Abstract
Antibody–drug conjugates (ADC) achieve targeted drug delivery to a tumor and have demonstrated clinical success in many tumor types. The activity and safety profile of an ADC depends on its construction: antibody, payload, linker, and conjugation method, as well as the number of payload drugs per antibody [drug-to-antibody ratio (DAR)]. To allow for ADC optimization for a given target antigen, we developed Dolasynthen (DS), a novel ADC platform based on the payload auristatin hydroxypropylamide, that enables precise DAR-ranging and site-specific conjugation. We used the new platform to optimize an ADC that targets B7-H4 (VTCN1), an immune-suppressive protein that is overexpressed in breast, ovarian, and endometrial cancers. XMT-1660 is a site-specific DS DAR 6 ADC that induced complete tumor regressions in xenograft models of breast and ovarian cancer as well as in a syngeneic breast cancer model that is refractory to PD-1 immune checkpoint inhibition. In a panel of 28 breast cancer PDXs, XMT-1660 demonstrated activity that correlated with B7-H4 expression. XMT-1660 has recently entered clinical development in a phase I study (NCT05377996) in patients with cancer.
Graphical Abstract
Introduction
Antibody–drug conjugates (ADCs) constitute a therapeutic modality that achieves targeted delivery to cells or tissues of interest by conjugating a payload drug to an antibody (1, 2). ADCs have been developed predominantly in oncology, and currently in the United States, there are 12 approved ADCs built on 8 distinct linker-payload platforms, all of which employ a cytotoxic payload. More recently, in early clinical development, there has been an increase in the number of ADCs for oncology based on payloads with noncytotoxic mechanisms of action, such as immune stimulation, as well as ADCs for therapeutic applications beyond oncology, such as inflammation and infectious disease (3–5).
B7-H4 (VTCN1) is a plasma membrane protein expressed in multiple tumor types, including breast cancer, ovarian cancer, and endometrial cancer, with limited expression in normal tissues (6–8). Accordingly, preclinical studies suggested that B7-H4 could be amenable to an ADC approach (8, 9) as well as to other therapeutic modalities (9, 10). B7-H4 has been reported to have an immune-suppressive function (11–14), and its expression in tumors has been associated with a lower frequency of tumor-infiltrating immune cells (7, 15). In a mouse tumor model, B7-H4 knockout was associated with increased antitumor immune response, reduced metastasis, and increased survival (16).
Strikingly, B7-H4 expression in tumors has minimal overlap with PD-L1 (also known as B7-H1) expression (17–19), which may reflect functional redundancy in inhibiting antitumor immunity. Consequently, in B7-H4+ tumors where PD-L1 expression is absent or low and PD-(L)1 immune checkpoint inhibitors may not be effective, a B7-H4 targeted agent may be an effective treatment option. The nonoverlapping expression profiles likely rooted in functional redundancy also provide a rationale for the potential benefit of combining B7-H4 and PD-L1 targeted therapies, to prevent tumors from escaping therapy by switching between these immunosuppressive ligands.
The pharmacology of an ADC has been shown to depend not only on the antibody, linker and payload, but also the method and antibody site of conjugation (20). An additional factor that impacts the pharmacology of ADCs is the number of payload drugs per antibody [drug-to-antibody ratio (DAR)], that balances distribution of the drug to individual cancer cells versus the tumor tissue. For instance, higher DAR might be required for cancer cell killing in cases of lower payload potency or lower target expression, yet in cases where payload dose (not antibody dose) defines the clinical dose level, higher DAR represents lower antibody dose, which could limit distribution throughout the tumor tissue in cases of large tumor size or poor tumor vascularization. Thus, the optimal DAR for a target is determined by target expression and internalization rate as well as tumor architecture and the characteristics of the ADC platform (21, 22). DAR and conjugation method can impact the safety profile, as illustrated by the clinical data of two MUC16-targeted ADCs that were based on the same vcMMAE linker-payload, one with stochastic conjugation DAR∼4 and the other with site-specific conjugation DAR∼2. The site-specific ADC had greater efficacy as well as significant ocular toxicity that was not observed with the stochastically conjugated ADC (23, 24). A major constraint of most ADC platforms is that the pharmacokinetic properties significantly deteriorate with higher DARs, which limits the ability to DAR-range and to identify the optimal DAR for a given target.
The Dolaflexin (DF) platform (25) overcame the historical challenges of achieving higher DAR due to compromised pharmacokinetics profiles (26). The hydrophilic scaffold and a charge compensating element offset the hydrophobic nature of the payload and thus enable high DAR without sacrificing desirable physicochemical and pharmacokinetic profiles (25). A distinguishing feature of the payload in the DF platform, auristatin F hydroxypropylamide (AF-HPA), is its controlled bystander effect. In general, bystander effect is the ability of an ADC to indirectly cause the death of antigen-negative cells; for instance, by the diffusion of payload delivered to antigen-positive cells into neighboring antigen-negative cells. In the case of DF, the ability of AF-HPA to diffuse into neighboring tumor cells is balanced by the intracellular conversion of AF-HPA to AF, a molecule that is highly active intracellularly and gets trapped in tumor cells because it is less membrane permeable and not a PgP substrate. In cytotoxicity assays, the potency of AF-HPA was within a range of ∼1 to 5 nmol/L across a panel of cancer cell lines, and AF is observed to be less potent likely due to reduced membrane permeability (25). The DF platform is the basis of XMT-1536 (UpRi), a NaPi2b-targeted ADC in clinical development; in the UPLIFT trial (NCT03319628), UpRi is administered at 36 mg/m2 (capped at 2.2 m2), which is the dose equivalent of 0.056 mg/kg payload (27). Notably, DF is not compatible with precise DAR-ranging or site-specific conjugation.
In this study, we defined an optimized B7-H4–targeted ADC by comparing ADCs with different DARs generated by different conjugation methods. We created a novel ADC platform, Dolasynthen (DS), in which the key features of DF were distilled into a fully synthetic framework that enables precise DAR ranging and site-specific conjugation. We then generated three B7-H4 ADCs with different drug ratios (DS DAR 2, DS DAR 6, and DF DAR 12) and compared them in preclinical studies to evaluate the impact of DAR on the pharmacologic profile. The site-specific DS DAR 6 ADC (XMT-1660) exhibited a superior preclinical profile and was selected for clinical development. Translational pharmacology studies with XMT-1660 demonstrated an expression-activity relationship that may inform clinical development.
Materials and Methods
Antibodies: The antibodies used were trastuzumab (anti-HER2); XMT-1535 (anti-NaPi2b; ref. 27); and XMT-1604 (anti–B7-H4). For OVCAR3, HBCx-24 and MX1 studies, the nonbinding control antibody was rituximab (binds human but not murine CD20), except for OV2423, CTG-1692, and mBR9013 studies, in which the nonbinding control was palivizumab (binds RSV).
The B7-H4 antibody XMT-1604 (CAS RN 2855971–15–0) was selected on the basis of the in vitro and in vivo characterization of 20 antibodies designed to overcome 3 developability liabilities identified with parental antibody 2F9 (U.S. patent no. 8,609,816): an unpaired cysteine, an aspartate isomerization sequence, and a methionine oxidation site. XMT-1604 exhibited comparable antigen binding properties and a lower polyspecificity score (Baculovirus particle ELISA) compared with parental 2F9; see U.S. Patent Application Publication US2022/0233707 A1. The sequence is provided in the Supplementary Material.
Syntheses of constructs
The DS scaffold-linker-payload (Constructs A and B in Supplementary Fig. S1) was the result of extensive optimization efforts including variation of the PEG motif and the presence or absence of a negative charge. The syntheses of constructs A-C used for conjugation in this study are described in the Supplementary Material and the synthesis for construct D was reported previously (25).
Syntheses of ADCs
DS ADC (Fig. 1A) was synthesized analogous to ADC1 where NaPi2b mAb was used instead of trastuzumab. The purified conjugate had a drug to trastuzumab ratio of 12.1 as determined by hydrolysis followed by RP-HPLC.
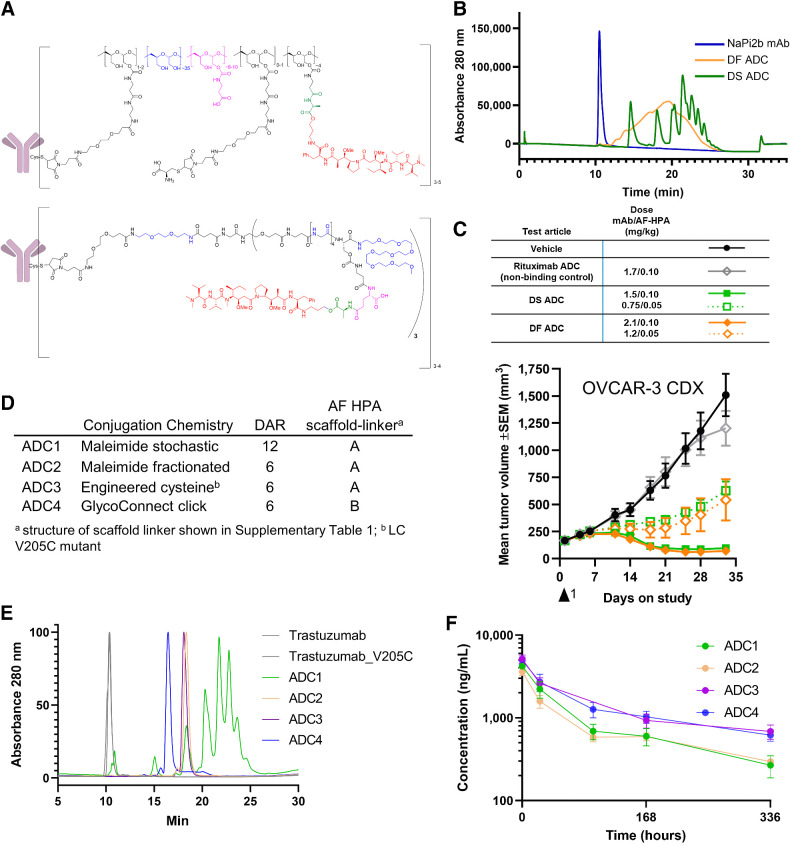
Figure 1.
