ABSTRACT
Current laparoscopic camera motion automation relies on rule-based approaches or only focuses on surgical tools. Imitation Learning (IL) methods could alleviate these shortcomings, but have so far been applied to oversimplified setups. Instead of extracting actions from oversimplified setups, in this work we introduce a method that allows to extract a laparoscope holder’s actions from videos of laparoscopic interventions. We synthetically add camera motion to a newly acquired dataset of camera motion free da Vinci surgery image sequences through a novel homography generation algorithm. The synthetic camera motion serves as a supervisory signal for camera motion estimation that is invariant to object and tool motion. We perform an extensive evaluation of state-of-the-art (SOTA) Deep Neural Networks (DNNs) across multiple compute regimes, finding our method transfers from our camera motion free da Vinci surgery dataset to videos of laparoscopic interventions, outperforming classical homography estimation approaches in both, precision by , and runtime on a CPU by .
KEYWORDS: Deep learning, homography estimation, laparoscopic surgery, image processing and analysis, visual data mining and knowledge discovery, virtual reality
1. Introduction
The goal in IL is to learn an expert policy from a set of expert demonstrations. IL has been slow to transition to interventional imaging. In particular, the slow transition of modern IL methods into automating laparoscopic camera motion is due to a lack state-action-pair data (Kassahun et al. 2016; Esteva et al. 2019). The need for automated laparoscopic camera motion (Pandya et al. 2014; Ellis et al. 2016) has, therefore, historically sparked research in rule-based approaches that aim to reactively centre surgical tools in the field of view (Agustinos et al. 2014; Da Col et al. 2020). DNNs could contribute to this work by facilitating SOTA tool segmentations and automated tool tracking (Garcia-Peraza-Herrera et al. 2017, 2021; Gruijthuijsen et al. 2021).
Recent research contextualises laparoscopic camera motion with respect to (w.r.t.) the user and the state of the surgery. DNNs could facilitate contextualisation, as indicated by research in surgical phase and skill recognition (Kitaguchi et al.2020). However, current contextualisation is achieved through handcrafted rule-based approaches (Rivas-Blanco et al. 2014, 2017), or through stochastic modelling of camera positioning w.r.t. the tools (Weede et al. 2011; Rivas-Blanco et al. 2019). While the former do not scale well and are prone to nonlinear interventions, the latter only consider surgical tools. However, clinical evidence suggests camera motion is also caused by the surgeon’s desire to observe tissue (Ellis et al. 2016). Non-rule-based, i.e. IL, attempts that consider both, tissue, and tools as source for camera motion are Ji et al. 2018; Su et al. 2020; Wagner et al. 2021, but they utilise an oversimplified setup, require multiple cameras or tedious annotations.
In current laparoscopic camera motion automation, DNNs merely solve auxiliary tasks. Consequentially, current laparoscopic camera motion automation is rule-based, and disregards tissue. While modern IL approaches could alleviate these issues, clinical data of laparoscopic surgeries remains unusable for IL. Therefore, SOTA IL attempts rely on artificially acquired data (Ji et al. 2018; Su et al. 2020; Wagner et al. 2021).
In this work, we aim to extract camera motion from videos of laparoscopic interventions, thereby creating state-action-pairs for IL. To this end, we introduce a method that isolates camera motion (actions) from object and tool motion by solely relying on observed images (states). To this end, DNNs are supervisedly trained to estimate camera motion while disregarding object, and tool motion. This is achieved by synthetically adding camera motion via a novel homography generation algorithm to a newly acquired dataset of camera motion free da Vinci surgery image sequences. In this way, object, and tool motion reside within the image sequences, and the synthetically added camera motion can be regarded as the only source, and therefore ground truth, for camera motion estimation. Extensive experiments are carried out to identify modern network architectures that perform best at camera motion estimation. The DNNs that are trained in this manner are found to generalise well across domains, in that they transfer to vast laparoscopic datasets. They are further found to outperform classical camera motion estimators.
2. Related work
Supervised deep homography estimation was first introduced in (DeTone et al. 2016) and got improved through a hierarchical homography estimation in (Erlik Nowruzi et al. 2017). It got adopted in the medical field in (Bano et al. 2020). All three approaches generate a limited set of homographies, only train on static images, and use non-SOTA VGG-based network architectures (Simonyan and Zisserman 2014).
Unsupervised deep homography estimation has the advantage to be applicable to unlabelled data, e.g. videos. It was first introduced in (Nguyen et al. 2018), and got applied to endoscopy in (Gomes et al. 2019). The loss in image space, however, can’t account for object motion, and only static scenes are considered in their works. Consequentially, recent work seeks to isolate object motion from camera motion through unsupervised incentives. Closest to our work are Le et al. (2020), where the authors generate a dataset of camera motion free image sequences. However, due to tool, and object motion, their data generation method is not applicable to laparoscopic videos, since it relies on motion free image borders. Zhang et al. (2020) provide the first work that does not need a synthetically generated dataset. Their method works fully unsupervised, but constraining what the network minimises, is difficult to achieve.
