Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive, currently untreatable Schwann cell–derived neoplasms with hyperactive mitogen-activated protein kinase and mammalian target of rapamycin signaling pathways. To identify potential therapeutic targets, previous studies used genome-scale shRNA screens that implicated the neuregulin-1 receptor erb-B2 receptor tyrosine kinase 3 (erbB3) in MPNST proliferation and/or survival. The current study shows that erbB3 is commonly expressed in MPNSTs and MPNST cell lines and that erbB3 knockdown inhibits MPNST proliferation and survival. Kinomic and microarray analyses of Schwann and MPNST cells implicate Src- and erbB3-mediated calmodulin-regulated signaling as key pathways. Consistent with this, inhibition of upstream (canertinib, sapitinib, saracatinib, and calmodulin) and parallel (AZD1208) signaling pathways involving mitogen-activated protein kinase and mammalian target of rapamycin reduced MPNST proliferation and survival. ErbB inhibitors (canertinib and sapitinib) or erbB3 knockdown in combination with Src (saracatinib), calmodulin [trifluoperazine (TFP)], or proviral integration site of Moloney murine leukemia kinase (AZD1208) inhibition even more effectively reduces proliferation and survival. Drug inhibition enhances an unstudied calmodulin-dependent protein kinase IIα phosphorylation site in an Src-dependent manner. The Src family kinase inhibitor saracatinib reduces both basal and TFP-induced erbB3 and calmodulin-dependent protein kinase IIα phosphorylation. Src inhibition (saracatinib), like erbB3 knockdown, prevents these phosphorylation events; and when combined with TFP, it even more effectively reduces proliferation and survival compared with monotherapy. These findings implicate erbB3, calmodulin, proviral integration site of Moloney murine leukemia kinases, and Src family members as important therapeutic targets in MPNSTs and demonstrate that combinatorial therapies targeting critical MPNST signaling pathways are more effective.
Graphical abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are highly aggressive Schwann cell–derived neoplasms that are common in individuals with the tumor susceptibility syndrome neurofibromatosis type 1 (NF1) as well as at sites of previous radiotherapy and sporadically in the general population.1 Patients with MPNSTs have a poor prognosis, largely because the radiotherapeutic and chemotherapeutic regimens currently used to treat MPNSTs are ineffective. Previous attempts to develop new, more effective therapies for MPNSTs have largely focused on the signaling pathways that are aberrantly activated in these neoplasms. All NF1-associated MPNSTs and plexiform neurofibromas (the benign precursors to MPNSTs) as well as most sporadic and radiation-induced MPNSTs carry null mutations of the NF1 gene.2,3 The NF1 gene encodes neurofibromin, a GTPase-activating protein that inactivates Ras. NF1-null mutations thus result in the activation of multiple classic Ras (H-Ras, N-Ras, and K-Ras) and R-Ras (R-Ras and R-Ras2) subfamily proteins in MPNSTs4,5 and the signaling pathways downstream of these small GTP-binding proteins. However, attempts to develop new treatments targeting Ras in neurofibromas and MPNSTs and key Ras-regulated signaling pathways in MPNSTs have been unsuccessful. For example, tipifarnib, a farnesyltransferase inhibitor that blocks a key post-translational modification of Ras proteins, is ineffective in children and young adults with plexiform neurofibromas.6,7 Although MAPK/ERK kinase (MEK) inhibitors, such as selumetinib, are effective against neurofibromas,8 these agents are ineffective against MPNSTs.8, 9, 10
This led to the possibility that agents targeting key upstream activators of Ras proteins might be effective against MPNSTs. We hypothesized that activation of one or more receptor tyrosine kinases (RTKs), occurring in conjunction with NF1 loss and additional driver mutations, is required for the activation of key signaling pathways that drive MPNST cell proliferation and survival and confer MEK inhibitor resistance. On querying all 58 human RTKs with pharmacologic and genome-scale shRNA screens, erbB kinases and insulin-like growth factor 1 receptor were consistently observed to be required for MPNST cell proliferation and combinatorial therapy with the broad-spectrum erbB inhibitor canertinib and the insulin-like growth factor 1 receptor inhibitor picropodophyllin abolished Ras hyperactivation in MPNST cells and more effectively inhibited xenograft growth (D.P. Jenkins, S.N. Brosius, A.M. Prechtl, S.T. Guest, J.F.L., S.H. Worley, A. Alers, B. Turner-Ivey, K. Armeson, E. Garrett-Mayer, T. Hull, S.L.C., unpublished data). The importance of erbB receptors is further underlined by a recent demonstration that erbB4 promotes MPNST proliferation and xenograft growth.11 However, ablating erbB4 expression had no effect on Ras hyperactivation and this RTK instead promoted MPNST growth by activating other non–Ras-regulated signaling proteins, such as STAT3, STAT5, and phospholipase Cγ. These observations led to the investigation of whether additional erbB receptors contributed to MPNST proliferation and survival.
The current genome-scale shRNA screens indicated that erb-B2 receptor tyrosine kinase 3 (erbB3) was the erbB kinase most consistently required for MPNST proliferation and/or survival. Consequently, this study tested the hypothesis that erbB3 promoted MPNST proliferation and survival by contributing to the activation of essential non–mitogen-activated protein kinase cytoplasmic signaling pathways. Whether erbB3 was widely expressed and activated in MPNST cell lines and surgically resected MPNSTs and what effect erbB3 knockdown had on MPNST proliferation and survival was investigated. The attempts to identify erbB3-regulated signaling cascades indicated that this RTK regulated calcium-dependent signaling pathways. This led to testing the possibility that simultaneous inhibition of erbB3 [by erbB3 knockdown or with broad-spectrum erbB inhibitors (canertinib and sapitinib)] and calmodulin signaling could additively inhibit MPNST proliferation and/or survival. The calmodulin inhibitor trifluoperazine (TFP) alters the activation of proviral integration site of Moloney murine leukemia (PIM) serine/threonine protein kinases, as indicated by kinomics analysis.12 Consequently, effectiveness of the PIM kinase inhibitor AZD1208 was tested, alone and in combination with erbB inhibitors. Paradoxically, TFP and AZD1208 enhanced the phosphorylation of calmodulin-dependent protein kinase IIα (CaMKII) and erbB3 and trifluoperazine promoted erbB3 phosphorylation by activating Src family kinases, which are upstream activators of STAT3 and STAT5. This, in turn, led to the testing of the effectiveness of treatment regimens that used the Src family kinase inhibitor saracatinib to inhibit erbB3 signaling, alone and in combination with trifluoperazine or AZD1208.
Materials and Methods
Antibodies and Reagents
See Table 1 for a complete list of antibodies, extraction reagents, and cellular labeling kits. The pan-erbB inhibitor canertinib (number C-1202) was from LC Laboratories (Woburn, MA). The epidermal growth factor receptor (EGFR)/erbB2/erbB3 [sapitinib (AZD8931); number S2192], pan-PIM kinase (AZD1208; number S7104), and Src family kinase [saracatinib (AZD0530); number S1006] inhibitors were purchased from Selleckchem (Houston, TX), whereas the calmodulin inhibitor trifluoperazine (number T8516) was obtained from Sigma Aldrich (St. Louis, MO).
Table 1.
Antibodies and Cell Stains
| Antibody target | Vender | Species | Catalog no. | Application |
|---|---|---|---|---|
| ErbB3 XP | Cell Signaling Technology (Danvers, MA) | Rabbit mAb | 12708 | IB, ICC-IF |
| ErbB3 | Santa Cruz Biotechnology (Dallas, TX) | Mouse mAb | sc-84155 | IHC |
| ErbB3 | Santa Cruz Biotechnology | Rabbit pAb | sc-285 | IHC |
| Phospho-ErbB3Tyr1328 | Cell Signaling Technology | Rabbit mAb | 14525 | IB |
| Phospho-ErbB3Tyr1289 | Cell Signaling Technology | Rabbit mAb | 4791 | IB |
| EGFR | Cell Signaling Technology | Rabbit mAb | 4267 | IB |
| ErbB2 | Cell Signaling Technology | Rabbit mAb | 4290 | IB |
| ErbB4 | Abcam (Cambridge, UK) | Rabbit mAb | 32375 | IB |
| STAT3 | Cell Signaling Technology | Mouse mAb | 9139 | IB |
| Phospho-STAT3Tyr705 | Cell Signaling Technology | Rabbit mAb | 2535 | IB |
| CaMKII | Cell Signaling Technology | Rabbit pAb | 3362 | IB |
| Phospho-CaMKIITyr231 | Cell Signaling Technology | Rabbit pAb | 3356 | IB |
| S6 ribosomal protein | Cell Signaling Technology | Mouse mAb | 2317 | IB |
| Phospho-S6Ser240/244 XP | Cell Signaling Technology | Rabbit mAb | 5364 | IB |
| Akt | Cell Signaling Technology | Mouse mAb | 2920 | IB |
| Phospho-AktSer473 XP | Cell Signaling Technology | Rabbit mAb | 4060 | IB |
| p44/42 MAPK (ERK 1/2) | Cell Signaling Technology | Rabbit mAb | 4696 | IB |
| Phospho-ERK1/2Thr202/Tyr204 | Cell Signaling Technology | Rabbit mAb | 4370 | IB |
| Cleaved caspase-3 | Cell Signaling Technology | Rabbit mAb | 9664 | IB |
| GAPDH | Cell Signaling Technology | Mouse mAb | 97166 | IB |
| Actinin | Cell Signaling Technology | Mouse mAb | 69758 | IB |
| Rb mAb IgG XP isotype control | Cell Signaling Technology | Rabbit | 3900 | ICC-IF |
| Alexa Fluor secondary antibodies | Life Technologies (Carlsbad, CA) | Alexa Fluor 568 anti-Rb IgG | A10042 | ICC-IF |
| ActinGreen 488 ReadyProbes | Invitrogen (Waltham, MA) | R37110 | ICC-IF | |
| Hoechst | Thermo Fisher Scientific (Waltham, MA) | 83570 | ICC-IF, Celigo | |
| NE-PER Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher Scientific | 78833 | IB | |
| IRDye secondary antibodies | LI-COR (Lincoln, NE) | IRDye 800CW, IRDye 680RD | IB | |
| LIVE/DEAD Cell Imaging Kit (488/570) | Thermo Fisher Scientific | R37601 | Celigo |
CaMKII, calmodulin-dependent protein kinase IIα; EGFR, epidermal growth factor receptor; ErbB, erythroblastic leukemia viral oncogene homolog B; ERK, extracellular signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IB, immunoblot; ICC-IF, immunocytochemistry-immunofluorescence; IHC, immunohistochemistry; mAb, monoclonal antibody; MAPK, mitogen-activated protein kinase; pAb, polyclonal antibody; Phospho, phosphorylated; Rb, retinoblastoma transcriptional corepressor.
Human MPNST Specimens and MPNST-Derived Cell Lines
Formalin-fixed, paraffin-embedded specimens of surgically resected human MPNSTs were retrieved from the files of the University of Alabama at Birmingham Department of Pathology and the Medical University of South Carolina Department of Pathology and Laboratory Medicine. The authors have previously described the sources of their human MPNST cell lines.11,13, 14, 15, 16 MPNST cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 10 μg/mL streptomycin, and 10 IU/mL penicillin. These cell lines were regularly tested for Mycoplasma infection. The morphology and doubling times of the lines were regularly evaluated, and cell line identities were validated by short tandem repeat analyses, as recommended by ATCC (Manassas, VA) Technical Bulletin 8.
Immunoblots
Cells were lysed with 1% boiling SDS, heated to 70°C for 10 minutes, briefly sonicated, and then clarified. These lysates were used to assess all targets except EGFR, erbB2, erbB3, phosphorylated erbB3, and erbB4; the authors used mammalian protein extraction reagent (MPER) lysis buffer to prepare lysates for assessing erbB kinase expression as they have found that MPER gives optimal results when immunoblotting for erbB receptors located in multiple distinct cellular compartments. For neuregulin-1α (NRG1α) and NRG1β stimulation immunoblots, cells were serum starved overnight and stimulated with vehicle [phosphate-buffered saline (PBS)/glycerol] or with the growth factors at 5, 10, 15, and 20 nmol/L in PBS/glycerol carrier-free diluent, and lysates were collected at 30 minutes. Protein concentrations were determined using a Pierce (St. Louis, MO) bicinchoninic acid protein assay kit. Equal amounts of protein were resolved on 7.5% or 4% to 20% gradient polyacrylamide gels and transferred to 0.45 μmol/L nitrocellulose membranes at 300 mA for 30 minutes utilizing Flashblot transfer buffer (Advansta, San Jose, CA; number R-03090-D50). Membranes were blocked with LI-COR (Lincoln, NE) Intercept–Tris-buffered saline blocking buffer and probed with primary antibodies overnight at 4°C. The next day, blots were washed three times in Tris-buffered saline with 0.1% Tween 20, and immunoreactivity was detected by incubating the blots with 1:20,000 secondary LI-COR IRDye 680– or IRDye 800–conjugated antibodies in LI-COR Tris-buffered saline with 0.1% Tween 20. Blots were imaged and analyzed on a LI-COR Odyssey Fc imaging system.
Cell Fractionation
An NE-PER Nuclear and Cytoplasmic Extraction Reagent kit (Thermo Fisher Scientific, Waltham, MA; number 78833) was used to isolate nuclear and cytoplasmic fractions from MPNST cells. Cells were trypsinized and pelleted by centrifugation for 5 minutes at 500 × g before pellets were washed one time with PBS. Pellets were resuspended, transferred to 1.5-mL tubes, and again pelleted by centrifugation at 500 × g for 3 minutes. Cell fractionation was then performed as recommended by the kit's manufacturer. Cellular fractions were immediately resolved onto 4% to 20% gradient gels for immunoblotting.
Immunocytochemistry
Cells were seeded onto 18-mm number 1.5 round coverslips (Electron Microscopy Sciences, Hatfield, PA; number 72222-01) in 12-well sterile tissue culture dishes at a density of 150,000 cells per coverslip and maintained at 37°C in a tissue culture incubator. At 70% to 75% log-phase confluency, cells were gently washed three times (5 minutes per wash) with 1× PBS, followed by fixation with 4% paraformaldehyde in PBS for 15 minutes at room temperature. Fixed cells were washed three times (5 minutes per wash) with 1× PBS. Cells were incubated for 10 minutes in 50 mmol/L ammonium chloride to quench residual aldehydes and then permeabilized for 10 minutes at room temperature with 0.3% Triton X-100 in PBS. Cells were washed three times (5 minutes per wash) with 1× PBS and then blocked with 5% goat serum for 1 hour at room temperature. Cells were incubated with primary antibodies diluted in 1% bovine serum albumin in PBS at 4°C overnight. Cells were washed three times (5 minutes per wash) with PBS containing 0.05% Triton X-100 to remove unbound primary antibody and then incubated with species appropriate Alexa Fluor 488– or Alexa Fluor 568–conjugated secondary antibodies in 1% bovine serum albumin in PBS for 1 hour at room temperature in the dark. Secondary incubations were followed with four 5-minute washes with PBS containing 0.05% Triton X-100. F-actin was stained using an ActinGreen-488 conjugated direct stain (Invitrogen, Waltham, MA; number R37110) for 30 minutes, followed by staining with Hoechst 33342 nuclear stain (0.04 μg/mL) for 10 minutes at room temperature in the dark. A 5-minute wash with water was performed before mounting the coverslips onto glass slides with ProLong Glass Antifade mounting media (Invitrogen; number P36980) overnight. Imaging was performed on a Zeiss (White Plains, NY) Imager M2 or Zeiss LS 980 with Airyscan 2 the following day.
