ABSTRACT
Prevotella copri is an abundant member of the human gastrointestinal microbiome, whose relative abundance has curiously been associated with positive and negative impacts on diseases, such as Parkinson’s disease and rheumatoid arthritis. Yet, the verdict is still out on the definitive role of P. copri in human health, and on the effect of different diets on its relative abundance in the gut microbiome. The puzzling discrepancies among P. copri studies have only recently been attributed to the diversity of its strains, which substantially differ in their encoded metabolic patterns from the commonly used reference strain. However, such strain differences cannot be resolved by common 16S rRNA amplicon profiling methods. Here, we scrutinize P. copri, its versatile metabolic potential, and the hypotheses behind the conflicting observations on its association with diet and human health. We also provide suggestions for designing studies and bioinformatics pipelines to better research P. copri.
Keywords: microbiome, diet, metabolism, genomic variation, pharmacomicrobiomics, toxicomicrobiomics, polysaccharide utilization loci, study design
Prevotella copri, a microbiome-derived bacterial species
Since the emergence of human microbiome studies, almost 20 years ago, the gut and oral microbe, Prevotella copri, has been repeatedly detected and mentioned in microbiome profiling studies. Owing to the vast distribution and variable abundance of this intriguing microbe, reports are contradictory about whether it is beneficial or detrimental to human health,1 to what extent its impact is on different conditions, and whether these impacts are causal or just associations (Figure 1).
Figure 1.
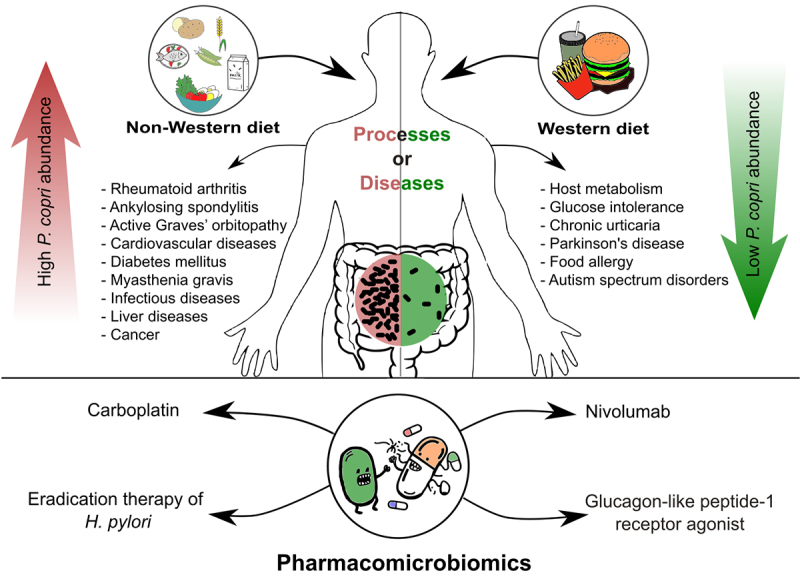
A schematic diagram summarizing the impact of Prevotella copri on the human host, with emphasis on the impact of diet, involvement in diseases, and pharmacomicrobiomic interactions. The figure was generated by Inkscape v. 1.3.
Prevotella copri is one of the main species of the Prevotella genus and is abundant in the human body, especially in the oral cavity and gastrointestinal tract. It is an anaerobic, Gram-negative, non-spore-forming bacterium.2 The name Prevotella (derived from the name of the French microbiologist, A. R. Prévot) was officially assigned to the genus in 1990 to distinguish “moderately saccharolytic, predominantly oral Bacteroides species” from other Bacteroides,3 whereas the species P. copri (from the ancient Greek Kopron = excrement or feces) was only taxonomically recognized in 2007, being among fecal Prevotella strains.4
Since the earliest human microbiome studies, the two genera Prevotella and Bacteroides have been observed to be inversely correlated, i.e., high abundance of P. copri was often associated with low abundance of Bacteroides. This observation led to a popular hypothesis of the presence of different microbiome ‘enterotypes’,5 and it was attributed to the fact that both genera belong to the same class and that they strive for the same nutrients.6–9
Nutrition, metabolism, and adaptation to the human gut
P. copri can flourish on a range of nutrients, among which are complex polysaccharides. Polysaccharide utilization loci (PULs), proposed by Bjursell et al.,10 are sets of genes that encode proteins required for the metabolism and transport of complex polysaccharides in prokaryotes. Their products allow the human gut microbiota to adjust to alterations in host development and host diet.10,11 In P. copri, such loci are common, and are both distinct and distinctive. In other terms, P. copri has unique PULs, which vary among different strains, and their gene products can distinctively break down a wide range of plant-derived (but not animal) polysaccharides in the human gut.12
Various studies suggested that Prevotella thrives on plant- or fiber-rich diets. These include diets commonly described as non-Western, Mediterranean, or rural African diet.13,14 The utilization of plant polysaccharides was compared between P. copri grown in culture media alone or with Candida spp. While glucose was observed to accelerate P. copri growth under no specific conditions, wheat arabinoxylan promoted P. copri growth only in a dose-dependent manner and at a slower rate. Starch and carboxymethyl cellulose did not promote P. copri growth.15
Wheat bran extract, enriched in arabinoxylan oligosaccharides, enhances the growth of Prevotella species, especially P. copri.15 The high abundance of P. copri did not correlate with weight loss in participants on an arabinoxylan-oligosaccharide diet.16 While wheat bran fermentation was suggested to cause the enrichment of fecal microbiota with P. copri in one study,17 a diet intervention of wheat peptides and the fucose-rich sulfated polysaccharide fucoidan reduced the relative abundance of P. copri and was proposed to alleviate chronic gastritis.18 Although dietary fiber might influence Prevotella, pinpointing a particular fiber ingredient that can enrich this versatile organism remains challenging.14
The Western diet is a high-fat, high-sugar, and low-fiber diet.19 It is characterized by a high content of processed food. Limiting advanced glycation end products, which are present in Western diets, significantly reduced the abundance of P. copri.20 However, intriguingly enough, a study demonstrated that a high relative abundance of P. copri was associated with high-protein and Western diet.21 The metabolic machinery of P. copri might be a great demonstration of its genomic and functional strain-level variability, which allows P. copri to easily adapt to the type of nutrients available in the gut: while some strains can degrade carbohydrates and fibers, others can biosynthesize branched-chain amino acids (BCAAs) from meat-based diets.22 Recently, a four-week study on rats identified Prevotella among the bacteria that slightly decrease with age but are selectively enriched in pomegranate-fed animals.23
Another intriguing finding concerning the metabolizing power of P. copri is its ability to detoxify superoxide radicals and tolerate reactive oxygen species, which may otherwise increase inflammation.24 In a study of the gut microbiome of cyclists, a high abundance of Prevotella was associated with amino acid and carbohydrate metabolism.25 The inconsistency in Prevotella’s response to diet may be due to the interplay between host genetics and microbe-microbe interactions.17 Environmental factors and host habits may also contribute to another dimension of variation: geographical variation.21
A major player in the gut ecosystem and its microbial composition is bile. Bile acids, the main components of bile – produced by hepatocytes, are important for fat digestion and act as antibacterial agents against most Gram-negative gut bacteria, such as Prevotella, Bacteroides, and Parabacteroides.26 Enteric bacteria, by definition, are able to survive bile solubility. Bacteria also modulate the bile production and composition. Deoxycholic acid and lithocholic acid, which are secondary bile acids, are only produced by the action of bacterial enzymes during enterohepatic circulation;27 therefore, the gut microbiota composition plays a role in controlling the bile salt pool in the gut.28 The role of P. copri in the metabolism of bile salts has been previously reported.29 The reduction in P. copri abundance after the administration of antibiotics in rats resulted in a lower ratio of primary to secondary bile salts.29 Consequently, dysbiosis can change bile acid proportions, which are important for the prevention of liver diseases, such as cholestasis, inflammation, fibrosis, and tumors.30,31
In sickness and health: the ups and downs of P. copri in the human host
Multiple incongruent conclusions were drawn from research on P. copri, which include, but are not limited to, its relative abundance in the human gut, its correlation with diet, its encoded metabolic pathways, and its effect on host metabolism and host health (Table 1). One example is the association between P. copri and inflammation and glucose intolerance. Upon metabolizing fibers, P. copri produces short-chain fatty acids (SCFAs), which protect the mucosal barrier and reduce the possibility of inflammation.13 In one study, the decline in SCFAs was associated with a low abundance of P. copri, and therefore suggested to cause inflammatory diseases, such as rheumatoid arthritis, type 1 diabetes (T1D), and T2D.13 Yet, other studies have associated a higher abundance of P. copri with new-onset rheumatoid arthritis,43 or with impaired glucose tolerance.44 The latter was explained as an association with the production of BCAAs by P. copri.44
Table 1.