Only (Le et al. 2020) and (Zhang et al. 2020) train DNNs on object motion invariant homography estimation. Contrary to their works, we train DNNs supervisedly. We do so by applying the data generation of DeTone et al. (2016) to image sequences rather than single images. We further improve their method by introducing a novel homography generation algorithm that allows to continuously generate synthetic homographies at runtime, and by using SOTA DNNs.
3. Materials and methods
3.1. Theoretical background
Two images are related by a homography if both images view the same plane from different angles and distances. Points on the plane, as observed by the camera from different angles in homogeneous coordinates are related by a projective homography (Malis and Vargas 2007)
| (1) |
Since the points and are only observed in the 2D image, depth information is lost, and the projective homography can only be determined up to scale . The distinction between projective homography and homography in Euclidean coordinates , with the camera intrinsics , is often not made for simplicity, but is nonetheless important for control purposes Huber, et al., 2021. The eight unknown parameters of can be obtained through a set of matching points = by rearranging (1) into
| (2) |
where holds the entries of as a column vector. The ninth constraint, by convention, is usually to set . Classically, is obtained through feature detectors but it may also be used as a means to parameterise the spatial transformation. Recent deep approaches indeed set as the corners of an image, and predict . This is also known as the four point homography
| (3) |
which relates to through (2), where .
3.2. Data preparation
Similar to (Le et al. 2020), we initially find camera motion free image sequences, and synthetically add camera motion to them. In our work, we isolate camera motion free image sequences from da Vinci surgeries, and learn homography estimation supervisedly. We acquire publicly available laparoscopic, and da Vinci surgery videos. An overview of all datasets is shown in Figure 1. Excluded are synthetic, and publicly unavailable datasets. Da Vinci surgery datasets, and laparoscopic surgery datasets require different pre-processing steps, which are described below.
Figure 1.
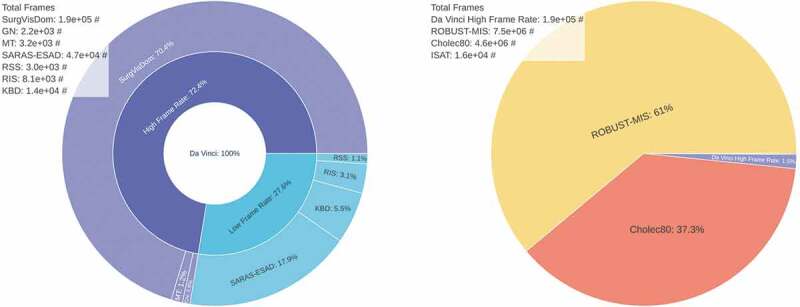
Da Vinci surgery and laparoscopic surgery datasets, referring to Section 3.2. Shown are relative sizes and the absolute number of frames. Da Vinci surgery datasets (left). Included are: SurgVisDom Zia, et al., 2021, GN Giannarou, et al., 2012, MT Mountney, et al., 2010, SARAS-ESAD Bawa, et al., 2020, KBD Hattab, et al., 2020, RIS Allan, et al., 2019, and RSS Allan, et al., 2020. They are often released at a low frame rate of 1fps for segmentation tasks. Much more laparoscopic surgery data is available (right). Included are ROBUST-MIS Maier-Hein, et al., 2020, Cholec80 Twinanda, et al., 2016, ISAT Bodenstedt, et al., 2018, and the HFR da Vinci dataset from (left) for reference.
3.2.1. Da Vinci surgery data pre-processing
Many of the da Vinci surgery datasets are designed for tool or tissue segmentation tasks, therefore, they are published at a frame rate of , see Figure 1 (left). We merge all high frame rate (HFR) datasets into a single dataset and manually remove image sequences with camera motion, which amount to of all HFR data. We crop the remaining data to remove status indicators, and scale the images to pixels, later to be cropped by the homography generation algorithm to a resolution of .
3.2.2. Laparoscopic surgery data pre-processing
Laparoscopic images are typically observed through a Hopkins telescope, which causes a black circular boundary in the view, see Figure 2. This boundary does not exist in da Vinci surgery recordings. For inference on the laparoscopic surgery image sequences, the most straightforward approach is to crop the view. To this purpose, we determine the centre and radius of the circular boundary, which is only partially visible. We detect it by randomly sampling points on the boundary. This is similar to work in (Münzer et al. 2013), but instead of computing an analytical solution, we fit a circle by means of a least squares solution through inversion of
| (4) |
Figure 2.