Development of DS and site-specific technology. A, DS ADCs (bottom structure) incorporate structural elements of the DF platform (top structure) within a fully synthetic, well-defined scaffold with a specific number of drugs per conjugated unit. B, The HIC of DS ADC indicates enhanced homogeneity over the DF ADC; C, Antitumor activity of DS and DF ADC following a single dose is comparable at equivalent drug dose and 2 dose levels; D, Trastuzumab ADCs made by four distinct approaches and two scaffold-linker payloads to generate DAR12 and DAR6 conjugates; E, HIC HPLC of trastuzumab ADCs; ADC3 and ADC4 show a fully homogeneous profile; F, Pharmacokinetics profile of ADC1–4 following a single intravenous bolus administration of ADC equivalent to a 0.199 mg/kg AF HPA dose to female CB.17 SCID mice bearing JIMT-1 human breast carcinoma xenograft tumors (6 mice per group) and samples collected at 10 min, 24 hours, 96 hours (ADC3 did not have the 96 hours timepoint due to operator error), 168 hours, and 336 hours. Graph depicts conjugated drug analyte concentration over the course of the study.
DF ADC (Fig. 1A) was synthesized as previously described for DF ADCs (25).
ADC1: Trastuzumab (2 mg, 0.014 μmol) was combined with TCEP-HCl (0.056 μmol) and 50 mmol/L HEPES, 1 mmol/L EDTA, pH 7 and TBS (249 μL, pH 7.6) to achieve a final antibody concentration of 5 mg/mL. The reaction continued for 90 minutes at 37°C. Then Construct A (0.535 mg, 0.084 μmol) in 50 mmol/L HEPES, 1 mmol/L EDTA, pH 7 was added to the reduced antibody and reaction was allowed to proceed for 60 minutes at 37°C. Reaction was quenched with L-cysteine (25 μg) in in 50 mmol/L HEPES, 1 mmol/L EDTA, pH 7. The crude product was purified by CHT Type II (Bio-Rad, P/N 7324756) loading with 10 mmol/L sodium phosphate, pH 6.5 and washing with the same buffer until UV absorbance at 214 nm returned to baseline. ADC was then eluted with 10 mmol/L sodium phosphate, 2 mol/L sodium chloride, pH 6.5. Eluted ADC was formulated by three rounds of ultrafiltration-dilution using a 30 kDa MWCO centrifugal filter (Millipore Sigma P/N UFC9030). The purified conjugate had a drug to trastuzumab ratio of 12.1 as determined by hydrolysis followed by RP-HPLC.
ADC2: To a solution of trastuzumab (40 mg, 0.275 μmol), in TEAA buffer (50 mmol/L, 1 mmol/L EDTA, pH 7, 0.831 mL) was added a solution of TCEP-HCl (0.118 mg, 0.413 μmol). A solution of Construct A (10.7 mg/mL, 1.65 mmol/L, prepared as described in Supplementary Material) in DMA was added and the resulting mixture was incubated for 1 hour at room temperature. L-cysteine (16 mg/mL, 132 mmol/L) was added, and the reaction mixture stirred for 30 minutes. The crude reaction mixture was purified by hydrophobic interaction chromatography (HIC) to give the desired conjugate (6.7 mg, 11% yield). The purified conjugate had a drug to trastuzumab ratio of 6.4 as determined by hydrolysis followed by RP-HPLC.
ADC3: To a solution of cysteine engineered trastuzumab LC V205C (30 mg, 0.21 μmol), in conjugation buffer (25 mmol/L HEPES, 25 mmol/L NaCl, 1 mmol/L EDTA, pH 8, 5.84 mL, 5.14 mg/mL) was added a solution of TCEP-HCl (0.573 mg, 2.1 μmol) and the resulting mixture was shaken for 4 hours at 37°C. The interchain disulfides were reoxidized by adding dehydroascorbic acid (dhAA) dissolved in reaction buffer (8.71 mg/mL, 50 mmol/L) and the mixture was rotated for 2 hours at room temperature. A solution of Construct A (6.4 mg/mL, 1 mmol/L) in DMSO was added and the resulting mixture was stirred for 1.5 hours at room temperature. The pH of the mixture was adjusted to ∼5.1 with 1 mol/L acetic acid and the crude product was purified by HPLC to give the desired conjugate (5.9 mg, 12% yield). The purified conjugate had a drug to trastuzumab ratio of 6.6 as determined by hydrolysis followed by RP-HPLC.
ADC4: Azido-modified trastuzumab was prepared as described above for the B7-H4 antibody. Azido-modified trastuzumab (16.5 mg, 0.113 μmol) in TBS was combined with Construct B in water (7.18 mg, 1.130 μmol) at a final antibody concentration of 15 mg/mL. Reaction proceeded overnight at 30°C. The crude product was purified by CHT Type II (Bio-Rad, P/N 7324756) loading with 10 mmol/L sodium phosphate, pH 6.5 and washing with the same buffer until UV absorbance at 214 nm returned to baseline. ADC was then eluted with 10 mmol/L sodium phosphate, 2 mol/L sodium chloride, pH 6.5. Eluted material was then polished with a Poros Ethyl HIC column (ThermoFisher P/N A36653) using a linear gradient from 25 mmol/L sodium phosphate, 1.5 mol/L ammonium sulfate, pH 7 to 25 mmol/L sodium phosphate, 10% acetonitrile, pH 7. Desired DAR 6 fractions were pooled and formulated by three rounds of ultrafiltration-dilution using a 30 kDa MWCO centrifugal filter (Millipore Sigma, P/N UFC9030). The purified conjugate had a drug to trastuzumab ratio of 5.9 as determined by hydrolysis followed by RP-HPLC.
Azido-modified B7-H4 antibody: To the B7-H4 antibody (12.71 mg, 0.088 μmol) in 50 mmol/L Tris-HCl, pH 7.6, was added in the following order: Endo SH (0.127 mg, 1 w-%), GalNAcT (0.64 mg, 5 w-%), UDP-azido sugar (1.34 mg, 2.12 μmol), and MnCl2 (1.18 mg, 9.4 μmol), to achieve a final antibody concentration of 13.5 g/L. The reaction was stirred at 30 rpm for 17 hours at 30°C. The crude azido-modified B7-H4 antibody was purified by Protein A chromatography and dialysis to give the azido-modified B7-H4 antibody (10.53 mg, 83% yield).
XMT-1660 B7-H4 DS DAR 6: Azido-modified B7-H4 antibody (10.03 mg, 0.070 μmol) in PBS, pH 7.2 and Construct B (CAS RN 2669084–60–8; structure in Supplementary Table S1; 4.25 mg, 0.67 μmol, prepared as described in Supplementary Materials) in water, were gently mixed, then left for 20 hours at 30°C without shaking or rocking. The crude product was purified by UF/DF and HIC to give XMT-1660 B7-H4 DS DAR 6 (5.85 mg, 58% yield; CAS RN 2855974–40–0), that had a DAR of 5.9 as determined by reduced RP-HPLC.
B7-H4 DS DAR 2: Azido-modified B7-H4 antibody (50 mg, 0.346 μmol) and Construct C (7.12 mg, 3.34 μmol prepared as described in Supplementary Material) instead of Construct B were used in Step 2 as described above. The purified B7-H4 DS DAR 2 (30.1 mg, 60% yield) had a DAR of 2.0 as determined by reduced RP-HPLC
B7-H4 DF DAR 12: To a solution of B7-H4 antibody (20 mg, 0.139 μmol) in TEAA buffer, pH 7 (4 mL) was added a solution of TCEP (0.0993 mg, 0.347 μmol) while stirring. The mixture was incubated for 1.5 hours at room temperature. The partially reduced B7-H4 antibody was then added to a vigorously stirred solution of Construct D (Supplementary Table S1; 18 mg, 1.807 μmol, prepared as described previously; ref. 25) in TEAA buffer, pH 6 (1.8 mL). The stirring was continued for 1 hour at room temperature. The reaction was quenched with an aqueous solution of cysteine (0.421 mg, 3.47 μmol) in TEAA buffer, pH 7 (0.084 mL). After stirring for 30 minutes at ambient temperature at pH 7.0, the reaction mixture was acidified to pH 5.8. The crude product was purified by WCX to give B7-H4 DF DAR 12 (10 mg, 50% yield), that had a DAR of 11.9 as determined by hydrolysis followed by RP-HPLC.
Control DS DAR 6 ADCs: Conjugate was synthesized as described for XMT-1660 B7-H4 DS DAR 6, except azido-modified Rituximab or Palivizumab antibody (131.4 mg, 0.91 μmol, prepared as described for B7-H4 antibody), was used in Step 2 instead of azido-modified B7-H4 antibody had a DAR of 5.9 as determined by reduced RP-HPLC.
Control DF DAR 12 was synthesized as described for B7-H4 DF DAR 12, except Rituximab antibody (100 mg, 6.99 μmol) was used instead of B7-H4 antibody. The purified conjugate (62.6 mg, 63% yield) had a DAR of 10.8 as determined by hydrolysis followed by RP-HPLC.
