligand (6-sulfation in GlcNAc) could maintain positional stabilities in the binding site of the Siglec-9, whereas the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal) appeared to form unfavorable electrostatic repulsions with the negatively charged D58 residue. After the mutation of D58S in the Siglec-9, the 6’-sulfo-sLacNAc ligand obtained positional stability in the binding site, which confirmed that the unfavorable electrostatic repulsions between the negatively charged D58 in the CC’ loop and the 6-sulfate in Gal exclude the recognition to 6’-sulfo-sLacNAc by the Siglec-9. In the Siglec-3 complexes, MD simulations showed that the both 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc (6-sulfation in Gal and GlcNAc, respectively) could maintain positional stability in the Siglec-3 binding site via strong interactions between sialic acid and the conserved R109. Different from the single binding pose observed in the experimentally measured structures for the Siglec-8 – 6’-sulfo-sLex complex and MD simulations for the Siglec-9 – 6-sulfo-sLacNAc complex, the sulfated ligands in the Siglec-3 complexes adopted two binding poses. Each of the poses permitted the formations of different interactions between the 6-sulfate groups and the amino acids in both G-strand and CC’ loop. Binding free energies computed with molecular mechanics generalized born surface area (MM-GBSA) methodology for 6’-sulfo-sLacNAc, 6’,6-disulfo-sLacNAc, and 6-sulfo-sLacNAc ligands agreed to their ranking of the experimentally measured binding affinities. Further point mutation of S58A in the CC’ loop confirmed the necessity for a two-state binding model. The mutation of S58A abolished the interaction between the 6-sulfate in Gal and the residues in the CC’ loop. Instead of disengaging from the Siglec-3 binding site as did the non-sulfated sLacNAc ligand, the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal) maintained a stable binding. These distinctive binding patterns observed in MD simulations suggested a novel binding state other than that observed in the Siglec-8 – 6’-sulfo-sLex complex is necessary. The mutation of I56R enhanced the populations for the sC state revealed the competing nature of the two states.
The present study examined the structural mechanisms for Siglec-8/9/3 recognizing the sLacNAc ligands with different 6-sulfations, in which the variety of the residues in the CC’ loop has a critical role by forming interactions with the sulfate groups. A novel two-state binding model was proposed to describe the bindings for the Siglec-3 complex, where the CC’ loop contains only neutral residues and form moderate interactions with the sulfate groups.
2. Method
2.1. Structure Preparation
The initial coordinates for the Siglec-8 – 6’-sulfo-sLex and Siglec-3 were obtained from the Protein Data Bank (PDB entry code: 2N7B [36] and 6D49 [9], respectively). The amino acid residues and the sialic acid residue from Chain A for Siglec-3 were extracted. The initial coordinates for the 6’-sulfo-sLacNAc ligand were obtained from the Siglec-8 – 6’-sulfo-sLex complex. In order to obtain the coordinates for the 6’-sulfo-sLacNAc ligand in Siglec-3, the ring atoms for the sialic acid residue in the 6’-sulfo-sLex ligand was superimposed with those for the sialic acid residue in the Siglec-3 binding site. The homology model for Siglec-9 was generated by the automated SWISS-MODEL server [58] with the experimentally measured structure of Siglec-8 as the template. The modeling for the Siglec-9 with AlphaFold2 [59] was carried out using Aphafold2_advanced python notebook that was running on Google Collaboratory cloud computing facilities with GPU hardware via ChimeraX (version 1.3) [60] interface from its amino acid sequence. Prior to AlphaFold2 predictions, HMMER3.3 [61] was used to compute the sequence alignments. Default settings for AlphaFold2 were employed, including max_msa_clusters and max_extra_msa for the number of randomly chosen sequence clusters provided to the AlphaFold2 neural network and the number of extra sequences used to compute additional summary statistics, respectively. Multiple databases were searched before utilizing all 5 neural networks for predicting structures without a given template, including Uniref90 (version: August 2021) [62], [63], smallBFD (version: 2018) [64], and Mgnify sequence database (version: 2018_12) [65]. The final structure with the top score was energy minimized in openMM [66] with AMBER force field before used in further modeling studies. The 6’-sulfo-sLacNAc ligand was positioned into the molecular models of the Siglec-9 by superimposing the Siglec-8 – 6’-sulfo-sLex complex structure with the models of Siglec-9 via their Cα atoms in the protein backbones. For consistency, the index of residues in this study followed those in 2N7B. The 6’-sulfo-sLacNAc ligand was generated with the tLEaP module in AMBER18 [67] after removing the coordinates for fucose from the 6’-sulfo-sLex ligand and changing the residue name for the GlcNAc residue accordingly. The sLacNAc ligand was, then, generated with the tLEaP module after removing the coordinates for the sulfate group from the 6’-sulfo-sLacNAc ligand and changing the residue name for the Gal residue accordingly. The 6’,6-disulfo-sLacNAc and 6-sulfo-sLacNAc ligands were generated by adding a sulfate group to the O6 position of GlcNAc in 6’-sulfo-sLacNAc and sLacNAc ligand, respectively, with tLEaP module [67]. The residue name for GlcNAc was changed accordingly and the partial charge for O6 atom in GlcNAc was increased by + 0.031 (http://legacy.glycam.org/docs/help/2014/04/04/sulfating-glycam-residues/). Force field parameters for carbohydrate molecules were taken from GLYCAM06 (version j) [68], and those for proteins were taken from AMBER18 (ff99sb) [69]. Counter sodium ions were added to neutralize each protein complex sing tLEaP module before they were solvated in a truncated octahedral box (8 Å buffer with TIP3P water model).
2.2. Simulation Setup
Energy minimization for the solvated complexes was performed in two steps under the canonical ensembles (nVT): 1. only the water molecules and counter ions were subjected to energy minimization (500 steps steepest descent followed by 24500 steps conjugate gradient), and the atoms in solute were restrained (100 kcal/mol∙Å2); 2. the energy minimization circle was repeated with restraints only applied to the Cα atoms on the protein backbone and ring atoms in the oligosaccharides. Subsequently, the solvated complexes were heated to 300 K over 50 ps under the nVT condition with restraint (10 kcal/mol∙Å2) ronly applied to the Cα atoms on the protein backbone. Then, the solvated system was equilibrated at 300 K under isothermal-isobaric ensembles (nPT) with a Berendsen thermostat [70] for 10 ns, during which all restraints were removed. The production run of the MD simulations for each Siglec complex were performed for 800 ns with the GPU implementation of PMEMD from AMBER18 software package [71]. The simulation time step in all MD simulations was set to 2 fs with the covalent bonds involving hydrogen atoms constrained using SHAKE algorithm [72]. A non-bonded cutoff of 8 Å was applied to van der Waals interaction energy calculations, and the particle mesh Ewald approximation was applied to the long-range electrostatic interaction energy calculations. Standard 1–4 nonbonded scaling factors for proteins (2.0/1.2) and carbohydrate molecules (1.0/1.0) were employed [68].
2.3. Extraction of structures for sG and sC states
The centers of the distributions for the φ/ψ angles of the glycosidic linkages in the oligosaccharide were selected from the centers of the gaussian function fitting curves to the histograms of the φ/ψ angles obtained from the MD simulation. The complex structure in the MD simulation whose φ/ψ angles in the glycosidic linkages for ligand were within 2° of the selected centers were extracted to represent the desired binding state.
2.4. Binding interaction energy and entropy calculations
The molecular mechanics-generalized born surface area (MM-GBSA) methodology [73] for calculating binding interaction energies and per-residue contributions were carried out on 8000 snapshots extracted evenly from 800 ns of MD simulations using a single trajectory methodology with the MMPBSA.py.MPI module in AMBER. The MM-GBSA energy for binding interactions were estimated as in Eq. [1]:
| (1) |
where is the gas-phase interaction energy between protein and ligand, including the electrostatic and the van der Waals energies, as the bonded energies were canceled under single trajectory methodolody. and are the polar and non-polar components of the desolvation free energies, respectively, approximated by generalized born model and solvent accessible surface area. The GB1OBC model (igb = 2) [74] and internal dielectric constant (εint) of 4.0 were applied in all MM-GBSA calculations [75], [76]. Quasi-harmonic (QH) entropies () contributed from rotations, transitions, and vibrations were calculated using the cpptraj module in AMBERTOOLS [77] and fit linearly as a function of inverse simulation period. The intercept with the Y-axis of the linear fitting function is the extrapolation of QH entropy to an infinite simulation period [78]. Conformational entropies associated with changes in the glycosidic torsion angle distributions that occur upon binding were computed using the Karplus – Kushick (KK) approach [79]. Calculations for MM-GBSA interaction energies and entropies were performed for the Siglec-3 complexes, including Siglec-3 – 6’-sulfo-sLacNAc, Siglec-3 – 6-sulfo-sLacNAc, and Siglec-3 – 6’,6-disulfo-sLacNAc complexes.
2.5. Adaptive steered molecular dynamics (ASMD) simulation
The starting geometries for the ASMD simulations were the representative structures for the Siglec-3 – 6’-sulfo-sLacNAc complex with the ligand in the different binding states [80], [81], [82]. The C2 atom of sialic acid was pulled with the Cζ atom of R109 in Siglec-3 fixed during the ASMD simulations. Each ASMD simulation was divided into 12 steps with a pulling distance of 1 Å for each step and an exceedingly slow constant velocity of 0.2 Å/ns, achieved with a harmonic force constant of 1.0 kcal/mol/Å. Every step was conducted with 25 independent trajectories to obtain statistically reliable results. All ASMD simulations were performed with the GPU implementation of PMEMD from AMBER18 software package [71]. Data collections were carried out with a time step of 2 fs under the nVT condition and 300 K using the Langevin thermostat [83]. Potential mean forces (PMF) along the pulling coordinate are calculated with scripts available on the AMBER tutorial website [84].