Controversial results in studies reporting on Prevotella copri. A selection of recent studies on P. copri, with emphasis on conflicting results.
| Condition | Conclusion | Ref. |
|---|---|---|
| Rheumatoid Arthritis (RA) vs. Osteoarthritis (OA) | P. copri tended to be more abundant in patients with RA, but with no statistical significance. | 32 |
| P. copri was significantly more abundant in the synovial fluid of patients with RA. | 33 | |
| RA vs. Healthy Controls | P. copri genes were not detected in healthy controls, but were abundant in patients with early onset RA. | 34 |
| RA vs. Healthy Controls | No difference in relative abundance of P. copri between controls and patients on treatment for RA. | 35 |
| Type 2 Diabetes (T2D) |
Patients with T2D suffered from intestinal dysbiosis, marked by an increase of relative P. copri abundance | 36 |
| Type 1 Diabetes (T1D) |
P. copri was one of the prevalent species in patients with T1D. | 37 |
| Chronic Urticaria (CU) |
At the species level, CU patients had significantly fewer P. copri amplicons than healthy controls. | 38 |
| Parkinson’s Disease (PD) |
Relative abundance of P. copri decreased in the PD cohort compared with the control group. | 13 |
| Chronic Liver Disease | P. copri was significantly more abundant in patients with advanced fibrosis and suggested to be used as a noninvasive marker for liver fibrosis. | 39 |
| Human Immunodeficiency Virus (HIV) |
In untreated adult patients with HIV-1, P. copri was more abundant than in control subjects. P. copri was positively associated with CD1c+ mDCs CD40 expression. | 40 |
| Gastric Cancer | P. copri was relatively more abundant in the gastric cancer cohort than the healthy controls. It was suggested as a risk factor for gastric cancer. | 41 |
| Food Allergy | No food allergies were reported in children whose mothers had P. copri in their gut. | 42 |
One of the key characteristics of P. copri is the expanded heterogeneity between strains.45 These divergent strains have been referred to as the “P. copri complex”.46 Compared with other microorganisms in the human gut, P. copri strains vary widely, perhaps because the organism is highly abundant, well-adapted, and widely distributed among humans. The diversity within the P. copri complex could be a clue to the P. copri paradox in health and disease states,43 and consequently, using just one reference strain of P. copri limits the exploration of the genomic and functional diversity of other P. copri strains.12 One study attributed discordance in P. copri research to strain differences, microbiome niche, host-microbiome metabolic interactions, and study design.47
In the next section, we discuss examples of studies associating relative P. copri abundance with human health and disease (Figure 1), highlighting how the conclusions on P. copri have been inconsistent and how challenging the research on P. copri is.
Examples in which P. copri is less abundant in disease
Host metabolism
To investigate the role of the human gut microbiome and diet in regulating human metabolism, Bäckhed et al. studied the effect of barely kernel-based bread (BKB) on glucose metabolism.8 In that study, responders were healthy individuals whose insulin and glucose levels improved with BKB consumption, while non-responders were those with least or no improvement in either insulin or glucose. P. copri was relatively more abundant in responders in whom complex polysaccharides were metabolized.8 When the microbiotas were transferred from responders and non-responders into two groups of germ-free mice, mice with responders’ microbiota had a higher relative abundance of Prevotella and enhanced glucose levels. These results suggested that Prevotella spp. may be beneficial to human health. Metagenomic analysis of fecal samples from responders and non-responders indicated that responders had a higher relative abundance of microbial genes encoding for complex polysaccharide fermentation. The latter finding was attributed to the capacity of Prevotella, specifically P. copri, to metabolize complex polysaccharides.48
Intestinal gluconeogenesis is a metabolic process that depends on succinate to aid in glucose homeostasis. Therefore, succinate may have metabolic advantages. Although succinate production by P. copri was shown to enhance glucose metabolism and insulin levels, succinate levels alone are insufficient to explain the beneficial role of P. copri in host glucose tolerance. Therefore, Prevotella was suggested to positively affect glucose levels independently of succinate production.8 An additional mechanism that may interpret glucose control by P. copri is increased bile acid metabolism and farnesoid X receptor signaling. In a study on Goto-Kakizaki rats that underwent vertical sleeve gastrectomy, the authors proposed P. copri as a plausible probiotic for T2D because of the abovementioned mechanisms.29 In a different study on the microbiome of 1,098 individuals, P. copri was found to have a higher relative abundance in individuals with good cardiometabolic markers, such as low visceral fat, high polyunsaturated fatty acids, low C-peptide, and lower postprandial glucose levels.49
Chronic urticaria
Chronic urticaria (CU) is a skin condition in which patients experience pruritus and sometimes swollen areas that persist for six weeks or longer without a clear cause.50 It might be due to the lack of balance in the immune system, which is tightly connected to the gut microbiota. A change in the diversity of the gut microbial community often results in a state of dysbiosis and may trigger inflammatory or allergic reactions.51–53 In a study that compared the gut microbiome of healthy individuals with CU, the relative abundance of P. copri, among other bacterial species, was significantly lower in patients with CU than in healthy individuals.38
Parkinson’s disease
Parkinson’s disease (PD) affects the central, autonomic, and enteric nervous systems. Accumulation and aggregation of the neuronal protein, α-synuclein, may contribute to PD propagation.54 In addition to a less diverse gut microbial community, the microbiome of patients with PD was found to contain less P. copri. Such microbiome imbalance was suggested to stimulate inflammation, Lewy body formation, and α-synuclein aggregation in nerve cells.55 Through the vagus nerve, these abnormal proteins can be transferred from the enteric system to the central nervous system.56 The past observation was further confirmed in a more elaborate study that used shotgun metagenomics:13 patients with early stage PD, who did not receive anti-Parkinson treatment, levodopa (L-DOPA), maintained a significantly lower relative abundance of P. copri in comparison with age-matched healthy controls.13
Autism spectrum disorders
The gut-brain axis is a term that describes the two-way relationship between the gastrointestinal tract and brain in humans. Accruing evidence has established a vital role for the gut microbiota in the communication between the gut and the brain. Consequently, intestinal dysbiosis may be involved in neurological disorders.57
A major example of the involvement of microbiota in disease through the gut-brain axis is autism spectrum disorder. According to the National Institute of Mental Health, autism spectrum disorders (ASD) are behavioral and communication disorders in which symptoms may appear in the early years of life.58 A comparison of children with ASD with typically cognitive children showed a decreased relative abundance of P. copri in the first group.59 Although diet is an important element affecting gut microbiota composition, Prevotella and P. copri-like operational taxonomic units were absent in children with autism suffering from gastrointestinal problems unrelated to their diet.60 Intriguingly, in another study, the relative abundance of P. copri was significantly higher in children with ASD than that in healthy controls.61