HIC: The hydrophobicity of the ADC was determined by HIC-HPLC on a Shimadzu Prominence HPLC system equipped with a DAD. A TSK gel butyl-NPR column (2.5 μm particle size) that was held at 35°C for these analyses. Mobile phase A was 1.5 mol/L ammonium sulfate, 25 mmol/L sodium phosphate, and pH 7.0, and mobile phase B was 25 mmol/L sodium phosphate, 10% isopropyl alcohol, and pH 7.0. Separations were performed with a 0% to 100% linear gradient of mobile phase B over 25 minutes. The flow rate was 1 mL/min. Sample injections ranged from ∼10 μg to 100 μg.
ELISA: B7-H4 proteins from human, monkey, rat, and mouse (R&D Systems 6576-B7–050, 10085-B7, 2154-B7 and Creative Biomart VTCN1–1519R) were coated onto the surface of wells of a 96-well plate by incubation with proteins (1 μg/mL) overnight at 4°C. Wells were then blocked with BSA (4% BSA in PBS) and then incubated with test article at fivefold serial dilutions for 2 hours with rocking at room temperature. Plates were washed 3 times in PBST. Wells were incubated with secondary anti-human IgG conjugated to HRP (0.16 μg/mL in PBST; Jackson ImmunoResearch, catalog no. 109–035–097) for 1 hour and washed 3 times. The HRP substrate, TMB (Bethyl Lab, catalog no. E102), was added and incubated for four minutes. The reaction was quenched with sulfuric acid (0.2 N, 100 μL). The absorbance at 450 nm was measured in a Spectramax M5 plate reader (Molecular Devices). EC50 values were determined with GraphPad Prism, RRID:SCR_002798 by four-parameter curve fitting.
Cell lines: MX-1 cells (RRID:CVCL_4774) (Creative Bioarray CSC-C0264) were cultured in DMEM:F12K with 10% FBS and 1% penicillin/streptomycin. CAMA-1 cells (RRID:CVCL_1115; ATCC HTB-21) were cultured in EMEM with 10% FBS and 1% penicillin/streptomycin. HEK293 cells (RRID:CVCL_0045; ATCC CRL-1573) and HEK293-B7-H4 cells (exogenous expression of human B7-H4) were cultured in EMEM with 10% FBS and 1% penicillin/streptomycin. HEK-293-B7-H4 cells were stable pools generated by transduction of B7-H4 lentivirus (G&P Biosciences, catalog no. LTV0745) and selection and maintenance in 3 μg/mL puromycin. Cell lines were authenticated using the CellCheck 9 Plus assay by IDEXX, which uses nine markers (eight STR and amelogenin) to verify identity by STR profiling. The assay also performs multiplex PCR to identify for interspecies contamination and Mycoplasma contamination. Cell lines were authenticated and mycoplasma-tested within 3 months of all experiments. Cell lines were cultured and studied for less than 3 months after being thawed; after 3 months, a new vial was thawed, tested, and used for experimentation.
T-cell proliferation assay: HEK293 or HEK293-B7-H4 cells were plated at a density of 50,000 cells per well in a 96-well plate and allowed to adhere overnight. Media was replaced with T cell media (Iscove's modified Dulbecco's medium with 10% FBS and 5% penicillin/streptomycin). Cells were incubated with 10 nmol/L antibody for 2 hours at 37°C, prior to the addition of CD3+ T cells. CD3+ cells were prepared as follows: frozen human PBMCs (2.5×107 cells; StemCell Technologies, Lot# 20017203C) were thawed and enriched for CD3+ T cells using EasySEP human T cell isolation kit (StemCell Technologies, 17951); CD3+ T cells were labeled with CellTrace Violet cell proliferation kit (CTV; ThermoFisher Scientific, #C34557) and adjusted to 2×106 cells/mL; 100,000 CTV-labeled T cells were added to each well containing test article-treated cells, stimulated with Immunocult Human CD3/CD28 T Cell Activator (StemCell Technologies, #10971), and incubated at 37°C in 5% CO2. After 4 days, T cell proliferation was assessed by the dilution of CTV-labeled T cells using flow cytometry. Cocultured cells were transferred to a U-bottom 96-well plate, washed with PBS, stained with LIVE/DEADTM Fixable Aqua dead cell staining dye (ThermoFisher Scientific, #L34966), and stained with fluorophore conjugated targeted or isotype control antibodies [FITC anti-human CD45 (BioLegend, catalog no. 304006, RRID:AB_314394) PE/Cy7 anti-human CD4 (BioLegend, catalog no. 357410, RRID:AB_2565662), PE anti-human CD8 (BioLegend, catalog no. 344705,AB_1953243)]. Cells were analyzed on a MACSQuant flow cytometer. Data analysis was performed using FlowJo, RRID:SCR_008520 software using the following hierarchical flow: (i) single gate, (ii) live cells, (iii) CD45+ cells, and (iv) diluted CTV (compared with unstimulated T cells). Percentage of proliferating CD4+ or CD8+ T cell were calculated using CD4+ combined with diluted CTV and CD8+ combined with diluted CTV, respectively.
Flow cytometry: Cells were detached with Accutase solution (Mediatech/Corning) before flow cytometry analysis. Cells were stained with test articles in DMEM with 6% goat serum on ice, washed, stained with secondary antibody, washed, and 5,000 cells per sample were analyzed on a flow cytometer. EC50 values were calculated with GraphPad Prism by four-parameter curve fitting.
Cytotoxicity: Cells were plated and allowed to adhere overnight. Cells were treated with serial dilutions of test article for 3 days. CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, G7570) was performed, and signal was measured with a SpectraMax M5 plate reader (Molecular Devices). Dose-response curves were generated with GraphPad Prism, and IC50 values were calculated by four-parameter curve fitting.
All animal experiments were approved by and performed in accordance with the Institutional Animal Care and Use Committee protocols at the following research facilities: Charles River Discovery Services (North Carolina, USA), Translational Drug Development, LLC (Arizona, USA), Champions Oncology (Maryland, USA), Crown Bioscience, Inc. (Taicang, P.R. China), Labcorp Early Development Laboratories Inc. (Indiana, USA), Charles River Laboratories, Inc. (Ohio, USA). All facilities are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The HBCx-24 patient-derived xenograft (PDX) experiment was performed at Xentech (Ervy, Fance) in accordance with French Legislation concerning the protection of laboratory animals and a currently valid license for experiments on vertebrate animals, issued by the French Ministry of Higher Education, Research and Innovation. Efficacy studies were performed at Translational Drug Development, LLC [OVCAR-3 cell line–derived xenograft (CDX)], Charles River Discovery Services (JIMT-1 CDX and MX-1 CDX), Crown Bioscience, Inc. (OV2423 PDX and mBR9013 syngeneic model), Champions Oncology, Inc. (CTG-1692 PDX and Breast PDX Trial), Rat Biodistribution study was performed at Charles River Laboratories, Inc. (RRID:SCR_003792), and NHP PK study was performed at Labcorp Early Development Laboratories Inc.
For efficacy studies, mice were randomized when tumors reached a mean of 100 to 150 mm3 for each model, except for the CTG-1692 PDX which was randomized at a mean of 150 to 300 mm3, and treated according to the doses, schedules, and routes shown in the figures. Tumors were measured by caliper twice weekly and tumor volumes were calculated using the formula: (a × b2 / 2) where ‘b’ is the smallest diameter and ‘a’ is the largest diameter. For OVCAR-3 (RRID:CVCL_0465), female athymic nude mice (HSD: Athymic Nude-Foxn1mu, Envigo, RRID:RGD_5508395) were subcutaneously inoculated in the right flank with 5×106 OVCAR-3 cells / mouse. For MX-1 (RRID:CVCL_4774), female athymic nude mice (Crl:NU(Ncr)-Foxn1mu, Charles River) were subcutaneously implanted in the right flank with 1 mm3 MX-1 tumor fragments. For JIMT-1 (RRID:CVCL_2077), female CB.17 SCID mice (Fox Chase SCID, C.B-17/Icr-Prkdcscid/IcrIcoCrl, Charles River Laboratories) were subcutaneously inoculated in the right flank with 1×107 JIMT-1 cells / mouse. For HBCx-24 and CTG-1692 PDX, female athymic nude mice (HSD: Athymic Nude-Foxn1mu, Envigo, RRID:RGD_5508395) were subcutaneously implanted in the right flank with tumor fragments. For OV2423, female BALB/c nude mice (BALB/cNj-Foxn1nu/Gpt, GemPharmatech) were subcutaneously implanted in the right flank with 2 to 3 mm³ tumor fragments. For mBR9013, female FVB/NJ mice were implanted in the right flank with 2 to 3 mm³ tumor fragments.
Breast PDX Panel: A panel of 30 breast cancer PDX models, (Champions Oncology) annotated by prior treatment history, and divided between TNBC and ER-positive subtypes, was implanted into athymic Nude-Foxn1nu mice (HSD: Athymic Nude-Foxn1mu, Envigo, RRID:RGD_5508395). When tumors reached an average volume of 150 to 300 mm3, animals (n = 3) were treated with a single intravenous administration of XMT-1660 4.71 mg/kg (antibody dose) / 0.15 mg/kg (payload dose) or saline vehicle. Tumor volumes were measured until the vehicle control group reached a mean tumor volume of 1,500 mm3 or day 28 post-dose. The outcome was reported as median best response (MBR): the volume of each individual tumor was compared with its volume on Day 0, with a best response value on a scale starting at −1 indicating 100% regression (with for example, −0.3 indicating 30% regression, 0 indicating no change, and + 0.5 indicating 50% increase); MBR was defined as the MBR of the 3 tumors evaluated for each model. At the endpoint, xenografts, or tumor beds (in the case of no palpable mass), were collected and formalin fixed, paraffin embedded (FFPE).