3. Results and discussion
3.1. Variety of amino acids in the CC’ loop
The sequences in the CC’ loop for Siglec-8/9/3 displayed a high degree of variety, especially in the range for residues 56–59 (Fig. 1). These different amino acid residues of the CC’ loop have been considered as the cause for binding specificities of several Siglecs [24], [85], [86], [87]. In the present study, residues 56 – 59 in Siglec-8/9/3 displayed distinctive electrostatic properties. Residue 56 in the Siglec-8 is an arginine residue, which contains a positively charged side chain, whereas residue 58 in the Siglec-9 is an aspartate, which contains a negatively charged side chain. Therefore, the CC’ loop in Siglec-8 and Siglec-9 displayed opposite electrostatic potentials (Fig. 1), suggesting that they could form different interactions with the negatively charged sulfate group in the ligand. The neutral S58 in the CC’ loop of Siglec-3 is different from both Siglec-8 and Siglec-9, which could possibly bring distinctive recognition modes to the sulfate groups in the oligosaccharides. In parallel, Siglec-8/9/3 showed distinctive binding preferences for sulfated oligosaccharides. Glycan microarray study revealed that 6’-sulfo-sLex and 6’-sulfo-sLacNAc (6-sulfation in Gal) are preferred ligands for Siglec-8, while Siglec-9 showed specificity towards 6-sulfo-sLex and 6-sulfo-sLacNAc (6-sulfation in GlcNAc) [30]. Recent studies [39] discovered that Siglec-3 could recognize sLacNAc motif with both 6’-sulfation and 6-sulfation (6-sulfation in Gal and GlcNAc, respectively). The present study employs molecular models for the Siglec-8/9/3 complexes to explore the impact of the variety of the amino acid residues in their CC’ loop to their binding interactions with the sulfated sLacNAc motifs.
3.2. The CC’ loop in the Siglec-8 recognizes 6-sulfation in Gal
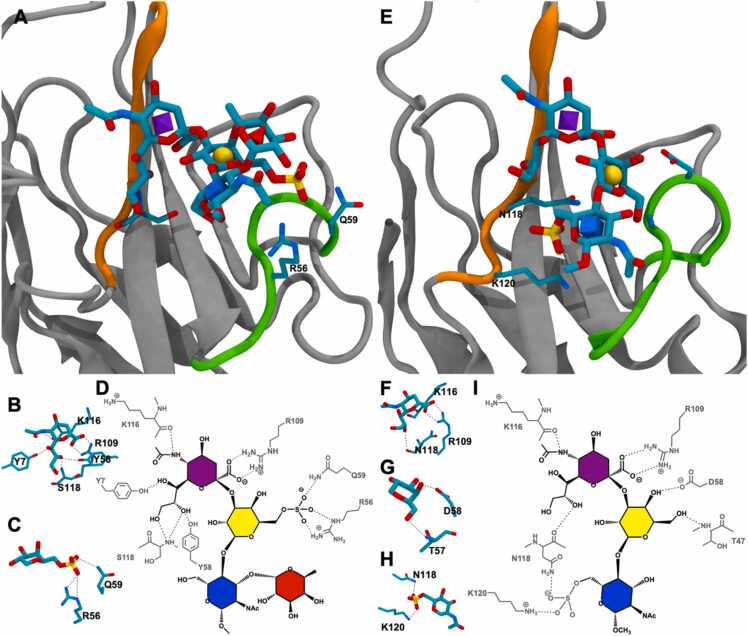
In the experimentally measured structure for the Siglec-8 – 6’-sulfo-sLex complex, the R56 in the CC’ loop formed direct interactions with the sulfate group at the O6-position of Gal and the Q59 flanked the 6-sulfate group in Gal to form a “clamp-like” configuration. The simultaneous interactions between the sulfate group and two residues in the CC’ loop have been considered to be origin of the binding specificity for the 6’-sulfo-sLex in the Siglec-8 [36] (Fig. 2). All 20 NMR-elucidated structures for the Siglec-8 – 6’-sulfo-sLex complex (entry code in protein databank: 2N7B) displayed a single pose of the ligand oligosaccharide, in which the glycosidic linkage conformation between Neu5Ac – Gal oriented the 6-position of the Gal residue toward the CC’ loop and permitted the interactions between the 6’-sulfo group and the residues in the CC’ loop. In the present study, three independent MD simulations were performed for the molecular model of the Siglec-8 – 6’-sulfo-sLex complex built from its NMR-elucidated structure, and the 6’-sulfo-sLex ligand in the MD simulations bound stably to the receptor (Fig. S1) in a single dominant conformation (Fig. S2), which was the single pose observed in its NMR-elucidated structures (φ/ψ, C2-C3-O3-C2’ and C3-O3-C2’-C3’, for Neu5Ac – Gal: 180°/240°). This dominant conformation was selected upon binding from three conformations observed for the 6’-sulfo-sLex ligand in the unbound state (Fig. S3). Furthermore, the preferred ring conformations for each monosaccharide in the 6’-sulfo-sLex ligand remained unchanged upon binding to the Siglec-8 (1C4 for Neu5Ac and Fuc; 4C1 for Gal and GlcNAc) (Fig. S4). Yet, the conformational preference for the ω angle (O5-C5-C6-O6) in Gal switched from tg (ω ∼ 180°) to gt (ω ∼ 60°) rotamer, as the gt rotamer could promote interactions for the 6’-sulfo group with R56 and Q59 on the CC’ loop (Fig. S5 A, D – F). Hydrogen bond analyses for MD simulations showed that Neu5Ac and 6’-sulfo group provided all stable interactions (Table 1 and Fig. 2), in which the experimentally observed key hydrogen bond interactions were preserved in MD simulations (Table 1). Stable CH-π interactions were observed for Neu5Ac with TYS7 and TRP117. The molecular model for the Siglec-8 – 6’-sulfo-sLex model in the present study reproduced the structural features obtained in experiments. Correspondingly, the MM-GBSA energy calculations reproduced the relative binding contributions observed from experimental mutagenesis studies [36], in which the sialic acid residue dominated the binding contribution and sulfated Galactose residue also made significant contributions (Table S1). The decompositions of the MM-GSBA energies showed that the non-polar contributions, including van der Waals and non-polar solvation energies, provided the majority of the favorable interaction energies, while the electrostatic and polar solvation energies almost cancelled each other. The distances between each monosaccharide and its interacting amino acid residue in the binding site were stable during MD simulations (Fig. S6), which suggested the non-polar contributions of the interactions were mostly stable despite the low occupancies for some of the hydrogen bond interactions. Furthermore, the mutation of R56A and Q59A in the CC’ loop reduced the binding affinity by 7.5 and 2 folds, respectively. Our previous study showed that the Siglec-8 (R56A) – 6’-sulfo-sLex complex had a less negative MM-GBSA energy than the endogenous Siglec-8 – 6’-sulfo-sLex complex by 1.7 kcal/mol [48]. The reductions observed in the binding affinities for the Siglec-8 (R56A) – 6’-sulfo-sLex and Siglec-8 (Q59A) – 6’-sulfo-sLex complexes confirmed the 6-sulfate group in Gal interacted with residues in the CC’ loop.
Fig. 2.
Binding sites for the Siglec-8 – 6’-sulfo-sLex (A) and Siglec-9 – 6-sulfo-sLacNAc (E) complexes from the representative structures in MD simulations. Proteins are shown in grey with the CC’ loop and the G-strand colored in green and orange, respectively. Monosaccharides are shown in licorice representation and their identities shown with 3D-SNFG nomenclature (GlcNAc, blue cube; Gal, yellow sphere; Neu5Ac, purple diamond) inside each ring [88]. Key residues in the CC’ loop and the G-strand are labeled accordingly. The hydrogen bond interactions represented in dashed lines in the Siglec-8 – 6’-sulfo-sLex complex for Neu5Ac (B) and 6’-sulfo group (C) and those in the Siglec-9 – 6-sulfo-sLacNAc complex for Neu5Ac (F), Gal (G), and 6-sulfo group (H). Schematic representations of binding sites of the Siglec-8 – 6’-sulfo-sLex (D) and Siglec-9 – 6-sulfo-sLacNAc (I) complexes showing hydrogen bonds as dashed lines.
Table 1.
Stable intermolecular hydrogen bond pairs observed in the MD simulations for the Siglec-8 – 6’-sulfo-sLex and the Siglec-9 – 6-sulfo-sLacNAc complexes.
| Ligand | Siglec-8 – 6'-sulfo-sLex | Siglec-9 – 6-sulfo-sLacNAc | |
|---|---|---|---|
| Carbohydrate - Protein Interactions | |||
| Neu5Ac | O1a | R109 - Nη1, 3.0 ± 0.1b(89)c, d | R109 - Nη1, 2.9 ± 0.1 (100)e |
| R109 - Nη2, 2.9 ± 0.1 (100)d, e | R109 - Nη2, 3.0 ± 0.1 (100)e | ||
| N5 | K116 - O, 2.9 ± 0.1(88)d | K116 - O, 2.9 ± 0.1 (90) | |
| O7 | Y7 - Oη, 3.0 ± 0.1 (34)d | --f | |
| O8 | Y58 - Oη, 3.1 ± 0.1(42) | N118 - O, 3.0 ± 0.1 (81) | |
| S118 - N, 3.1 ± 0.1 (65)d | |||
| O9 | S118 - N, 3.0 ± 0.1 (33)d | -- | |
| Gal | O4 | -- | D58 - Oδ, 2.8 ± 0.1 (100)a, e |
| O6 | -- | T57 - N, 3.1 ± 0.1 (65) | |
| Sulfate - Protein Interactions | |||
| 6’-sulfo (6-sulfation in Gal) | Oa | R56 - Nη2, 3.0 ± 0.1 (76)d | |
| R56 - Nε, 2.9 ± 0.1 (59)d | |||
| Q59 - Nε2, 3.0 ± 0.1 (58)d | |||
| 6-sulfo (6-sulfation in GlcNAc) | Oa | -- | N118 - Nδ, 3.0 ± 0.1 (74) |
| -- | K120 - Nζ, 2.9 ± 0.1 (64) | ||
when multiple hydrogen bonds are formed between multiple equivalent heavy atoms, the occupancy of the interaction listed is the sum of all the individual hydrogen bonds and the distance is the average of all the individual hydrogen bonds.
distance in Å.
percentage (%) based on a distance between non-hydrogen atoms of less than 3.5 Å.
also observed in experimentally measured structure (PDB ID: 2N7B).
the occupancy of the interactions between multiple equivalent heavy atoms, calculated as the sum of all the individual hydrogen bonds, is greater than 100%.
no stable interactions observed.
Collectively, both the present and previous experimental and molecular modeling studies for the Siglec-8 – 6’-sulfo-sLex complex showed that the positively charged residues on the CC’ loop locked the ligand in a single conformation, by forming strong interactions with the 6-sulfate in Gal.