Food allergy
In the Barwon Infant Study in Australia, the microbiota of mothers and their infants was analyzed, and the infants were tested for different types of food allergies. Remarkably, infants whose mothers carried P. copri during pregnancy were at lower risk for food allergy, especially mothers who were on a high-fiber high-fat diet.42 Suggested mechanisms behind the observed P. copri-associated maternal protection against food allergy development in infants include succinate production, maternal IgG bound to epitopes from P. copri, and P. copri endotoxin inhibiting Toll-like receptor 4 signaling during the development of the fetal immune system.42 Moreover, P. copri was reported to be more abundant in healthy controls than in individuals with multiple food allergies, and this observation was explained by SCFA production in healthy controls.62
Examples of P. copri enrichment in disease conditions
Rheumatoid arthritis
The Centers for Disease Control and Prevention define rheumatoid arthritis (RA) as an inflammatory autoimmune disorder that gradually destructs joint tissues. While the exact reasons behind RA remain unknown, genetic variations between individuals are insufficient to explain RA.63 Through distinguishable mechanisms, researchers have advocated the human intestinal microbiota as a vital player in joint diseases, especially in the state of dysbiosis.32,33,43
Accruing studies on P. copri strongly associate its putative pro-inflammatory characteristics with the development of RA. In a study of patients with new-onset untreated rheumatoid arthritis (NORA), P. copri was relatively more abundant in patients with NORA than in healthy controls.43 In the same study, a mouse model of gut inflammation demonstrated that P. copri augmented chemical colitis, and mice showed more severe disease. The authors suggested that limiting the spread of P. copri might halt the incidence or progression.43 A recent study demonstrated the prevalence of P. copri in the early stages of RA. The authors compared healthy controls with the naïve patients with RA and those receiving disease-modifying antirheumatic drugs (DMARDs).34 To explore the association between early RA and P. copri, Maeda and coworkers inoculated germ-free SKG mice with fecal microbiota obtained from patients with RA and treated them with zymosan, which triggers the disease.63 Severe arthritis in SKG mice was explained by an increase in intestinal T-helper 17 cells. In addition, lymphocytes and P. copri-stimulated dendritic cells responded to the arthritis-related autoantigen 60S ribosomal protein L23a by producing interleukin-17.63
The interactions between P. copri and the immune system, and how these interactions may reflect RA progression have been investigated.64,65 During an RA flare of a chronic patient on DMARDs, a peripheral blood mononuclear cell sample contained a P. copri HLA-DR-presented peptide. This peptide was predicted to be derived from a P. copri 27-kDa protein and is believed to stimulate an immunogenic response via T-helper 1 cells. Both the epitope and the whole P. copri organism were able to trigger host antibody responses, which varied between IgA and IgG responses that were only specific to patients with RA rather than other types of arthritis, or to healthy controls. Based on this, one may predict local and systemic immune responses to either P. copri or its T-cell epitope. Surprisingly, P. copri 16S rRNA genes were detected in the synovial fluid of patients with NORA and chronic RA, albeit at low abundance.64 Based on these findings, the authors of that study hypothesized that P. copri might continue to circulate from the intestines to the joints, carried by macrophages, during the disease course.64 A comparison of the microbiome composition in the synovial fluid of patients with RA with that of patients with osteoarthritis (OA) demonstrated a higher relative abundance of P. copri in patients with RA. This finding further supports the unique relationship between the organism and RA.33 A recent study reported a correlation between anti-P. copri P27 antibodies with autoantibodies found in RA patients and proposed a potential causal role for P. copri in RA evolution and synovitis.66
Even though the above studies suggested P. copri as a key microbe associated with RA, a study on patients receiving treatment for RA found no such association:35 Neither the abundance of P. copri nor the family Prevotellaceae was significantly different between patients with RA and healthy controls.35 Methotrexate (MTX) is one of the main therapeutic agents used for the treatment of RA, and its mechanism of action depends on the inhibition of dihydrofolate reductase (DHF). Enzymes involved in tetrahydrofolate biosynthesis were found to be much less expressed in the P. copri-enriched microbiome. Therefore, oral MTX had better bioavailability in patients with NORA having P. copri in their microbiome, because of the lower competition between the host and microbiota DHF.43 Metagenomic analysis of the microbiomes of healthy individuals vs. patients with NORA delineated specific open reading frames (ORF) in each cohort. At least two ORFs of the nuo operon, coding for components of NADH:ubiquinone oxidoreductase, were associated with healthy status. In contrast, particular ORFs encoding components of an ATP-binding cassette iron transporter were NORA-specific. These distinct ORFs were proposed to serve as microbiome markers to differentiate between healthy and diseased individuals.43 Yet, defining an enterotype as a biomarker for RA has remained an unachieved goal, at least until 2019.67
The discrepant conclusions of the above studies, if we assume they were performed rigorously, endorse the existence of dissimilar strains of P. copri with variable genetic makeup and phenotypes, or dissimilar regulatory programs for P. copri strains under different conditions.68 Studies started exploring this strain dissimilarity to define signature genes associated with “RA-pathogenic” strains of P. copri; those genes belong to the accessory – rather than core – genomes of these strains.69
Ankylosing spondylitis
Ankylosing spondylitis (AS) is a type of chronic inflammatory arthritis that mainly affects the spine and causes rigidity and fusion of the vertebrae.70 In a study on Chinese patients with AS and healthy controls, multiple Prevotella species were relatively more abundant in the patient cohort. Among these, P. copri, P. melaninogenica, and Prevotella sp. C561.71 When the fecal microbiota of healthy controls was compared with that of untreated patients with AS and a subset of those patients after treatment, the relative abundance of P. copri was found to be higher in patients with AS, but lower after treatment with DMARDs or TNF-alpha inhibitors.72
Active graves’ orbitopathy
Graves’ orbitopathy (GO) is an autoimmune disorder that threatens the sight of patients with Graves’ disease.73 Thyrotropin receptor autoantibody (TRAb) is recommended by the Thyroid Association guidelines as a marker for Graves’ disease and for GO management.74 In patients with GO, who had high levels of TRAb, the abundance of the family Prevotellaceae, and P. copri species, in particular, in the gut microbial community was correlated with TRAb levels. However, the exact mechanism by which P. copri influences TRAb levels has not been reported.75