RNA was extracted from FFPE samples using the Qiagen RNeasy FFPE kit according to the manufacturer's instructions. Samples were equalized on the basis of nanodrop reading and cDNA produced using the Thermofisher SuperScript IV VILO Master Mix with exDNase Enzyme. Gene expression assays were set up with the TaqMan Fast Advanced Master Mix. ABI assay Hs01552471_g1 was used for VTCN1. Hs99999903_m1 ACTB and Hs03929097_g1 GAPDH were used as endogenous controls. Expression data was analyzed as ΔΔ Ct of the average of animals in each vehicle treated group (generally n = 3) referencing a universal RNA control.
In the PDX panel, IHC to detect B7-H4 expression was performed on a single vehicle treated animal from each model. Briefly, tissues were sectioned at 4 μ onto positively charged slides and dried overnight. Using the Leica BOND III platform, sections were baked, dewaxed and subjected to antigen retrieval (LEICA BOND III ER1+ Proteinase K). The primary B7-H4 antibody (Abcam ab209242) was used at a concentration of 0.2 μg/mL (prepared in DAKO/Agilent diluent S3022). Signal was detected using the Leica BOND Polymer Refine system/DAB chromogen. Slides were evaluated by light microscopy, and tumor proportion score (TPS = percent positive at any intensity) of membrane reactivity were calculated. TPS ranged from 0 to 100 and was calculated on the basis of membrane immunoreactivity; cytoplasmic reactivity, if noted, was not included in the score.
For B7-H4 IHC on other samples (not in the PDX panel): Deparaffinization of FFPE samples was completed with multiple changes of xylenes and alcohols at decreasing concentrations. Manual antigen unmasking was done using heat induced epitope retrieval with an electronic pressure cooker. Slides were immersed in citrate buffer at pH 6.0 (Vector Laboratories, catalog no. H-3301–250), heated to 99°C and incubated for 20 minutes. Following a 20-minute cooldown period, samples were blocked with Dual Endogenous Enzyme Block (Agilent Technologies, catalog no. S200389–2) to quench potential endogenous peroxidase activity. Slides were then sequentially incubated with rabbit monoclonal anti–B7-H4 (clone EPR20236, Abcam, catalog no. ab209242; final dilution 1:2,000) primary antibody for 30 minutes, followed by an HRP-labeled anti-rabbit secondary antibody (Agilent Technologies, catalog no. K400311–2) for an additional 30 minutes. Chromogenic detection was initiated using liquid DAB+ (Agilent Technologies catalog no. K346811–2) for 5 minutes followed by hematoxylin counterstaining (Abcam, catalog no. ab220365) for 10 seconds. Completed slides were dehydrated in increasing concentrations of alcohols, cleared in xylenes and mounted in non-aqueous mounting media (Leica Microsystems, catalog no. 3801730). All staining steps were performed manually using a standard humidity chamber and several rinses with 1× TBST (Boston Bioproducts, catalog no. MSPP-IBB181X) followed each incubation step to remove excess reagents. Images were captured using the Olympus cellSens Entry 1.17 microscope camera (Olympus Corporation, Japan).
A study was conducted in male Sprague Dawley rats to investigate the biodistribution of XMT-1660 vs. a Control DS DAR 6 ADC. All animals were administered with a single intravenous dose of 9.0/∼0.3 mg/kg (mAb / AF-HPA) and n = 3/group were euthanized at each timepoint ranging from 24 hours to 672 hours post dose. Kidney, Liver and Lung tissues were sampled for bioanalysis of released drug.
A study in naïve cynomolgus monkeys was conducted to evaluate B7-H4 DS DAR 2 and XMT-1660 at a matched payload dose of 0.09 mg/kg administered via 45-minute intravenous infusion (1 male/1 female per group). Pharmacokinetic data for total mAb, Conjugated drug and free payload (AF-HPA) are shown.
Data availability
The data generated in this study are available in the manuscript and the Supplementary Data.
Results
Development of the DS platform
With the goal of retaining the key features of DF while achieving the ability for precise DAR ranging and site-specific conjugation, we designed a fully synthetic scaffold comprised of peptide-based units, hydrophilicity-endowing units, and a branching unit that allows for modulation of hydrophilicity and AF-HPA payload multiplicity per antibody-linked unit. This effort yielded the DS platform, with the key features of DF translated to DS as follows (Fig. 1A): (i) the hydrophilic polymer backbone (blue) was mimicked by short peptide sequences, PEG2 or PEG8 moieties; (ii) the β-alanine moiety (purple) was replaced by a glutamic acid that allows for precise charge balance of the AF-HPA payload; (iii) the alanine ester linker (green) is maintained; (iv) the number of AF-HPA payloads per scaffold is defined at 3 by a tris-hydroxymethyl-methylamine core, in this case generating a trimer, and (v) the platform is amenable to various bioconjugation chemistries including maleimide and click by variation of the terminal bioconjugation-enabling moiety. DS can be based on a trimer (constructs A and B, Supplementary Table S1) or a monomer (construct C, Supplementary Table S1). The constructs used in this report were synthesized and characterized as described in the Supplementary Material.
To compare the physicochemical and pharmacologic properties of DF and DS ADCs, we first generated stochastic (not site-specific) DF and DS ADCs by the partial reduction of native interchain disulfides of a tool anti-NaPi2b IgG1 antibody (27). The resulting ADCs had DAR 10.5 (DF) and DAR 12.1 (DS) which were deemed reasonably comparable for this purpose. The ADCs exhibited a striking difference in physicochemical properties, as showcased by HIC (Fig. 1B): the DS ADC (green) displayed discrete peaks, an indication of enhanced homogeneity, while the DF ADC (orange) showed a narrow range of lipophilicity and no distinguishable individual species (25), consistent with the heterogeneous nature of the DF polymeric platform. As expected, both ADCs display a shift in lipophilicity compared with unconjugated mAb (blue), which is attributable to AF-HPA (Fig. 1B). Antigen binding was comparable for the ADCs as well as the unconjugated antibody (Supplementary Fig. S1A). Similarly, cytotoxicity against cancer cells with endogenous expression of the antigen was comparable for the DF and DS ADCs (Supplementary Fig. S1B). The ADCs also exhibited comparable activity in vivo when compared at equal payload dose in a tumor xenograft study (Fig. 1C). Taken together, these results demonstrate that DS retains the pharmacologic profile of DF.
We next designed a set of experiments to evaluate the impact of conjugation technology on the in vivo performance of DS ADCs. A set of ADCs was synthesized by conjugation of DS trimer constructs A and B to trastuzumab (Fig. 1D). ADC1 was generated with stochastic (not site-specific) conjugation and thus was analogous to the NaPi2b DS ADC described above and served to bridge to the earlier experiments; ADC2 was generated with maleimide conjugation at partially reduced native cysteines and was fractionated by chromatography to be enriched in DAR 6 species (28); ADC3 was based on an engineered trastuzumab variant with an LC V205C mutation (20) that afforded a homogeneous ADC; and ADC4 was synthesized by remodeling the N-linked glycan and introducing a sugar containing an azido group that is amenable to strain-promoted azide-alkyne cycloaddition (“click chemistry”) previously described as GlycoConnect (29). HIC analysis indicated that ADC2, ADC3, and ADC4 were largely homogeneous, while ADC1 displayed heterogeneity as expected (Fig. 1E). ADC4 had the shortest HIC retention time, which in some contexts has been associated with enhanced pharmacokinetics (30). All 4 ADCs demonstrated comparable binding to HER2 peptide and comparable cytotoxicity against a breast cancer cell line with endogenous moderate HER2 expression (Supplementary Fig. S2; Supplementary Table S2).
In contrast to the in vitro results, the comparison of the 4 ADCs in vivo revealed significant differentiation. First, in a HER-2 expressing JIMT-1 mouse tumor xenograft study with a single administration at equal payload doses, the site-specific ADC3 and ADC4 outperformed the stochastic ADC1, and to a lesser extent the fractionated stochastic ADC2 (Supplementary Fig. S3). Second, in pharmacokinetics analysis at equal payload doses in tumor-bearing mice, the site-specific ADC3 and ADC4 showed significantly increased exposures over ADC1 and ADC2 (Fig. 1F; Supplementary Table S2). Thus, site-specific DS ADCs exhibited favorable pharmacokinetics and pharmacology over stochastic DS ADCs, and the cysteine engineering and glycan remodeling approaches yielded ADCs with comparable profiles. Taken together, these data demonstrate that the novel DS platform retains the pharmacology profile of DF and is amenable to site-specific conjugation, thus achieving the design criteria described above.
Antibody does not block B7-H4 function
The DS platform was then incorporated into the discovery and optimization of a B7-H4–targeted ADC for clinical development. In lieu of engineering cysteines, the GlycoConnect technology was chosen for the clinical candidates. The key desired attribute of the antibody was specific, potent binding to B7-H4 in human and one or more nonclinical species (to facilitate nonclinical assessments). The selected antibody (XMT-1604) is a human IgG1 kappa monoclonal antibody that specifically binds to human B7-H4 with sub nanomolar affinity and exhibits comparable binding profiles to recombinant B7-H4 from human, cynomolgus monkey, mouse, and rat (Supplementary Table S3).