RNA Interference
Sigma (St. Louis, MO) shRNA lentiviral vectors targeting erbB3 (TRCN0000199993, TRCN0000010344, and TRCN0000009835) and a nontargeting control (SHC002) were obtained from the Medical University of South Carolina Hollings Cancer Center shRNA Technology Shared Resource. HEK293T cells were transfected to generate lentivirus using Lipofectamine 2000 (Thermo Fisher Scientific; number 11668019) and MISSION shRNA Lentiviral Packaging Mix (Sigma; product number SHP001) in accordance with the MISSION shRNA user manual. Transfections were performed in antibiotic-free Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 10 μg/mL streptomycin, and 10 IU/mL penicillin. Viral supernatant was harvested 24 and 48 hours after transfection, filtered through 0.45 μmol/L filters, and stored in 500-μL aliquots at −80°C.
MPNST cells plated in 2.5 mL of antibiotic-free Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 10 μg/mL streptomycin, and 10 IU/mL penicillin in 60-mm dishes were reverse transduced at 40% confluency with 250 μL of viral supernatant and polybrene (final concentration, 10 μg/mL). Puromycin selection was initiated 48 hours after infection and continued for 72 hours before validating knockdowns via immunoblot analysis.
Celigo Imaging Cytometer Assays
Cells were detached using Corning CellStripper (Thermo Fisher Scientific) and counted using a LUNA-FL Dual Fluorescence Cell Counter (Logos Biosystems, Annandale, VA). Cells were seeded at a density of 1200 cells per well in black-walled 96-well plates (Corning Inc., Corning, NY; number 3603). For assessment of direct cell number, each condition was performed in triplicate, and end point reads were performed 1, 3, 5, and 7 days after plating. Cells were stained with Hoechst 33342 (5 μg/mL) and incubated for 30 minutes at 37°C. Plates were read on a Celigo high-throughput imaging cytometer (Redwood City, CA) utilizing the direct cell count for total cell number option with exposure times of 100,000 milliseconds. Reads were analyzed with Celigo software version 2.1 and exported into Excel version 2304 (Microsoft, Redmond WA) and GraphPad Prism version 9.5.1 (Boston, MA) for statistical analyses. For cell viability analyses, cells were seeded as described above and were stained at end point reads with calcein AM (1 μmol/L) and propidium iodide (1 μmol/L) for 15 minutes at 37°C. Cells were then imaged on a Celigo cytometer using the Live + Dead software option. Images were analyzed using Fiji ImageJ version 2.9.0 (https://downloads.imagej.net/fiji/releases/2.9.0).
High-Density Oligonucleotide Microarray Hybridization, Scanning, and Analysis
Cultures of wild-type Schwann cells were prepared from the sciatic nerves of adult Sprague-Dawley rats, per previously described methods.17 Rat Schwann cell cultures were initially maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 10 μg/mL streptomycin, and 10 IU/mL penicillin supplemented with 50 nmol/L NRG1β and 2 μmol/L forskolin. For microarray experiments, Schwann cells were plated on poly-l-lysine/laminin–coated plates and allowed to reach 70% confluency. To avoid the effects of Schwann cell mitogens present in serum, cells were incubated in Schwann cell–defined medium17 for 24 hours before NRG1 stimulation. This medium was then replaced with Schwann cell–defined medium containing 10 nmol/L NRG1β or vehicle, and cells were incubated for an additional 24 hours. Cells were then harvested, and total RNA was isolated from them using TRIzol reagent (Life Technologies, Grand Island, NY), as recommended by the manufacturer.
Double-stranded cDNA was synthesized from 20 μg of total RNA using the SuperScript Choice System (Life Technologies) and a high-performance liquid chromatography–purified oligo-dT primer containing an adjacent T7 RNA polymerase promoter (GENSET Corp., La Jolla, CA). Biotinylated riboprobes were transcribed from cDNA templates using an Enzo BioArray High Yield Transcript Labeling Kit (Affymetrix, Inc., Santa Clara, CA). Biotinylated cRNA was purified using RNeasy columns, fragmented, and hybridized to rat genome U34A oligonucleotide arrays (Affymetrix, Inc.) at 45°C in 100 mmol/L 2-(N-morpholino)ethanesulfonic acid (MES; pH 7.4)/1 mol/L NaCl/20 mmol/L EDTA/0.01% Tween-20 containing 50 pmol/L Affymetrix control oligonucleotide B2, 1.5 pmol/L Escherichia coli bioB RNA, 5 pmol/L E. coli bioC RNA, 25 pmol/L E. coli bioD RNA, and 100 pmol/L E. coli Cre recombinase RNA. After 16 hours of hybridization, arrays were washed and stained with streptavidin-phycoerythrin. Hybridization signals were amplified by labeling stained arrays with a biotinylated goat anti-streptavidin antibody (Vector Laboratories, Burlingame, CA) and restaining with streptavidin-phycoerythrin (Affymetrix, Inc., EUKGE-WS2 version 4 protocol). Arrays were scanned twice at 570 nmol/L using an Affymetrix GeneArray scanner. Data sets from Affymetrix U34A microarrays were processed using Affymetrix Microarray Suite 5.0 (Santa Clara, CA). Total fluorescent intensities for all arrays were normalized to a fixed value of 150. Average differences were calculated as the mean of hybridization signal intensities for perfect match oligonucleotides subtracted from intensities for mismatch oligonucleotides (oligonucleotides differing from perfect match oligonucleotides at a single, centrally placed position); outliers were excluded from calculations of average differences. The CIs for the presence or absence of a particular mRNA were calculated for each transcript using a matrix-based decision algorithm; these calculations were performed using the default settings in Microarray Suite software. Hybridization efficiency was evaluated by determining whether the E. coli bioB, bioC, bioD, and cre mRNAs spiked into the hybridization mixture were detectable; in all array experiments reported in this study, all four control transcripts were detected. To assess the quality of the transcribed RNA target, signals were determined for oligonucleotide probe sets corresponding to the 3′, middle, and 5′ portions of glyceraldehyde-3-phosphate dehydrogenase and β-actin mRNAs, and the 3′/5′ ratio for these signals was calculated. In the array experiments included in this study, the 3′/5′ ratios for glyceraldehyde-3-phosphate dehydrogenase and β-actin mRNAs were ≤2, indicating that cDNAs used for in vitro transcription were near full length and thus efficiently transcribed.
Normalization based on loess approach was performed to make array intensities comparable for group comparison. These normalized values were filtered using the two-sample t-test (cutoff, P < 0.05 in different genotypes). The selected genes were then examined to identify subgroups of genes based on similar Gene Ontology annotation patterns.
Kinomics Analysis
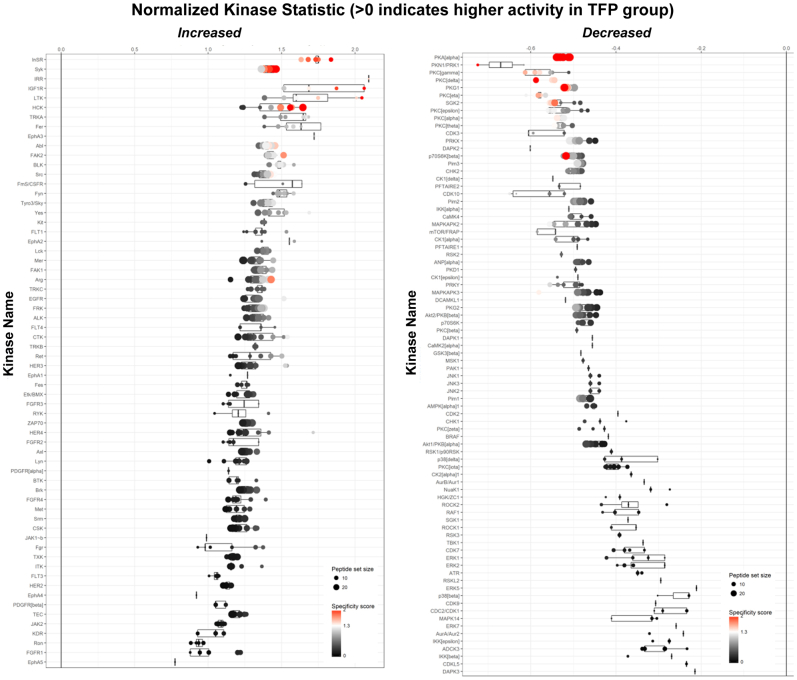
The authors have previously described the performance of kinomics analyses in ST88-14 MPNST cells challenged with trifluoperazine.12 These assays were performed at the University of Alabama at Birmingham Kinome Core using a Pam Station 12 automatic kinomics instrument (PamGene International BV, Hertogenbosch, the Netherlands), as previously described.18, 19, 20, 21 Automated array pumping of lysates and detection reagents, as well as imaging over multiple cycles, and exposure times of fluorescein isothiocyanate–immunolabeled phosphorylated target peptides were conducted using Evolve software version 3.0 (PamGene) on both phosphotyrosine (part 86312) and phosphoserine/phosphothreonine (part 87102) arrays. Images were gridded, and kinetic phosphorylation intensities were integrated and further analyzed with BioNavigator software version 6.3. Comparative (trifluoperazine versus vehicle) peptide phosphorylation data were used to identify trifluoperazine-responsive kinases using BioNavigator Upkin PamApps (PamGene; phosphotyrosine and phosphoserine/phosphothreonine version 6.0). Trifluoperazine-altered kinases (with a PamApp-generated sensitivity plus specificity score >1.5) were uploaded (July 20, 2022) to GeneGo MetaCore (Clarivate, London, UK) to generate an AutoExpand network model of trifluoperazine-altered kinases.
Results
Most Human MPNSTs and MPNST-Derived Cell Lines Express ErbB3
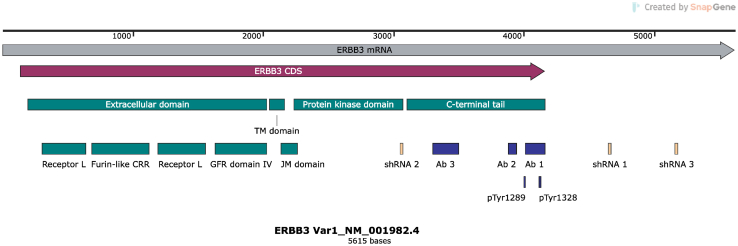
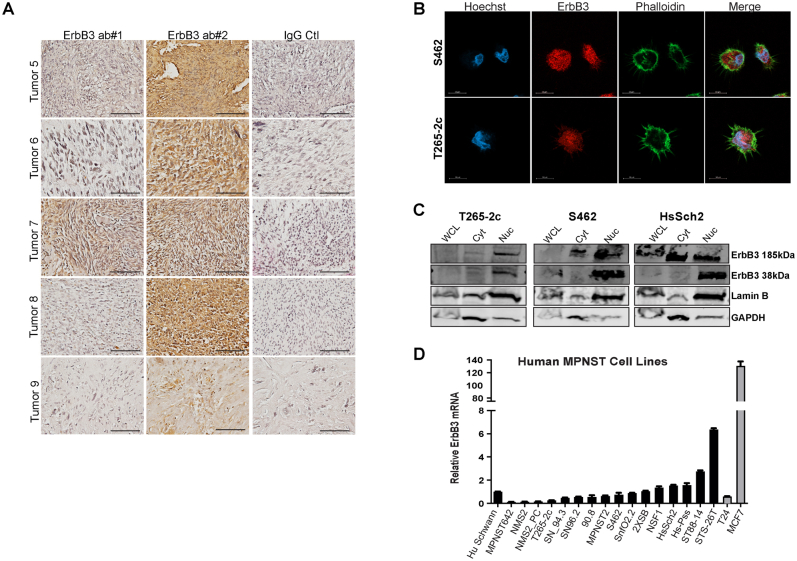
A previous study using the anti-erbB3 IgM monoclonal antibody RTJ.1 demonstrated that four of five (80%) of the surgically resected MPNSTs stained were immunoreactive for erbB3.15 To expand on and confirm this, immunohistochemistry on nine more surgically resected MPNSTs was performed using both a rabbit polyclonal antibody recognizing an epitope at the carboxy terminus of erbB3 and 5A12, and a mouse IgG1 monoclonal antibody that recognizes an epitope spanning amino acids 1250 to 1270 in the 1342–amino acid full-length protein. Supplemental Figure S1 provides a schematic showing the sites targeted by the antibodies and shRNAs used in this study. Seven of the nine tumors (78%) had moderate to high levels of erbB3 immunoreactivity with both antibodies (Figure 1A and Supplemental Figure S2A), which is consistent with the previous observations with the RTJ.1 monoclonal antibody.15 The distribution of immunoreactivity detected with both antibodies was also highly similar; although there was some tumor-to-tumor variability, both the rabbit polyclonal antibody and the 5A12 mouse monoclonal antibody detected nuclear, cytoplasmic, and membranous erbB3 immunoreactivity.
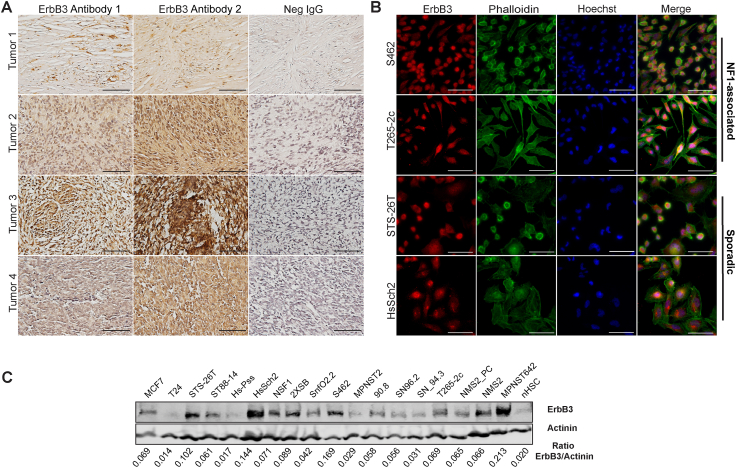
Figure 1.