Diabetes mellitus
Despite studies describing the role of P. copri in improving glucose homeostasis, contradicting evidence suggests that the same microbe may be associated with insulin resistance.44,76 In individuals with insulin resistance, BCAA levels were high, according to metabolomic analysis. These levels were attributed, at least in part, to the overabundance of P. copri in the gut microbiome of these patients.44 Functional metagenomic analysis reflected the ability of P. copri to synthesize BCAAs, lipopolysaccharides, and tryptophan. Mice developed insulin resistance and a high BCAAs level when orally administered P. copri.44 A different study confirmed the enrichment of P. copri in patients with T2D and suggested P. copri as a potential biomarker for the development of this important human disease.76
The outer membrane of P. copri contains lipopolysaccharide (LPS). Pooled LPS from the microbiome of healthy individuals, especially from members of the order Bacteroidales, was found to play a role in tolerating the host immune system.77 The surge of LPS in the blood results in a state of metabolic endotoxemia, which results in an inflammatory reaction and insulin resistance.77 A positive correlation between BCAAs and LPS in the serum was concluded.36 Patients with T2D, especially those on metformin only (rather than metformin and glibenclamide), had a higher relative abundance of P. copri than healthy controls. An oral antidiabetic regimen was suggested to influence P. copri, and a functional food was proposed to decrease its abundance.36
Because the intestinal walls are more permeable in presence of Gram-negative bacteria, a host inflammatory response is expected with P. copri. This may explain the poor glucose management reported in patients with T1D, as P. copri was more abundant in diabetic patients than in healthy participants.37
Two other studies have reported a higher relative abundance of P. copri in adult and pediatric patients with T1D.78,79
Cancer
The link between P. copri and different types of cancer remains inexplicit. P. copri may have a protective effect against colorectal cancer, as demonstrated by tumor size shrinkage in rats.80 While some studies have reported P. copri to be one of the causes of colorectal cancer and a useful diagnostic microbial biomarker for colorectal cancer,81,82 P. copri was reportedly more abundant in the fecal microbiomes of healthy individuals than those of patients with colorectal cancer.83,84 In a study combining wet-lab investigation and meta-analysis of fecal microbiome data of patients with melanoma, P. copri was more abundant in patients with stage I and stage II melanoma than in healthy individuals.85 While P. copri had a lower abundance in gastric tumor and peritumor microenvironments compared with normal tissue,86 patients with gastric cancer had a higher relative abundance of P. copri than healthy controls.41 Moreover, in a study performed specifically to determine the risk of gastric cancer in the Korean population, participants whose gastric microbiome had P. copri were found to be at higher risk of developing gastric cancer than those with P. copri-free microbiome.41 A set of microorganisms in the salivary microbiome, including P. copri, were shortlisted as biomarkers for oropharyngeal and hypopharyngeal cancers.87
Cardiovascular diseases
To delve into the role of the gut microbiota in cardiovascular diseases with comparable risk factors, researchers investigated the gut microbiome profiles of patients with coronary artery disease and valve calcification. P. copri is a key microbe, particularly in patients with valve calcification. Additionally, P. copri was positively associated with high levels of low-density lipoprotein, which is a risk factor for cardiovascular diseases.88 This finding contradicts the positive correlation reported between the abundance of P. copri and high-density lipoprotein cholesterol in another study.89 In addition, P. copri is significantly more abundant in the gut microbiota of patients with acute cerebral infarction than in healthy individuals.90
Liver diseases
When lipid droplets accumulate in 5% or more of liver cells, patients develop nonalcoholic fatty liver disease (NAFLD). When accompanied by inflammation, NAFLD evolves into a more aggressive disease state called nonalcoholic steatohepatitis (NASH). NASH may lead to liver fibrosis and, eventually liver cirrhosis. P. copri positively correlated with more severe liver fibrosis in children.24 A reasonable justification for this correlation is the highly significant increase in the expression of the microbiome-derived inflammatory products. For example, intermediate biomolecules involved in LPS biosynthesis and flagellar assembly discriminated between healthy obese children and children with chronic liver disease, particularly NASH. Despite opposing findings on the role of P. copri in liver diseases, P. copri may represent a noninvasive predictor of liver fibrosis, particularly in the state of microbiome dysbiosis.24,39
As stated above, liver health status is strongly associated with bile-microbiome interactions. In primary sclerosing cholangitis, a liver disease coupled with inflammation in the bile ducts, P. copri was negatively correlated with the disease, especially in patients with concurrent inflammatory bowel disease.91 Enriching the gut microbiota with P. copri was shown to decrease cholestasis and liver fibrosis.92 Conversely, in stage-four hepatitis C, P. copri was more abundant than in healthy volunteers;93 this observation was suggested to be associated with inflammation and high Th17 and IL-17 levels.93 Likewise, a recent metagenomic study associated P. copri with NASH in subjects with obesity and attributed this risk for NASH to lower abundance of butyrate-producing pathways and possible higher intestinal permeability.94
Myasthenia gravis (MG)
Liu et al. reported a correlation between MG development and dysbiosis in pediatric patients. An increase in the relative abundance of P. copri and other microbes in patients with myasthenia gravis causes a reduction in SCFA production. This reduction seems to play a crucial role in MG development.95
Infectious diseases
Listeria monocytogenes
Listeriosis is a foodborne illness caused by the Gram-positive bacterium, Listeria monocytogenes. It accounts for massive outbreaks and, in some cases, can lead to severe illnesses, such as sepsis and meningitis. The intestinal microbiota protects against Listeria monocytogenes colonization of the gut and against its transfer to the bloodstream.96 However, P. copri might decrease the thickness of the mucosal barriers, thus enhancing their permeability to Listeria and exacerbating inflammation,97 most likely because Prevotella synthesizes sulfatase enzymes that degrade the intestinal mucosa’s mucin lining.98 In contrast, the bacteriocin Lmo2776, produced by Listeria monocytogenes, has an inhibitory effect on the growth of P. copri and may decrease its abundance, avoiding excessive inflammation.97
Bacteremia
The first case of blood infection with P. copri was reported in an elderly patient with heart failure. Gut microbiome analysis showed that P. copri was the most abundant microorganism in the gut, and was thus suggested as a biomarker for failure of the intestinal barrier.2 As stated above, dysbiosis may increase gut permeability and translocate some members of the gut microbial community to the blood.