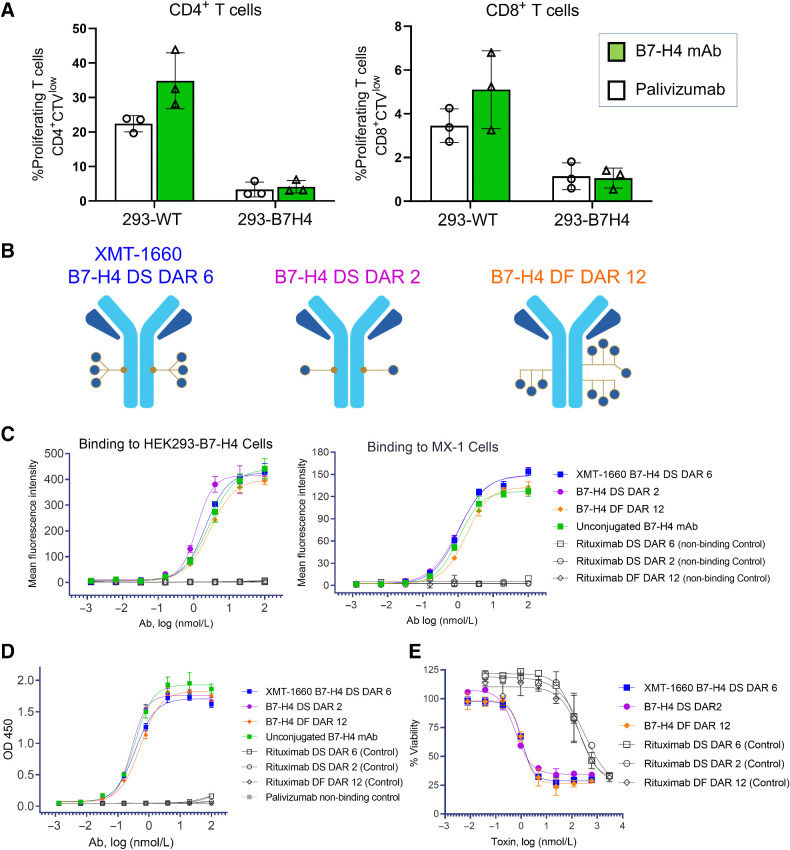
Certain anti–B7-H4 antibodies have been reported to inhibit the T-cell suppressive activity of B7-H4 (13, 31). We reasoned that an antibody without function-blocking activity would avoid any unnecessary safety liability and would not compromise ADC activity, which is typically driven by antigen binding and payload delivery. To confirm that our selected antibody did not exhibit this type of functional activity, wildtype HEK293 (HEK293-WT) cells or HEK293-B7-H4 cells (engineered to exogenously express B7-H4) were incubated with the B7-H4–targeted antibody XMT-1604 or a nonbinding control antibody prior to the addition of Cell Trace Violet (CTV)-labeled CD3+ T cells. T cell proliferation was assessed using flow cytometry measuring the dilution of CTV-labeled T cells. Consistent with the reported immune inhibitory function of B7-H4, the HEK293-B7-H4 cells reduced the proliferation of both CD4+ T cells (3.4% proliferating compared with 22.4% with HEK293-WT) and CD8+ T cells (1.1% compared with 3.5% with HEK293-WT) (Fig. 2A). The B7-H4 antibody did not block the effect of HEK293-B7-H4 cells on CD4+ T cells (4.1% compared with 3.4% with control antibody) or CD8+ T cells (1.1% compared with 1.1% with control antibody) (Fig. 2A). These data indicate that the B7-H4 antibody does not exhibit function-blocking activity.
Figure 2.
In vitro characterization of B7-H4 ADCs. A, The anti–B7-H4 antibody does not block the functional activity of B7-H4 ligand. HEK293 or HEK293-B7-H4 cells were incubated with 10 nmol/L antibody (B7-H4 or nonbinding control) prior to the addition of CellTrace Violet-labeled CD3+ T cells and CD3/CD28 T-cell Activator. T cell proliferation was determined after 4-day incubation. Data represent the mean (± standard deviation) of triplicate samples. B, Schematics of the B7-H4–targeted ADCs evaluated in this study: site-specific DS DAR 2 and DAR 6, and DF DAR 12. C, Binding of ADCs to HEK293-B7-H4 cells and MX-1 breast cancer cells. Flow cytometry analysis was performed with B7-H4 ADCs, unconjugated antibody, and nonbinding control ADCs. D, Binding of ADCs to recombinant human B7-H4 protein. ELISA was performed with B7-H4 ADCs, unconjugated antibody, and non-binding control ADCs. E, B7-H4 ADCs elicit target-dependent cytotoxicity. HEK293-B7-H4 cells were incubated with test article for 3 days, and viability was measured with CellTiter-Glo.
In vitro characterization of B7-H4 targeting ADCs
The DS platform enables the comparison of ADCs with different DARs, allowing for the evaluation of ADCs with differing DARs and the selection of the DAR with the most desirable profile. To this end, we generated B7-H4 ADCs with DAR 2 and DAR 6 (site-specific DS) and DAR 12 (DF; Fig. 2B; Supplementary Fig. S4). All three ADCs exhibited potent, specific binding to HEK293-B7-H4 exogenously expressing cells and MX-1 triple negative breast cancer endogenously expressing cells, as assessed by flow cytometry (Fig. 2C). The binding profiles of the ADCs were comparable with one another and to the unconjugated mAb, which indicated that the bioconjugation process did not impact binding (Fig. 2C). The EC50 values for binding were 1.2–2.8 nmol/L for HEK293-B7-H4 and 1.1–1.7 nmol/L for MX-1. Consistent with the cell binding results, all three ADCs and unconjugated mAb exhibited comparable, potent, and specific binding profiles to recombinant B7-H4 protein by ELISA (Fig. 2D). The control ADCs did not exhibit appreciable binding in these assays. Together these results demonstrate that the binding profiles of all three ADCs were comparable with one another and were not impacted by bioconjugation.
The cytotoxic activity of the ADCs was evaluated on HEK293-B7-H4 cells. All three ADCs elicited potent, target-dependent cytotoxicity, with IC50 values of 0.6, 1, and 1.1 nmol/L payload for DS DAR 2, DS DAR 6, and DF DAR 12 ADCs, respectively (Fig. 2E). The potency of the B7-H4 ADCs was >200-fold higher than that of the nonbinding control ADCs (Fig. 2E). These results indicate that all three B7-H4 ADCs exhibited comparable, potent, and specific cytotoxicity. Taken together, the in vitro data of all three B7-H4 ADCs did not reveal any significant differences among their activity profiles. The DAR 6 ADC (XMT-1660) was later tested on CAMA-1 breast cancer cells which have endogenous expression of B7-H4, and demonstrated specific binding and cytotoxicity, with IC50 = 0.052 nmol/L payload (Supplementary Fig. S5).
In vivo comparison of B7-H4 ADCs
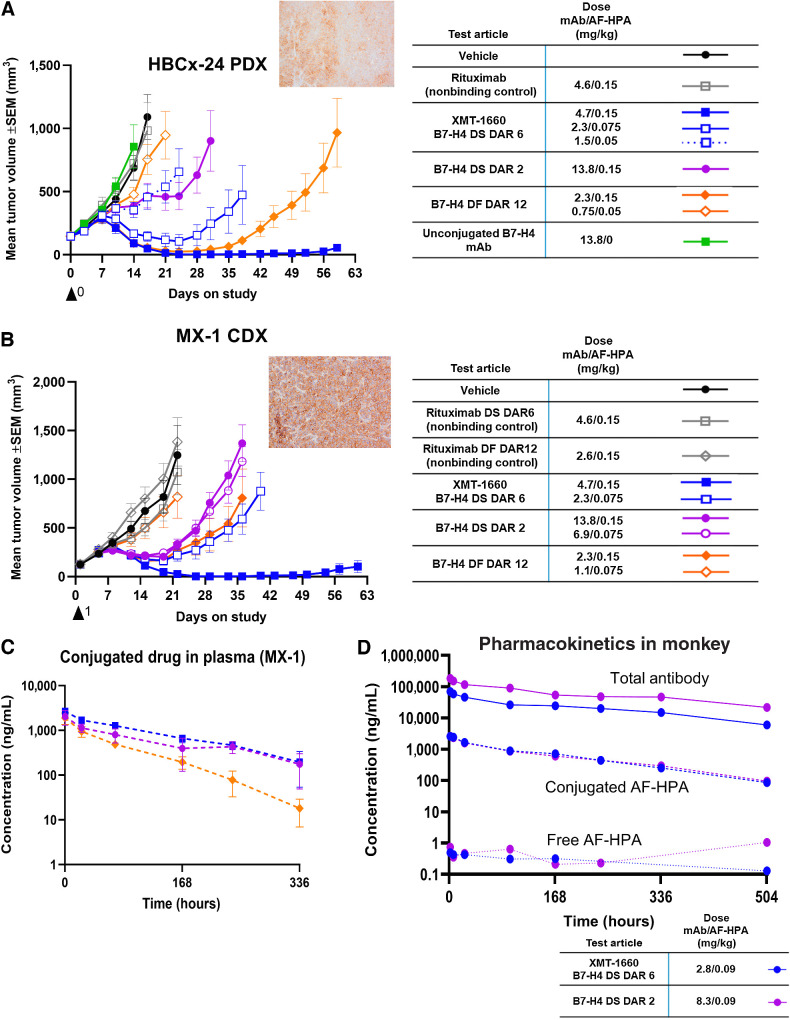
We next compared the B7-H4 ADCs head-to-head in two breast cancer models with endogenous B7-H4 expression: the MX-1 CDX model and the HBCx-24 PDX model. To allow for a more robust interpretation of the results, these studies included at least 2 dose levels of each ADC as well as nonbinding control ADCs. In both models, the DS DAR 6 ADC (XMT-1660) induced complete tumor regression after a single administration and significantly outperformed the DAR 2 and DF DAR 12 ADCs (Fig. 3A and B; Supplementary Fig. S6). Analyzing the data across dose levels indicated that XMT-1660 was >2-fold more potent than the DAR 2 ADC and 1.5–2-fold more potent than the DAR 12 ADC on a payload basis (tumor responses are shown in Supplementary Table S4). Comparing the activity at equal payload dose is likely the clinically relevant approach because payload dose (not antibody dose) typically defines the clinical dose for ADCs. Notably, the DAR 2 ADC underperformed the DAR 6 ADC despite being administered at a 3-fold higher antibody dose, which could enhance tumor distribution. This finding could be explained by not reaching the threshold of payload concentration in the cancer cell to achieve efficient cell killing due to saturation of the B7-H4 antigens on the cancer cell surface. Neither the nonbinding control ADCs nor the unconjugated antibody exhibited antitumor activity, which confirmed the target-dependent, payload-dependent basis of the observed effect. The plasma pharmacokinetics analysis of conjugated drug in MX-1 tumor-bearing animals showed the enhanced plasma exposure and the slower clearance of the site-specific DS ADCs relative to the DF ADC (Fig. 3C; Supplementary Fig. S7), consistent with the data obtained during platform development (Fig. 1F).