ErbB3 is variably expressed in human malignant peripheral nerve sheath tumors (MPNSTs) and MPNST-derived cell lines. A: Representative images of erbB3 immunostaining of four different surgically resected MPNSTs. Panels show images of sections stained with two different erbB3-specific antibodies recognizing two different epitopes within the erbB3 protein. Negative (Neg) IgG panels show representative images of sections stained with nonimmune control antibody. Images taken with a 40× objective lens. B: Immunocytochemistry with an antibody recognizing a third distinct epitope in erbB3 shows ubiquitous erbB3 expression in four MPNST-derived cell lines. Images taken with a 63× oil objective showing neurofibromatosis type 1 (NF1)–associated (S462 and T265-2c) and sporadic (STS-26T and HsSch2) MPNST cells stained with anti-erbB3 antibody (red), the lectin phalloidin (green; recognizes actin), and Hoechst 33342 (blue). C: Immunoblot analyses demonstrating the relative levels of erbB3 protein in whole cell lysates from 16 human MPNST-derived cell lines, nonneoplastic human Schwann cells (nHSCs), MCF-7 breast carcinoma cells, and T24 bladder carcinoma cells. Actinin was used as a loading control. Scale bars: 100 μm (A); 20 μm (B).
Subcellular distribution of erbB3 was further assessed by immunocytochemistry with the rabbit D22C5 XP monoclonal antibody, which recognizes an epitope in the carboxy terminus surrounding amino acid 1100 (Supplemental Figure S1), on two NF1-associated MPNST cell lines (S462 and T265-2c) and two sporadic MPNST cell lines (STS-26T and HsSch2). Ubiquitous staining was observed in the cytoplasm and perinuclear/nuclear areas of the cells, by both conventional immunofluorescence microscopy (Figure 1B) and confocal imaging utilizing Airyscan technology (Supplemental Figure S2B). This pattern of immunoreactivity was not evident when the primary antibody was replaced with a nonimmune control or staining was performed with secondary antibody only (Supplemental Figure S3). erbB3 nuclear expression was further confirmed via cellular fractionation, followed by immunoblot analyses (Supplemental Figure S2C). Interestingly, this antibody recognized a prominent band in the nuclear fractions at 38 kDa, which likely corresponds to a known truncated form of erbB3 derived from an mRNA with a shortened reading frame (ErbB3 sequence), which is freely available to the public and was obtained from the National Center for Biotechnology Information's website (GenBank; https://www.ncbi.nlm.nih.gov/gene/?term=BC002706.2; accession number BC002706.2).
Real-time PCR and immunoblot analyses with rabbit anti-erbB3 monoclonal antibody D22C5 assessed erbB3 mRNA and protein expression in a panel of 16 NF1-associated and sporadic human MPNST cell lines relative to the level of erbB3 expression in non-transformed human Schwann cells (nHSCs). Cell lines with low (T24 bladder carcinoma cells) and high (MCF7 breast adenocarcinoma cells) erbB3 expression were used as controls. Five MPNST cell lines (NSF1, HcSch2, Hs-Pss, ST88-14, and STS-26T) had ERBB3 mRNA expression higher than that of nHSCs, whereas the other 11 MPNST cell lines had ERBB3 mRNA expression that was similar or lower than that of nHSCs (Supplemental Figure S2D). In contrast, immunoblot analyses of these same cell lines showed that erbB3 protein expression in 13 of the 16 MPNST cell lines was higher than in nHSCs, with the remaining three MPNST cell lines (Hs-Pss, MPNST2, and SN94.3) having erbB3 protein expression similar to that seen in nHSCs (Figure 1C). Notably, erbB3 protein levels in seven of the MPNST cell lines (STS-26T, HsSch2, NSF1, 2XSB, S462, NMS1, and MPNST642) were equivalent to or higher than the levels evident in MCF7 cells. The differences between the levels of erbB3 mRNA and protein expression in most of these MPNST cell lines suggests that post-transcriptional regulatory mechanisms are responsible for increased erbB3 protein expression in MPNST cells.
ErbB3 Is Required for the Proliferation and Survival of MPNST Cells
Whether erbB3 is required for the proliferation and viability of MPNSTs was investigated next. To address this, lentiviral vectors expressing three different erbB3 shRNAs or a nontargeting shRNA control were transduced into two NF1-associated MPNST lines (S462 and T265-2c cells) and two sporadic MPNST cell lines (STS-26T and HsSch2 cells). Two of the erbB3 shRNAs effectively reduced expression of the 185-kDa form of erbB3 in all four MPNST cell lines relative to the nontargeting control (Figure 2A). The two shRNAs that effectively knocked down the 185-kDa form of erbB3 similarly reduced expression of the 38-kDa erbB3 nuclear variant identified in their cellular fractionation experiments (Figure 2B).
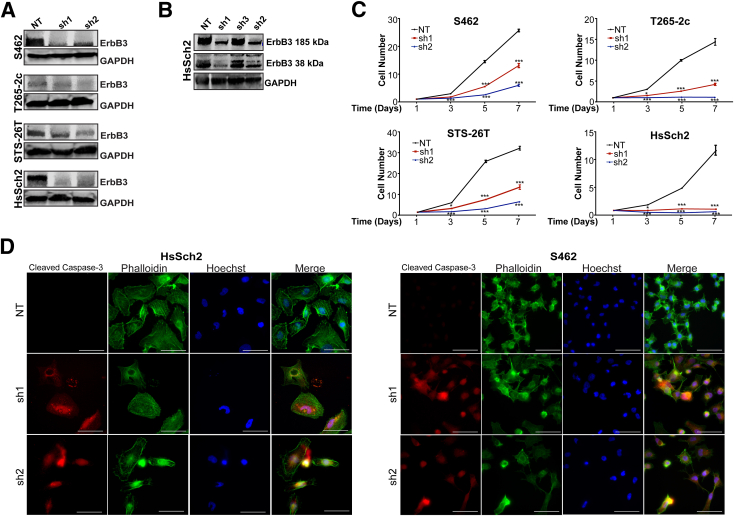
Figure 2.
ErbB3 loss decreases cellular proliferation and increases cell death in malignant peripheral nerve sheath tumors (MPNSTs). A: Immunoblots demonstrating reduced erbB3 expression in four MPNST cell lines transduced with two different lentiviral vectors expressing erbB3 shRNAs (sh1 and sh2) or a nontargeting control shRNA (NT). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. B: Immunoblot demonstrating reduction of the 185- and 38-kDa forms of erbB3 in cells transduced with sh1 and sh2 erbB3 shRNAs (but not sh3) compared with cells transduced with NT. C: Cells with stable erbB3 knockdown were seeded at optimized densities for each cell line, and direct cell counts were performed at 1, 3, 5, and 7 days later. ErbB3 knockdown decreases cell numbers relative to NT controls beginning 3 days after plating; statistical significance represents the comparison of each erbB3 shRNA to the nontargeting vector. Only those data with statistical significance are designated. D: Immunostaining for activated (cleaved) caspase-3 in HsSch2 and S462 MPNST cells transduced with sh1 and sh2 erbB3 shRNAs or a nontargeting control shRNA. Cleaved caspase-3 staining (red) is increased in cells with erbB3 knockdown. The actin cytoskeleton is counterstained with phalloidin (green), and nuclei are labeled with Hoechst 33342 (blue). Images taken with a 63× oil immersion objective. ∗P ≤ 0.05, ∗∗∗P ≤ 0.001. Scale bars = 20 μm (D).
Stable knockdown of erbB3 expression significantly reduced the number of cells in all four cell lines, as indicated by a high-throughput imaging cytometer used to analyze cell proliferation over the course of 7 days (Figure 2C). Prominent morphologic changes in surviving cells following erbB3 knockdown as well as increased granular debris in the background were observed, indicative of an increase in cell death (Supplemental Figure S4). To confirm whether this decrease reflected, at least in part, reduced cellular survival, caspase-3 activation in sporadic (HsSch2) and NF1-associated (S462) MPNST cell lines transduced with the erbB3 shRNAs or the nontargeting control was examined. erbB3 knockdown induced an increase in cleaved caspase-3 in both lines, whereas the control shRNA did not (Figure 2D). This indicates that erbB3 knockdown reduces both the proliferation and the viability of HsSch2 and S462 cells.
ErbB3 Activates Multiple Known Pathways in MPNST Cells and Nonneoplastic Schwann Cells as Well as Novel Calcium/Calmodulin-Regulated Signaling Pathways
Although it is well established that erbB3 activates the phosphatidylinositol 3-kinase/Akt and extracellular signal-regulated kinase (ERK) signaling pathways, little is known regarding other signaling pathways that are regulated by erbB3.22 To identify additional downstream effectors of erbB3 signaling, normal adult rat Schwann cells (which express erbB3, but not erbB4) were stimulated with vehicle or 10 nmol/L NRG1β for 24 hours. Microarray analyses were performed to identify genes potentially regulated by erbB3. In addition to an up-regulation of genes known to be downstream of phosphatidylinositol 3-kinase, several transcripts encoding effectors activated by calmodulin were up-regulated. Gene Ontology tools used on these microarray data identified several pathways that were enriched in NRG1β-stimulated Schwann cells, including pathways involved in growth factor receptor binding, cytoskeletal organization, transcription factor interactions, and calcium-calmodulin signaling (Supplemental Figure S5).23, 24, 25 Transcripts encoding CaMKIIα were prominently and significantly up-regulated in the calcium-calmodulin signaling pathway.
To determine whether NRG1 also activated some of the effectors in MPNST cells that were up-regulated in their Schwann cell microarray analyses, HsSch2 cells, the MPNST line with the highest level of erbB3 expression, were challenged with vehicle or a range of concentrations (5, 10, 15, and 20 nmol/L) of NRG1α or NRG1β for 30 minutes. Immunoblots of the lysates of these cells were probed with antibodies recognizing phosphorylated and non-phosphorylated forms of the proteins of interest. The effect of NRG1α and NRG1β on the phosphorylation of erbB3 at residue Y1289, which is a well-known p85/phosphatidylinositol 3-kinase binding site (Figure 3A), was examined first. Both NRG1α and NRG1β promoted erbB3 phosphorylation at Y1289 in a concentration-dependent manner, with NRG1β more robustly stimulating erbB3 phosphorylation at this residue. The effect of these NRG1 isoforms on the phosphorylation of Y1328, a site in erbB3 that is capable of binding Src homology 2 domain-containing-transforming protein 2 (SHC) domains and that has been predicted to be involved in calcium-mediated signaling (http://elm.eu.org/instances/LIG_PTB_Phospho_1/P21860/1322, last accessed April 3, 2023) was examined next. Both NRG1α and NRG1β promoted the erbB3-Y1328 phosphorylation in a concentration-dependent manner (Figure 3A), with NRG1β more robustly inducing phosphorylation at this site. The effects of NRG1α and NRG1β on the phosphorylation of Akt at S473 and ERK (Y202 and Y204) paralleled the effects of these growth factors on erbB3 phosphorylation.
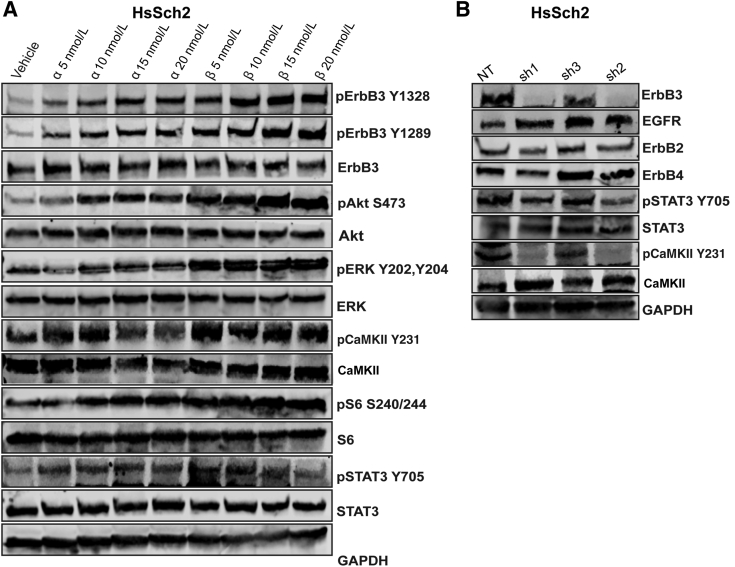
Figure 3.
ErbB3 signaling pathways involve activation of known target effectors as well as novel calcium-calmodulin–mediated effectors. A: Immunoblot analyses of HsSch2 malignant peripheral nerve sheath tumor cells challenged with vehicle (0.1% dimethyl sulfoxide) or neuregulin-1α (NRG1α) or NRG1β (5, 10, 15, or 20 nmol/L) for 30 minutes. Cell lysates were probed with the indicated monoclonal antibodies. NRG1α and NRG1β both increase the phosphorylation of erbB3, several known downstream effectors of erbB3 [Akt S473, extracellular signal-regulated kinase (ERK) Y202/204, S6 S240/244, and STAT3 Y705], as well as the calcium-calmodulin effector calmodulin-dependent protein kinase IIα (CaMKII) Y231 in a concentration-dependent manner. B: Stable erbB3 knockdown has little or no effect on the expression of other erbB3 family members but decreases STAT3 Y705 and CaMKII Y231. EGFR, epidermal growth factor receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NT, nontargeting control shRNA; p, phosphorylated; sh1/2, erbB3 shRNAs.
The effect of NRG1 on the phosphorylation of CaMKII, STAT3, and S6 kinase, three signaling molecules encoded by transcripts that were NRG1β responsive in their microarray analyses, was examined next. Both NRG1α and NRG1β promoted the phosphorylation of CaMKII at residue Y231. Unlike that for erbB3, Akt, and ERK, however, CaMKII Y231 phosphorylation was highly sensitive at the lowest (5 nmol/L) concentration of NRG1 tested (Figure 3A). NRG1α and NRG1β exerted concentration-dependent effects on the phosphorylation of S6 kinase at S240 and S244, with NRG1β having a greater effect than NRG1α. NRG1α and NRG1β additionally promoted the phosphorylation of STAT3 at residue Y705. As with CaMKII, the highest levels of STAT3 phosphorylation were already evident in cells challenged with 5 and 10 nmol/L concentrations of NRG1α and NRG1β (Figure 3A).
After establishing that stimulation of HsSch2 MPNST cells with NRG1α and NRG1β enhanced the phosphorylation of CaMKII and STAT3, whether the basal phosphorylation of these signaling proteins was erbB3 dependent was investigated next. For this, unstimulated HsSch2 cells were transduced with three shRNAs targeting erbB3 or a nontargeting control. Its effect on the phosphorylation of CaMKII and STAT3 72 hours after beginning puromycin selection was examined. Knockdown of erbB3 expression with the two most potent shRNAs (sh1 and sh2) reduced the phosphorylation of STAT3 at Y705 (Figure 3B). Strikingly, these same two shRNAs nearly completely abolished the phosphorylation of CaMKII at residue Y231. In both instances, erbB3 knockdown had virtually no effect on the total levels of CaMKII and STAT3 protein. To determine whether these changes might reflect a loss of expression of other erbB kinases, the level of expression of EGFR, erbB2, and erbB4 was assessed in cells transduced with the erbB3 shRNAs. Knockdown of erbB3 had, at most, minimal effects on the levels of erbB2 and erbB4 while slightly increasing the expression of EGFR (Figure 3B).