3.3. The CC’ loop in the Siglec-9 recognizes 6-sulfation in GlcNAc
As the experimentally measured structures for the Siglec-9 complexes are currently unavailable, the initial geometry for Siglec-9 were generated through comparative modeling or other structure predicting techniques. In the present study, the homology model with the experimentally measured structure for the Siglec-8 as the template and the AlphaFold2 with only the sequence of the Siglec-9 were used to predict the initial structure for Siglec-9, simultaneously. When the structures predicted from two fundamentally different algorithms with and without template structures, and their MD simulations showed structural and dynamic similarities, the reliability of the molecular model for Siglec-9 would be built. The modeled structures for the Siglec-9, including the side chain conformation for the conserved R109 residue essential for binding to the sialic acid residue, from two independent approaches shared a high degree of similarity (Fig. S7). Three independent MD simulations for the molecular models of the Siglec-9 – 6-sulfo-sLacNAc, in which the protein structures were generated by the homology model and AlphaFold2 predictions, were carried out. The Siglec-9 – 6-sulfo-sLacNAc complexes generated with both approaches showed similar behaviors in their MD simulations. The positional RMSD for the ring atom in the ligand trisaccharide along the trajectories of MD simulations for the Siglec-9 – 6-sulfo-sLacNAc complex were generally under 5 Å in all six MD simulations (Fig. S1). The hydrogen bond interactions were also similar in six MD simulations (Table S2 and Table S3). Thus, the reliability and confidence for the molecular model of Siglec-9 were built.
Siglec-9 displayed significant binding preference for sLacNAc motif with 6-sulfation in GlcNAc, such as 6-sulfo-sLacNAc and 6-sulfo-sLex [30], unlike the specificity to the 6-sulfation in Gal (6’-sulfo-sLacNAc and 6’-sulfo-sLex) found in Siglec-8. Such preference in Siglec-9 could be used to validate the molecular model for the Siglec-9 complexes. The 6-sulfo-sLacNAc ligand maintained stable interactions in the binding site of Siglec-9, as the positional RMSD values for the ligand were generally under 5 Å in all six MD simulations. In contrast, those for the ligand in the Siglec-9 – 6’-sulfo-sLacNAc complex exceeded 20 Å (Fig. S8 and Fig. S9), which suggested the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal) disengaged from the Siglec-9 binding site. Both observations from independent MD simulations were in consistent with the binding specificities of Siglec-9 observed in glycan array screenings [30]. Thus, the molecular model for Siglec-9 was validated for representing its binding specificity to the 6-sulfo-sLacNAc ligand over the 6’-sulfo-sLacNAc ligand, and will be used to understand the impact from residues in the CC’ loop.
The ligand in the Siglec-9 – 6-sulfo-sLacNAc complex in MD simulations displayed a single dominant conformation that was similar to that observed in the Siglec-8 – 6’-sulfo-sLex complex (Fig. S2), indicating that the trisaccharide in the 6-sulfo-sLacNAc adopted the same conformation as that in the Siglec-8 – 6’-sulfo-sLex complex. Similar to the binding of the 6’-sulfo-sLex to the Siglec-8, the dominant conformation of 6-sulfo-sLacNAc in the Siglec-9 complex was also selected upon binding from three conformations observed in its unbound state. Furthermore, the preferred ring conformations for each monosaccharide in the ligand remained unchanged upon binding to the Siglec-9 (1C4 for Neu5Ac; 4C1 for Gal and GlcNAc) (Fig. S10). The preference of ω angle in GlcNAc also remained unchanged upon binding, where the gg rotamer (ω ∼ 300°) could promote interactions for the 6-sulfate group (Fig. S11). As the 6-sulfations occurred in different positions in the 6’-sulfo-sLex and 6-sulfo-sLacNAc ligands, these two sulfate groups were seen in MD simulations to interact with residues in the CC’ loop and G-strand, respectively (Fig. 2). The hydrogen bond analysis (Table 1) showed that the 6-sulfate group in GlcNAc formed stable interactions with N118 and K120 in the G-strand, whereas the 6-sulfate group in Gal interacted with R56 and Q59 in the CC’ loop in the Siglec-8 complex. It should also be noted that the essential interactions between the sialic acid residue and the conserved R109 were indeed observed in the MD simulations for the Siglec-9 – 6-sulfo-sLacNAc complex in all six independent MD simulations (Table 1). The MM-GBSA energies for the Siglec-9 – 6-sulfo-sLacNAc complex were similar to those for the Siglec-8 – 6’-sulfo-sLex complex, in which the sialic acid residue dominated the energy contributions and the sulfated GlcNAc also made significant contribution (Table S4 and S5). The non-polar contributions also provided the majority of the favorable interaction energies and the distances between each monosaccharide and the amino acids in the binding site were also stable during MD simulations (Fig. S6). In the Siglec-9 – 6’-sulfo-sLacNAc complex, the close proximity between the 6-sulfation in Gal and the negatively charged side chain of D58 residue in the CC’ loop observed in molecular models generated by both approaches for the Siglec-9 suggested that the unfavorable van der Waals (vdW) and electrostatic interactions could exclude the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal). For further validations, a point mutation of D58S was introduced into the molecular models for the Siglec-9 – 6’-sulfo-sLacNAc complex to remove these unfavorable interactions. The positional RMSD values for the ring atoms of the ligand in the Siglec-9 (D58S) – 6’-sulfo-sLacNAc complex were generally under 10 Å (Fig. S8 and Fig. S9), indicating the stability for the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal) was achieved after removing the negatively charged residues and eliminating unfavorable repulsions between the 6-sulfate group in Gal and the CC’ loop. Thus, the theoretical point mutation of D58S confirmed that Siglec-9 not recognizing 6’-sulfo-sLacNAc was caused by the unfavorable interactions between the 6-sulfation in Gal and the negatively charged residues on the CC’ loop.
Collectively, Siglec-9 formed stable interactions to the 6-sulfo-sLacNAc ligand via interactions with the sialic acid residue and 6-sulfo group. Unlike the 6’-sulfo group in the 6’-sulfo-sLex ligand interacting with the CC’ loop in the Siglec-8, the 6-sulfo group in the Siglec-9 – 6-sulfo-sLacNAc complex interacted with residues in the G-strand. The oligosaccharides in both ligands adopted the same conformation. The negatively charged residues in the CC’ loop also prevented the binding for the 6’-sulfation in the Gal with the Siglec-9.
3.4. Two-state binding model for the Siglec-3 complexes
Unlike the Siglec-8 and Siglec-9, Siglec-3 has been found to bind both 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc (6-sulfation in Gal and GlcNAc, respectively) ligands [39]. The neutral S58 residue on the CC’ loop of Siglec-3, different from the charged residues in the Siglec-8 and Siglec-9, may not be sufficient to lock the sulfate group at the 6-position in Gal by forming strong and favorable interactions, or reject it by forming unfavorable repulsions. In order to study the impact of the neutral residues on the CC’ loop to the binding of sulfate moieties in ligand, molecular models for the Siglec-3 – 6’-sulfo-sLacNAc, Siglec-3 – 6-sulfo-sLacNAc, and Siglec-3 – 6’,6-disulfo-sLacNAc complexes were generated, with which MD simulations were performed.
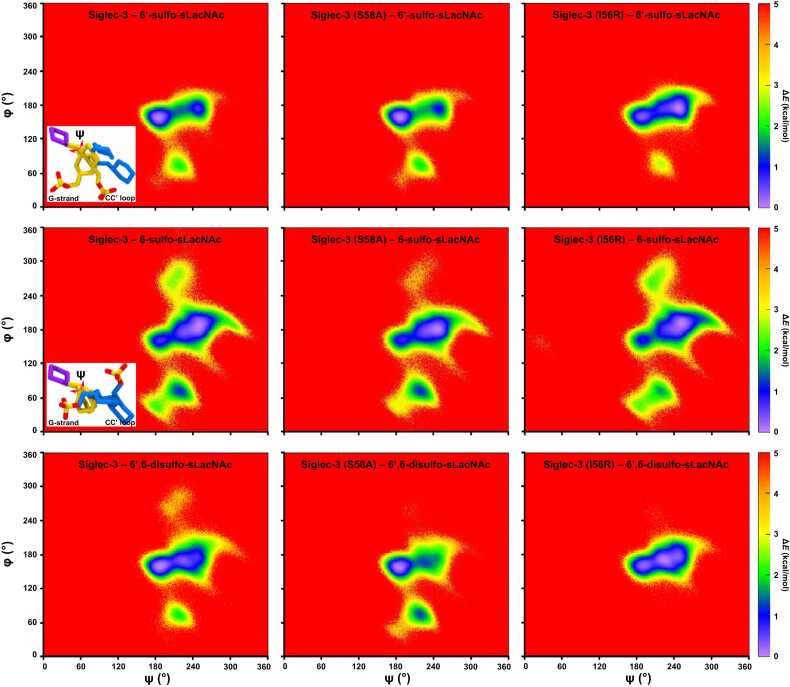
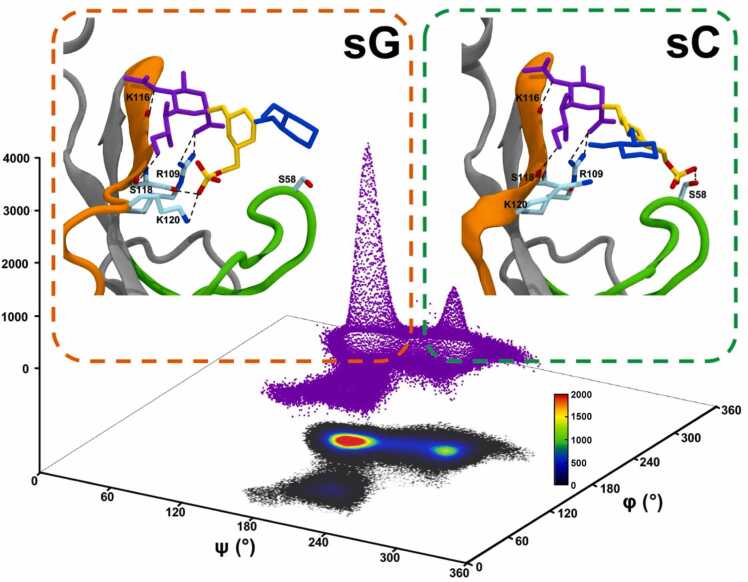
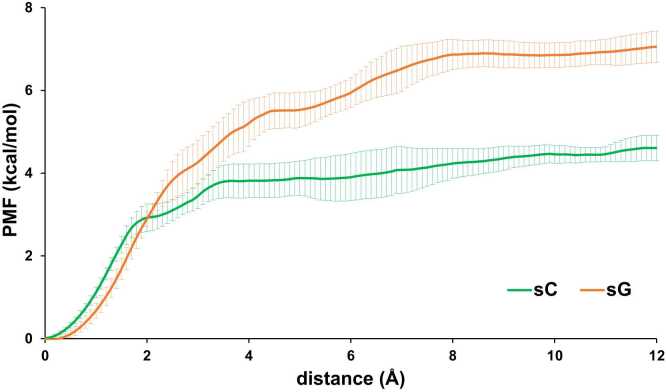
All three sulfated ligands maintained positional stability in the binding site of Siglec-3, as the positional RMSD values for the ring atoms in all three ligands were all generally under 10 Å (Fig. S12). Each monosaccharide in all three ligands maintained stable interactions with the amino acids in the binding site during MD simulations (Fig. S13). Furthermore, the preferred ring conformations for each monosaccharide in three ligands were maintained upon binding to the Siglec-3 (1C4 for Neu5Ac; 4C1 for Gal and GlcNAc) (Fig. S10, S14, and S15). The 2D-RMSD for the pyranose ring atoms in the ligand showed that the ligand adopted two distinct conformations (Fig. S16), one near 2 Å and the other near 5 Å. Furthermore, the frequent transitions between two conformations indicated that all three ligands were able to easily switch from one conformation to the other while maintaining positional stabilities in the binding site of Siglec-3. A closer monitoring of the ligand conformations in MD simulations showed that these two distinctive states were resulted by the ψ angle (C3-O3-C2’-C3’) for Neu5Acα2–3 Gal. As seen in Fig. 3, the ψ angle for Neu5Acα2–3 Gal adopted two conformations, one near 180° and the other near 250°. The φ angle (C2-C3-O3-C2’, 150–180°) for Neu5Acα2–3 Gal (Fig. 3) and the glycosidic linkage for Gal-GlcNAc adopted a single conformation (φ, C4-O4-C1’-C2’, near 170° and ψ, C3-C4-O4-C1’, near 120°) (Fig. S17). The conformations for the Neu5Acα2–3 Gal motif observed in the MD simulations were also found in the co-crystal complex structures in protein databank [89], [90], [91], [92], [93]. Subsequently, the sulfate moieties at different locations in three all ligands interacted with different regions of Siglec-3 (Fig. 4). In the Siglec-3 – 6’-sulfo-sLacNAc complex, when the ψ angle for the Neu5Acα2–3 Gal was near 250°, the 6-position of Gal was close the CC’ loop; thus, in this state, namely sC state, the 6’-sulfo group in Gal interacted with the residues in the CC’ loop. The conformation of the 6’-sulfo-sLacNAc ligand in this state is similar to the 6’-sulfo-sLex ligand in the experimentally measured structure of the Siglec-8 complex. When the ψ angle for the Neu5Acα2–3 Gal was near 180°, the 6-position of Gal was close the G-strand; thus, in this state, namely sG state, the 6’-sulfo group in Gal interacted with the residues in the G-strand of the Siglec-3. Furthermore, the sG state appeared to be a lower energy conformation with a higher population in the MD simulation than the sC state (Fig. 3 and Fig. 4). To validate this binding preference observed for the sG state in the Siglec-3 – 6’-sulfo-sLacNAc complex, ASMD simulations for the complex with the ligand starting from each binding state were performed three independent times. Both PMF energy profiles displayed similar shapes, and showed that the ligand dissociating in the sG state required more work than it in the sC state (Fig. 5). Furthermore, the PMF curve for sG state reached plateau with a longer distance, which suggested the dissociation of the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal) in the sG state required a longer process than that in the sC state, thus, confirmed that the 6’-sulfo-sLacNAc ligand in the sG state adopted a more optimized binding position.