Traveler’s diarrhea
To investigate the relationship between the gut microbiome and travelers’ diarrhea, a large study team analyzed the gut microbiome of 43 participants before and after their travel to tropical regions. P. copri was more abundant in participants who had diarrhea.99 In a symptomatic infection by Entamoeba histolytica, the abundance of P. copri was reported to be higher than that in asymptomatic infection by the same parasite.100,101 Yet, explaining the association between P. copri and traveler’s diarrhea is quite confusing because there has been no consensus in the literature on whether P. copri may cause or protect against it.100,102,103 This discrepancy can be attributed to the vast differences between the probable strains of P. copri.
Human immunodeficiency virus
Chronic activation of the innate immune system, and thus chronic inflammation, may lead to a worse prognosis for human immunodeficiency virus (HIV). A possible rationale for chronic inflammation is microbial translocation from the gut to the circulatory system together with LPS production. High expression of colonic myeloid dendritic cells (CD40 cells) was positively correlated with HIV viral load and P. copri abundance in patients with AIDS, compared with healthy individuals. Similarly, P. copri prompts the maturation of myeloid dendritic cells to produce inflammatory cytokines in vitro.40 Not only does P. copri have this signature in naïve HIV patients, but it is also more abundant in children with HIV and on antiretroviral therapy.104 Even during treatment, antiretroviral therapy did not protect against increased intestinal permeability, as indicated by the positively correlated soluble CD14, a marker for microbial translocation, with the higher abundance of P. copri in the treated cohort.104
Pharmacomicrobiomics and P. copri
Pharmacomicrobiomics studies mutual interactions between the human microbiome and medications.105,106 Prevotella is a gut microorganism with several documented pharmacomicrobiomic interactions. Carboplatin is an anticancer agent that may cause inflammation of the intestinal membrane in some patients. It also impacts the composition and diversity of the gut microbiota. In a mouse experiment, the severity of carboplatin-induced mucositis increased with a high abundance of P. copri.107 Moreover, when mice were pretreated with metronidazole, a drug with antifungal and selective antimicrobial activity against anaerobes, the intensity of the mucosal damage decreased as P. copri abundance was reduced. In light of these observations, targeting P. copri was suggested as a potential feasible strategy to relieve carboplatin-induced mucositis.107
Eradication therapy for Helicobacter pylori has been found to increase high-density lipoprotein levels, which are negatively associated with cardiovascular disease risk.89 This observation was attributed to disturbance in gut microbiome composition. Intriguingly, the abundance of P. copri, unlike several other members of the phylum Bacteroidetes, was positively correlated with high-density lipoprotein cholesterol after eradication therapy.89
P. copri affects the human response to nivolumab, which is an immunotherapeutic agent (anti-programmed cell death-1) used to treat many types of cancer.108 The activity of nivolumab was measured by the number of unique memory CD8+ T cells and natural killer cells after drug use. Patients with non-small cell lung carcinoma receiving nivolumab were classified as responders or non-responders according to the number of immune cells produced. Responders had more diverse gut microbiomes before and during drug treatment than non-responders did. P. copri was one of the most abundant microorganisms in the responders’ microbiomes. This was explained by the beneficial role of P. copri, which synergizes with nivolumab’s action.109
Glucagon-like peptide-1 receptor agonist (GLP-1 RA) is an antidiabetic drug that induces variable responses in individuals. When the gut microbiome composition was compared between responders and non-responders by 16S rRNA amplicon sequencing, P. copri had higher relative abundance in non-responders, which has been taken as evidence for a negative correlation between P. copri abundance and glycemic control with GLP-1 RA.110
The laborious P. copri
This section discusses why studying and reaching conclusions on P. copri is toilsome work. This includes both the study design and data analysis.
Study design
Sampling methods
Fecal samples are the most common type of samples used in gut microbiome research. Fecal sampling is noninvasive, inexpensive, and relatively easy. However, fecal samples do not reflect inherent alterations in the microbiota along the gastrointestinal tract and may be more susceptible to environmental contamination than other tissues. Other sampling techniques, such as mucosal biopsy, intestinal fluid collection, and rectal swabs, have been reviewed elsewhere.111
Suitable storage conditions for fecal samples play a major role in obtaining reliable research results. These conditions, if not well maintained and kept consistent, may introduce variability across studies. The anaerobic nature of P. copri adds another dimension to the complexity of its sample collection and storage. The cultivation of noncommercial P. copri strains is prohibitive because of their intolerance to oxygen and slow growth.
Confounding factors
Most microbiome studies focused on the association between the human microbiome and disease state. Researchers aspire to find straightforward interrelations between microbes and illnesses, so that alternative therapies can be introduced to patients. Nevertheless, numerous confounding factors can stand against this goal. For instance, host-microbiome and microbiome-microbiome interactions are copious, and fluctuate between individuals and within the same individual. Geographical location has striking consequences on an individual’s lifestyle, eating habits, and even microbiome evolution. Although researchers invest considerable efforts to design controlled microbiome studies and minimize confounding variables, recruiting healthy individuals or patients with similar microbiota for microbiome studies can be challenging. Other than strain variations, an additional explanation for the vastly contradictory results between the above studies is the variability in confounding factors; thus, we have indicated experimental approaches and analysis strategies in selected studies (Table 2).
Table 2.
Different statistical and bioinformatics approaches used in studying P. copri.
| Condition | Approach | Analysis | Observation | Ref. |
|---|---|---|---|---|
| NORA, Chronic RA | 16S rRNA | GraphPad Prism 6 | ↑ abundance in synovial fluid | 64 |
| RA vs. healthy | V3-V5 16S rRNA regions | IM TORNADO/LEfSe | No difference in abundance | 35 |
| AS vs. RA and Behcet’s Disease | Metagenomic shotgun sequencing | In-house pipeline | ↑ abundance in AS cohort ↓ abundance in patients with AS receiving TNFi or DMARDs rather than NSAIDs only |
72 |
| T1D | V3-V4 16S rRNA regions | QIIME | ↑ prevalence in patients with T1D | 37 |
| T2D | Whole genome sequencing | R packages | P. copri enhanced glucose tolerance | 29 |
| PD | V3-V4 16S rRNA region | QIIME/ANOSIM/STAMP software | ↑ abundance in control cohort | 55 |
| Active Graves’ Orbitopathy | V4 16S rRNA region | Co-occurrence Network Analysis. Weighted Gene Coexpression Network Analysis |
↑ P. copri with ↑levels of thyrotropin receptor antibody. | 75 |
| HIV | V3-V4 16S rRNA regions | QIIME/LEfSE | ↑ P. copri in children with perinatally transmitted HIV and on ART | 104 |
| Bacteremia | V3, V4, V6 16S rRNA regions | MicrobAT system/BLAST | ↑ P. copri in blood and intestine of patient with heart failure. | 2 |
| Acute Cerebral Infarction | V4 16S rRNA region | QIIME/PLS-DA | ↑ P. copri in disease cohort | 90 |
| Myasthenia Gravis | Wholegenome shotgun sequencing | In-house bioinformatics pipeline. | ↑ P. copri in pediatric patients with MG | 95 |
↑ = increase; ↓ = decrease; Ref = reference.