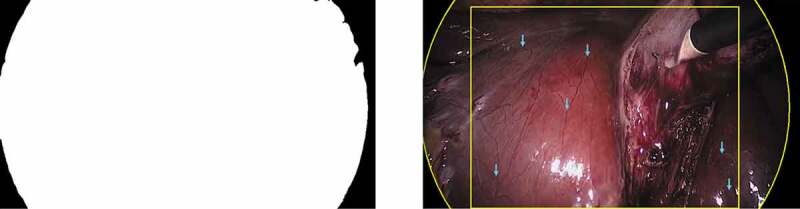
Cholec80 dataset pre-processing, referring to Section 3.2.2. The black boundary circle is automatically detected through fitting a circle to the binary segmentation. The binary segmentation mask is shown on the left. The circular boundary fitting and static landmarks (blue arrows) are shown on the right. Landmarks are manually annotated and tracked over time.
where the circle’s centre is , and its radius is . We then crop the view centrally around the circle’s centre, and scale it to a resolution of . An implementation is provided on GitHub.1
3.2.3. Ground truth generation
One can simply use the synthetically generated camera motion as ground truth at train time. For inference on the laparoscopic dataset, this is not possible. We therefore generate ground truth data by randomly sampling image sequences with frames each from the Cholec80 dataset. In these image sequences, we find characteristic landmarks that are neither subject to tool, nor to object motion, see Figure 2 (right). Tracking of these landmarks over time allows one to estimate the camera motion in between consecutive frames through (2).
3.3. Deep homography estimation
In this work we exploit the static camera in da Vinci surgeries, which allows us to isolate camera motion free image sequences. The processing pipeline is shown in Figure 3.
Figure 3.
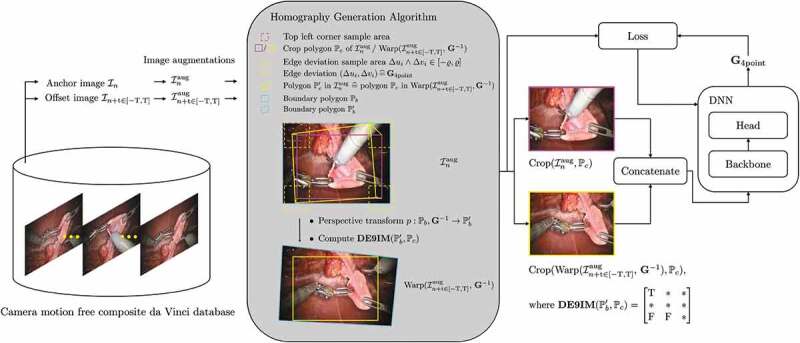
Deep homography estimation training pipeline, referring to Section 3.3. Image pairs are sampled from the HFR da Vinci surgery dataset. The homography generation algorithm then adds synthetic camera motion to the augmented images, which is regressed through a backbone DNN.
Image pairs are sampled from image sequences of the HFR da Vinci surgery dataset of Figure 1. An image pair consists of an anchor image , and an offset image . The offset image is sampled uniformly from and interval around the anchor. The HFR da Vinci surgery dataset is relatively small, compared to the laparoscopic datasets, see Figure 1 (right). Therefore, we apply image augmentations to the sampled image pairs. They include transform to greyscale, horizontal, and vertical flipping, cropping, change in brightness, and contrast, Gaussian blur, fog simulation, and random combinations of those. Camera motion is then added synthetically to the augmented image via the homography generation algorithm from Section 3.4. A DNN, with a backbone, then learns to predict the homography between the augmented image, and the augmented image with synthetic camera motion at time step .
3.4. Homography generation algorithm
In its core, the homography generation algorithm is based on the works of DeTone et al. 2016. However, where DeTone et al. crop the image with a safety margin, our method allows to sample image crops across the entire image. Additionally, our method computes feasible homographies at runtime. This allows us to continuously generate synthetic camera motion, rather then training on a fixed set of precomputed homographies. The homography generation algorithm is summarised in Alg. 1, and visualised in Figure 3.
Initially, a crop polygon is generated for the augmented image . The crop polygon is defined through a set of points in the augmented image , which span a rectangle. The top left corner is randomly sampled such that the crop polygon resides within the image border polygon , hence , where , and are the height and width of the crop, and the border polygon, respectively. Following that, a random four point homography (3) is generated by sampling edge deviations . The corresponding inverse homography is used to warp each point of the border polygon to . Finally, the Dimensionally Extended 9-Intersection Model (Clementini et al. 1994) is used to determine whether the warped polygon contains , for which we utilise the Python library Shapely.2 If the thus found intersection matrix satisfies
| (5) |
the homography is returned, otherwise a new four point homograpy is sampled. Therein, indicates that the intersection matrix may hold any value, and indicate that the intersection matrix must be true or false at the respective position. In the unlikely case that no homography is found after maximum rollouts, the identity is returned. Once a suitable homography is found, a crop of the augmented image is computed, as well as a crop of the warped augmented image at time , . This keeps all computationally expensive operations outside the loop.
| Algorithm 1: Homography generation algorithm, referring to Section 3.4. |
|---|
| Randomly sample crop polygon of desired shape in ; |
| while rollouts maximum rollouts do |
| Randomly sample , where ; |
| Perspective transform boundary polygon ; |
| Compute intersection matrix ; |
| if then |
| return, ; |
| end |
| Increment rollouts; |
| end |
| return, ; |
4. Experiments
We train DNNs on a train split of the HFR da Vinci surgery dataset from Figure 1. The test split is referred to as test set in the following. Inference is performed on the ground truth set from Section 3.2.3. We compute the Mean Pairwise Distance (MPD) of the predicted value for from the desired one. We then compute the Cumulative Distribution Function (CDF) of all MPDs. We evaluate the CDF at different thresholds , e.g. of all homography estimations are below a MPD of . We additionally evaluate the compute time on a GeForce RTX 2070 GPU, and a Intel Core i7-9750 H CPU.