Fig. 3.
The heatmaps of the φ/ψ (φ: C2-C3-O3-C2’, ψ: C3-O3-C2’-C3’) angles for the glycosidic linkage between the Neu5Ac and galactose residues in the wild type and mutated Siglec-3 complexes. Energy differences (ΔE) were calculated based on number of frames in different conformations at 298 K and the most sampled conformation in each complex was the employed as the reference. The representative structures of the 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc ligands in different states while binding to Siglec-3 were added. Monosaccharides are shown in licorice representation and colored according to the SNFG nomenclature [41], [42].
Fig. 4.
The two-state model for the 6’-sulfo-sLacNAc ligand binding to Siglec-3. In each labeled state, proteins are shown in grey with the CC’ loop and the G-strand colored in green and orange, respectively. Monosaccharides are shown in licorice representation and colored according to the SNFG nomenclature [41], [42]. Stable hydrogen bond interactions are shown with dashed lines with involved amino acid residues labeled and shown in licorice representation. The distributions of the φ/ψ angles for the glycosidic linkage between the sialic acid and galactose residues are shown beneath the structures. The 3D representation for the number of the structures sampled for both states in three independent MD simulations is shown in purple dots.
Fig. 5.
PMF calculated for the Siglec-3 – 6’-sulfo-sLacNAc complex with the ligand in the sG and sC states.
Structures for the Siglec-3 – 6’-sulfo-sLacNAc complex with the ligand in each state were extracted from three independent MD simulations to analyze the structural features for both states. The sialic acid residue in both states displayed strong and stable interactions with the conserved R109 (Table 2), similar to those seen in the Siglec-8 and Siglec-9 complexes. In the sC state, the 6-sulfate group in Gal interacted with S58 in the CC’ loop, as stable hydrogen bond interactions were observed in structures collected for the sC state. In the sG state, the sulfate group in Gal interacted with S118 and K120 in the G-strand (Fig. 4 and Table 2) and these interactions were also seen along the entire course of MD simulations with lower occupancies (Table S6).
Table 2.
Stable intermolecular hydrogen bond pairs observed in the structures selected for sG and sC states from the MD simulations for the Siglec-3 – 6’-sulfo-sLacNAc, Siglec-3 – 6-sulfo-sLacNAc, and Siglec-3 – 6’,6-disulfo-sLacNAc complexes.
| Ligand – Atom |
Protein – Atom | Siglec-3 – 6’-sulfo-sLacNAc |
Siglec-3 – 6’,6-disulfo-sLacNAc |
Siglec-3 – 6-sulfo-sLacNAc |
||||
|---|---|---|---|---|---|---|---|---|
| sG state | sC state | sG state | sC state | sG state | sC state | |||
| Neu5Ac | O1a | R109, Nη1 | 2.9 ± 0.2 (100)b, c, d | 2.9 ± 0.2 (100)d | 2.9 ± 0.2 (100)d | 2.9 ± 0.2 (100)d | 2.9 ± 0.3 (100)d | 2.9 ± 0.2 (100)d |
| R109, Nη2 | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | ||
| N5 | K116, O | 2.9 ± 0.2 (90) | 2.9 ± 0.2 (91) | 2.9 ± 0.2 (93) | 2.9 ± 0.2 [98] | 3.0 ± 0.2 (93) | 2.9 ± 0.1 [95] | |
| O9 | S118, O | 3.0 ± 0.2 (21) | 2.9 ± 0.2 (59) | 2.9 ± 0.2 (20) | 2.9 ± 0.2 (45) | 2.9 ± 0.2 (29) | 2.9 ± 0.2 (40) | |
| 6’-sulfo | O | S58, Oγ | --e | 2.8 ± 0.2 (92) | -- | 2.8 ± 0.2 (79) | -- | -- |
| S118, Oγ | 2.8 ± 0.2 (92) | -- | 2.7 ± 0.2 (66) | -- | -- | -- | ||
| K120, Nζ | 3.0 ± 0.2 (48) | -- | 3.0 ± 0.2 (45) | -- | -- | -- | ||
| 6-sulfo | O | S118, Oγ | -- | -- | -- | -- | -- | 2.9 ± 0.3 (47) |
| K120, Nζ | -- | -- | -- | 2.9 ± 0.2 (50) | -- | 2.9 ± 0.2 (33) | ||
when multiple hydrogen bonds are formed between multiple equivalent heavy atoms, the occupancy of the interaction listed is the sum of all the individual hydrogen bonds and the distance is the average of all the individual hydrogen bonds.
distance in Å.
percentage (%) based on a distance between non-hydrogen atoms of less than 3.5 Å.
the occupancy of the interactions between multiple equivalent heavy atoms, calculated as the sum of all the individual hydrogen bonds, is greater than 100%.
no stable interactions observed.
The two states were also adopted by the 6-sulfo-sLacNAc ligand in the Siglec-9 complex in three independent MD simulations (sG state: φ/ψ for Neu5Ac – Gal, 155°/185°; sC state: φ/ψ for Neu5Ac – Gal, 175°/250°; Gal – GlcNAc for both states: φ/ψ – 170°/120°). However, unlike the 6’-sulfo-sLacNAc ligand in the Siglec-3 complex, the sC state was a lower energy state instead of the sG state (Fig. 3). The extracted structures from three independent MD simulations from different states showed that the 6-position in GlcNAc was close to the G-strand when the ligand was in the sC state. The 6-sulfo group in GlcNAc interacted with S118 and K120 residues in the G-strand of Siglec-3 (Table 2). Yet, no stable interactions were observed for the 6-sulfo group when the ligand was in the sG state, since the 6-position of the GlcNAc residue was not facing either the CC’ loop or the G-strand.
Similarly, the two states adopted by the 6’,6-disulfo-sLacNAc ligand were similar to those observed in the 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc ligands (sG state: φ/ψ for Neu5Ac – Gal, 160°/188°; sC state: φ/ψ for Neu5Ac – Gal, 170°/227°; Gal – GlcNAc for both states: φ/ψ – 170°/120°). Furthermore, both sulfate groups in the 6’,6’-disulfo-sLacNAc ligand displayed exceedingly similar behaviors to the corresponding sulfate groups in the 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc ligands. In sC state, the sulfate group in Gal of the 6’,6-disulfo-sLacNAc ligand formed stable interactions with the S58 residue in the CC’ loop; in sG state, it interacted with S118 and K120 residues in the G-strand. The sulfate group in GlcNAc only form stable interactions with the residues in the G-strand while the oligosaccharide was in the sC state. It is also worth noting that the 6-sulfate (6-sulfo in GlcNAc) moiety in the 6-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands preferred the gg rotamer, which is the same as that in the Siglec-9 – 6-sulfo-sLacNAc complex. The gg rotamer allowed the 6-sulfate to form more interactions with residues on the G-strand in both Siglec-9 and Siglec-3 complexes (Fig. S11, C-E). Inversely, the 6’-sulfate (6-sulfo in Gal) moiety in the 6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands preferred gt and tg rotamers while binding to Siglec-3. In the sC state, the 6’-sulfate in the gt rotamer was closer to the S58 in the CC’ loop of Siglec-3 (Fig. S5, G-I). In the sG state, the tg rotamer permitted the 6’-sulfate positioning between the S118 and K120 in the G-strand of Siglec-3 (Fig. S5, J-L). Thus, the 6’-sulfate in Siglec-3 complexes preferred both gt and tg rotamers, which is different from the dominant gt rotamer in the Siglec-8 – 6’-sulfo-sLex complex (Fig. S5, D-F).