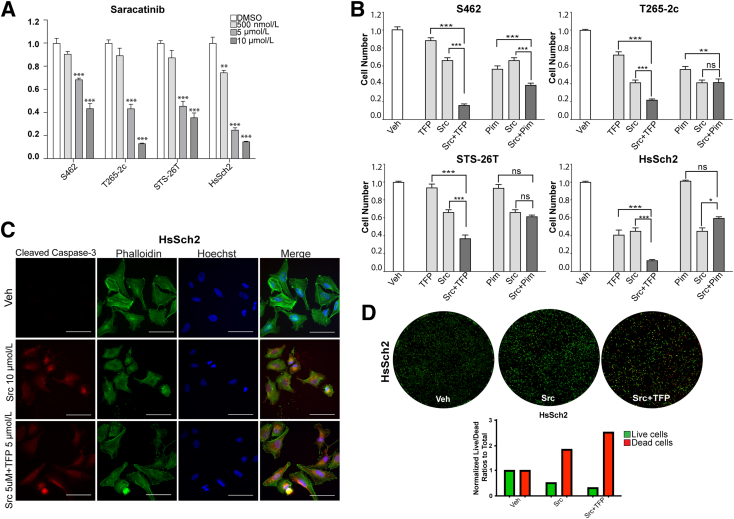
Pan-ErbB, Calmodulin, and PIM Kinase Inhibitors Impair MPNST Cell Proliferation and/or Survival
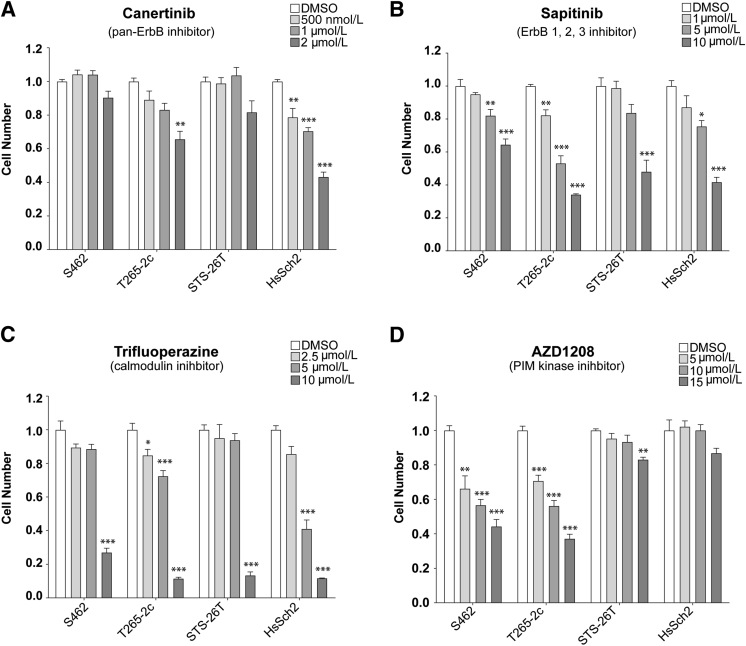
Prior kinomics studies examining the effects of the calmodulin inhibitor trifluoperazine on ST88-14 MPNST cells12 indicated that the activity of PIM1 and PIM3, two constitutively active members of a family of calmodulin-dependent protein kinase–related proteins26 downstream of STAT3/5 signaling, was inhibited by trifluoperazine. This suggested that inhibitors of PIM kinases may also inhibit the proliferation and/or survival of MPNST cells. This prompted the comparison of the relative effectiveness of erbB inhibitors, trifluoperazine, and the pan-PIM kinase inhibitor AZD1208. Two NF1-associated (S462 and T265-2c) and two sporadic (STS-26T and HsSch2) MPNST cell lines were challenged with vehicle, the pan-erbB inhibitor canertinib, the EGFR/erbB2/erbB3 inhibitor sapitinib, trifluoperazine, or AZD1208. Each line was first challenged with vehicle or a range of concentrations of each inhibitor. The relative cell numbers were determined 1, 3, 5, and 7 days after beginning treatment (Supplemental Figure S6), and low, middle, and high concentrations to be used in subsequent experiments were identified. Canertinib did not significantly reduce the number of S462 and STS-26T cells even at the highest concentration tested (2 μmol/L) (Figure 4A). T265-2c and HsSch2 cells differed in their relative sensitivity to canertinib, with HsSch2 cells being more sensitive. In contrast, significant reductions in cell numbers were identified in all four MPNST lines challenged with 1 to 10 μmol/L sapitinib, although these lines differed in their relative sensitivity to this agent (Figure 4B). Similarly, cell numbers were reduced in all four MPNST cell lines treated with trifluoperazine with marked reductions in cell numbers seen at the highest concentration tested (Figure 4C). Both NF1-associated (S462 and T265-2c) MPNST cell lines also demonstrated significant reductions in cell numbers when challenged with AZD1208 (Figure 4D). In contrast, the two sporadic MPNST cell lines were relatively insensitive to AZD1208, with STS-26T cells showing a small, but significant, reduction in cell numbers only at the highest concentration tested and HsSch2 cells showing no significant response at any of the concentrations tested.
Figure 4.
ErbB, calmodulin, and proviral integration site of Moloney murine leukemia (PIM) inhibitors reduce cell numbers in malignant peripheral nerve sheath tumor (MPNST) cultures. Normalized direct cell counts of four MPNST cell lines after 7 days of exposure to vehicle [dimethyl sulfoxide (DMSO)], canertinib, sapitinib, trifluoperazine, or AZD1208. A: Effects of the pan-erbB inhibitor canertinib (500 nmol/L, 1 μmol/L, or 2 μmol/L concentrations) on cell numbers after 7 days of treatment. B: Effects of the epidermal growth factor receptor/erbB2/erbB3 inhibitor sapitinib (1, 5, or 10 μmol/L concentrations) on cell numbers after 7 days of treatment. C: Effects of the calmodulin inhibitor trifluoperazine (2.5, 5, or 10 μmol/L concentrations) on cell numbers after 7 days of treatment. D: Effects of the pan-PIM kinase inhibitor AZD1208 (5, 10, or 15 μmol/L concentrations) on cell numbers after 7 days of treatment. Statistical significance represents the comparison of each monotherapy to the vehicle treated. Only those data with statistical significance are designated. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001.
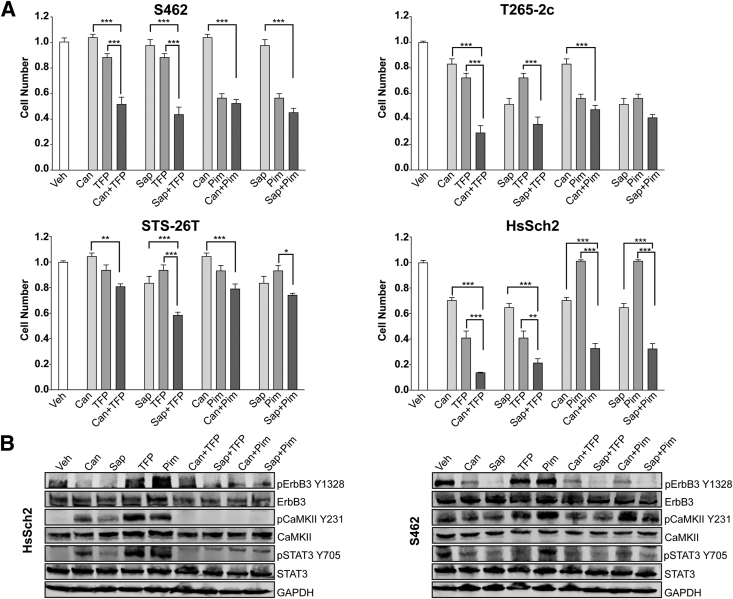
Whether combinatorial treatment with the erbB inhibitors and trifluoperazine or AZD1208 would more effectively reduce MPNST cell numbers compared with treatment with each individual agent was examined next. For this, direct cell counts of NF1-associated (S462 and T265-2c) were compared with sporadic (STS-26T and HsSch2) MPNST cells in cultures challenged with 1 μmol/L canertinib or 5 μmol/L sapitinib alone or in combination with 5 μmol/L trifluoperazine or 7.5 μmol/L AZD1208 (Figure 5A). In S462 and T265-2c cells, combining either canertinib or sapitinib with trifluoperazine at the middle dosages of each drug more effectively reduced cell numbers than either the erbB inhibitor or trifluoperazine alone at the highest dosages of the drugs. In STS-26T cells, although the combination of canertinib and trifluoperazine was not significantly differently from trifluoperazine alone, the combination of sapitinib and trifluoperazine did significantly reduce cell numbers compared with either sapitinib or trifluoperazine alone. In contrast, combinatorial therapies that used AZD1208 with canertinib or sapitinib did not produce results significantly different from those observed with the most effective monotherapy in S462, T265-2c, and STS-26T cells. In the high erbB3-expressing HsSch2 cell line, however, all four combinatorial therapies were superior to monotherapy with any of the four agents.
Figure 5.
Treatment with a combination of erbB inhibitors with trifluoperazine (TFP) or AZD1208 variably affects cell numbers and the phosphorylation of key signaling molecules. A: Normalized direct cell counts of four malignant peripheral nerve sheath tumor cell lines 48 hours after exposure to vehicle (Veh; 0.1% dimethyl sulfoxide), 1 μmol/L canertinib (Can), 5 μmol/L sapitinib (Sap), 5 μmol/L TFP, 7.5 μmol/L AZD1208, or combinations of these agents; all combinatorial treatments used half the concentration of each drug used in the monotherapy treatments. Statistical significance represents the comparison of each monotherapy (canertinib or trifluoperazine) to the drug combination. Only those data with statistical significance are designated. B: Immunoblot analyses of lysates of cells treated for 48 hours with the indicated monotherapies or combination therapies. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001. CaMKII, calmodulin-dependent protein kinase IIα; p, phosphorylated; Pim, proviral integration site of Moloney murine leukemia.
To verify that the combinatorial therapies were having the expected effects on the phosphorylation of their targets, lysates of NF1-associated (S462) and sporadic (HsSch2) MPNST cells were immunoblotted and probed with antibodies recognizing phosphorylated erbB3, CaMKII, or STAT3, a protein previously shown to be activated by erbB receptors.11 As expected, canertinib and sapitinib, both alone and as part of combinatorial therapies, reduced erbB3-Y1328 phosphorylation (Figure 5B). Surprisingly, however, both trifluoperazine and AZD1208 modestly increased erbB3 phosphorylation in both lines. Furthermore, canertinib, sapitinib, trifluoperazine, and AZD1208 unexpectedly increased the phosphorylation of CaMKII at residue Y231 in HsSch2 cells and to a lesser extent in S462 cells. In contrast, all four combinatorial therapies abolished CaMKII phosphorylation in HsSch2 cells, but not in S462 cells. The effects of these agents on STAT3 phosphorylation (residue Y705) also differed between the two lines. In HsSch2 cells, the effects of the monotherapies and combinatorial therapies on STAT3 phosphorylation paralleled the effects of these agents on CaMKII phosphorylation. In S462 cells, however, AZD1208 had no effect on STAT3 phosphorylation, whereas canertinib, sapitinib, trifluoperazine, and all four combinatorial therapies reduced STAT3 phosphorylation relative to the vehicle control.
ErbB3 Knockdown in Combination with Trifluoperazine Promotes Cell Death
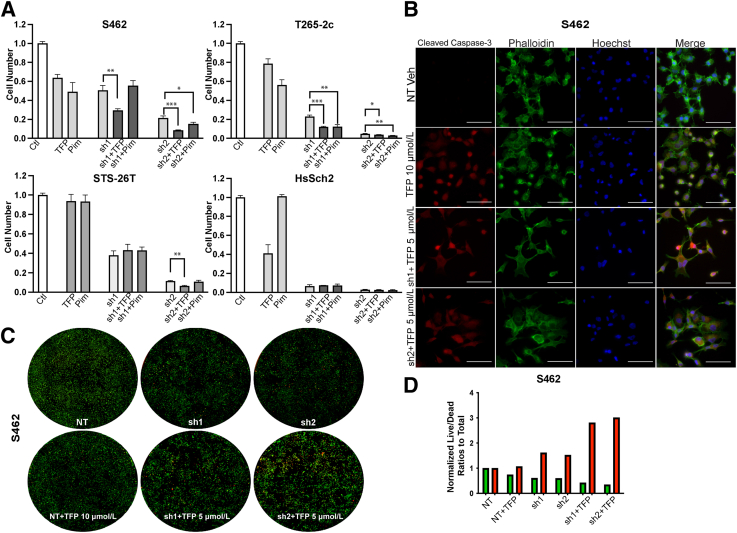
Although canertinib and sapitinib inhibit erbB3, they also inhibit the action of other erbB kinases. Consequently, the possibility that the effects noted with erbB inhibitors reflect canertinib or sapitinib actions on an erbB receptor other than erbB3 could not be ruled out. Unfortunately, although several monoclonal antibodies targeting erbB3 are in development, inhibitors that solely target erbB3 are not yet commercially available. To determine whether inhibition of erbB3 in combination with trifluoperazine or AZD1208 had an enhanced effect on MPNST cell proliferation and/or survival, the drug treatment was combined drug treatment with erbB3 loss using two NF1-associated and two sporadic MPNST cell lines transduced with erbB3 shRNAs or a nontargeting control. After allowing adequate time for erbB3 knockdown, these cells were challenged with vehicle, trifluoperazine, or AZD1208 and the number of cells present were determined after 7 days of treatment (Figure 6A). In the NF1-associated S462 cell line, TFP treatment combined with erbB3 knockdown reduced cell numbers to a greater extent than erbB3 knockdown or trifluoperazine treatment alone. However, treatment with AZD1208 did not further reduce the number of S462 cells transduced with erbB3 shRNAs. In contrast, the addition of both trifluoperazine and AZD1208 to cultures of T265-2c expressing erbB3 shRNAs led to a decrease in the total number of viable cells. In STS-26T cells, neither trifluoperazine nor AZD1208 treatment of cells with erbB3 knockdown produced statistically significant reductions in cell numbers compared with the erbB3 sh1 alone; however, the combination of sh2 and trifluoperazine did produce a further significant decrease in cell numbers. In HsSch2 cells, trifluoperazine and ADZ1208 also failed to further reduce cell numbers relative to the erbB3 shRNAs alone. However, because both shRNAs potently reduced cell numbers, the profound knockdown of erbB3 in this line may have masked additional effects induced by trifluoperazine or ADZ1208.
Figure 6.