4.1. Backbone search
In this experiment, we aim to find the best performing backbone for homography estimation. Therefore, we run the same experiment repeatedly with fixed hyperparameters, and varying backbones. We train each network for epochs, with a batch size of , using the Adam optimiser with a learning rate of . The edge deviation is set to , and the sequence length to .
4.2. Homography generation algorithm
In this experiment, we evaluate the homography generation algorithm. For this experiment we fix the backbone to a ResNet-34, and train it for epochs, with a batch size of , using the Adam optimiser with a learning rate of 1e-3. Initially, we fix the sequence length to , and train on different edge deviations . Next, we fix the edge deviation to , and train on different sequence lengths , where a sequence length of corresponds to a static pair of images.
5. Results
5.1. Backbone search
The results are listed in Table 1. It can be seen that the deep methods generally outperform the classical methods on the test set. There is a tendency that models with more parameters perform better. On the ground truth set, this tendency vanishes. The differences in performance become independent of the number of parameters. Noticeably, many backbones still outperform the classical methods across all thresholds on the ground truth set, and low compute regime models also run quicker on CPU than comparable classical methods. E.g. we find that EfficientNet-B0, and RegNetY-400MF run at , and on a CPU, respectively. Both outperform SURF & RANSAC in homography estimation, which runs at .
Table 1.
Results referring to Section 5.1. All methods are tested on the da Vinci HFR test set, indicated by , and the Cholec80 inference set, indicated by . Best, and second best metrics are highlighted with bold character. Improvements in precision and compute time are given w.r.t. SURF & RANSAC
| Name | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VGG-style | ||||||||||
| ResNet-18 | ||||||||||
| ResNet-34 | ||||||||||
| ResNet-50 | ||||||||||
| EfficientNet-B0 | ||||||||||
| EfficientNet-B1 | ||||||||||
| EfficientNet-B2 | ||||||||||
| EfficientNet-B3 | ||||||||||
| EfficientNet-B4 | ||||||||||
| EfficientNet-B5 | ||||||||||
| RegNetY-400MF | ||||||||||
| RegNetY-600MF | ||||||||||
| RegNetY-800MF | ||||||||||
| RegNetY-1.6GF | ||||||||||
| RegNetY-4.0GF | ||||||||||
| RegNetY-6.4GF | ||||||||||
| SURF & RANSAC | N/A | N/A | N/A | |||||||
| SIFT & RANSAC | N/A | N/A | N/A | |||||||
| ORB & RANSAC | N/A | N/A | N/A |
5.2. Homography generation algorithm
Given that ResNet-34 performs well on the ground truth set, and executes fast on the GPU, we run the homography generation algorithm experiments with it. It can be seen in Figure 4 (left), that the edge deviation is neglectable for inference. In Figure 4 (right), one sees the effects of the sequence length on the inference performance. Notably, with , corresponding to static image pairs, the SURF & RANSAC homography estimation outperforms the ResNet-34. For the other sequence lengths, ResNet-34 outperforms the classical homography estimation. The CDF for the best performing combination of parameters, with , and , is shown in Figure 5. Our method generally outperforms SURF & RANSAC. The advantage of our method becomes most apparent for a . Even the identity outperforms SURF & RANSAC for large MPDs. This aligns with the qualitative observation that motion is often overestimated by SURF & RANSAC, which is shown in Figure 6. An exemplary video is provided.3
Figure 4.
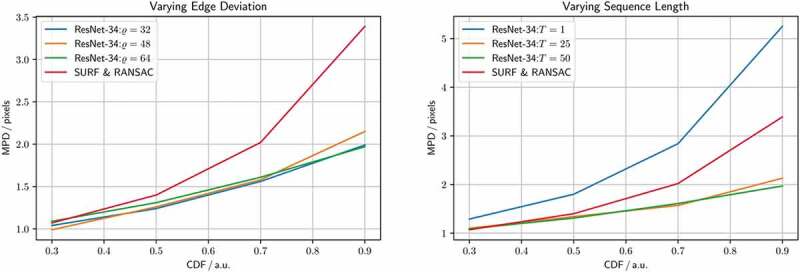
Homography generation optimisation, referring to Section 5.2. Shown is a ResNet-34 homography estimation for different homography generation configurations, and a SURF & RANSAC homography estimation for reference. Varying edge deviation , and fixed sequence length (left). Varying sequence length , and fixed edge deviation (right).
Figure 5.