Notably, the sulfate group at the 6-position of Gal in both 6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands could interact with the CC’ loop and the G-strand; the sulfate group at the 6-position of GlcNAc in both 6-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands could only interact with the G-strand. The two sulfate groups in the 6’,6-disulfo-sLacNAc ligand displaying the same behaviors as the ones in the 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc ligands suggested that the sulfate groups at different positions of the sLacNAc ligand for the Siglec-3 complex form interactions independently from each other, and their interactions with different regions of the Siglec-3 relied on the poses of the oligosaccharide.
3.5. Energies of two binding states for the Siglec-3 complexes
A recent study [39] discovered that the 6-sulfation in Gal and GlcNAc residues could enhance the binding for sLacNAc motif with the Siglec-3 (6,6’-disulfo-sLacNAc: 6’-sulfo-sLacNAc: 6-sulfo-sLacNAc: sLacNAc = 28: 8: 2: 1 in binding affinities). Theoretical interaction energies were computed for the Siglec-3 complexes using the MM-GBSA approximation, in which the classical mechanical force field provides the energies associated with electrostatic and van der Waals interactions, and the generalized Born model provides an estimate of the desolvation free energy. An advantage of the MM-GBSA approach is the ability to decompose the total interaction energies into the contributions from monosaccharide and amino acid in each complex (Table 3). Structures employed for each energy calculation were evenly extracted from simulation trajectories, containing conformations for both sC and sG states.
Table 3.
Per-residue interaction energies and entropic penalties (in kcal/mol) for Siglec-3 – 6’-sulfo-sLacNAc, Siglec-3 – 6’,6-disulfo-sLacNAc, and Siglec-3 – 6-sulfo-sLacNAc complexes in three independent replicas of MD simulations.
| replica 1 | replica 2 | replica 3 | Average | |
|---|---|---|---|---|
| Siglec-3 – 6'-sulfo-sLacNAc | ||||
| Per-residue MM-GBSA energies | ||||
| Neu5Ac | -9.0 ± 1.7 (65%) | -8.9 ± 1.6 (65%) | -8.8 ± 2.2 (63%) | -8.9 |
| Gal | -2.0 ± 0.7 (14%) | -1.8 ± 0.6 (13%) | -2.0 ± 0.7 (14%) | -1.9 |
| GlcNAc | -1.2 ± 0.8 (9%) | -1.2 ± 0.9 (9%) | -1.2 ± 0.9 (9%) | -1.2 |
| 6'-SO3 | -1.7 ± 0.8 (12%) | -1.8 ± 0.8 (13%) | -1.9 ± 0.8 (14%) | -1.8 |
| MM-GBSA | -13.9 ± 2.1 | -13.8 ± 2.1 | -13.9 ± 2.6 | -13.8 |
| Entropic penalties | ||||
| -TΔSRTV (all) | 8.1 | 8.6 | 8.7 | 8.5 |
| -TΔSqC | 1.5 | 1.5 | 1.3 | 1.4 |
| -TΔS | 9.6 | 10.1 | 10.0 | 9.9 |
| Binding free energies | ||||
| ΔGbinding | -4.2 | -3.6 | -3.9 | -3.9 |
| Siglec-3 – 6',6-disulfo-sLacNAc | ||||
| Per-residue MM-GBSA energies | ||||
| Neu5Ac | -9.4 ± 1.5 (65%) | -9.5 ± 1.6 (65%) | -9.5 ± 1.5 (65%) | -9.5 |
| Gal | -1.9 ± 0.6 (13%) | -1.8 ± 0.7 (12%) | -1.8 ± 0.7 (13%) | -1.8 |
| GlcNAc | -0.9 ± 0.7 (7%) | -1.0 ± 0.7 (7%) | -1.0 ± 0.7 (7%) | -1.0 |
| 6'-SO3 | -1.8 ± 0.8 (12%) | -1.8 ± 0.8 (12%) | -1.8 ± 0.8 (12%) | -1.8 |
| 6-SO3 | -0.5 ± 0.6 (3%) | -0.6 ± 0.7 (4%) | -0.5 ± 0.5 (3%) | -0.5 |
| MM-GBSA | -14.5 ± 2.0 | -14.6 ± 2.1 | -14.6 ± 2.1 | -14.6 |
| Entropic penalties | ||||
| -TΔSRTV (all) | 8.1 | 8.1 | 8.7 | 8.3 |
| -TΔSqC | 1.5 | 1.6 | 1.2 | 1.4 |
| -TΔS | 9.6 | 9.7 | 9.9 | 9.7 |
| Binding free energies | ||||
| ΔGbinding | -5.0 | -4.9 | -4.8 | -4.9 |
| Siglec-3 – 6-sulfo-sLacNAc | ||||
| Per-residue MM-GBSA energies | ||||
| Neu5Ac | -9.6 ± 1.3 (68%) | -9.6 ± 1.3 (68%) | -9.6 ± 1.3 (70%) | -9.6 |
| Gal | -1.6 ± 0.9 (11%) | -1.6 ± 0.9 (11%) | -1.9 ± 1.0 (14%) | -1.7 |
| GlcNAc | -1.4 ± 0.8 (10%) | -1.4 ± 0.8 (10%) | -1.1 ± 0.9 (9%) | -1.3 |
| 6-SO3 | -1.5 ± 0.9 (11%) | -1.5 ± 0.9 (11%) | -1.0 ± 0.8 (7%) | -1.3 |
| MM-GBSA | -14.1 ± 2.0 | -14.1 ± 2.0 | -13.6 ± 2.1 | -13.9 |
| Entropic penalties | ||||
| -TΔSRTV (all) | 10.2 | 11.1 | 10.7 | 10.7 |
| -TΔSqC | 1.0 | 0.8 | 0.7 | 0.8 |
| -TΔS | 11.2 | 11.9 | 11.4 | 11.5 |
| Binding free energies | ||||
| ΔGbinding | -2.9 | -2.2 | -2.3 | -2.4 |
The per-residue decomposition of the MM-GBSA energies showed that, similar to Siglec-8 and Siglec-9 complexes, the sialic acid residue contributed more than half of the total interaction energies in all three ligands (Table 3). The dominant energy contribution from the sialic acid residue (63 – 65%) agreed with its essential role in the Siglec-3 – ligand interactions. The Gal residue in all three ligands contributed similarly to the binding interactions (13 – 14%); the GlcNAc residue in all three ligands also contributed similarly (9%). Since the trisaccharide in all three ligands contributed similarly to the binding energies, the differences in the binding affinities of three sulfated ligands with the Siglec-3 were caused by their differences in sulfation patterns. The 6’-sulfo groups (6-sulfation in Gal) in 6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands both contributed − 1.8 kcal/mol, while the 6-sulfo groups (6-sulfation in GlcNAc) in 6-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands contributed − 1.3 and − 0.5 kcal/mol, respectively.
The total MM-GBSA energies suggested that the 6’,6-disulfo-sLacNAc ligand had stronger binding strength than the 6’-sulfo-sLacNAc and 6-sulfo-sLacNAc ligands, which could be resulted from the contributions from two sulfate groups. The 6’-sulfo group in Gal displayed higher contributions than the 6-sulfo group in GlcNAc, which agreed to the observations that 6’-sulfo group in Gal could form interactions with the Siglec-3 in both sC and sG states while the 6-sulfo group in GlcNAc could only form interactions while the ligand was in the sC state. For the same reason, the 6-sulfo group in GlcNAc in the 6’,6-disulfo-sLacNAc ligand had a much smaller contribution than the 6’-sulfo group in Gal. The entropic penalties for the 6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands were comparable and both smaller than that for the 6-sulfo-sLacNAc ligand. The predicted binding free energies for these three ligands (−4.9 kcal/mol for 6’,6-disulfo-sLacNAc, −3.9 kcal/mol for 6’-sulfo-sLacNAc, −2.4 kcal/mol for 6-sulfo-sLacNAc) agreed to the ranking measured from the electrospray ionization – MS assay (6,6’-disulfo-sLacNAc: 6’-sulfo-sLacNAc: 6-sulfo-sLacNAc: sLacNAc = 28: 8: 2: 1), from which the results are consistent with other approaches such as isothermal titration calorimetry [94]. It should be noted that a value of 4.0 for the internal dielectric constant was applied in the MM-GBSA energy calculations. Values for internal dielectric constant greater than 1 have been proposed to account the charge polarization upon ligand binding, which is not included in the atomic partial charge model. The choice of the internal dielectric constant value has been shown to be somewhat system dependent, and values of 2–4 have been proposed for polar or charged binding site [76]. Although the sLacNAc ligand showed measurable binding to Siglec-3, its affinity appeared to be lower than the other three sulfated ligands. Correspondingly, it is not surprising that the non-sulfated sLacNAc ligand was observed to be positional unstable in the binding site, as the positional RMSD values for the ring atoms in the trisaccharide were over 20 Å after 400 ns of all three independent MD simulations (Fig. S18). As a stable trajectory is required in order to estimate meaningful interaction energies, the binding energy calculations were not performed for the Siglec-3 – sLacNAc complex.
In summary, the two-state model represented the structural and energetic binding features of the 6’-sulfo-sLacNAc, 6-sulfo-sLacNAc, and 6’,6-disulfo-sLacNAc ligands in the Siglec-3 complexes. Different binding poses for the trisaccharide led to different interactions for the sulfate groups, hence different binding strengths for different sulfated sLacNAc ligands. The two-state binding interactions were only observed in the Siglec-3 complexes with sulfated sLacNAc ligand, but not in the Siglec-8 or Siglec-9 complexes, suggesting that the interactions formed by the sulfate groups with the neutral residues in the CC’ loop of Siglec-3 may not be sufficient enough to hold the ligand in a single binding pose.
3.6. Point Mutagenesis in the CC’ loop of Siglec-3
In order to further validate and study the two-state model, two point-mutations, S58A and I56R, were introduced to the CC’ loop of Siglec-3. S58 in wild-type Siglec-3 could form stable interactions with the 6’-sulfo group in Gal residue; thus, the S58A could abolish these interactions and make the sC state unstable. On the other hand, I56R could enhance the interactions between the CC’ loop and the sulfate group and make the sC state easier to access.