Figure 3.
In vivo profile comparison of B7-H4 ADCs. A, Antitumor activity of B7-H4 ADCs in HBCx-24 PDX model. Inset, B7-H4 IHC. B, Antitumor activity of B7-H4 ADCs in MX-1 breast cancer cell line model. Inset, B7-H4 IHC. C, Plasma levels of conjugated drug in MX-1 tumor-bearing animals following a single administration of ADC at 0.15 mg/kg payload. Refer to legend in part B. D, Plasma levels of three analytes in cynomolgus monkeys following a single administration of ADC at 0.09 mg/kg payload. Blue, XMT-1660 DAR 6 DS; purple, DAR 2 DS.
The DAR 6 ADC (XMT-1660) and the DAR 2 ADC were evaluated at equal payload dose in a repeat-dose study in cynomolgus monkeys. Both ADCs exhibited comparable pharmacokinetics profiles, with comparable clearance rates and high stability in circulation, as indicated by the extremely low quantities of free payload (Fig. 3D). Moreover, the ADCs exhibited comparable safety profiles which were consistent with AF-HPA–based ADCs (27). On the basis of the totality of the nonclinical data, XMT-1660 was determined to have a superior profile and thus considered to be the optimized B7-H4 ADC for advancement into clinical development.
Antitumor activity in ovarian tumors and PD-1 refractory breast tumor
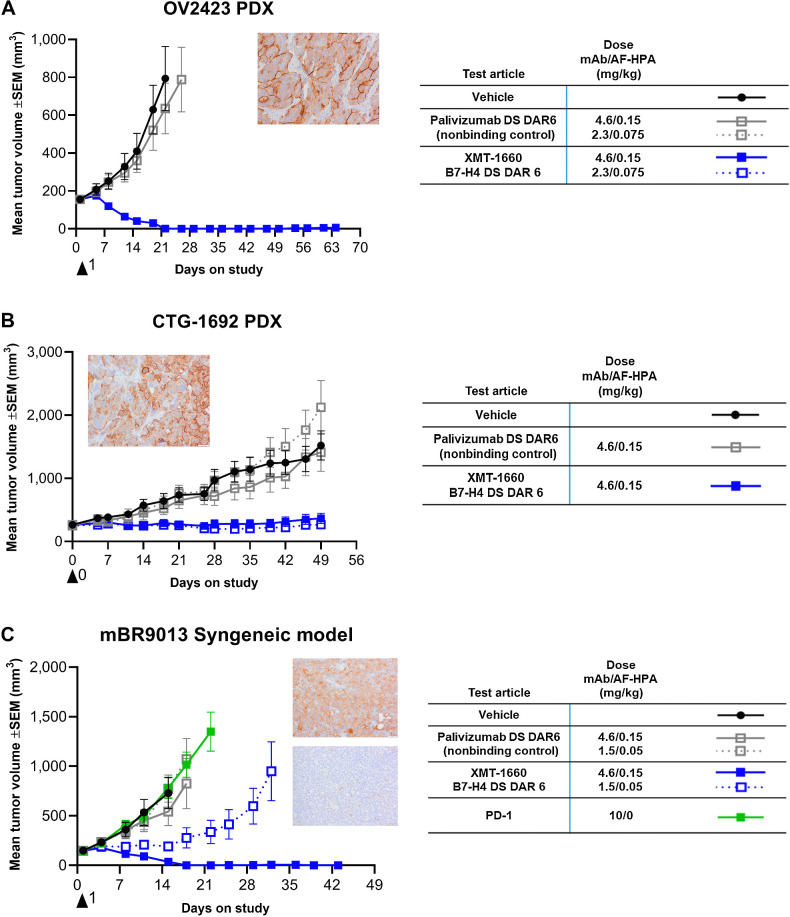
Additional pharmacology studies were conducted with XMT-1660 to inform clinical development strategy. Because the expression of B7-H4 is prominent in ovarian cancer, the antitumor activity of XMT-1660 was evaluated in ovarian cancer PDX models. XMT-1660 exhibited robust antitumor activity in OV2423 and CTG-1692 PDX models (Supplementary Table S4), which express B7-H4 (Fig. 4A and B; Supplementary Fig. S6). The control ADC was not active in these models, confirming the target-dependent basis of the activity.
Figure 4.
XMT-1660 activity in ovarian tumors and PD-1 refractory breast tumor. A, Antitumor activity of XMT-1660 in OV2423 ovarian cancer PDX. Inset, B7-H4 IHC. B, Antitumor activity of XMT-1660 in CTG-1692 ovarian cancer PDX. Inset, B7-H4 IHC. C, Antitumor activity of XMT-1660 in mBR9013 MMTV-ERBB2-derived syngeneic tumor in FVB/NJ. Anti–PD-1 immune checkpoint inhibitor was also evaluated (dosed BIWx3 (IP)). Inset, B7-H4 (top) and PD-L1 (bottom) IHC.
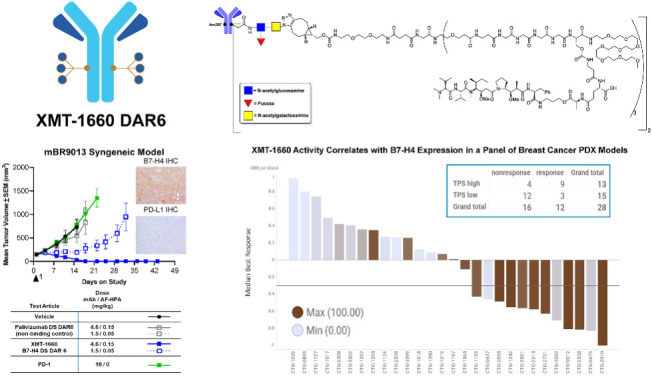
On the basis of the nonoverlapping expression patterns of B7-H4 and PD-L1 in primary tumors, we sought a tumor model in immune-competent animals to enable the evaluation of PD-(L)1 immune checkpoint inhibitor as well as that of XMT-1660. The mBR9013 breast cancer model, which was derived from a mouse mammary tumor virus (MMTV)-ERBB2 transgenic mouse that expresses rat HER2 under the direction of the MMTV promoter, was determined to be suitable because it exhibits endogenous expression of murine B7-H4 and is propagated in immune competent FVB/NJ mice (Fig. 4C; Supplementary Fig. S6); PD-L1 expression was observed on a small percentage of tumor cells (Fig. 4C). and also, on antigen presenting cells. Importantly, XMT-1660 is fully cross-reactive with murine B7-H4 (Supplementary Table S3). Strikingly, XMT-1660 induced complete tumor regression in mBR9013 after a single administration, whereas the anti–PD-1 antibody had no effect (Fig. 4C; Supplementary Table S4). These results suggest that XMT-1660 could be effective in patients who are refractory or resistant to immune checkpoint inhibitors.
In vivo activity in a panel of breast cancer PDX
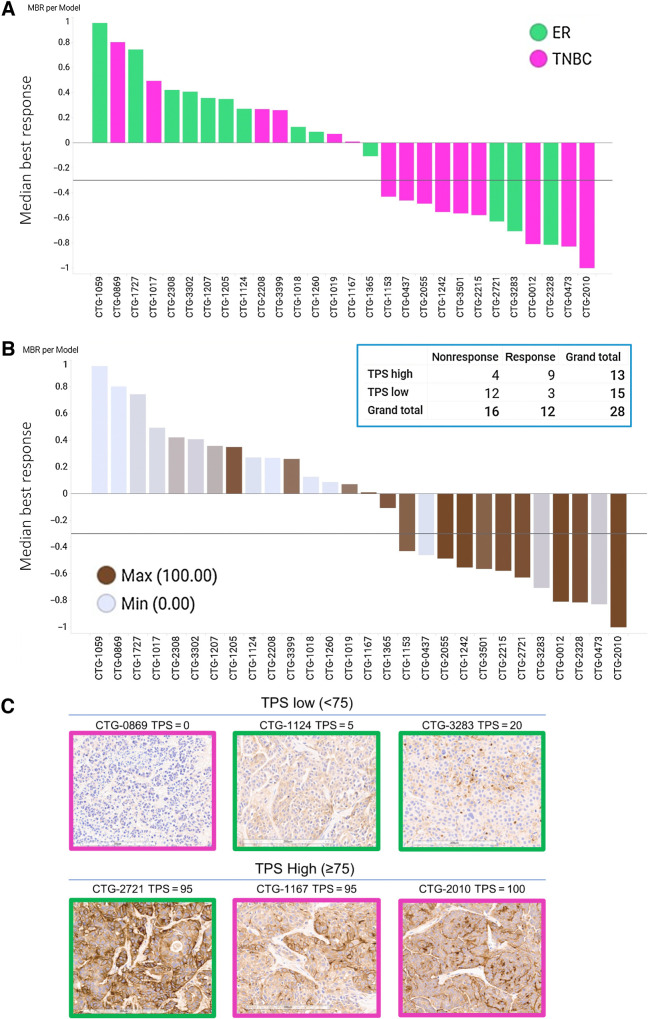
To gain an understanding of the determinants of antitumor activity of XMT-1660, an unselected panel of 30 breast cancer PDX models was simultaneously evaluated for expression of B7-H4 and in vivo activity of XMT-1660. Two PDX models were excluded from the analysis due to uncharacteristic growth patterns or tumor phenotypes that disallowed confident interpretation of the data. The outcome was expressed for each tumor model in terms of MBR: the median change in tumor volume among the 3 animals for that tumor model. In the panel, 12/28 (43%) breast cancer PDX models achieved MBR between −1 and −0.3 (representing at least 30% tumor shrinkage) following a single administration of 4.7/0.15 mg/kg (antibody/payload) XMT-1660; tumor volume reductions were seen in 13/28 (46%) models (Fig. 5A; Supplementary Fig. S8). Of the 12 PDX in which MBR ≤ −0.3 tumor reduction was achieved, 10 were models derived from patients who had received prior therapies. Achieving MBR ≤ −0.3 was more frequent in TNBC models 9/15 (60%) compared with ER-positive models 3/13 (23%), (green: ER+; pink: TNBC).