Sample size
A major limitation in drawing strong conclusions is the sample size of the study. While a large sample size evidently increases the power of a study and may limit the influence of confounding factors, researchers are sometimes limited by resources, funding, compliance of participants, and the nature of the disease under study.
In Table 3, we compile a representative set of studies on P. copri, with careful delineation of their type, sample size, and conclusions about P. copri abundance. While the study designs can be more comprehensive, we focused on samples in which P. copri was detected.
Table 3.
Sample type and size in selected studies on involvement of P. copri in human diseases.
| Condition | Samples | Sample Size* | Abundance in Disease State | Ref |
|---|---|---|---|---|
| RA vs. OA | Synovial tissue Synovial fluid |
Total: 183 patients RA [n = 125] OA [n = 58] |
↑ | 33 |
| RA vs. healthy | Fecal | Total: 31 early RA [n = 17] controls [n = 14] |
↑ | 63 |
| RA vs. healthy | Fecal | Total: 72 Control [n = 32] RA [n = 40] |
No difference | 35 |
| NORA vs. healthy | Fecal | 114 stool samples [RA and control] | ↑ | 43 |
| T2D | Fecal | Total: 80 Healthy [n = 22] T2D [n = 58] |
↑ | 36 |
| T1D | Fecal | Total: 48 Healthy [n = 28] T1D [n = 20] |
↑ | 37 |
| CU | Fecal | Total: 20 Healthy [n = 10] CU [n = 10] |
↓ | 38 |
| Primary sclerosing cholangitis-inflammatory bowel disease (PSC-IBD) |
Fecal | Total: 106 PSC [n = 11] PSC-IBD [n = 32] UC [n = 32] HCT [n = 31] |
↓ | 91 |
| PD | Fecal | Total: 155 PD [n = 89] Controls [n = 66] |
↓ | 55 |
| NAFLD | Fecal | Total: 124 NAFLD [n = 87] Obese [n = 37] |
↑ | 24 |
| ASD | Fecal | Total: 44 children ASD [n = 23] neurotypical [n = 21] |
↓ | 59 |
| MG | Fecal | Total: 99 pediatrics MG [n = 53] Healthy [n = 46] |
↑ | 95 |
*In these studies, additional samples were sometimes analyzed; however, the number given only refers to samples in which P. copri was detected.
↑ = increase; ↓ = decrease; Ref = reference.
Animal models
To validate the observations made on human samples and to establish causality, researchers use animal models in which they inoculate the microbe of interest, mostly after depletion of the normal microbiota of the animal, and often after reconstituting the microbiota with certain organisms. They may test for immune responses, as in P. copri studies.
Even though animal models, especially mice, can serve this purpose, the influence of differences in physiology is substantial. In addition, the murine and human microbiomes vary widely.68 Yet, the greatest strength of animal models is their ability to establish causality, which is usually practically and ethically not feasible in humans. Once an association with a certain organism is established, this organism can be added to the animal microbiome in different amounts, and at different time points. Such analysis can identify whether the studied phenotype is a consequence of the addition of the organism and whether it is dose dependent.
In silico analysis
Most microbiome studies apply 16S rRNA amplicon sequencing to characterize the human microbiome community. Although efficient, sequencing variable regions of the 16S rRNA gene might be less specific to discern differences between species and is unable to discern strain differences. Species differentiation has been made possible by full 16S rRNA gene sequencing with long-read sequencing strategies, such as nanopore112 or single molecule real-time (SMRT) sequencing.113 Still, amplicon sequencing cannot describe the functional potential of a significant microbe, notably the substantial proportion of functions encoded by the non-core genome of a particular species or genus. Researchers often seek to explain why a certain microbe is either positively or negatively correlated with the disease; shotgun metagenomic sequencing provides the data needed for comprehensive functional analysis that may guide the understanding of the association between microbes and diseases.
Shotgun metagenomic sequencing, with sufficient sequencing depth, allows de novo assembly to unveil new taxa or new strains that may share a similar core genome but a variable accessory genome. Comparative genomics clarifies strain variations at the single-nucleotide level, genomic rearrangement, and loss or gain of genes, plasmids, or operons. This can hugely shape the microorganism’s metabolizing potential, its interaction with the host, its coping with the host immune system, and its dynamic communication with other microbes in the same niche.43,45
Exploring the complexity of P. copri’s gene pool is only at its early stages. Genomic and phylogenomic studies are accruing to estimate the actual diversity of this human gut-adapted organism. For example, Nii and coworkers69 attempted to address this question with reference to the involvement of P. copri in disease (specifically RA),69 while Lo Presti and colleagues took a phylogenetic approach to link specific clades to the pathogenicity of bowel diseases.114
Because of this immense genomic repertoire, so-called reference strains used in disease-association studies are not always informative because strains vary geographically, and because, while reference strains are historically more convenient or first to study, they are not necessarily a true representative of a species, if such a ‘representative strain’ concept ever exists. This reference strain dilemma powerfully accounts for the many discrepancies in research findings regarding P. copri.14 As reviewed above, the same biological mechanism carried out by P. copri could contribute to two opposing conclusions, while two mutually exclusive metabolic pathways carried by P. copri have been proven in different studies.1,12,22 In one person, a single strain of a species prevails in the microbiome, either stochastically or according to the host lifestyle, and other host and environmental factors that structure the microbiome composition.45 Prevotella, in general, and P. copri, in particular, happened to be highly abundant115 and easily detectable microbiome members (since the emergence of culture-independent techniques); thus, they become ‘usual suspects’ behind several studied phenotypes.
Reviewing multiple publications reveals how variable the sequencing approach, and bioinformatic and statistical analyses are between studies (Table 2). We assume that these might inflict an extra layer of variability on the conclusions of the P. copri studies.
A glimpse into the future
A call for best practices in microbiome analysis is as crucial as the rapid advancement in sequencing technologies, and analysis tools and programs. The best practices reinforce the reliability and reproducibility of microbiome studies. The best microbiome analysis practices can pave the way for well-founded meta-analyses, more research hypotheses, and more solid conclusions. Best laboratory practices combined with established and transparent standard operating procedures for analysis can ensure high-quality research and reduce technical errors.
Updating the available databases with curated reference genome sequences of the different strains of P. copri will guarantee a better understanding of its association with disease and health and create less confusion. Developing computational tools and algorithms that enhance strain-level resolution will accelerate and empower correlation studies. Focusing on strain-level analyses, whether through genome sequencing of cultured isolates, single-cell genomics, or metagenome-assembled genome sequencing, will help estimate the actual diversity of the P. copri complex and the breadth of its genomic repertoire. Using comparative genomics and phylogenomics will allow a precise definition of the core/pan genome and an estimation of the number of key clades/types of this versatile species, respectively. Machine learning algorithms will develop various classifiers to determine signature gene sets, regulons, and hub genes involved in specific association with, or protection from, different disease conditions.