In the MD simulations for the Siglec-3 (S58A) complexes with three sulfate ligands, 6’-sulfo-sLacNAc, 6-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc, all three ligands maintained stable bindings, as the positional RMSD values for the ring atoms in the trisaccharides were under 10 Å (Fig. S19). Furthermore, the glycosidic linkages in three ligands also populated in the same conformations as those observed in the wild-type Siglec-3 complexes (Fig. 3 and Fig. S17). However, the hydrogen bond analyses showed that the key interactions between the 6’-sulfo group in Gal (6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc) failed to form stable interactions with the residues in the CC’ loop in the MD simulations (Table 4). Thus, the mutation of S58A in the CC’ loop abolished the interactions previously observed while the ligand was in the sC state and made the sC state less stable, but failed to destabilize the binding for the sulfate sLacNAc ligands. If the sulfated sLacNAc ligands could only adopt the sC state that was observed for the 6’-sulfo-sLex ligand in the Siglec-8 complex, the S58A mutation would abolish the interactions between the 6’-sulfo group in Gal and the CC’ loop, which would make the trisaccharide in the sulfated sLacNAc ligands solely responsible for the 6’-sulfo-sLacNAc ligand binding to Siglec-3. If so, the sulfated sLacNAc ligand would display similar interactions to the sLacNAc ligand in the Siglec-3 complex, which could lead to unstable bindings observed in the Siglec-3 – sLacNAc complex.
Table 4.
Stable intermolecular hydrogen bond pairs observed in the structures selected for sG and sC states from the MD simulations for the Siglec-3 (S58A) – 6’-sulfo-sLacNAc, Siglec-3 (S58A) – 6-sulfo-sLacNAc, and Siglec-3 (S58A) – 6’,6-disulfo-sLacNAc complexes.
| Ligand – Atom |
Protein – Atom | Siglec-3 (S58A) – 6’-sulfo-sLacNAc |
Siglec-3 (S58A) – 6’,6-disulfo-sLacNAc |
Siglec-3 (S58A) – 6-sulfo-sLacNAc |
||||
|---|---|---|---|---|---|---|---|---|
| sG state | sC state | sG state | sC state | sG state | sC state | |||
| Neu5Ac | O1a | R109, Nη1 | 2.9 ± 0.2 (100)b, c, d | 2.9 ± 0.3 (100)d | 2.9 ± 0.2 (100)d | 2.9 ± 0.3 (100)d | 2.9 ± 0.2 (100)d | 2.9 ± 0.2 (100)d |
| R109, Nη2 | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | ||
| N5 | K116, O | 2.9 ± 0.2 (93) | 2.9 ± 0.2 (92) | 2.9 ± 0.2 [95] | 2.9 ± 0.2 [97] | 3.0 ± 0.2 (90) | 2.9 ± 0.2 (93) | |
| O9 | S118, O | 3.0 ± 0.2 (18) | 2.9 ± 0.2 (63) | 3.0 ± 0.2 (18) | 2.9 ± 0.2 (42) | 2.9 ± 0.2 (26) | 2.9 ± 0.2 (33) | |
| 6’-sulfo | O | A58 | --e | -- | -- | -- | -- | -- |
| S118, Oγ | 2.7 ± 0.2 [97] | -- | 2.7 ± 0.2 (93) | -- | -- | -- | ||
| K120, Nζ | 3.0 ± 0.2 (55) | -- | 3.0 ± 0.2 (57) | -- | -- | -- | ||
| 6-sulfo | O | K120, Nζ | -- | -- | -- | 2.9 ± 0.2 (32) | -- | 2.9 ± 0.2 (42) |
when multiple hydrogen bonds are formed between multiple equivalent heavy atoms, the occupancy of the interaction listed is the sum of all the individual hydrogen bonds and the distance is the average of all the individual hydrogen bonds.
distance in Å.
percentage (%) based on a distance between non-hydrogen atoms of less than 3.5 Å.
the occupancy of the interactions between multiple equivalent heavy atoms, calculated as the sum of all the individual hydrogen bonds, is greater than 100%.
no stable interactions observed.
Therefore, these observations suggested that besides the sC state, a novel conformation existed for the sulfated sLacNAc ligand in the Siglec-3 complexes. Other than disengaging from the binding site, the stable bindings for the 6’-sulfo-sLacNAc ligand in the Siglec-3 (S58A) complex confirmed the existence of a novel binding pose (sG state) in addition to the sC state and the necessity of a two-state binding model. Furthermore, the populations of structures in the sG states for the 6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands appeared to be enhanced in the Siglec-3 (S58A) complexes, comparing to those in the wild-type complexes (Fig. 3 and Fig. S17). Without the interactions formed between the sulfate group in Gal and the CC’ loop in the sC state, the sG state from the two-state binding model gained a higher population.
The mutation of I56R, in contrast, could enhance the bindings for the sulfate group in Gal with the CC’ loop by introducing a positively charged side chain. Three sulfated sLacNAc ligands were observed to maintain stable bindings to the Siglec-3 (I56R), as the positional RMSD values for the ring atoms in the ligand were also generally under 10 Å (Fig. S20). The glycosidic linkages in three sulfated sLacNAc ligands also populated in the same conformations as those observed in the wild-type Siglec-3 complexes (Fig. 3 and Fig. S17). Furthermore, the hydrogen bond analyses showed that the sulfate group in Gal could form stable hydrogen bond interactions with R56 in addition to S58 (Table 5). Notably, the population for the sC state was enhanced and became the dominant binding pose for both 6’-sulfo-sLacNAc and 6’,6-disulfo-sLacNAc ligands, in contrast to the dominant sG state observed in the wild-type Siglec-3 complexes (Fig. 3 and Fig. S17). Such strong interactions between the 6’-sulfo group in Gal and the CC’ loop were also observed in the Siglec-8 – 6’-sulfo-sLex complex, in which R56 residue in the CC’ loop of Siglec-8 was seen to form key interactions with the 6’-sulfo group in Gal and lock the oligosaccharide in a single binding pose. Therefore, the enhancement of the sC state after the mutation of I56R suggested a balanced competition existed between the sC and sG states.
Table 5.
Stable intermolecular hydrogen bond pairs observed in the structures selected for sG and sC states from the MD simulations for the Siglec-3 (I56R) – 6’-sulfo-sLacNAc, Siglec-3 (I56R) – 6-sulfo-sLacNAc, and Siglec-3 (I56R) – 6’,6-disulfo-sLacNAc complexes.
| Ligand – Atom |
Protein – Atom | Siglec-3 (I56R) – 6’-sulfo-sLacNAc |
Siglec-3 (I56R) – 6’,6-disulfo-sLacNAc |
Siglec-3 (I56R) – 6-sulfo-sLacNAc |
||||
|---|---|---|---|---|---|---|---|---|
| sG state | sC state | sG state | sC state | sG state | sC state | |||
| Neu5Ac | O1a | R109, Nη1 | 2.9 ± 0.2 (100)b, c, d | 2.9 ± 0.3 (100)d | 2.9 ± 0.2 (100)d | 2.9 ± 0.3 (100)d | 2.9 ± 0.2 (100)d | 2.9 ± 0.2 (100)d |
| R109, Nη2 | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | 3.0 ± 0.2 (100)d | ||
| N5 | K116, O | 2.9 ± 0.2 (83) | 2.9 ± 0.1 (92) | 2.9 ± 0.2 (87) | 2.9 ± 0.2 [97] | 3.0 ± 0.2 (68) | 2.9 ± 0.2 (84) | |
| O9 | S118, O | 2.9 ± 0.2 (14) | 2.9 ± 0.2 (57) | 2.9 ± 0.2 (17) | 2.9 ± 0.2 (42) | 2.9 ± 0.2 (25) | 2.9 ± 0.2 (27) | |
| 6’-sulfo | O | R56, Nε | 3.0 ± 0.2 (26) | 3.0 ± 0.2 (26) | 3.0 ± 0.2 (37) | 3.0 ± 0.2 (68) | --e | -- |
| R56, Nη2 | 3.0 ± 0.2 (21) | 3.0 ± 0.2 (22) | 3.0 ± 0.2 (23) | 2.9 ± 0.2 (51) | -- | -- | ||
| S58, Oγ | 2.7 ± 0.2 (35) | 2.8 ± 0.2 [96] | 2.7 ± 0.2 (41) | 2.8 ± 0.2 (91) | -- | -- | ||
| 6-sulfo | O | R56, Nε | -- | -- | -- | -- | -- | 3.0 ± 0.2 (24) |
| R56, Nη2 | -- | -- | -- | -- | -- | 2.9 ± 0.2 (18) | ||
| S58, Oγ | -- | -- | -- | -- | -- | 2.7 ± 0.2 (46) | ||
| K120, Nζ | -- | -- | -- | 2.9 ± 0.2 (49) | -- | 2.9 ± 0.2 (12) | ||
when multiple hydrogen bonds are formed between multiple equivalent heavy atoms, the occupancy of the interaction listed is the sum of all the individual hydrogen bonds and the distance is the average of all the individual hydrogen bonds.
distance in Å.
percentage (%) based on a distance between non-hydrogen atoms of less than 3.5 Å.
the occupancy of the interactions between multiple equivalent heavy atoms, calculated as the sum of all the individual hydrogen bonds, is greater than 100%.
no stable interactions observed.
Collectively, the mutations for the residues in the CC’ loop confirmed the necessity of the two-state binding model and the competing natures of the two states.
4. Discussion
In the present study, we confirmed with theoretical models and calculations that the sequence variety of the CC’ loops of the Siglec-8/9/3 could determine the recognitions of ligand containing sulfated sLacNAc motifs. Siglec-8 contains a positively charged R56 residue in its CC’ loop and is able to lock the 6’-sulfo-sLex ligand (6-sulfation in Gal) in a single binding pose as shown in its experimentally measured co-complex structure. In contrast, the Siglec-9 contains a negatively charged D58 residue, and the theoretical mutagenesis in the present study confirmed that the unfavorable electrostatic repulsions between D58 and the sulfate group in Gal excluded the recognition for the 6’-sulfo-sLacNAc ligand. Different from the charged residues in the CC’ loop in the Siglec-8 and Siglec-9, the Siglec-3 has neutral residues in the CC’ loop. The molecular models for the Siglec-3 complexes showed that the sulfated ligands adopted a two-state binding model, in which the glycosidic linkage for Neu5Ac – Gal populated into two distinctive conformations and orientated the sulfate groups in Gal and GlcNAc to form different interactions with the CC’ loop and G-strand in the Siglec-3. The ranking from the MM-GBSA energies calculated for the Siglec-3 complexes agreed to the experimentally measured affinities, which validated the molecular models for the Siglec-3 complexes. In the two-state binding model, the sulfate groups in Gal and GlcNAc behaved independently, and their interactions with the Siglec-3 depended on the binding pose for the oligosaccharide in the two-state model. The theoretical mutagenesis study confirmed the necessity of the two-state binding model and the competing natures of the two states. The mutation of S58A abolished the interactions for the sulfate group in Gal with the CC’ loop, but the 6’-sulfo-sLacNAc ligand (6-sulfation in Gal) still maintained a stable binding, confirming that there should be a novel binding pose in addition to the one observed in the Siglec-8 – 6’-sulfo-sLex and the Siglec-9 – 6-sulfo-sLacNAc complexes. The mutation of I56R added a charged side chain to the CC’ loop and promoted the interactions for the sulfate group in Gal with the CC’ loop, and consequently enhanced the distributions of the sC state for both 6’-sulfo-sLacNAc (6-sulfation in Gal) and 6’,6-disulfo-sLacNAc (6-sulfations in both Gal and GlcNAc) ligands, indicating the competing natures between the two states.