Effect of erbB3 knockdown in combination with trifluoperazine (TFP) or AZD1208 treatment on cell numbers. A: Normalized direct cell counts of four malignant peripheral nerve sheath tumor cell lines transduced with erbB3 shRNAs (sh1 or sh2) or a nontargeting control (Ctl) and then treated for 48 hours with 5 μmol/L TFP or 7.5 μmol/L AZD1208. Statistical significance represents the comparison of each shRNA (sh1 or sh2) to the drug with shRNA combination. Only those data with statistical significance are designated. B: Immunostains for activated (cleaved) caspase-3 in S462 cells transduced with erbB3 shRNAs or a nontargeting control and then treated for 48 hours with 5 μmol/L TFP or 7.5 μmol/L AZD1208. Cells transduced with a nontargeting shRNA and then treated with vehicle (Veh) were used as a control. C: Representative wells of S462 cells transduced with erbB3 shRNAs or a nontargeting control, treated for 48 hours with 5 μmol/L TFP and then exposed to calcein AM to label live cells (green) and BOBO-3 iodide (red) to label dead cells. D: Green (live) and red (dead) separated channel measurements analyzed with particle count Fiji ImageJ version 2.9.0 analyses in S462 cells transduced with erbB3 shRNAs (sh1 and sh2) or NT, treated with vehicle or 10 μmol/L TFP alone and then exposed to calcein AM and BOBO-3 iodide. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001. Scale bars = 20 μm (B). NT, nontargeting control shRNA; Pim, proviral integration site of Moloney murine leukemia.
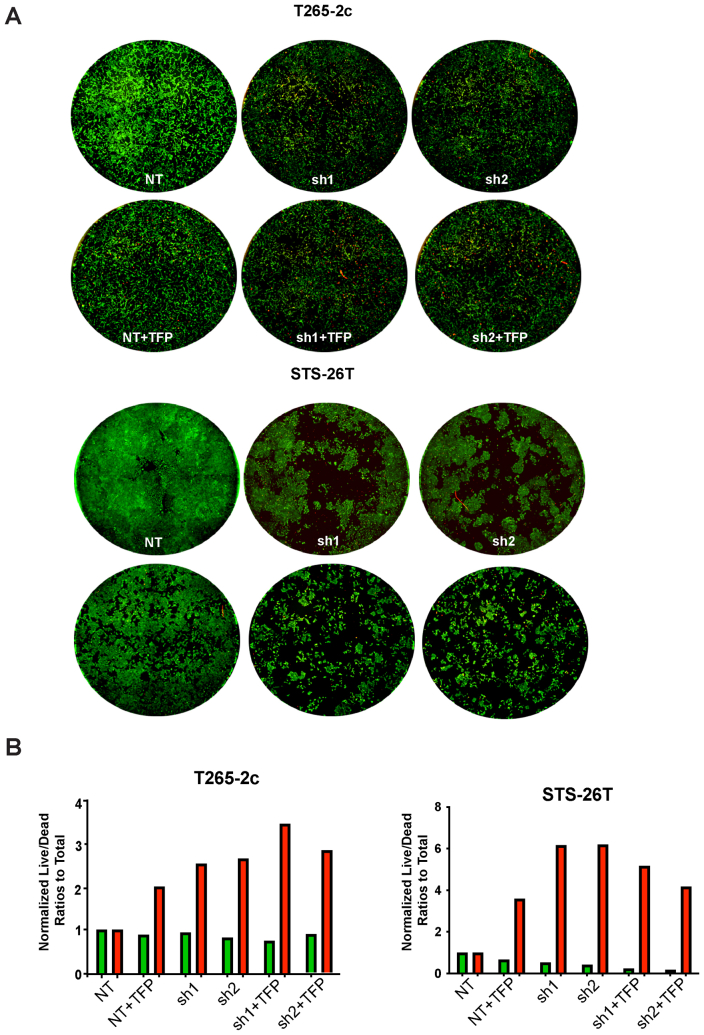
Because the combination of erbB3 knockdown and trifluoperazine treatment produced the greatest reduction in cell numbers in the NF1-associated S462 and T265-2c lines, whether this reduction in cell numbers reflected, at least in part, increased cell death was investigated next. S462 cells with erbB3 or nontargeting shRNAs were transduced, treated with trifluoperazine, and immunostained for cleaved caspase-3 (Figure 6B). The combination of erbB3 knockdown and 5 μmol/L trifluoperazine markedly enhanced the number of cleaved caspase-3 immunoreactive cells compared with the nontargeting control or cells treated with 10 μmol/L trifluoperazine alone. Live/dead assays were performed to quantify the effect of combined erbB3 knockdown and trifluoperazine treatment. For this, cells were exposed to calcein AM and propidium iodide staining, and the calcein and propidium iodide signals were then quantified using a Celigo imaging cytometer. Examination of the stained wells demonstrated a marked decrease in the number of viable cells and an increase in dead cells (Figure 6C). Although the number of dead cells was increased by the erbB3 shRNAs relative to the nontargeting control and trifluoperazine alone, erbB3 knockdown in combination with trifluoperazine treatment even more potently induced cell death (Figure 6D). Similar results were obtained with T265-2c and STS-26T cells (Supplemental Figure S7).
Trifluoperazine Activates Multiple Src Family Kinases, which Results in Enhanced ErbB3 Phosphorylation
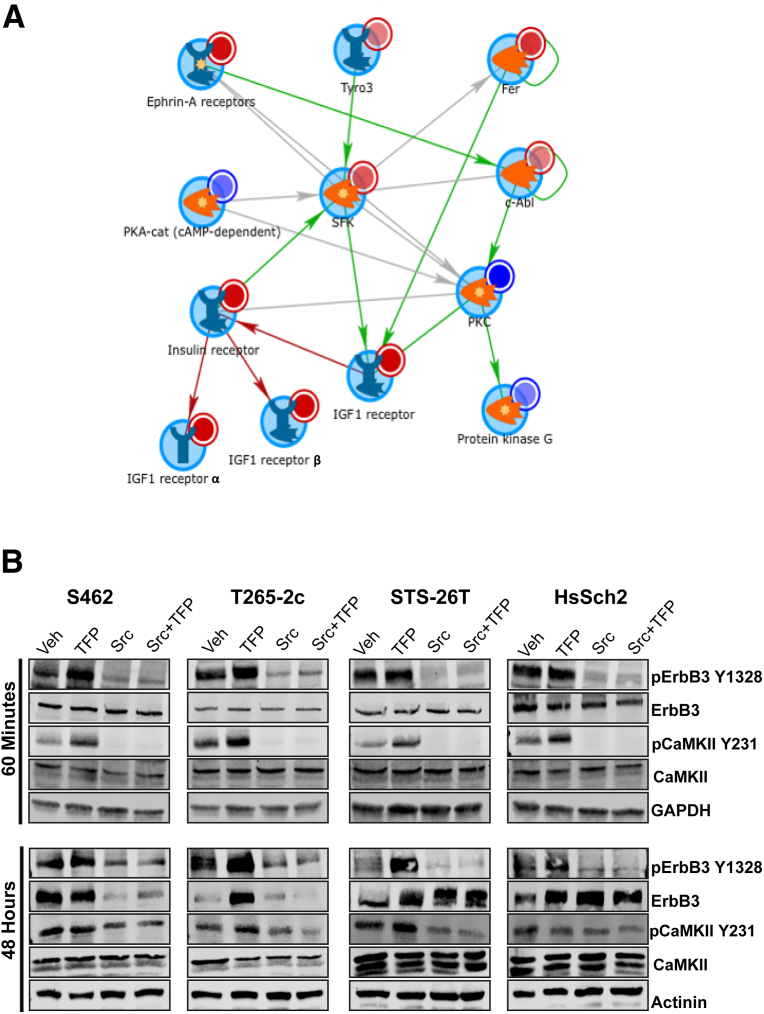
Trifluoperazine significantly decreases the proliferation and survival of MPNST cells in vivo and in vitro,12,13 effects that are associated with the inactivation of multiple kinases.12 However, the observation that trifluoperazine enhances erbB3 phosphorylation suggested that this drug likely also activates one or more kinases, which leads to enhanced phosphorylation of erbB3 and other proteins, such as CaMKII. To identify kinases whose activity is increased by trifluoperazine treatment, previously generated kinomic data sets were re-analyzed using updated algorithms that have become available since their original publication. This re-analysis identified multiple kinases that demonstrated increased or decreased activity following trifluoperazine treatment (Supplemental Figure S8). In keeping with previous observations, trifluoperazine treatment of ST88-14 MPNST cells decreased the activity of kinases regulating mammalian target of rapamycin–, ERK-, and calmodulin-mediated signaling. However, as indicated in Table 2, trifluoperazine increased the activity of the nonreceptor tyrosine kinase Syk, two RTKs (the insulin-like growth factor 1 receptor and insulin receptor-related protein), and five members of the Src family of kinases (Src, Yes, Fyn, HCK, and BLK). Network modeling with MetaCore was used to define how the trifluoperazine-sensitive kinases mapped within known annotated networks. This MetaCore analysis identified an Src family kinase–centric network (Figure 7A) that connected with the insulin-like growth factor 1 receptor. As this network contained most of the kinases whose activity was enhanced by trifluoperazine treatment, this suggested that one or more Src family members might be required for the increased phosphorylation of CaMKII-Y231 and erbB3-Y1328.
Table 2.
Kinase Signaling Increased by Trifluoperazine Relative to Vehicle Treatment
| Kinase | Uniprot ID | Protein type | Mean final score | Mean kinase statistic |
|---|---|---|---|---|
| Syk | P43405 | NRTK | 2.512 | 1.416 |
| IRR | P14616 | RTK | 2.510 | 2.093 |
| IGF1R | P08069 | RTK | 2.494 | 1.756 |
| HCK | P08631 | SFK | 2.051 | 1.445 |
| BLK | P51451 | SFK | 1.680 | 1.484 |
| Fyn | P06241 | SFK | 1.623 | 1.503 |
| Src | P12931 | SFK | 1.588 | 1.386 |
| Yes | P07947 | SFK | 1.535 | 1.465 |
Scores generated with the algorithms indicated in Materials and Methods denote the likelihood that the indicated kinase is differentially affected. Bold entries are Src family kinases (SFK).
BLK, B lymphoid tyrosine kinase; Fyn, FYN oncogene related to SRC, FGR, YES; HCK, hematopoietic cell kinase, Src family tyrosine kinase; ID, identifier; IGF1R, insulin-like growth factor 1 receptor; IRR, insulin receptor-related protein; NRTK, non-receptor tyrosine kinase; RTK, receptor tyrosine kinase; Src, v-Src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog; Syk, spleen associated tyrosine kinase; Yes, Yamaguchi sarcoma viral oncogene homolog 1.
Figure 7.
Kinomics, MetaCore, and Src inhibitor analyses implicating Src family kinases in basal and trifluoperazine-induced erbB3 and calmodulin-dependent protein kinase IIα (CaMKII) phosphorylation. A: MetaCore analysis of the data shown identifies an Src-centric network in which the insulin-like growth factor 1 (IGF1) receptor and several Src family kinases show increased activity in response to trifluoperazine (TFP) treatment. B: Immunoblot analyses of four malignant peripheral nerve sheath tumor cell lines treated for with vehicle (Veh), 10 μmol/L TFP, 10 μmol/L saracatinib (Src), or preloaded for 60 minutes with 5 μmol/L saracatinib and then treated with 5 μmol/L trifluoperazine (Src + TFP). Cells were harvested 60 minutes and 48 hours after beginning treatment. Lysates were probed with antibodies recognizing erbB3-Y1328 phosphorylation, total erbB3, CaMKII-Y231 phosphorylation, or total CaMKII. Actinin was used as a loading control. car, catalytic; c-Abl, cellular Abelson murine leukemia viral oncogene homolog 1; Fer, FER tyrosine kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; p, phosphorylated; PKA, protein kinase A; PKC, protein kinase C; SFK, Src family kinase; Tyro3, TYRO3 protein tyrosine kinase.
To determine whether Src family kinase activity is required for the increased erbB3 and CaMKII phosphorylation observed in trifluoperazine-treated MPNST cells, two NF1-associated (S462 and T265-2c) and two sporadic (STS-26T and HsSch2) MPNST cell lines were treated either with vehicle or a 5 μmol/L concentration of saracatinib, which inhibits the Src family of kinases, for 60 minutes (an initial preload with saracatinib). Vehicle-treated cells were then challenged for another 60 minutes or 48 hours with vehicle, 10 μmol/L trifluoperazine, or 10 μmol/L saracatinib monotherapy treatments, whereas saracatinib preloaded cells were challenged with 5 μmol/L trifluoperazine. Lysates prepared from the cells were immunoblotted before being probed with antibodies recognizing the proteins of interest. As in previous experiments, a 60-minute treatment with trifluoperazine enhanced the phosphorylation of erbB3-Y1328 and CaMKII-Y231 in all four cell lines, while having no effect on the total levels of erbB3 and CaMKII (Figure 7B). However, saracatinib markedly reduced erbB3 phosphorylation in these lines and completely abolished CaMKII phosphorylation. After 48 hours of treatment, saracatinib similarly reduced levels of ErbB3-Y1328. Furthermore, saracatinib treatment also led to reduced levels of total erbB3 protein in S462 and T265-2c cells. Similar reductions in erbB3 and CaMKII phosphorylation were evident in cells treated with both saracatinib and trifluoperazine. In contrast, saracatinib, alone and in combination with trifluoperazine, also reduced levels of erbB3-Y1328. However, in these sporadic MPNST cell lines, 48 hours of treatment with trifluoperazine, saracatinib, or trifluoperazine plus saracatinib increased the amount of total erbB3 present.
Combinatorial Treatment with Trifluoperazine and Saracatinib Reduces MPNST Cell Numbers and Enhances Cell Death Compared with Monotherapy with Either Agent
Although the above mentioned results indicated that erbB3 knockdown effectively inhibited the proliferation and survival of the four MPNST cell lines tested, agents that specifically target erbB3 are not commercially available. However, the observation that the Src inhibitor saracatinib effectively inhibited erbB3 phosphorylation suggested that this agent could have effects on MPNST cells similar to those produced by erbB3 knockdown. To test this possibility, S462, T265-2c, STS-26T, and HsSch2 cells were treated with a range of concentrations of saracatinib and quantified cell numbers 1, 3, 5, and 7 days after beginning treatment (Supplemental Figure S6). After establishing the concentration range of saracatinib that was potentially effective, these same cell lines were challenged with vehicle or 500 nmol/L, 5 μmol/L, or 10 μmol/L saracatinib for 7 days and the number of cells present were determined using a Celigo imaging cytometer. Saracatinib reduced cell numbers in all four MPNST cell lines in a concentration-dependent manner (Figure 8A).
Figure 8.