To grasp the sophistication of the ongoing dynamics between the host and its microbiome and within the microbiome itself, one must implement multi-omics analyses and avoid conclusions based on geographically, spatially, or temporally confined studies. With more geographically representative microbiomes sequenced, and with the complexity and heterogeneity of omics data generated, machine learning can be implemented to incorporate data multidimensionality into context, thus distinguishing real disease associations from diet- or geography-based confounders. With all of the above implemented, perhaps one day, primary care physicians will request a routine microbiome analysis through which they can intervene noninvasively, by removing, adding, or modifying specific organisms to improve the quality of human life.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Abbreviations
AS: Ankylosing spondylitis; ASD: Autism spectrum disorders; BCAAs: Branched-chain amino acids; BKB: Barley kernel-based bread; CU: Chronic urticaria; DHF: Dihydrofolate reductase; DMARDs: Disease-modifying antirheumatic drugs; GLP-1 RA: Glucagon-like peptide-1 receptor agonist; GO: Graves’ orbitopathy; HIV: Human immunodeficiency virus; L-DOPA: l-3,4-dihydroxyphenylalanine or Levodopa; LPS: Lipopolysaccharides; MG: Myasthenia gravis; MTX: Methotrexate; NAFLD: Nonalcoholic fatty liver disease; NASH: Nonalcoholic steatohepatitis; NORA: New-onset untreated rheumatoid arthritis; OA: Osteoarthritis; PD: Parkinson’s disease; PSC-IBD: Primary sclerosing cholangitis-inflammatory bowel disease; RA: Rheumatoid arthritis; SCFAs: Short-chain fatty Acids; T1D: Type 1 diabetes; T2D: Type 2 diabetes; TRAb: Thyrotropin receptor autoantibody
References
- 1.Claus SP. The Strange Case of Prevotella copri: Dr. Jekyll or Mr. Hyde? Cell Host & Microbe. 2019;26(5):577–18. doi: 10.1016/j.chom.2019.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Posteraro P, De Maio F, Menchinelli G, Palucci I, Errico FM, Carbone M, Sanguinetti M, Gasbarrini A, Posteraro B. First bloodstream infection caused by Prevotella copri in a heart failure elderly patient with Prevotella-dominated gut microbiota: a case report. Gut Pathog. 2019;11(1):44. doi: 10.1186/s13099-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah HN, Collins DM. Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol. 1990;40(2):205–208. doi: 10.1099/00207713-40-2-205. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi H, Shibata K, Sakamoto M, Tomita S, Benno Y. Prevotella copri sp. nov. And Prevotella stercorea sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2007;57(5):941–946. doi: 10.1099/ijs.0.64778-0. [DOI] [PubMed] [Google Scholar]
- 5.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9(1):e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Franke T, Deppenmeier U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol Microbiol. 2018;109(4):528–540. doi: 10.1111/mmi.14058. [DOI] [PubMed] [Google Scholar]
- 10.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281(47):36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 11.White BA, Lamed R, Bayer EA, Flint HJ. Biomass utilization by gut microbiomes. Annu Rev Microbiol. 2014;68(1):279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 12.Fehlner-Peach H, Magnabosco C, Raghavan V, Scher JU, Tett A, Cox LM, Gottsegen C, Watters A, Wiltshire-Gordon JD, Segata N, et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe. 2019;26(5):680–90.e5. doi: 10.1016/j.chom.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedarf JR, Hildebrand F, Coelho LP, Sunagawa S, Bahram M, Goeser F, Bork P, Wüllner U. Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson’s disease patients. Genome Med. 2017;9(1):39. doi: 10.1186/s13073-017-0428-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol. 2016;13(2):69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 15.Pareek S, Kurakawa T, Das B, Motooka D, Nakaya S, Rongsen-Chandola T, Goyal N, Kayama H, Dodd D, Okumura R, et al. Comparison of Japanese and Indian intestinal microbiota shows diet-dependent interaction between bacteria and fungi. NPJ Biofilms Microbiomes. 2019;5(1):37. doi: 10.1038/s41522-019-0110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen L, Sørensen CV, Wøhlk FU, Kjølbæk L, Astrup A, Sanz Y, Hjorth MF, Benítez-Páez A. Microbial enterotypes beyond genus level: Bacteroides species as a predictive biomarker for weight change upon controlled intervention with arabinoxylan oligosaccharides in overweight subjects. Gut Microbes. 2020;12(1):1847627. doi: 10.1080/19490976.2020.1847627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Paepe K, Verspreet J, Courtin CM, Van de Wiele T. Microbial succession during wheat bran fermentation and colonisation by human faecal microbiota as a result of niche diversification. ISME J. 2020;14(2):584–596. doi: 10.1038/s41396-019-0550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kan J, Cheng J, Xu L, Hood M, Zhong D, Cheng M, Liu Y, Chen L, Du J. The combination of wheat peptides and fucoidan protects against chronic superficial gastritis and alters gut microbiota: a double-blinded, placebo-controlled study. Eur J Nutr. 2020;59(4):1655–1666. doi: 10.1007/s00394-019-02020-6. [DOI] [PubMed] [Google Scholar]
- 19.Rakhra V, Galappaththy SL, Bulchandani S, Cabandugama PK. Obesity and the Western diet: how we got here. Mo Med. 2020;117:536–538. [PMC free article] [PubMed] [Google Scholar]
- 20.Yacoub R, Nugent M, Cai W, Nadkarni GN, Chaves LD, Abyad S, Honan AM, Thomas SA, Zheng W, Valiyaparambil SA, et al. Advanced glycation end products dietary restriction effects on bacterial gut microbiota in peritoneal dialysis patients; a randomized open label controlled trial. PLoS One. 2017;12(9):e0184789. doi: 10.1371/journal.pone.0184789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Filippis F, Pasolli E, Tett A, Tarallo S, Naccarati A, De Angelis M, Neviani E, Cocolin L, Gobbetti M, Segata N, et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25(3):444–53.e3. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Metwaly A, Haller D. Strain-level diversity in the gut: The P. copri case. Cell Host Microbe. 2019;25(3):349–350. doi: 10.1016/j.chom.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Attia H, ElBanna SA, Khattab RA, Farag MA, Yassin AS, Aziz RK. Integrating microbiome analysis, metabolomics, bioinformatics, and histopathology to elucidate the protective effects of pomegranate juice against benzo-alpha-pyrene-induced colon pathologies. Int J Mol Sci. 2023;24(13):24. doi: 10.3390/ijms241310691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, Bross C, Durelle J, Goyal NP, Hamilton G, et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology. 2019;157(4):1109–1122. doi: 10.1053/j.gastro.2019.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen LM, Bautista EJ, Nguyen H, Hanson BM, Chen L, Lek SH, Sodergren E, Weinstock GM. Community characteristics of the gut microbiomes of competitive cyclists. Microbiome. 2017;5(1):98. doi: 10.1186/s40168-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng X, Huang F, Zhao A, Lei S, Zhang Y, Xie G, Chen T, Qu C, Rajani C, Dong B, et al. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017;15(1):120. doi: 10.1186/s12915-017-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Urdaneta V, Casadesús J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med. 2017;4:163. doi: 10.3389/fmed.