The present study demonstrated with molecular models the sequence variety of the CC’ loop could determine the recognition and binding of the sulfated oligosaccharides for several members of the Siglec family. The two-state binding model provided insights into the structural mechanisms for their recognitions.
CRediT authorship contribution statement
Yucheng Wang: Investigation, Formal analysis, Writing – original draft. Yujie Peng: Formal analysis, Visualization. Rui Long: Formal analysis. Peiting Shi: Investigation. Yinghao Zhang: Investigation. De-Xin Kong: Conceptualization, Funding acquisition. Jinshui Zheng: Conceptualization. Xiaocong Wang: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Dr. Hui Liu at Huazhong Agricultural University for insightful discussions. This work was supported by the National Natural Science Foundation of China (No. 21977033) and the Fundamental Research Funds for the Central Universities (Project 2662022XXYJ003).
Supporting Information
The Supporting Information is available online.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.08.014.
Appendix A. Supplementary material
Supplementary material.
.
Data Availability
AMBER18 is available at www.ambermd.org. The MD simulation trajectory files are available upon request. Figures were created with VMD (www.ks.uiuc.edu/Research/vmd/) and gnuplot.
References
- 1.Varki A., Angata T. Siglecs – the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 2.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 3.McMillan S.J., Crocker P.R. CD33-related sialic-acid-binding immunoglobulin-like lectins in health and disease. Carbohydr Res. 2008;343:2050–2056. doi: 10.1016/j.carres.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Lunemann J.D., von Gunten S., Neumann H. Targeting sialylation to treat central nervous system diseases. Trends Pharmacol Sci. 2021;42:998–1008. doi: 10.1016/j.tips.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Murugesan G., Weigle B., Crocker P.R. Siglec and anti-Siglec therapies. Curr Opin Chem Biol. 2021;62:34–42. doi: 10.1016/j.cbpa.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 6.O'Reilly M.K., Paulson J.C. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–248. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Gunten, S., and Bochner, B.S. (2008) Basic and Clinical Immunology of Siglecs. in Year in Immunology 2008 (Rose, N. R. ed.). pp 61–82. [DOI] [PMC free article] [PubMed]
- 8.Gonzalez-Gil A., Schnaar R.L. Siglec Ligands. Cells. 2021;10:1260. doi: 10.3390/cells10051260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miles L.A., Hermans S.J., Crespi G.A.N., Gooi J.H., Doughty L., Nero T.L., et al. Small molecule binding to alzheimer risk factor CD33 promotes Aβ phagocytosis. iScience. 2019;19:110–118. doi: 10.1016/j.isci.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenza M.P., Atxabal U., Oyenarte I., Jiménez-Barbero J., Ereño-Orbea J. Current Status on Therapeutic Molecules Targeting Siglec Receptors. Cells. 2020;9:2691. doi: 10.3390/cells9122691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim J., Sari-Ak D., Bagga T. Siglecs as therapeutic targets in cancer. Biology. 2021;10:1178. doi: 10.3390/biology10111178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magesh S., Ando H., Tsubata T., Ishida H., Kiso M. High-affinity ligands of siglec receptors and their therapeutic potentials. Curr Med Chem. 2011;18:3537–3550. doi: 10.2174/092986711796642580. [DOI] [PubMed] [Google Scholar]
- 13.O'Sullivan J.A., Chang A.T., Youngblood B.A., Bochner B.S. Eosinophil and mast cell Siglecs: From biology to drug target. J Leukoc Biol. 2020;108:73–81. doi: 10.1002/JLB.2MR0120-352RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Sullivan J.A., Carroll D.J., Bochner B.S. Glycobiology of eosinophilic inflammation: contributions of siglecs, glycans, and other glycan-binding proteins. Front Med. 2017;4:12. doi: 10.3389/fmed.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crocker P.R., Kelm S., Dubois C., Martin B., McWilliam A.S., Shotton D.M., et al. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 1991;10:1661–1669. doi: 10.1002/j.1460-2075.1991.tb07689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman S.D., Kelm S., Barber E.K., Crocker P.R. Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood. 1995;85:2005–2012. [PubMed] [Google Scholar]
- 17.Blixt O., Collins B.E., van den Nieuwenhof I.M., Crocker P.R., Paulson J.C. Sialoside specificity of the siglec family assessed using novel multivalent probes - identification of potent inhibitors of myelin-associated glycoprotein. J Biol Chem. 2003;278:31007–31019. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- 18.Kiwamoto T., Kawasaki N., Paulson J.C., Bochner B.S. Siglec-8 as a drugable target to treat eosinophil and mast cell-associated conditions. Pharmacol Ther. 2012;135:327–336. doi: 10.1016/j.pharmthera.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J.Q., Nicoll G., Jones C., Crocker P.R. Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J Biol Chem. 2000;275:22121–22126. doi: 10.1074/jbc.M002788200. [DOI] [PubMed] [Google Scholar]
- 20.Brinkman-Van der Linden E.C.M., Varki A. New aspects of siglec binding specificities, including the significance of fucosylation and of the sialyl-Tn epitope. J Biol Chem. 2000;275:8625–8632. doi: 10.1074/jbc.275.12.8625. [DOI] [PubMed] [Google Scholar]
- 21.Collins B.E., Kiso M., Hasegawa A., Tropak M.B., Roder J.C., Crocker P.R., et al. Binding specificities of the sialoadhesin family of I-type lectins - Sialic acid linkage and substructure requirements for binding of myelin-associated glycoprotein, Schwann cell myelin protein, and sialoadhesin. J Biol Chem. 1997;272:16889–16895. doi: 10.1074/jbc.272.27.16889. [DOI] [PubMed] [Google Scholar]
- 22.Moebius J.M., Widera D., Schmitz J., Kaltschmidt C., Piechaczek C. Impact of polysialylated CD56 on natural killer cell cytotoxicity. BMC Immunol. 2007;8:13. doi: 10.1186/1471-2172-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angata T., Kerr S.C., Greaves D.R., Varki N.M., Crocker P.R., Varki A. Cloning and characterization of human Siglec-11 - A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. 2002;277:24466–24474. doi: 10.1074/jbc.M202833200. [DOI] [PubMed] [Google Scholar]
- 24.Yamaji T., Teranishi T., Alphey M.S., Crocker P.R., Hashimoto Y. A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to alpha 2,8-disialyl and branched alpha 2,6-sialyl residues - A comparison with Siglec-9. J Biol Chem. 2002;277:6324–6332. doi: 10.1074/jbc.M110146200. [DOI] [PubMed] [Google Scholar]
- 25.Campanero-Rhodes M.A., Childs R.A., Kiso M., Komba S., Le Narvor C., Warren J., et al. Carbohydrate microarrays reveal sulphation as a modulator of siglec binding. Biochem Biophys Res Commun. 2006;344:1141–1146. doi: 10.1016/j.bbrc.2006.03.223. [DOI] [PubMed] [Google Scholar]
- 26.Kimura N., Ohmori K., Miyazaki K., Izawa M., Matsuzaki Y., Yasuda Y., et al. Human B-lymphocytes express alpha 2-6-sialylated 6-sulfo-N-acetyllactosamine serving as a preferred ligand for CD22/Siglec-2. J Biol Chem. 2007;282:32200–32207. doi: 10.1074/jbc.M702341200. [DOI] [PubMed] [Google Scholar]
- 27.Muthana S.M., Campbell C.T., Gildersleeve J.C. Modifications of glycans: biological significance and therapeutic opportunities. ACS Chem Biol. 2012;7:31–43. doi: 10.1021/cb2004466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gama C.I., Tully S.E., Sotogaku N., Clark P.M., Rawat M., Vaidehi N., et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 29.Bochner B.S., Alvarez R.A., Mehta P., Bovin N.V., Blixt O., White J.R., et al. Glycan array screening reveals a candidate ligand for Siglec-8. J Biol Chem. 2005;280:4307–4312. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]
- 30.Yu H.F., Gonzalez-Gil A., Wei Y.D., Fernandes S.M., Porell R.N., Vajn K., et al. Siglec-8 and Siglec-9 binding specificities and endogenous airway ligand distributions and properties. Glycobiology. 2017;27:657–668. doi: 10.1093/glycob/cwx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floyd H., Ni J., Cornish A.L., Zeng Z.Z., Liu D., Carter K.C., et al. Siglec-8 - A novel eosinophil-specific member of the immunoglobulin superfamily. J Biol Chem. 2000;275:861–866. doi: 10.1074/jbc.275.2.861. [DOI] [PubMed] [Google Scholar]
- 32.Youngblood B.A., Leung J., Falahati R., Williams J., Schanin J., Brock E.C., et al. Discovery, Function, and Therapeutic Targeting of Siglec-8. Cells. 2020;10:19. doi: 10.3390/cells10010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bochner B.S. Siglec-8 on human eosinophils and mast cells, and Siglec-F on murine eosinophils, are functionally related inhibitory receptors. Clin Exp Allergy. 2009;39:317–324. doi: 10.1111/j.1365-2222.2008.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nutku E., Aizawa H., Hudson S.A., Bochner B.S. Ligation of Siglec-8: a selective mechanism for induction of human eosinophil apoptosis. Blood. 2003;101:5014–5020. doi: 10.1182/blood-2002-10-3058. [DOI] [PubMed] [Google Scholar]
- 35.Jia Y., Yu H.F., Fernandes S.M., Wei Y.D., Gonzalez-Gil A., Motari M.G., et al. Expression of ligands for Siglec-8 and Siglec-9 in human airways and airway cells. J Allergy Clin Immunol. 2015;135:799–810. doi: 10.1016/j.jaci.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Propster J.M., Yang F., Rabbani S., Ernst B., Allain F.H.T., Schubert M. Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8. Proc Natl Acad Sci USA. 2016;113:E4170–E4179. doi: 10.1073/pnas.1602214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lajaunias F., Dayer J.M., Chizzolini C. Constitutive repressor activity of CD33 on human monocytes requires sialic acid recognition and phosphoinositide 3-kinase-mediated intracellular signaling. Eur J Immunol. 2005;35:243–251. doi: 10.1002/eji.200425273. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez B., Escalona Z., Uenishi H., Toki D., Revilla C., Yuste M., et al. Molecular and functional characterization of porcine Siglec-3/CD33 and analysis of its expression in blood and tissues. Dev Comp Immunol. 2015;51:238–250. doi: 10.1016/j.dci.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Jung J., Enterina J.R., Bui D.T., Mozaneh F., Lin P.-H., Nitin, et al. Carbohydrate sulfation as a mechanism for fine-tuning siglec ligands. ACS Chem Biol. 2021;16:2673–2689. doi: 10.1021/acschembio.1c00501. [DOI] [PubMed] [Google Scholar]