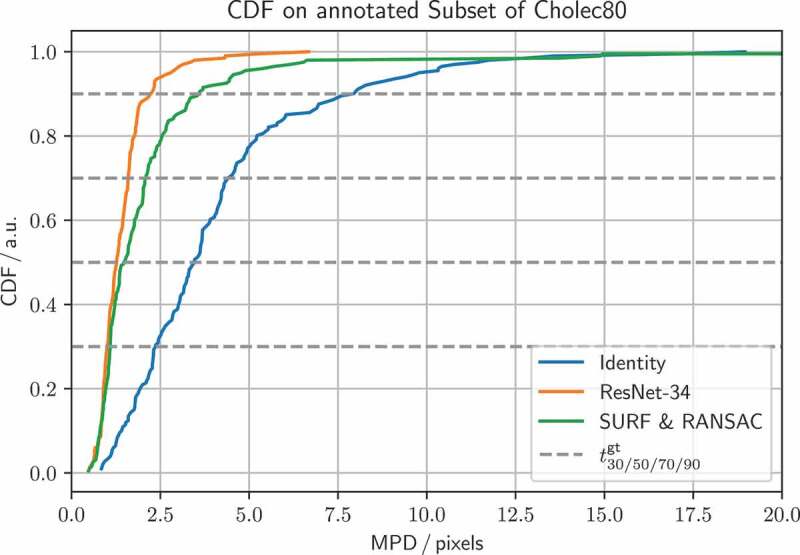
CDF for SURF & RANSAC, and ResNet-34, trained with a sequence length , and edge deviation , referring to Section 5.2. The identity is added for reference. CDF thresholds for the SURF & RANSAC are , and for the ResNet-34 . ResNet-34 generally performs better, and has no outliers.
Figure 6.
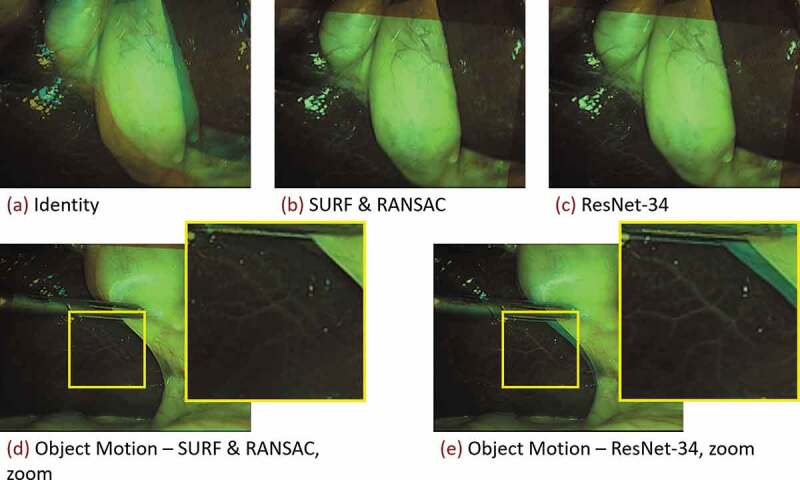
Classical homography estimation using a SURF feature detector under RANSAC outlier rejection, and the proposed deep homography estimation with a ResNet-34 backbone, referring to Section 5.2. Shown are blends of consecutive images from a resampled Cholec80 exemplary sequence (Twinanda et al. 2016). Decreasing the framerate from originally to , increases the motion in between consecutive frames. (Top row) Homography estimation under predominantly camera motion. Both methods perform well. (Bottom row) Homography estimation under predominantly object motion. Especially in the zoomed images it can be seen that the classical method (d) misaligns the stationary parts of the image, whereas the proposed method (e) aligns the background well.
6. Discussion
In this work we supervisedly learn homography estimation in dynamic surgical scenes. We train our method on a newly acquired, synthetically modified da Vinci surgery dataset and successfully cross the domain gap to videos of laparoscopic surgeries. To do so, we introduce extensive data augmentation and continuously generate synthetic camera motion through a novel homography generation algorithm.
In Section 5.1, we find that, despite the domain gap for the ground truth set, DNNs outperform classical methods, which is indicated in Table 1. The homography estimation performance proofs to be independent of the number of model parameters, which indicates an overfit to the test data. The independence of the number of parameters allows to optimise the backbone for computational requirements. E.g. a typical laparoscopic setup runs at , the classical method would thus already introduce a bottleneck at . On the other hand, EfficientNet-B0, with , and RegNetY-400MF, with , introduce no latency, and could be integrated into systems without GPU.
In Section 5.2, we find that increasing the edge deviation has no effect on the homography estimation, see Figure 4 (left). This is because the motion in the ground truth set does not exceed the motion in the training set. In Figure 4 (right), we further find how training DNNs on synthetically modified da Vinci surgery image sequences enables our method to isolate camera from object and tool motion, validating our method. In Figure 5, it is demonstrated that ResNet-34 generally outperforms SURF & RANSAC. This shows that generating camera motion synthetically through homographies, which approximates the surgical scene as a plane, does not pose an issue.
The object, and tool motion invariant camera motion estimation allows one to extract a laparoscope holder’s actions from videos of laparoscopic interventions, which enables the generation of image-action-pairs. In future work, we will generate image-action-pairs from laparoscopic datasets and apply IL to them. Describing camera motion (actions) by means of a homography is grounded in recent research for robotic control of laparoscopes (Huber et al. 2021). This work will therefore support the transition towards robotic automation approaches. It might further improve augmented reality, and image mosaicing methods in dynamic surgical environments.