Figure 5.
XMT-1660 activity across a panel of breast cancer PDX. A, Waterfall plot of MBR among n = 3 per PDX model. Pink bars, TNBC. Green bars, ER+ breast cancer. B, The data from part A shown with B7-H4 IHC score in each model. Inset, responsive (MBR ≤ −0.3) and nonresponsive tumors when categorized by B7-H4 IHC as low TPS (< 75) or high TPS (≥75). C, Representative B7-H4 IHC images from pink (TNBC) and green (ER+) PDX models with low TPS (<75) and high TPS (≥75).
Higher B7-H4/VTCN1 RNA values (shown as ΔΔCt of the average of animals) were associated with greater response in this sample set (Supplementary Fig. S9). Similarly, higher B7-H4 IHC scores were associated with response (Fig. 5B; IHC images shown in Fig. 5C). IHC analysis yielded a TPS for each sample, which indicates the % of tumor cells expressing membranous B7-H4 regardless of expression level. Notably, 9/13 TPS high tumors responded to XMT-1660 achieving MBR ≤ −0.3 (Fig. 5B). Employing a cut-off of TPS 75 (TPS high ≥ 75, TPS low < 75) identified 9/12 (75%) responding models while 12/15 (80%) models with TPS low did not achieve MBR ≤ −0.3 (Fig. 5B). Overall, these results indicate that the antitumor activity of XMT-1660 is associated with the expression level of B7-H4, as expected, and suggest that, in the clinic, it may be possible to prospectively enrich for responders based on tumor expression.
To assess the biodistribution of the ADC, we dosed XMT-1660 and a nonbinding control DAR6 ADC in Male Sprague Dawley Rats at 9 mg/kg antibody dose and measured released payload in liver, kidney, and lung (Supplementary Fig. S10). These data demonstrate the comparable distribution of released drug between XMT-1660 and nonbinding control ADC, which suggests, based on this limited study, that the B7-H4-directed antibody component of XMT-1660 did not impact biodistribution. Moreover, no accumulation of the released drug was observed.
Discussion
In this study we defined and characterized XMT-1660, a DAR-optimized B7-H4 ADC for the treatment of cancer. We developed a novel ADC platform, DS, to enable DAR optimization and site-specific conjugation while retaining the salient features of DF. We then applied both platforms to evaluate B7-H4 ADCs with DAR 2, 6, and 12. To our knowledge, no prior publication has described the evaluation of ADCs with a 6-fold range of DAR to optimize an ADC clinical candidate.
The features of DF that enable high DAR ADCs with favorable physicochemical and pharmacokinetic profiles—a hydrophilic scaffold and charge compensation—are advantageous in the construction of ADCs with low DAR as well as high DAR. In creating DS, we optimized a synthetic platform that retains these features while providing the capability to DAR-range. In this study we present the characterization of DAR 2, 6, and 12 ADCs. DS is amenable to various methods of site-specific conjugation, and our results in this study and elsewhere support the hypothesis that site-specific conjugation (32) can improve the therapeutic window (20) of ADCs, likely by improving the pharmacokinetics profile. The results in this study, though limited to two site-specific conjugation methods, suggest that the advantages of site-specific conjugation with DS do not require a particular conjugation site or technology.
The optimal DAR for a given target may depend on factors such as the target expression level on tumor cells, the extent of heterogeneity of target expression across the tumor, the overall sensitivity of the tumor cells (e.g., of a particular tumor type) to the payload, and the tumor architecture parameters such as vascular density and interstitial pressure (33). Consequently, the optimal DAR may vary among targets. It is noted that a confounding factor in comparing the pharmacology of ADCs with different DARs is the inability to simultaneously compare them at equal antibody dose and payload dose: ADCs with different DARs that are dosed at the same payload dose necessarily have different antibody doses, and vice versa. In general, as more targets are evaluated on these platforms in the preclinical and clinical settings, the datasets could enable a more predictive, less empirical approach to identifying the optimal ADC design.
In the case of B7-H4, the site-specific DS DAR 6 ADC (XMT-1660) exhibited superior in vivo antitumor activity and noninferior in vitro, pharmacokinetics, and toxicology profiles relative to the DS DAR 2 and DF DAR 12 ADCs. XMT-1660 exhibited superior antitumor activity over the other ADCs in both tumor models tested (a CDX and a PDX). We based our comparisons on equal payload dose because in clinical studies with ADCs, the payload dose (not antibody dose) typically determines the clinical dose level. Thus XMT-1660 was selected for advancement into clinical development.
Our in vivo studies demonstrate the dose-dependent, target-dependent antitumor activity of XMT-1660 in several models of breast cancer and ovarian cancer; tumor regressions were achieved after a single administration in all tumor models (Figs. 3 and 4). XMT-1660 also has potential for the treatment of other tumor types that express B7-H4, such as endometrial cancer. An important clinically relevant finding is the antitumor activity observed in PDX models established from previously treated tumors (Fig. 5). As expected, based on the targeted delivery mediated by an ADC, the activity of XMT-1660 correlated with target expression (Fig. 5B; Supplementary Fig. S9), which suggests that in the clinic, it may be possible to prospectively enrich for responders based on the expression of B7-H4. Interestingly, in the PDX panel, the TNBC models exhibited higher B7-H4 expression and greater antitumor activity than the ER models; future work will be needed to determine whether or not this finding is also observed in primary human samples.
The minimal overlap of B7-H4 and PD-L1 expression in tumors has important implications for the clinical development of XMT-1660. It is possible that XMT-1660 could be effective against tumors that do not respond to PD-(L)1 immune checkpoint inhibitors, for example due to their lack of PD-L1+ expression. Indeed, XMT-1660 induced tumor regression in a syngeneic breast cancer model that was refractory to anti–PD-1 (Fig. 4C). Moreover, the combination of XMT-1660 and an immune checkpoint inhibitor may be beneficial to preclude a tumor's evasion of therapy by switching from expression of one antigen to the other. Particularly in this setting, the use of an antibody that does not block the immune-suppressive function of B7-H4 (Fig. 2A) may reduce the likelihood of exacerbating immune-related toxicity induced by the checkpoint inhibitor.
In summary, we have developed novel ADC platforms, demonstrated the importance of DAR optimization during ADC discovery, and defined and characterized XMT-1660, a B7-H4–targeted DS DAR 6 ADC that is currently in a phase I clinical study (NCT05377996).
Supplementary Material
Supplementary Materials
Acknowledgments
The authors gratefully acknowledge contributions from Kelly Lancaster, Annika Yau, Dmitriy Gumerov, Mark Nazzaro, and Barrett Nehilla.
This article is featured in Selected Articles from This Issue, p. 997
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
D. Toader reports a patent for US20220233707 pending, a patent for US20210220477 pending, and a patent for US20230021500 pending. S.P. Fessler reports personal fees and nonfinancial support from Mersana Therapeutics; personal fees from Forma Therapeutics; and personal fees and nonfinancial support from Matchpoint Therapeutics outside the submitted work. C.N. Chin reports a patent for B7-H4-Targeted Antibody–Drug Conjugates and Methods of Use Thereof pending. J.R. Duvall reports personal fees from Mersana Therapeutics; and personal fees from The Broad Institute outside the submitted work. M.V. Kozytska reports a patent for WO2018098269A2 pending and a patent for US20230043447A1 pending. R. Mosher reports other support from Mersana Therapeutics outside the submitted work; and I was formerly an employee of Mersana Therapeutics; I am currently an employee of Kymera Therapeutics. M. Damelin reports a patent for US20220233707 pending and a patent for US20210220477 pending. T.B. Lowinger reports other support from Mersana Therapeutic outside the submitted work; in addition, T.B. Lowinger has a patent for US8808679B2 issued to Mersana, a patent for US11135307B2 issued to Mersana, a patent for US20230043447A1 pending to Mersana, a patent for US20230021500A1 pending to Mersana, a patent for US20210220477A1 pending to Mersana, and a patent for US20220233707A1 pending to Mersana. No disclosures were reported by the other authors.