Saracatinib reduces malignant peripheral nerve sheath tumor (MPNST) cell numbers and viability, and a combination of saracatinib and trifluoperazine (TFP) even more potently reduces cell numbers and viability. A: Normalized direct cell counts of four MPNST cell lines treated with vehicle [Veh; dimethyl sulfoxide (DMSO)] or saracatinib (500 nmol/L, 5 μmol/L, and 10 μmol/L concentrations). Statistical significance represents the comparison of each monotherapy to the vehicle treated. Only those data with statistical significance are designated. B: Normalized direct cell counts of four MPNST cell lines treated with vehicle, 5 μmol/L TFP, 5 μmol/L saracatinib (Src), 7.5 μmol/L AZD1208 [proviral integration site of Moloney murine leukemia (Pim)], or the combinations of these drugs. Statistical significance represents the comparison of each monotherapy (trifluoperazine or saracatinib) to the drug combination. C: HsSch2 cells were treated with vehicle, 10 μmol/L saracatinib (Src), or a combination of 5 μmol/L saracatinib and 5 μmol/L trifluoperazine. Cells were then immunostained for cleaved caspase-3 (red) and counterstained with phalloidin (labels actin cytoskeleton) and Hoechst 33342 (nuclear dye). Images taken with a 63× oil immersion objective. D: Top panels: Representative wells of HsSch2 cells treated for 48 hours with Veh, 10 μmol/L saracatinib (Src), or a combination of 5 μmol/L saracatinib and 5 μmol/L trifluoperazine and then exposed to calcein AM to label live cells (green) and BOBO-3 iodide (red) to label dead cells. Bottom panel: Green (live) and red (dead) separated channel measurements were analyzed with particle count Fiji ImageJ version 2.9.0 analyses (bar graph). ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001. Scale bars = 20 μm (C). Ns, not significant.
Trifluoperazine decreased cell numbers in all four MPNST lines (Figure 4C), AZD1208 decreased the proliferation of both NF1-associated MPNST lines (Figure 4D), and both trifluoperazine and AZD1208 increased erbB3 phosphorylation (Figure 5B). Therefore, the next set of experiments were conducted to determine whether combining Src inhibition via saracatinib with the calmodulin inhibitor trifluoperazine or the PIM kinase inhibitor AZD1208 would more effectively decrease cell numbers compared with monotherapy with any of these three agents. For these experiments, two NF1-associated MPNST lines (S462 and T265-2c) and two sporadic MPNST lines (STS-26T and HsSch2) were challenged with vehicle, trifluoperazine alone (TFP), saracatinib alone (Src), ADZ1208 alone (PIM), saracatinib plus trifluoperazine, or a combination of saracatinib and AZD1208. Seven days after initiating therapy, the viable cell numbers in each condition were determined using a Celigo imaging cytometer. In all four cell lines, the reduction in cell numbers in response to the Src inhibitor saracatinib in combination with the calmodulin inhibitor TFP was significantly higher than seen in cells challenged with saracatinib or trifluoperazine alone, which was higher than that seen with vehicle-treated cells (Figure 8B). In contrast, responses to combined treatment with saracatinib (Src) and AZD1208 (PIM) were more variable. In S462 cells, the combination of saracatinib and trifluoperazine reduced cell numbers that were significantly lower than treatment with saracatinib or trifluoperazine alone. However, in T265-2c, STS-26T, and HsSch2 cells, the combination of saracatinib and AZD1208 did not reduce cell numbers beyond those obtained with the most effective monotherapy (Figure 4).
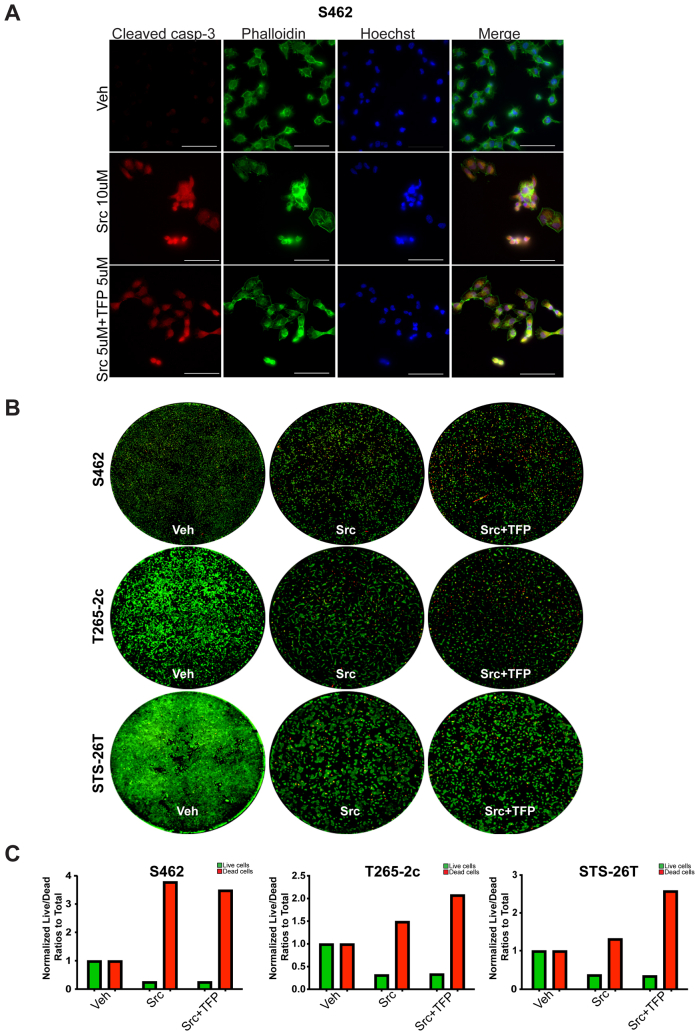
Whether treatment with saracatinib or a combination of saracatinib and trifluoperazine reduced cell numbers, at least in part, by increasing cell death was investigated next. HsSch2 MPNST cells were challenged with vehicle, 10 μmol/L saracatinib, or 5 μmol/L saracatinib in combination with 5 μmol/L trifluoperazine and were immunostained with an antibody recognizing cleaved caspase-3 (Figure 8C). Both 10 and 5 μmol/L saracatinib in combination with 5 μmol/L trifluoperazine markedly increased immunoreactivity for cleaved caspase-3. On the basis of this observation, live/dead assays were performed on HsSch2 cells treated with saracatinib or saracatinib plus trifluoperazine. Both saracatinib and the combination of saracatinib and trifluoperazine significantly increased the number of dead cells relative to that observed in vehicle-treated controls (Figure 8D). Furthermore, cell death in cultures treated with both saracatinib and trifluoperazine was significantly higher than death in cultures exposed to saracatinib alone. Similar findings were obtained from T265-2c and STS-26T cells (Supplemental Figure S9).
Discussion
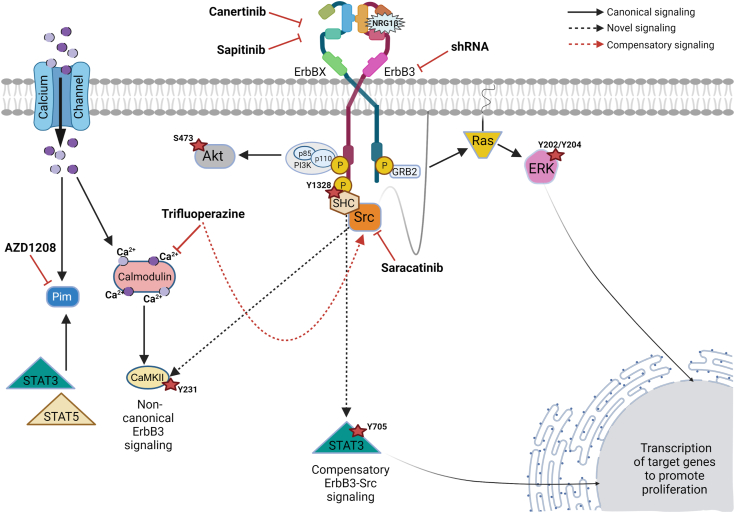
ErbB3 is a powerful oncogenic driver that promotes proliferation, invasion, and drug resistance in several types of human cancers.27 In keeping with the initial results of the genome-scale shRNA screens, the current study shows that erbB3 is commonly expressed in MPNSTs, it is overexpressed in many MPNST cell lines, it is essential for the proliferation and survival of human MPNST cells, and erbB inhibitors, such as canertinib and sapitinib, and erbB3 knockdown inhibit MPNST proliferation and survival. Microarray, kinomic, and immunoblot analyses provided evidence that erbB3 regulates calcium-calmodulin signaling effectors, that the calmodulin inhibitor trifluoperazine and the PIM kinase inhibitor AZD1208 inhibit MPNST proliferation and survival, and that combining erbB3 inhibition, via either pharmacologic inhibition of erbB receptors or erbB3 knockdown, with trifluoperazine inhibits MPNST proliferation and survival even further. Kinomics data showed that trifluoperazine activates multiple Src family kinases (Src, Fyn, Yes, HCK, and BLK); inhibition of these kinases with saracatinib potently reduces erbB3 and CaMKII phosphorylation and blocks trifluoperazine-induced phosphorylation of erbB3 and CaMKII. As with erbB3 knockdown, saracatinib potently inhibited MPNST proliferation and survival; and combined treatment with saracatinib and trifluoperazine was superior to monotherapy with either of these agents. The current study thus identified erbB3, calmodulin, PIM kinases, and Src family kinases as important therapeutic targets in MPNSTs. However, it also raised important new questions, such as how erbB3 protein levels are regulated in MPNST cells, the functions of the different erbB3 isoforms present in MPNST cells, the factors responsible for the variability that we observed in responses to some of our candidate therapeutic agents and combinatorial therapies, and how signaling molecules, such as CaMKII, are phosphorylated in MPNST cells.
We14,15 and others28 have reported that the expression of erbB3 protein is increased in plexiform neurofibromas, MPNSTs, and MPNST cell lines. In contrast, the expression of erbB3 mRNA is decreased in MPNSTs.29 To resolve this apparent contradiction, erbB3 immunoreactivity was examined in surgically resected MPNSTs using two antibodies targeting different epitopes on the erbB3 receptor. In keeping with the previous finding that 80% of the surgically resected MPNSTs we stained were immunoreactive for erbB3 using the RTJ.1 monoclonal antibody,15 both a rabbit anti-erbB3 polyclonal antibody and the 5A12 mouse monoclonal antibody stained 78% of the MPNSTs examined and all three antibodies produced highly similar patterns of staining. A fourth antibody, the rabbit D22C5 XP monoclonal antibody, produced the same pattern of staining in two NF1-associated MPNST cell lines and two sporadic MPNST cell lines. Furthermore, the immunoblot analyses of 16 human MPNST cell lines showed that erbB3 protein levels in 13 of the 16 MPNST lines were higher than those in nonneoplastic Schwann cells. These findings thus support previous reports that erbB3 protein expression is commonly elevated in human MPNSTs. Intriguingly, however, the real-time PCR analyses also demonstrated that erbB3 mRNA levels in 10 of these same 16 human MPNST cell lines were lower than those observed in nonneoplastic Schwann cells. The findings regarding erbB3 protein and transcript expression in MPNSTs are thus consistent with all of the previous studies cited. Given the disparity between the levels of erbB3 protein and mRNA expression in MPNST cell lines, it seems likely that the increased erbB3 protein expression evident in MPNST cells is the result of a post-transcriptional regulatory mechanism.
In addition to the expected membranous immunoreactivity, erbB3 immunoreactivity was also observed in the cytoplasm and nucleus of MPNST cells. These observations were in keeping with the cell fractionation experiments, which also identified erbB3 protein in cytoplasmic and nuclear fractions. The presence of the 185-kDa form of erbB3 in the cytoplasm is consistent with the previous demonstration that erbB proteins are regulated by endocytosis and that erbB3 is constitutively internalized, after which its degradation or stabilization is dependent on the level of protein kinase C activity.30 Nuclear localization of erbB3 protein has also been reported in several types of human cancer22,31,32 and was recently shown to be negatively regulated by NRG1.33 Although some intranuclear forms of erbB3 may result from proteolytic cleavage of full-length erbB3 protein, others are truncated forms of erbB3 that are translated from one of several different alternatively spliced mRNAs,34, 35, 36 resulting in the generation of proteins that vary in mass from 38 to 90 kDa. A 38-kDa erbB3-related protein predominantly present in the nuclear fraction was identified in MPNST cells. That the levels of this 38-kDa species were markedly reduced in MPNST cells transduced with erbB3 shRNAs strongly supports the identification of this protein as a truncated form of erbB3. Because the epitope recognized by this erbB3 antibody is in the C-terminal tail of the erbB3 protein and the two shRNAs that abolish its expression target sequences encoding the extreme C-terminal region of the tyrosine kinase domain and in the 3′ untranslated region, respectively, it is likely that the 38-kDa erbB3 species encompasses the carboxy terminal region of the protein rather than the extracellular domain. At present, the role of this 38-kDa species is not known. This question will be of considerable interest in future studies.
The demonstration that erbB3 knockdown profoundly reduced MPNST cell proliferation and survival in two NF1-associated and two sporadic MPNST cell lines identified erbB3 as an important therapeutic target in MPNSTs. This was further corroborated by the demonstration that treatment with the pan-erbB inhibitor canertinib and the EGFR/erbB2/erbB3 inhibitor sapitinib also reduced cell numbers. However, it is notable that canertinib and sapitinib differed in their relative ability to reduce MPNST cell numbers, with sapitinib being universally more effective. The reason behind this is unclear at present. One possibility is that this reflects the differential affinity of these drugs for erbB3 and erbB4; sapitinib targets erbB3 with a much lower half-maximal inhibitory concentration (IC50), whereas canertinib has a lower IC50 for erbB4. Canertinib is a pan-erbB inhibitor (inhibits EGFR, erbB2, erbB3, and erbB4) while sapatinib only inhibits EGFR, erbB2, and erbB3.11,15 Another possibility is that a differential effect of these drugs on erbB3/erbB4 heterodimers might explain their varying effectiveness. However, because erbB4 also promotes MPNST cell proliferation,11 canertinib rather than sapitinib could be the more effective agent. Other possibilities include differences in the half-life of these two drugs, a varying ability of the MPNST cells to metabolize canertinib and sapitinib, and differences in the ability of canertinib and sapitinib to be taken up by MPNST cells. These observations underline the need for future studies comparing the effectiveness of the multiple erbB inhibitors that are currently available in several cell lines and animal models.
Calmodulin inhibitors, such as trifluoperazine and tamoxifen, inhibit MPNST cell proliferation and survival in vitro and in xenografted mice.12,13 These studies, together with the finding that basal and NRG1β-stimulated erbB3 activity promotes the phosphorylation of CaMKII, prompted testing of the effectiveness of trifluoperazine and the PIM kinase inhibitor AZD1208. Trifluoperazine markedly reduced cell numbers and survival in all four of the MPNST lines tested and AZD1208 was effective against the NF1-associated S462 and T265-2c MPNST cell lines, but not the two sporadic MPNST cell lines. In keeping with the observation that erbB3 activity modulates calcium-regulated signaling pathways, a combination of trifluoperazine with either erbB inhibitor (canertinib or sapitinib) reduced cell numbers to a greater extent than monotherapy in most of the cell lines tested. In contrast, the combination of AZD1208 with either (canertinib or sapitinib) was more effective only in HsSch2 cells. The variable responsiveness of MPNST cells to AZD1208, alone or as part of a combinatorial therapy with erbB inhibitors, suggests that there may be a distinct subset of MPNSTs that are sensitive to PIM kinase inhibitors. At present, PIM inhibitor sensitivity cannot be linked to a specific molecular subtype of MPNSTs because such molecular subtypes have not yet been defined for these neoplasms.