Biographies
Martin received his BSc and MSc in Physics from the University of Heidelberg. For his Master’s Thesis he implemented an optimal control scheme for dynamically balanced locomotion of humanoid robots and automated high level commands to said control with neural networks. He joined the RViM Lab in 2019 to develop data driven visual servo algorithms for surgical robots to assist surgeons in minimally invasive surgery. His main interests are optimal control, artificial intelligence and medical imaging. He is supervised by Dr. Christos Bergeles and Prof. Tom Vercauteren.
Seb Ourselin is Head of the School of Biomedical Engineering & Imaging Sciences, King’s College London; dedicated to the development, translation and clinical application of medical imaging, computational modelling, minimally invasive interventions and surgery. In collaboration with Guys & St Thomas’ NHS Foundation Trust (GSTT), he is leading the establishment of the MedTech Hub, located at St Thomas’ campus. The vision of the MedTech Hub is to create a unique infrastructure that will develop health technologies including AI, medical devices, workforce and operational improvements that will be of global significance. Previously, he was based at UCL where he formed and led numerous activities including the UCL Institute of Healthcare Engineering, the EPSRC Centre for Doctoral Training in Medical Imaging and Wellcome EPSRC Centre for Surgical and Interventional Sciences. He is co-founder of Brainminer, an academic spin-out commercialising machine learning algorithms for brain image analysis. Their clinical decision support system for dementia diagnosis, DIADEM, obtained CE marking. Over the last 15 years, he has raised over £50M as Principal Investigator and has published over 420 articles (over 20,000 citations, h-index 73). He is/was an associate editor for IEEE Transactions on Medical Imaging, Journal of Medical Imaging, Nature Scientific Reports, and Medical Image Analysis. He has been active in conference organisation (12 international conferences as General or Program Chair) and professional societies (APRS, MICCAI). He was elected Fellow of the MICCAI Society in 2016.
Christos Bergeles received the Ph.D. degree in Robotics from ETH Zurich, Switzerland, in 2011. He was a postdoctoral research fellow at Boston Children’s Hospital, Harvard Medical School, Massachusetts, and the Hamlyn Centre for Robotic Surgery, Imperial College, United Kingdom. He was an Assistant Professor at the Wellcome/EPSRC Centre for Interventional and Surgical Sciences (which he co-founded) at University College London. He is now a Senior Lecturer (Associate Professor) at King's College London, leading the Robotics and Vision in Medicine Lab. Dr. Bergeles received the Fight for Sight Award in 2014, and the ERC Starting Grant in 2016. His main research area is image-guided micro-surgical robotics.
Tom Vercauteren is Professor of Interventional Image Computing at King’s College London where he holds the Medtronic / Royal Academy of Engineering Research Chair in Machine Learning for Computer-assisted Neurosurgery. He is also co-founder and Chief Scientific Officer at Hypervision Surgical, a spin-out focusing on intraoperative hyperstral imaging. From 2014 to 2018, he was Associate Professor at UCL where he acted as Deputy Director for the Wellcome / EPSRC Centre for Interventional and Surgical Sciences (2017-18). From 2004 to 2014, he worked for Mauna Kea Technologies, Paris where he led the research and development team designing image computing solutions for the company’s CE- marked and FDA-cleared optical biopsy device. His work is now used in hundreds of hospitals worldwide. He is a Columbia University and Ecole Polytechnique graduate and obtained his PhD from Inria in 2008. Tom is also an established open-source software supporter.
Funding Statement
This work was supported by core and project funding from the Wellcome/EPSRC [Wellcome / EPSRC Centre for Interventional and Surgical Sciences WT203148/Z/16/Z; NS/A000049/1; WT101957; NS/A000027/1]. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 101016985 (FAROS project).
Notes
Disclosure statement
TV is supported by a Medtronic/RAEng Research Chair [Royal Academy of Engineering RCSRF1819\7\34]. SO and TV are are co-founders and shareholders of Hypervision Surgical. TV holds shares from Mauna Kea Technologies.
References
- Agustinos A, Wolf R, Long JA, Cinquin P, Voros S. 2014. Visual servoing of a robotic endoscope holder based on surgical instrument tracking. In: 5th IEEE RAS/EMBS International Conference on Biomedical Robotics and Biomechatronics São Paulo, Brazil. IEEE. p. 13–18. [Google Scholar]
- Allan M, Kondo S, Bodenstedt S, Leger S, Kadkhodamohammadi R, Luengo I, Fuentes F, Flouty E, Mohammed A, Pedersen M, et al. 2020. 2018 robotic scene segmentation challenge. arXiv preprint arXiv: 200111190.
- Allan M, Shvets A, Kurmann T, Zhang Z, Duggal R, Su YH, Rieke N, Laina I, Kalavakonda N, Bodenstedt S, et al. 2019. 2017 robotic instrument segmentation challenge. arXiv preprint arXiv: 190206426.