Authors' Contributions
D. Toader: Conceptualization, resources, data curation, supervision, writing–original draft, writing–review and editing. S.P. Fessler: Conceptualization, supervision, methodology, project administration. S.D. Collins: Conceptualization, resources, formal analysis, supervision, visualization, writing–review and editing. P.R. Conlon: Investigation, methodology. R. Bollu: Supervision, investigation, methodology. K.C. Catcott: Investigation, methodology. C.N. Chin: Conceptualization. A. Dirksen: Conceptualization, supervision. B. Du: Investigation, methodology. J.R. Duvall: Supervision, methodology. S. Higgins: Methodology, writing–review and editing. M.V. Kozytska: Investigation, methodology. K. Bellovoda: Investigation, methodology. C. Faircloth: Investigation, methodology. D. Lee: Conceptualization, supervision. F. Li: Supervision, validation, writing–original draft, project administration. L. Qin: Investigation, methodology. C. Routhier: Investigation, methodology. P. Shaw: Investigation, methodology. C.A. Stevenson: Investigation, methodology. J. Wang: Investigation, methodology. P. Wongthida: Investigation, methodology. E. Ter-Ovanesyan: Investigation, methodology. E. Ditty: Investigation, methodology. S.P. Bradley: Investigation, methodology. L. Xu: Conceptualization, formal analysis, supervision. M. Yin: Conceptualization, supervision, methodology. A.V. Yurkovetskiy: Conceptualization, resources, supervision. R. Mosher: Conceptualization, data curation, formal analysis, supervision, visualization, writing–review and editing. M. Damelin: Conceptualization, resources, data curation, formal analysis, supervision, validation, writing–original draft. T.B. Lowinger: Conceptualization, resources, formal analysis, supervision, writing–review and editing.
References
- 1. Beck A, Goetsch L, Dumontet C, Corvaïa N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat Rev Drug Discovery 2017;16:315–37. [DOI] [PubMed] [Google Scholar]
- 2. Drago JZ, Modi S, Chandarlapaty S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat Rev Clin Oncol 2021;18:327–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, et al. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 2015;527:323–8. [DOI] [PubMed] [Google Scholar]
- 4. Han A, Olsen O, D'Souza C, Shan J, Zhao F, Yanolatos J, et al. Development of novel glucocorticoids for use in antibody–drug conjugates for the treatment of inflammatory diseases. J Med Chem 2021;64:11958–71. [DOI] [PubMed] [Google Scholar]
- 5. Dragovich PS. Antibody–drug conjugates for immunology. J Med Chem 2022;65:4496–9. [DOI] [PubMed] [Google Scholar]
- 6. Salceda S, Tang T, Kmet M, Munteanu A, Ghosh M, Macina R, et al. The immunomodulatory protein B7-H4 is overexpressed in breast and ovarian cancers and promotes epithelial cell transformation. Exp Cell Res 2005;306:128–41. [DOI] [PubMed] [Google Scholar]
- 7. Miyatake T, Tringler B, Liu W, Liu SH, Papkoff J, Enomoto T, et al. B7-H4 (DD-O110) is overexpressed in high risk uterine endometrioid adenocarcinomas and inversely correlated with tumor T-cell infiltration. Gynecol Oncol 2007;106:119–27. [DOI] [PubMed] [Google Scholar]
- 8. Leong SR, Liang W-C, Wu Y, Crocker L, Cheng E, Sampath D, et al. An Anti–B7-H4 antibody–drug conjugate for the treatment of breast cancer. Mol Pharm 2015;12:1717–29. [DOI] [PubMed] [Google Scholar]
- 9. Iizuka A, Nonomura C, Ashizawa T, Kondou R, Ohshima K, Sugino T, et al. A T-cell–engaging B7-H4/CD3-bispecific Fab-ScFv antibody targets human breast cancer. Clin Cancer Res 2019;25:2925–34. [DOI] [PubMed] [Google Scholar]
- 10. Smith JB, Lanitis E, Dangaj D, Buza E, Poussin M, Stashwick C, et al. Tumor regression and delayed onset toxicity following B7-H4 CAR T-cell therapy. Mol Ther 2016;24:1987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei J, Loke P, Zang X, Allison JP. Tissue-specific expression of B7x protects from CD4 T cell–mediated autoimmunity. J Exp Med 2011;208:1683–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azuma T, Zhu G, Xu H, Rietz AC, Drake CG, Matteson EL, et al. Potential role of decoy B7-H4 in the pathogenesis of rheumatoid arthritis: a mouse model informed by clinical data. PLoS Med 2009;6:E1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prasad DV, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T-cell activation. Immunity 2003;18:863–73. [DOI] [PubMed] [Google Scholar]
- 14. Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, et al. B7-H4, a molecule of the B7 family, negatively regulates T-cell immunity. Immunity 2003;18:849–61. [DOI] [PubMed] [Google Scholar]
- 15. Mugler KC, Singh M, Tringler B, Torkko KC, Liu W, Papkoff J, et al. B7-H4 expression in a range of breast pathology: correlation with tumor T-cell infiltration. Appl Immunohistochem Mol Morphol 2007;15:363–70. [DOI] [PubMed] [Google Scholar]
- 16. Abadi YM, Jeon H, Ohaegbulam KC, Scandiuzzi L, Ghosh K, Hofmeyer KA, et al. Host B7x promotes pulmonary metastasis of breast cancer. J Immunol 2013;190:3806–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L, Dong J, Li Z, Chen Y, Zhang Y. The B7H4-PDL1 classifier stratifies immuno-phenotype in cervical cancer. Cancer Cell Int 2021;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altan M, Kidwell KM, Pelekanou V, Carvajal-Hausdorf DE, Schalper KA, Toki MI, et al. Association of B7-H4, PD-L1, and tumor infiltrating lymphocytes with outcomes in breast cancer. NPJ Breast Cancer 2018;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, et al. Differential expression and significance of PD-L1, IDO-1, and B7-H4 in human lung cancer. Clin Cancer Res 2017;23:370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol 2008;26:925–32. [DOI] [PubMed] [Google Scholar]
- 21. Thurber GM, Wittrup D, Mechanistic KA. Compartmental model for total antibody uptake in tumors. J Theor Biol 2012;314:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thurber GM, Schmidt MM, Wittrup KD. Factors determining antibody distribution in tumors. Trends Pharmacol Sci 2008;29:57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu J, Burris H, Wang JS, Barroilhet L, Gutierrez M, Wang Y, et al. An open-label phase I dose-escalation study of the safety and pharmacokinetics of DMUC4064A in patients with platinum-resistant ovarian cancer. Gynecol Oncol 2021;163:473–80. [DOI] [PubMed] [Google Scholar]
- 24. Liu JF, Moore KN, Birrer MJ, Berlin S, Matulonis UA, Infante JR, et al. Phase I study of safety and pharmacokinetics of the Anti-MUC16 antibody–drug conjugate DMUC5754A in patients with platinum-resistant ovarian cancer or unresectable pancreatic cancer. Ann Oncol 2016;27:2124–30. [DOI] [PubMed] [Google Scholar]
- 25. Yurkovetskiy AV, Bodyak ND, Yin M, Thomas JD, Clardy SM, Conlon PR, et al. Dolaflexin: a novel antibody–drug conjugate platform featuring high drug loading and a controlled bystander effect. Mol Cancer Ther 2021;20:885–95. [DOI] [PubMed] [Google Scholar]
- 26. Hamblett KJ, Senter PD, Chace DF, Sun MMC, Lenox J, Cerveny CG, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody–drug conjugate. Clin Cancer Res 2004;10:7063–70. [DOI] [PubMed] [Google Scholar]
- 27. Bodyak ND, Mosher R, Yurkovetskiy AV, Yin M, Bu C, Conlon PR, et al. The Dolaflexin-based antibody–drug conjugate XMT-1536 targets the solid tumor lineage antigen SLC34A2/NaPi2b. Mol Cancer Ther 2021;20:896–905. [DOI] [PubMed] [Google Scholar]
- 28. Thompson P, Fleming R, Bezabeh B, Huang F, Mao S, Chen C, et al. Rational design, biophysical and biological characterization of site-specific antibody-- tubulysin conjugates with improved stability, efficacy and pharmacokinetics. J Control Release 2016;236:100–16. [DOI] [PubMed] [Google Scholar]
- 29. van Geel R, Wijdeven MA, Heesbeen R, Verkade JMM, Wasiel AA, van Berkel SS, et al. Chemoenzymatic conjugation of toxic payloads to the globally conserved n-glycan of native MAbs provides homogeneous and highly efficacious antibody–drug conjugates. Bioconjug Chem 2015;26:2233–42. [DOI] [PubMed] [Google Scholar]
- 30. Lyon RP, Bovee TD, Doronina SO, Burke PJ, Hunter JH, Neff-LaFord HD, et al. Reducing hydrophobicity of homogeneous antibody-- drug conjugates improves pharmacokinetics and therapeutic index. Nat Biotechnol 2015;33:733–5. [DOI] [PubMed] [Google Scholar]
- 31. Dangaj D, Lanitis E, Zhao A, Joshi S, Cheng Y, Sandaltzopoulos R, et al. Novel recombinant human B7-H4 antibodies overcome tumoral immune escape to potentiate T-cell antitumor responses. Cancer Res 2013;73:4820–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agarwal P, Bertozzi CR. Site-specific antibody–drug conjugates: the nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjug Chem 2015;26:176–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baxter LT, Jain RK. Transport of fluid and macromolecules in tumors. I. role of interstitial pressure and convection. Microvasc Res 1989;37:77–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Data Availability Statement
The data generated in this study are available in the manuscript and the Supplementary Data.