As noted above, trifluoperazine and AZD1208 enhanced or sustained the phosphorylation of erbB3-Y1328 and CaMKII-Y231. Because trifluoperazine also inhibits dopamine receptor D2 and dopamine receptor D2 is known to transactivate Src and EGFR, altered dopamine receptor D2 signaling may be one of several factors potentially involved in regulating these phosphorylation events. Also, the functional role of the phosphorylation of residue Y231 in CaMKII and Y1328 in erbB3 has not been established. However, the current findings suggest these sites modulate downstream signaling intensity or induce cellular stress responses that promote survival because these modifications occur at lower drug levels that do not induce the death of HsSch2 cells. In addition, NRG1- and erbB3-dependent activation of STAT3, an Src and Janus kinase (Jak) effector, was observed under serum-deprived conditions. Conversely, an increase in Src signaling was observed following trifluoperazine treatment that was associated with increased phosphorylated (p) STAT3-Y705 levels in HsSch2 cells. This is consistent with the finding that the phosphorylation of erbB3-Y1328 correlated with pSTAT3-Y705 levels under serum growth conditions. Canertinib and sapitinib treatment inhibited erbB3 phosphorylation and reduced, not completely ablated, pCaMKII and pSTAT3 phosphorylation. However, in S462 cells, different pCaMKII kinetics were observed with basal phosphorylation of this site that did not respond to combinatorial treatment, which differs from that observed with HsSch2 cells. Interestingly, S462 cells have copy number gains of the gene encoding CaMKII, which may contribute to this discrepancy (J.F.L., A.M. Prechtl, S.L.C., unpublished data).
The current findings shed light on the significance of understudied phosphorylation sites in both erbB3 (Y1328) and CaMKII (Y231). erbB3-Y1328 phosphorylation was constitutively, yet positively, responsive to NRG1β treatment. erbB3-Y1328 phosphorylation is associated Shc–growth factor receptor bound protein 2 (Grb2) binding, which is enhanced by NRG1β treatment.37 Herein, mutational analysis indicated that this site did not bind p85 and activate phosphatidylinositol 3-kinase signaling but instead regulated proliferation coupling to mitogen-activated protein kinase. CaMKII binds to ERK and promotes its phosphorylation and nuclear translocation.38 What is known about Y231-CaMKII phosphorylation is that this site is located within the kinase domain, and that CaMKII activity is regulated by a balance between activating and inhibitory autophosphorylation events that are unique for each of the four CaMKII isoforms (Thr286 and Thr305/306 for the α isoform).39 Paradoxically, the current study showed that trifluoperazine and AZD1208 sustained or enhanced erbB3-Y1328 and CaMKII-Y231 phosphorylation, whereas the erbB inhibitors reduced phosphorylation. Phosphorylation of Y1328 in erbB3 generates a docking site for SHC, which can, in turn, activate Src family kinases; saracatinib abolishes the phosphorylation of CaMKII-Y231, which suggests that erbB3-mediated activation of Src kinases is responsible for this CaMKII phosphorylation event (explained later). erbB inhibitors may be altering the erbB3 heterodimeric pairs because of their distinct binding ability for each erbB and the fact that trifluoperazine and AZD1208 are targeting downstream of these receptors. This is consistent with erbB3-Y1328 phosphorylation being constitutive. It is possible that in cancer cells with hyperactive Ras-ERK signaling, this allows CaMKII to be resistant to calmodulin and PIM inhibitors by coupling to ERK for sustained activation or transactivation.
NRG1 and its erbB3 receptor possibly induced CaMKII-Y231 phosphorylation via a novel calmodulin-independent mechanism. Trifluoperazine prevents the activation of calcium-calmodulin–related kinases, such as CaMKII, by blocking a calmodulin-dependent conformational change that allows the autophosphorylation of the kinase domain at T286 and CaMKII activation. However, because CaMKII is a multifunctional serine/threonine kinase, phosphorylation at tyrosine 231 is inconsistent with this being the result of an autophosphorylation event mediated by CaMKII itself. This is underlined by the fact that trifluoperazine treatment prevents CaMKII activation. On the other hand, the kinomics analyses indicated that trifluoperazine activated multiple Src family kinases that function as nonreceptor tyrosine kinases. Furthermore, the Src family inhibitor saracatinib abolished the baseline Y231 phosphorylation of CaMKII and prevented the trifluoperazine-induced increases in Y231 phosphorylation seen after 1 hour of trifluoperazine treatment. Given this, it seems likely that one or more Src family kinases mediate CaMKII-Y231 phosphorylation (Figure 9), which is in the CaMKII kinase domain. The effect(s) of Y231 phosphorylation on CaMKII activity and whether this site serves as a docking site for CaMKII interactions with one or more other proteins remains to be determined.
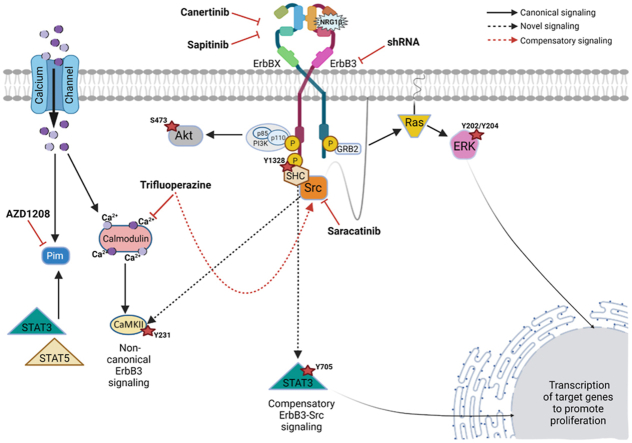
Figure 9.
Model of signaling pathways and therapeutic targets in malignant peripheral nerve sheath tumor cells downstream of erbB3. ErbBX refers to epidermal growth factor receptor, erbB2, or erbB4 heterodimeric pair complex. Dotted black lines indicate potentially novel signaling pathways. The red dotted line identifies the erbB3 compensatory pathway identified in this article. CaMKII, calmodulin-dependent protein kinase IIα; ERK, extracellular signal-regulated kinase; GRB2, growth factor receptor bound protein 2; NRG1, neuregulin-1; PI3K, phosphatidylinositol 3-kinase; Pim, proviral integration site of Moloney murine leukemia; SHC, Src homology 2 domain-containing transforming protein 1.
In summary, the current study identified erbB3, calmodulin, PIM kinases, and Src family kinases as important therapeutic targets in MPNSTs. As several drugs targeting these proteins are already in use clinically, it is possible that these findings can soon be moved from bench to bedside for the treatment of these highly aggressive spindle cell neoplasms. The current findings also indicate that combinatorial therapies targeting these proteins are promising, and future studies will test their effectiveness in animal models. A schematic of our understanding of the highly complex signaling network in which erbB3 operates is presented in Figure 9. However, these findings also raise some important new questions that need to be addressed in future studies. Although outside of the scope of this study, future work and expansion of study to determine what functions are performed by the truncated intranuclear form of erbB3 and to identify the post-transcriptional mechanisms by which erbB3 protein levels are elevated in MPNSTs may yield more insight into therapeutic target(s) in treatment of this extremely aggressive disease. It is also clear that our understanding of the mechanisms regulating CaMKII activity and interactions is incomplete. Given the critically important role that CaMKs play in the biology of multiple cell types and human cancers, clarifying the mechanisms involved in CaMKII regulation will have important implications that go well beyond understanding the biology of MPNSTs as well as further investigation into the erbB-calcium/CaMKII–mediated signaling contributing to the pathogenesis of the disease.
Footnotes
Supported by National Institute of Neurological Diseases and Stroke grants R01 NS048353 and R01 NS109655 (S.L.C.), the National Cancer Institute grant R01 CA122804 (S.L.C.), the Department of Defense grants X81XWH-09-1-0086 and W81XWH-12-1-0164 (S.L.C.), and the Children's Tumor Foundation grant 2020-01-005 (L.E.B.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2023.05.016.
Supplemental Data
Supplemental Figure S1.
ERBB3 mRNA features and location of sites targeted by shRNAs and encoding epitopes recognized by antibodies (Abs). Schematic of the ERBB3 mRNA (gray) indicating the coding region (red) and sequences encoding protein domains in erbB3 (green), and epitopes recognized by different erbB3 and phosphorylated erbB3 antibody (blue) targeting regions. Sites targeted by shRNAs are indicated in yellow. The black line at the top indicates locations measured in base pairs. The erbB3 sequence, NM_001982.4, is freely available to the public and was obtained from the National Center for Biotechnology Information's website (https://www.ncbi.nlm.nih.gov/gene/?term=erbb3). CDS, coding sequence; CRR, cysteine-rich region; GFR, growth factor region; JM, juxtamembrane; TM, transmembrane.
Supplemental Figure S2.
ErbB3 expression in surgically resected malignant peripheral nerve sheath tumors (MPNSTs) and MPNST cell lines. A: ErbB3 immunoreactivity in five surgically resected human MPNSTs assessed with two antibodies recognizing different epitopes in the protein. IgG Ctl indicates control immunostains in which the primary antibody has been replaced with nonimmune IgG. Images taken with a 40× objective lens. B: Confocal Airyscan 63× oil objective imaging demonstrating perinuclear and nuclear (Nuc) localization of erbB3 in MPNST cells. Blue channel (Hoechst; nuclei), red channel (erbB3), and green channel (phalloidin) are shown. C: Immunoblot analyses of nuclear and cytoplasmic (Cyt) fractions collected from three MPNST cell lines probed for erbB3, demonstrating the presence of full-length erbB3 (185 kDa; present in both nucleus and cytoplasm) and a truncated 38-kDa form of the protein (predominantly located in the nucleus). Lysates were also probed with antibodies recognizing the nuclear protein lamin B and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). D: Real-time PCR analysis showing relative levels of ERBB3 mRNA expression using nonisoform discriminatory PCR primers; mRNA levels are shown relative to nontransformed human Schwann cells as well as known low (T24 bladder cancer) and high (MCF-7 breast cancer) erbB3-expressing control cell lines. GAPDH mRNA was used as the normalization control. Scale bars: 100 μm (A); 20 μm (B). WCL, whole cell lysate.
Supplemental Figure S3.
Immunostaining controls. Immunostaining controls in HsSch2 and S462 malignant peripheral nerve sheath tumor cells in which the primary antibody was replaced with a nonimmune rabbit (Rb) IgG control or staining was performed with secondary antibody only. Cells have been counterstained with phalloidin, which labels the actin cytoskeleton; and Hoechst 33,342, which labels the nucleus. Scale bars = 20 μm.
Supplemental Figure S4.
Morphologic changes in malignant peripheral nerve sheath tumor cells following erbB3 knockdown. Phase-contrast images taken with an EVOS (Thermo Fisher Scientific, Waltham, MA) 10× objective, demonstrating decreased cell number 9 days after erbB3 knockdown compared with nontargeting shRNA control (NT). Note the changes in cell morphology and increased cellular debris in background. The identities of the cell lines are indicated to the left of each row of images. Scale bars = 400 μm. Sh1/2, erbB3 shRNAs.
Supplemental Figure S5.
Analyses of Schwann cell microarray data. Pathway and Gene Ontology (GO) utilizing EnrichR analyses of transcripts with expression that is altered in rat Schwann cells after 24 hours of treatment with 10 nmol/L neuregulin-1β. Pathways enriched included cytokine activity, growth factor receptor binding, growth factor activity, transcription factor protein/protein interactions, including STAT3, and calcium-mediated signaling pathways. These analyses also identified several cellular compartments involved with this signaling. The blue coloring represents those data sets that met significance (P < 0.5); the asterisks represent those data sets that still met significance after Benjamini-Hochberg correction.
Supplemental Figure S6.
Cell numbers in malignant peripheral nerve sheath tumor (MPNST) cultures 1 to 7 days after exposure to varying concentrations of canertinib, sapitinib, trifluoperazine, AZD1208, or saracatinib. Celigo cellular proliferation assay measurement of 7-day drug treatment for four MPNST cell lines. Biological replicates were normalized, and SEM was calculated. Veh, vehicle.
Supplemental Figure S7.
Live/dead analyses of malignant peripheral nerve sheath tumor cells transduced with erbB3 shRNAs (sh1 and sh2) and then challenged with vehicle or trifluoperazine (TFP). A: Representative images of live/dead T265-2c and STS-26T cells, as imaged on a Celigo high-throughput imager showing 48-hour nontargeting shRNA (NT) vehicle-treated controls and erbB3 knockdowns (sh1 and sh2) with and without 5 μmol/L TFP treatment. B: Live/dead staining for calcein AM (live cells; green) and BOBO-3 iodide (dead cells; red) analyses in T265-2c and STS-26T cells. Normalized live/dead cell ratios compared with total cell counts were analyzed with Fiji ImageJ version 2.9.0 direct particle measurement.
Supplemental Figure S8.
Kinomics analyses of neurofibromatosis type 1–associated ST88-14 malignant peripheral nerve sheath tumor cells treated with trifluoperazine (TFP). The activity of numerous kinases is decreased by trifluoperazine treatment (black/gray signals), with a smaller number of kinases demonstrating increased activity in response to trifluoperazine treatment (red/pink signals). Src, saracatinib treated.
Cell viability analyses of malignant peripheral nerve sheath tumor cells with saracatinib alone or saracatinib plus trifluoperazine (TFP). A: Immunocytochemistry for cleaved caspase-3 (casp-3; red staining) in S462 cells treated with vehicle (Veh), 10 μmol/L saracatinib (Src), or 5 μmol/L saracatinib in combination with 5 μmol/L trifluoperazine. The actin cytoskeleton was stained with phalloidin (green), and nuclei were labeled with Hoechst 33,342 (blue). Images taken with a 63× oil immersion objective. B: Representative images of wells containing S462, T265-2c, and STS-26T cells after performing live/dead assays, as imaged on a Celigo high-throughput imager. The illustrated images are of cells treated for 48 hours with vehicle, 10 μmol/L saracatinib, or 5 μmol/L saracatinib plus 5 μmol/L trifluoperazine. C: Live/dead assays [staining for calcein AM (live cells; green) and BOBO-3 iodide (dead cells; red)] were performed on S462, T265-2c, and STS-26T cells. Normalized live/dead cell ratios compared with total cell counts were analyzed with Fiji ImageJ version 2.9.0 direct particle measurement. Scale bars = 20 μm (A).