- Bano S, Vasconcelos F, Tella-Amo M, Dwyer G, Gruijthuijsen C, Vander Poorten E, Vercauteren T, Ourselin S, Deprest J, Stoyanov D.. 2020. Deep learning-based fetoscopic mosaicking for field-of-view expansion. Int J Comput Assist Radiol Surg. 15(11):1807–1816. doi: 10.1007/s11548-020-02242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa VS, Singh G, KapingA F, Leporini A, Landolfo C, Stabile A, Setti F, Muradore R, Oleari E, Cuzzolin F, et al. 2020. Esad: endoscopic surgeon action detection dataset. arXiv preprint arXiv: 200607164.
- Bodenstedt S, Allan M, Agustinos A, Du X, Garcia-Peraza-Herrera L, Kenngott H, Kurmann T, Müller-Stich B, Ourselin S, Pakhomov D, et al. 2018. Comparative evaluation of instrument segmentation and tracking methods in minimally invasive surgery. arXiv preprint arXiv: 180502475.
- Clementini E, Sharma J, Egenhofer MJ.. 1994. Modelling topological spatial relations: strategies for query processing. Comput Graph. 18(6):815–822. doi: 10.1016/0097-8493(94)90007-8. [DOI] [Google Scholar]
- Da Col T, Mariani A, Deguet A, Menciassi A, Kazanzides P, De Momi E. 2020. Scan: system for camera autonomous navigation in robotic-assisted surgery. In: 2020 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) Las Vegas, NV, USA. IEEE. p. 2996–3002. [Google Scholar]
- DeTone D, Malisiewicz T, Rabinovich A. 2016. Deep image homography estimation. arXiv preprint arXiv: 160603798.
- Ellis RD, Munaco AJ, Reisner LA, Klein MD, Composto AM, Pandya AK, King BW. 2016. Task analysis of laparoscopic camera control schemes. Int J Med Robot Compt Assist Surgery. 12(4):576–584. doi: 10.1002/rcs.1716. [DOI] [PubMed] [Google Scholar]
- Erlik Nowruzi F, Laganiere R, Japkowicz N. 2017. Homography estimation from image pairs with hierarchical convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision Workshops Venice, Italy. p. 913–920. [Google Scholar]
- Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. 2019. A guide to deep learning in healthcare. Nat Med. 25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- Garcia-Peraza-Herrera LC, Fidon L, D’Ettorre C, Stoyanov D, Vercauteren T, Ourselin S. 2021. Image compositing for segmentation of surgical tools without manual annotations. IEEE Trans Med Imaging. 40(5):1450–1460. doi: 10.1109/TMI.2021.3057884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Peraza-Herrera LC, Li W, Fidon L, Gruijthuijsen C, Devreker A, Attilakos G, Deprest J, Vander Poorten E, Stoyanov D, Vercauteren T, et al. 2017. Toolnet: holistically-nested real-time segmentation of robotic surgical tools. In: 2017 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) Vancouver, BC, Canada. IEEE. p. 5717–5722. [Google Scholar]
- Giannarou S, Visentini-Scarzanella M, Yang GZ. 2012. Probabilistic tracking of affine-invariant anisotropic regions. IEEE Trans Pattern Anal Mach Intell. 35(1):130–143. doi: 10.1109/TPAMI.2012.81. [DOI] [PubMed] [Google Scholar]
- Gomes S, Valério MT, Salgado M, Oliveira HP, Cunha A. 2019. Unsupervised neural network for homography estimation in capsule endoscopy frames. Procedia Comput Sci. 164:602–609. doi: 10.1016/j.procs.2019.12.226. [DOI] [Google Scholar]
- Gruijthuijsen C, Garcia-Peraza-Herrera LC, Borghesan G, Reynaerts D, Deprest J, Ourselin S, Vercauteren T, Poorten EV. 2021. A utonomous robotic endoscope control based on semantically rich instructions. arXiv preprint arXiv: 210702317.
- Hattab G, Arnold M, Strenger L, Allan M, Arsentjeva D, Gold O, Simpfendörfer T, Maier-Hein L, Speidel S. 2020. Kidney edge detection in laparoscopic image data for computer-assisted surgery. Int J Comput Assist Radiol Surg. 15(3):379–387. doi: 10.1007/s11548-019-02102-0. [DOI] [PubMed] [Google Scholar]
- Huber M, Mitchell JB, Henry R, Ourselin S, Vercauteren T, Bergeles C. 2021. Homography-based visual servoing with remote center of motion for semi-autonomous robotic endoscope manipulation. arXiv preprint arXiv: 211013245.