- 40.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varki A., Cummings R.D., Aebi M., Packer N.H., Seeberger P.H., Esko J.D., et al. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neelamegham S., Aoki-Kinoshita K., Bolton E., Frank M., Lisacek F., Lütteke T., et al. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 2019;29:620–624. doi: 10.1093/glycob/cwz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woods R.J. Predicting the structures of glycans, glycoproteins, and their complexes. Chem Rev. 2018;118:8005–8024. doi: 10.1021/acs.chemrev.8b00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.González-Cuesta M., Ortiz Mellet C., García Fernández J.M. Carbohydrate supramolecular chemistry: beyond the multivalent effect. Chem Commun. 2020;56:5207–5222. doi: 10.1039/d0cc01135e. [DOI] [PubMed] [Google Scholar]
- 46.Kamiya Y., Yagi-Utsumi M., Yagi H., Kato K. Structural and molecular basis of carbohydrate-protein interaction systems as potential therapeutic targets. Curr Pharm Des. 2011;17:1672–1684. doi: 10.2174/138161211796355074. [DOI] [PubMed] [Google Scholar]
- 47.Agostino M., Yuriev E., Ramsland P. A computational approach for exploring carbohydrate recognition by lectins in innate immunity. Front Immunol. 2011;2:23. doi: 10.3389/fimmu.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Hanes M.S., Cummings R.D., Woods R.J. Computationally guided conversion of the specificity of E-selectin to mimic that of Siglec-8. Proc Natl Acad Sci USA. 2022;119 doi: 10.1073/pnas.2117743119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Attrill H., Imamura A., Sharma R.S., Kiso M., Crocker P.R., van Aalten D.M.F. Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J Biol Chem. 2006;281:32774–32783. doi: 10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- 50.Xu F., Bandara A., Akiyama H., Eshaghi B., Stelter D., Keyes T., et al. Membrane-wrapped nanoparticles probe divergent roles of GM3 and phosphatidylserine in lipid-mediated viral entry pathways. Proc Natl Acad Sci USA. 2018;115:E9041–E9050. doi: 10.1073/pnas.1804292115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barb A.W., Wang X., Prestegard J.H. Refolded recombinant Siglec5 for NMR investigation of complex carbohydrate binding. Protein Expr Purif. 2013;88:183–189. doi: 10.1016/j.pep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhunia A., Jayalakshmi V., Benie A.J., Schuster O., Kelm S., Rama Krishna N., et al. Saturation transfer difference NMR and computational modeling of a sialoadhesin–sialyl lactose complex. Carbohydr Res. 2004;339:259–267. doi: 10.1016/j.carres.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 53.Forgione R.E., Di Carluccio C., Guzmán-Caldentey J., Gaglione R., Battista F., Chiodo F., et al. Unveiling molecular recognition of sialoglycans by human Siglec-10. iScience. 2020;23 doi: 10.1016/j.isci.2020.101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Carluccio C., Crisman E., Manabe Y., Forgione R.E., Lacetera A., Amato J., et al. Characterisation of the dynamic interactions between complex N-glycans and human CD22. ChemBioChem. 2020;21:129–140. doi: 10.1002/cbic.201900295. [DOI] [PubMed] [Google Scholar]
- 55.Yamakawa N., Yasuda Y., Yoshimura A., Goshima A., Crocker P.R., Vergoten G., et al. Discovery of a new sialic acid binding region that regulates Siglec-7. Sci Rep. 2020;10:8647. doi: 10.1038/s41598-020-64887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prescher H., Frank M., Gütgemann S., Kuhfeldt E., Schweizer A., Nitschke L., et al. Design, synthesis, and biological evaluation of small, high-affinity Siglec-7 ligands: toward novel inhibitors of cancer immune evasion. J Med Chem. 2017;60:941–956. doi: 10.1021/acs.jmedchem.6b01111. [DOI] [PubMed] [Google Scholar]
- 57.Soares C.O., Grosso A.S., Ereno-Orbea J., Coelho H., Marcelo F. Molecular recognition insights of sialic acid glycans by distinct receptors unveiled by NMR and molecular modeling. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.727847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eddy S.R. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 63.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steinegger M., Söding J. Clustering huge protein sequence sets in linear time. Nat Commun. 2018;9:2542. doi: 10.1038/s41467-018-04964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richardson L., Allen B., Baldi G., Beracochea M., Bileschi, Maxwell L., et al. MGnify: the microbiome sequence data analysis resource in 2023. Nucleic Acids Res. 2022;51:D753–D759. doi: 10.1093/nar/gkac1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A., et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Case, D.A. , Ben-Shalom, I.Y. , Brozell, S.R. , Cerutti, D.S. , Cheatham, I. , T.E., Cruzeiro, V. W. D.,.... et al. (2018) AMBER18.
- 68.Kirschner K.N., Yongye A.B., Tschampel S.M., Gonzalez-Outeirino J., Daniels C.R., Foley B.L., et al. GLYCAM06: a generalizable Biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Case D.A., Cheatham T.E., Darden T., Gohlke H., Luo R., Merz K.M., et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., Haak J.R. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 71.Gotz A.W., Williamson M.J., Xu D., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vangunsteren W.F., Berendsen H.J.C. Algorithms for macromoleculr dynamics and constraint dynamics. Mol Phys. 1977;34:1311–1327. [Google Scholar]
- 73.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S.H., Chong L., et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 74.Onufriev A., Bashford D., Case D.A. Modification of the generalized Born model suitable for macromolecules. J Phys Chem B. 2000;104:3712–3720. [Google Scholar]
- 75.Krishnamurthy V.R., Sardar M.Y.R., Ying Y., Song X., Haller C., Dai E., et al. Glycopeptide analogues of PSGL-1 inhibit P-selectin in vitro and in vivo. Nat Commun. 2015;6:6387. doi: 10.1038/ncomms7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hou T.J., Wang J.M., Li Y.Y., Wang W. Assessing the performance of the molecular mechanics/poisson boltzmann surface area and molecular mechanics/generalized born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem. 2011;32:866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 78.Sood A., Gerlits O.O., Ji Y., Bovin N.V., Coates L., Woods R.J. Defining the specificity of carbohydrate–protein interactions by quantifying functional group contributions. J Chem Inf Model. 2018;58:1889–1901. doi: 10.1021/acs.jcim.8b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karplus M., Kushick J.N. Method for estimating the configurational entropy of macromolecules. Macromolecules. 1981;14:325–332. [Google Scholar]
- 80.Ozer G., Keyes T., Quirk S., Hernandez R. Multiple branched adaptive steered molecular dynamics. J Chem Phys. 2014;141 doi: 10.1063/1.4891807. [DOI] [PubMed] [Google Scholar]
- 81.Ozer G., Quirk S., Hernandez R. Adaptive steered molecular dynamics: validation of the selection criterion and benchmarking energetics in vacuum. J Chem Phys. 2012;136 doi: 10.1063/1.4725183. [DOI] [PubMed] [Google Scholar]
- 82.Ozer G., Valeev E.F., Quirk S., Hernandez R. Adaptive Steered molecular dynamics of the long-distance unfolding of neuropeptide Y. J Chem Theory Comput. 2010;6:3026–3038. doi: 10.1021/ct100320g. [DOI] [PubMed] [Google Scholar]
- 83.Loncharich R.J., Brooks B.R., Pastor R.W. Langevin dynamics of peptides – the frictional dependence of isomerization rates of N-acetylalanyl-N’-methylamide. Biopolymers. 1992;32:523–535. doi: 10.1002/bip.360320508. [DOI] [PubMed] [Google Scholar]
- 84.Ozer G., Quirk S., Hernandez R. Thermodynamics of decaalanine stretching in water obtained by adaptive steered molecular dynamics simulations. J Chem Theory Comput. 2012;8:4837–4844. doi: 10.1021/ct300709u. [DOI] [PubMed] [Google Scholar]
- 85.Alphey M.S., Attrill H., Crocker P.R., van Aalten D.M.F. High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J Biol Chem. 2003;278:3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 86.Zhuravleva M.A., Trandem K., Sun P.D. Structural implications of Siglec-5-mediated sialoglycan recognition. J Mol Biol. 2008;375:437–447. doi: 10.1016/j.jmb.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Angata T. Specificity of glycan recognition by Siglecs. Trends Glycosci Glycotechnol. 2006;18:197–208. [Google Scholar]
- 88.Thieker D.F., Hadden J.A., Schulten K., Woods R.J. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 2016;26:786–787. doi: 10.1093/glycob/cww076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pronker M.F., Lemstra S., Snijder J., Heck A.J.R., Thies-Weesie D.M.E., Pasterkamp R.J., et al. Structural basis of myelin-associated glycoprotein adhesion and signalling. Nat Commun. 2016;7:13584. doi: 10.1038/ncomms13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Neu U., Hengel H., Blaum B.S., Schowalter R.M., Macejak D., Gilbert M., et al. Structures of merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Imberty A., Gautier C., Lescar J., Perez S., Wyns L. An unusual carbohydrate binding site revealed by the structures of two Maackia amurensis lectins complexed with sialic acid-containing oligosaccharides. J Biol Chem. 2000;275:17541–17548. doi: 10.1074/jbc.M000560200. [DOI] [PubMed] [Google Scholar]
- 92.Lu X.S., Qi J.X., Shi Y., Wang M., Smith D.F., Heimburg-Molinaro J., et al. Structure and receptor binding specificity of hemagglutinin H13 from avian influenza A virus H13N6. J Virol. 2013;87:9077–9085. doi: 10.1128/JVI.00235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seiradake E., Henaff D., Wodrich H., Billet O., Perreau M., Hippert C., et al. The cell adhesion molecule "CAR'' and sialic acid on human erythrocytes influence adenovirus in vivo biodistribution. PLOS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kitova E.N., El-Hawiet A., Schnier P.D., Klassen J.S. Reliable determinations of protein–ligand interactions by direct ESI-MS measurements. are we there yet? J Am Soc Mass Spectrom. 2012;23:431–441. doi: 10.1007/s13361-011-0311-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Data Availability Statement
AMBER18 is available at www.ambermd.org. The MD simulation trajectory files are available upon request. Figures were created with VMD (www.ks.uiuc.edu/Research/vmd/) and gnuplot.