References
- 1.Longo J.F., Weber S.M., Turner-Ivey B.P., Carroll S.L. Recent advances in the diagnosis and pathogenesis of neurofibromatosis type 1 (NF1)-associated peripheral nervous system neoplasms. Adv Anat Pathol. 2018;25:353–368. doi: 10.1097/PAP.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee W., Teckie S., Wiesner T., Ran L., Prieto Granada C.N., Lin M., Zhu S., Cao Z., Liang Y., Sboner A., Tap W.D., Fletcher J.A., Huberman K.H., Qin L.X., Viale A., Singer S., Zheng D., Berger M.F., Chen Y., Antonescu C.R., Chi P. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M., Wang Y., Jones S., Sausen M., McMahon K., Sharma R., Wang Q., Belzberg A.J., Chaichana K., Gallia G.L., Gokaslan Z.L., Riggins G.J., Wolinksy J.P., Wood L.D., Montgomery E.A., Hruban R.H., Kinzler K.W., Papadopoulos N., Vogelstein B., Bettegowda C. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brossier N.M., Prechtl A.M., Longo J.F., Barnes S., Wilson L.S., Byer S.J., Brosius S.N., Carroll S.L. Classic ras proteins promote proliferation and survival via distinct phosphoproteome alterations in neurofibromin-null malignant peripheral nerve sheath tumor cells. J Neuropathol Exp Neurol. 2015;74:568–586. doi: 10.1097/NEN.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber S.M., Brossier N.M., Prechtl A., Barnes S., Wilson L.S., Brosius S.N., Longo J.F., Carroll S.L. R-Ras subfamily proteins elicit distinct physiologic effects and phosphoproteome alterations in neurofibromin-null MPNST cells. Cell Commun Signal. 2021;19:95. doi: 10.1186/s12964-021-00773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widemann B.C., Dombi E., Gillespie A., Wolters P.L., Belasco J., Goldman S., Korf B.R., Solomon J., Martin S., Salzer W., Fox E., Patronas N., Kieran M.W., Perentesis J.P., Reddy A., Wright J.J., Kim A., Steinberg S.M., Balis F.M. Phase 2 randomized, flexible crossover, double-blinded, placebo-controlled trial of the farnesyltransferase inhibitor tipifarnib in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Neuro Oncol. 2014;16:707–718. doi: 10.1093/neuonc/nou004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Widemann B.C., Salzer W.L., Arceci R.J., Blaney S.M., Fox E., End D., Gillespie A., Whitcomb P., Palumbo J.S., Pitney A., Jayaprakash N., Zannikos P., Balis F.M. Phase I trial and pharmacokinetic study of the farnesyltransferase inhibitor tipifarnib in children with refractory solid tumors or neurofibromatosis type I and plexiform neurofibromas. J Clin Oncol. 2006;24:507–516. doi: 10.1200/JCO.2005.03.8638. [DOI] [PubMed] [Google Scholar]
- 8.de Blank P.M.K., Gross A.M., Akshintala S., Blakeley J.O., Bollag G., Cannon A., Dombi E., Fangusaro J., Gelb B.D., Hargrave D., Kim A., Klesse L.J., Loh M., Martin S., Moertel C., Packer R., Payne J.M., Rauen K.A., Rios J.J., Robison N., Schorry E.K., Shannon K., Stevenson D.A., Stieglitz E., Ullrich N.J., Walsh K.S., Weiss B.D., Wolters P.L., Yohay K., Yohe M.E., Widemann B.C., Fisher M.J. MEK inhibitors for neurofibromatosis type 1 manifestations: clinical evidence and consensus. Neuro Oncol. 2022;24:1845–1856. doi: 10.1093/neuonc/noac165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattingly R.R., Kraniak J.M., Dilworth J.T., Mathieu P., Bealmear B., Nowak J.E., Benjamins J.A., Tainsky M.A., Reiners J.J., Jr. The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor PD184352 (CI-1040) selectively induces apoptosis in malignant schwannoma cell lines. J Pharmacol Exp Ther. 2006;316:456–465. doi: 10.1124/jpet.105.091454. [DOI] [PubMed] [Google Scholar]
- 10.Jessen W.J., Miller S.J., Jousma E., Wu J., Rizvi T.A., Brundage M.E., Eaves D., Widemann B., Kim M.O., Dombi E., Sabo J., Hardiman Dudley A., Niwa-Kawakita M., Page G.P., Giovannini M., Aronow B.J., Cripe T.P., Ratner N. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. 2013;123:340–347. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longo J.F., Brosius S.N., Black L., Worley S.H., Wilson R.C., Roth K.A., Carroll S.L. ErbB4 promotes malignant peripheral nerve sheath tumor pathogenesis via Ras-independent mechanisms. Cell Commun Signal. 2019;17:74. doi: 10.1186/s12964-019-0388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosius S.N., Turk A.N., Byer S.J., Longo J.F., Kappes J.C., Roth K.A., Carroll S.L. Combinatorial therapy with tamoxifen and trifluoperazine effectively inhibits malignant peripheral nerve sheath tumor growth by targeting complementary signaling cascades. J Neuropathol Exp Neurol. 2014;73:1078–1090. doi: 10.1097/NEN.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byer S.J., Eckert J.M., Brossier N.M., Clodfelder-Miller B.J., Turk A.N., Carroll A.J., Kappes J.C., Zinn K.R., Prasain J.K., Carroll S.L. Tamoxifen inhibits malignant peripheral nerve sheath tumor growth in an estrogen receptor-independent manner. Neuro Oncol. 2011;13:28–41. doi: 10.1093/neuonc/noq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert J.M., Byer S.J., Clodfelder-Miller B.J., Carroll S.L. Neuregulin-1 beta and neuregulin-1 alpha differentially affect the migration and invasion of malignant peripheral nerve sheath tumor cells. Glia. 2009;57:1501–1520. doi: 10.1002/glia.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stonecypher M.S., Byer S.J., Grizzle W.E., Carroll S.L. Activation of the neuregulin-1/ErbB signaling pathway promotes the proliferation of neoplastic Schwann cells in human malignant peripheral nerve sheath tumors. Oncogene. 2005;24:5589–5605. doi: 10.1038/sj.onc.1208730. [DOI] [PubMed] [Google Scholar]
- 16.Longo J.F., Brosius S.N., Znoyko I., Alers V.A., Jenkins D.P., Wilson R.C., Carroll A.J., Wolff D.J., Roth K.A., Carroll S.L. Establishment and genomic characterization of a sporadic malignant peripheral nerve sheath tumor cell line. Sci Rep. 2021;11:5690. doi: 10.1038/s41598-021-85055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frohnert P.W., Stonecypher M.S., Carroll S.L. Lysophosphatidic acid promotes the proliferation of adult Schwann cells isolated from axotomized sciatic nerve. J Neuropathol Exp Neurol. 2003;62:520–529. doi: 10.1093/jnen/62.5.520. [DOI] [PubMed] [Google Scholar]
- 18.Ibrahim A.N., Yamashita D., Anderson J.C., Abdelrashid M., Alwakeal A., Estevez-Ordonez D., Komarova S., Markert J.M., Goidts V., Willey C.D., Nakano I. Intratumoral spatial heterogeneity of BTK kinomic activity dictates distinct therapeutic response within a single glioblastoma tumor. J Neurosurg. 2019;133:1683–1694. doi: 10.3171/2019.7.JNS191376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar D.S., Chakravarthi B., Robinson A.D., Anderson J.C., Agarwal S., Balasubramanya S.A.H., Eich M.L., Bajpai A.K., Davuluri S., Guru M.S., Guru A.S., Naik G., Della Manna D.L., Acharya K.K., Carskadon S., Manne U., Crossman D.K., Ferguson J.E., Grizzle W.E., Palanisamy N., Willey C.D., Crowley M.R., Netto G.J., Yang E.S., Varambally S., Sonpavde G. Therapeutically actionable PAK4 is amplified, overexpressed, and involved in bladder cancer progression. Oncogene. 2020;39:4077–4091. doi: 10.1038/s41388-020-1275-7. [DOI] [PubMed] [Google Scholar]
- 20.Akhand S.S., Chen H., Purdy S.C., Liu Z., Anderson J.C., Willey C.D., Wendt M.K. Fibroblast growth factor receptor facilitates recurrence of minimal residual disease following trastuzumab emtansine therapy. NPJ Breast Cancer. 2021;7:5. doi: 10.1038/s41523-020-00213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen H., Huang F., Zhang X., Ojo O.A., Li Y., Trummell H.Q., Anderson J.C., Fiveash J., Bredel M., Yang E.S., Willey C.D., Chong Z., Bonner J.A., Shi L.Z. Selective suppression of melanoma lacking IFN-gamma pathway by JAK inhibition depends on T cells and host TNF signaling. Nat Commun. 2022;13:5013. doi: 10.1038/s41467-022-32754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Black L.E., Longo J.F., Carroll S.L. Mechanisms of receptor tyrosine-protein kinase ErbB-3 (ERBB3) action in human neoplasia. Am J Pathol. 2019;189:1898–1912. doi: 10.1016/j.ajpath.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., Jeon M., Ma'ayan A. Gene set knowledge discovery with enrichr. Curr Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayer K.U., Schulman H. CaM kinase: still inspiring at 40. Neuron. 2019;103:380–394. doi: 10.1016/j.neuron.2019.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandullo-Sanchez L., Ocana A., Pandiella A. HER3 in cancer: from the bench to the bedside. J Exp Clin Cancer Res. 2022;41:310. doi: 10.1186/s13046-022-02515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedrich R.E., Nornberg L.K.N., Hagel C. ERBB2 and ERBB3 growth factor receptors, neuregulin-1, CD44 and Ki-67 proliferation index in neurofibromatosis type 1-associated peripheral nerve sheath tumors. Anticancer Res. 2022;42:2327–2340. doi: 10.21873/anticanres.15712. [DOI] [PubMed] [Google Scholar]
- 29.Levy P., Vidaud D., Leroy K., Laurendeau I., Wechsler J., Bolasco G., Parfait B., Wolkenstein P., Vidaud M., Bieche I. Molecular profiling of malignant peripheral nerve sheath tumors associated with neurofibromatosis type 1, based on large-scale real-time RT-PCR. Mol Cancer. 2004;3:20. doi: 10.1186/1476-4598-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietrich M., Malik M.S., Skeie M., Bertelsen V., Stang E. Protein kinase C regulates ErbB3 turnover. Exp Cell Res. 2019;382 doi: 10.1016/j.yexcr.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Almadori G., Coli A., De Corso E., Mele D.A., Settimi S., Di Cintio G., Brigato F., Scannone D., Carey T.E., Paludetti G., Lauriola L., Ranelletti F.O. Nuclear HER3 expression improves the prognostic stratification of patients with HER1 positive advanced laryngeal squamous cell carcinoma. J Transl Med. 2021;19:408. doi: 10.1186/s12967-021-03081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Bacco F., Orzan F., Erriquez J., Casanova E., Barault L., Albano R., D'Ambrosio A., Bigatto V., Reato G., Patane M., Pollo B., Kuesters G., Dell'Aglio C., Casorzo L., Pellegatta S., Finocchiaro G., Comoglio P.M., Boccaccio C. ERBB3 overexpression due to miR-205 inactivation confers sensitivity to FGF, metabolic activation, and liability to ERBB3 targeting in glioblastoma. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109455. [DOI] [PubMed] [Google Scholar]
- 33.Tagliaferro M., Rosa P., Bellenchi G.C., Bastianelli D., Trotta R., Tito C., Fazi F., Calogero A., Ponti D. Nucleolar localization of the ErbB3 receptor as a new target in glioblastoma. BMC Mol Cell Biol. 2022;23:13. doi: 10.1186/s12860-022-00411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S.H., Cheng C.J., Lee Y.C., Ye X., Tsai W.W., Kim J., Pasqualini R., Arap W., Navone N.M., Tu S.M., Hu M., Yu-Lee L.Y., Logothetis C.J. A 45-kDa ErbB3 secreted by prostate cancer cells promotes bone formation. Oncogene. 2008;27:5195–5203. doi: 10.1038/onc.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasan R., Leverton K.E., Sheldon H., Hurst H.C., Sarraf C., Gullick W.J. Intracellular expression of the truncated extracellular domain of c-erbB-3/HER3. Cell Signal. 2001;13:321–330. doi: 10.1016/s0898-6568(01)00155-3. [DOI] [PubMed] [Google Scholar]
- 36.Lee H., Maihle N.J. Isolation and characterization of four alternate c-erbB3 transcripts expressed in ovarian carcinoma-derived cell lines and normal human tissues. Oncogene. 1998;16:3243–3252. doi: 10.1038/sj.onc.1201866. [DOI] [PubMed] [Google Scholar]
- 37.Vijapurkar U., Cheng K., Koland J.G. Mutation of a Shc binding site tyrosine residue in ErbB3/HER3 blocks heregulin-dependent activation of mitogen-activated protein kinase. J Biol Chem. 1998;273:20996–21002. doi: 10.1074/jbc.273.33.20996. [DOI] [PubMed] [Google Scholar]
- 38.Cipolletta E., Monaco S., Maione A.S., Vitiello L., Campiglia P., Pastore L., Franchini C., Novellino E., Limongelli V., Bayer K.U., Means A.R., Rossi G., Trimarco B., Iaccarino G., Illario M. Calmodulin-dependent kinase II mediates vascular smooth muscle cell proliferation and is potentiated by extracellular signal regulated kinase. Endocrinology. 2010;151:2747–2759. doi: 10.1210/en.2009-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya M., Lee Y.K., Muratcioglu S., Qiu B., Nyayapati P., Schulman H., Groves J.T., Kuriyan J. Flexible linkers in CaMKII control the balance between activating and inhibitory autophosphorylation. Elife. 2020;9:e53670. doi: 10.7554/eLife.53670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell viability analyses of malignant peripheral nerve sheath tumor cells with saracatinib alone or saracatinib plus trifluoperazine (TFP). A: Immunocytochemistry for cleaved caspase-3 (casp-3; red staining) in S462 cells treated with vehicle (Veh), 10 μmol/L saracatinib (Src), or 5 μmol/L saracatinib in combination with 5 μmol/L trifluoperazine. The actin cytoskeleton was stained with phalloidin (green), and nuclei were labeled with Hoechst 33,342 (blue). Images taken with a 63× oil immersion objective. B: Representative images of wells containing S462, T265-2c, and STS-26T cells after performing live/dead assays, as imaged on a Celigo high-throughput imager. The illustrated images are of cells treated for 48 hours with vehicle, 10 μmol/L saracatinib, or 5 μmol/L saracatinib plus 5 μmol/L trifluoperazine. C: Live/dead assays [staining for calcein AM (live cells; green) and BOBO-3 iodide (dead cells; red)] were performed on S462, T265-2c, and STS-26T cells. Normalized live/dead cell ratios compared with total cell counts were analyzed with Fiji ImageJ version 2.9.0 direct particle measurement. Scale bars = 20 μm (A).