- Ji JJ, Krishnan S, Patel V, Fer D, Goldberg K. 2018. Learning 2d surgical camera motion from demonstrations. In: 2018 IEEE 14th International Conference on Automation Science and Engineering (CASE) Munich, Germany. IEEE. p. 35–42. [Google Scholar]
- Kassahun Y, Yu B, Tibebu AT, Stoyanov D, Giannarou S, Metzen JH, Vander Poorten E. 2016. Surgical robotics beyond enhanced dexterity instrumentation: a survey of machine learning techniques and their role in intelligent and autonomous surgical actions. Int J Comput Assist Radiol Surg. 11(4):553–568. doi: 10.1007/s11548-015-1305-z. [DOI] [PubMed] [Google Scholar]
- Kitaguchi D, Takeshita N, Matsuzaki H, Takano H, Owada Y, Enomoto T, Oda T, Miura H, Yamanashi T, Watanabe M, et al. 2020. Real-time automatic surgical phase recognition in laparoscopic sigmoidectomy using the convolutional neural network-based deep learning approach. Surg Endosc. 34(11):4924–4931. doi: 10.1007/s00464-019-07281-0. [DOI] [PubMed] [Google Scholar]
- Le H, Liu F, Zhang S, Agarwala A. 2020. Deep homography estimation for dynamic scenes. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Seattle, WA, USA. p. 7652–7661. [Google Scholar]
- Maier-Hein L, Wagner M, Ross T, Reinke A, Bodenstedt S, Full PM, Hempe H, Mindroc-Filimon D, Scholz P, Tran TN, et al. 2020. Heidelberg colorectal data set for surgical data science in the sensor operating room. arXiv preprint arXiv: 200503501. [DOI] [PMC free article] [PubMed]
- Malis E, Vargas M. 2007. Deeper understanding of the homography decomposition for vision-based control [dissertation]. INRIA. [Google Scholar]
- Mountney P, Stoyanov D, Yang GZ. 2010. Three-dimensional tissue deformation recovery and tracking. IEEE Signal Process Mag. 27(4):14–24. doi: 10.1109/MSP.2010.936728. [DOI] [Google Scholar]
- Münzer B, Schoeffmann K, Böszörmenyi L. 2013. Detection of circular content area in endoscopic videos. In: Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems Lubbock, TX, USA. IEEE. p. 534–536. [Google Scholar]
- Nguyen T, Chen SW, Shivakumar SS, Taylor CJ, Kumar V. 2018. Unsupervised deep homography: a fast and robust homography estimation model. IEEE Robot Autom Lett. 3(3):2346–2353. doi: 10.1109/LRA.2018.2809549. [DOI] [Google Scholar]
- Pandya A, Reisner LA, King B, Lucas N, Composto A, Klein M, Ellis RD. 2014. A review of camera viewpoint automation in robotic and laparoscopic surgery. Robotics. 3(3):310–329. doi: 10.3390/robotics3030310. [DOI] [Google Scholar]
- Rivas-Blanco I, Estebanez B, Cuevas-Rodriguez M, Bauzano E, Muñoz VF. 2014. Towards a cognitive camera robotic assistant. In: 5th IEEE RAS/EMBS International Conference on Biomedical Robotics and Biomechatronics São Paulo, Brazil. IEEE. p. 739–744. [Google Scholar]
- Rivas-Blanco I, López-Casado C, Pérez-del Pulgar CJ, Garcia-Vacas F, Fraile J, Muñoz VF. 2017. Smart cable-driven camera robotic assistant. IEEE Trans Hum Mach Syst. 48(2):183–196. doi: 10.1109/THMS.2017.2767286. [DOI] [Google Scholar]
- Rivas-Blanco I, Perez-del Pulgar CJ, López-Casado C, Bauzano E, Muñoz VF. 2019. Transferring know-how for an autonomous camera robotic assistant. Electronics. 8(2):224. doi: 10.3390/electronics8020224. [DOI] [Google Scholar]
- Simonyan K, Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv: 14091556.
- Su YH, Huang K, Hannaford B. 2020. Multicamera 3d reconstruction of dynamic surgical cavities: autonomous optimal camera viewpoint adjustment. In: 2020 International Symposium on Medical Robotics (ISMR) Atlanta, GA, USA. IEEE. p. 103–110. [Google Scholar]
- Twinanda AP, Shehata S, Mutter D, Marescaux J, De Mathelin M, Padoy N. 2016. Endonet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans Med Imaging. 36(1):86–97. doi: 10.1109/TMI.2016.2593957. [DOI] [PubMed] [Google Scholar]
- Wagner M, Bihlmaier A, Kenngott HG, Mietkowski P, Scheikl PM, Bodenstedt S, Schiepe-Tiska A, Vetter J, Nickel F, Speidel S, et al. 2021. A learning robot for cognitive camera control in minimally invasive surgery. Surg Endosc. 35(1):1–10. doi: 10.1007/s00464-020-08131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weede O, Mönnich H, Müller B, Wörn H. 2011. An intelligent and autonomous endoscopic guidance system for minimally invasive surgery. In: 2011 IEEE International Conference on Robotics and Automation Shanghai, China. IEEE. p. 5762–5768. [Google Scholar]
- Zhang J, Wang C, Liu S, Jia L, Ye N, Wang J, Zhou J, Sun J. 2020. Content-aware unsupervised deep homography estimation. In: European Conference on Computer Vision. Springer Glasgow, UK. p. 653–669. [Google Scholar]
- Zia A, Bhattacharyya K, Liu X, Wang Z, Kondo S, Colleoni E, van Amsterdam B, Hussain R, Hussain R, Maier-Hein L, et al. 2021. Surgical visual domain adaptation: Results from the miccai 2020 surgvisdom challenge.