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Péan N, Le Lay A, Brial F, Wasserscheid J, Rouch C, Vincent M, Myridakis A, Hedjazi L, Dumas M-E, Grundberg E, et al. Dominant gut Prevotella copri in gastrectomised non-obese diabetic Goto–Kakizaki rats improves glucose homeostasis through enhanced FXR signalling. Diabetologia. 2020;63(6):1223–1235. doi: 10.1007/s00125-020-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell CB, Cowen AE. Bile salt metabolism. II. Bile salts and disease. Aust NZ J Med. 1977;7(6):587–595. doi: 10.1111/j.1445-5994.1977.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 31.Chiang JYL, Ferrell JM. Bile acid metabolism in liver pathobiology. Gene Expr. 2018;18(2):71–87. doi: 10.3727/105221618X15156018385515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JY, Mannaa M, Kim Y, Kim J, Kim GT, Seo YS. Comparative analysis of fecal microbiota composition between rheumatoid arthritis and osteoarthritis patients. Genes (Basel). 2019;10(10):10. doi: 10.3390/genes10100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Chen B, Li S, Yang L, Zhu D, Wang Y, Wang H, Wang T, Shi B, Gai Z, et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci Rep. 2018;8(1):14305. doi: 10.1038/s41598-018-32675-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, Taneja V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8(1):43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina-Vera I, Sanchez-Tapia M, Noriega-López L, Granados-Portillo O, Guevara-Cruz M, Flores-López A, Avila-Nava A, Fernández ML, Tovar AR, Torres N, et al. A dietary intervention with functional foods reduces metabolic endotoxaemia and attenuates biochemical abnormalities by modifying faecal microbiota in people with type 2 diabetes. Diabetes Metab. 2019;45(2):122–131. doi: 10.1016/j.diabet.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Higuchi BS, Rodrigues N, Gonzaga MI, Paiolo JCC, Stefanutto N, Omori WP, Pinheiro DG, Brisotti JL, Matheucci E, Mariano VS, et al. Intestinal dysbiosis in autoimmune diabetes is correlated with poor glycemic control and increased interleukin-6: a pilot study. Front Immunol. 2018;9:1689. doi: 10.3389/fimmu.2018.01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu T, Chen Y, Guo Y, Sun J, Shen W, Yuan M, Zhang S, He P, Jiao X. Altered gut microbiota diversity and composition in chronic urticaria. Dis MarkersDis Markers. 2019;2019:1–11. doi: 10.1155/2019/6417471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong TS, Katzka W, Lagishetty V, Luu K, Hauer M, Pisegna J, Jacobs JP. A microbial signature identifies advanced fibrosis in patients with chronic liver disease mainly due to NAFLD. Sci Rep. 2020;10(1):2771. doi: 10.1038/s41598-020-59535-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, Smith DM, Landay AL, McManus MC, Robertson CE, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 2016;9(1):24–37. doi: 10.1038/mi.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunathilake MN, Lee J, Choi IJ, Kim YI, Ahn Y, Park C, Kim J. Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci Rep. 2019;9(1):13589. doi: 10.1038/s41598-019-50054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuillermin PJ, O’Hely M, Collier F, Allen KJ, Tang MLK, Harrison LC, Carlin JB, Saffery R, Ranganathan S, Sly PD, et al. Maternal carriage of Prevotella during pregnancy associates with protection against food allergy in the offspring. Nat Commun. 2020;11(1):1452. doi: 10.1038/s41467-020-14552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 45.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27(4):626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, Armanini F, Manghi P, Bonham K, Zolfo M, et al. The Prevotella copri complex comprises four distinct clades underrepresented in Westernized populations. Cell Host Microbe. 2019;26(5):666–79.e7. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benítez-Páez A, Kjølbæk L, Gómez Del Pulgar EM, Brahe LK, Astrup A, Matysik S, Schött H-F, Krautbauer S, Liebisch G, Boberska J, et al. A multi-omics approach to unraveling the microbiome-mediated effects of arabinoxylan oligosaccharides in overweight humans. mSystems. 2019;4(4):4. doi: 10.1128/mSystems.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodd D, Mackie RI, Cann IK. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol Microbiol. 2011;79(2):292–304. doi: 10.1111/j.1365-2958.2010.07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, Leeming E, Gibson R, Le Roy C, Khatib HA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27(2):321–332. doi: 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachdeva S, Gupta V, Amin SS, Tahseen M. Chronic urticaria. Indian J Dermatol. 2011;56(6):622–628. doi: 10.4103/0019-5154.91817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong DK, Mitchell SB, Barrett JS, Shepherd SJ, Irving PM, Biesiekierski JR, Smith S, Gibson PR, Muir JG. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J Gastroenterol Hepatol. 2010;25(8):1366–1373. doi: 10.1111/j.1440-1746.2010.06370.x. [DOI] [PubMed] [Google Scholar]
- 52.Michail S, Durbin M, Turner D, Griffiths AM, Mack DR, Hyams J, Leleiko N, Kenche H, Stolfi A, Wine E, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18(10):1799–1808. doi: 10.1002/ibd.22860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palm NW, de Zoete MR, Flavell RA. Immune–microbiota interactions in health and disease. Clin Immunol. 2015;159(2):122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stefanis L. α-synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2(2):a009399. doi: 10.1101/cshperspect.a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrov VA, Saltykova IV, Zhukova IA, Alifirova VM, Zhukova NG, Dorofeeva YB, Tyakht AV, Kovarsky BA, Alekseev DG, Kostryukova ES, et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull Exp Biol Med. 2017;162(6):734–737. doi: 10.1007/s10517-017-3700-7. [DOI] [PubMed] [Google Scholar]
- 56.Braak H, de Vos RA, Bohl J, Del Tredici K. Gastric α-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neurosci Lett. 2006;396(1):67–72. doi: 10.1016/j.neulet.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Suganya K, Koo BS. Gut–brain axis: Role of gut microbiota on neurological disorders and how probiotics/prebiotics beneficially modulate microbial and immune pathways to improve brain functions. Int J Mol Sci. 2020;21(20):21. doi: 10.3390/ijms21207551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.National Institutes of Mental Health NIH . Autism spectrum disorder. National Institutes of Health; 2018. [Google Scholar]
- 59.Kang DW, Ilhan ZE, Isern NG, Hoyt DW, Howsmon DP, Shaffer M, Lozupone CA, Hahn J, Adams JB, Krajmalnik-Brown R, et al. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe. 2018;49:121–131. doi: 10.1016/j.anaerobe.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, Krajmalnik-Brown R. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8(7):e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou R, Xu F, Wang Y, Duan M, Guo M, Zhang Q, Zhao H, Zheng H. Changes in the gut microbiota of children with autism spectrum disorder. Autism Res. 2020;13(9):1614–1625. doi: 10.1002/aur.2358. [DOI] [PubMed] [Google Scholar]
- 62.Goldberg MR, Mor H, Magid Neriya D, Magzal F, Muller E, Appel MY, Nachshon L, Borenstein E, Tamir S, Louzoun Y, et al. Microbial signature in IgE-mediated food allergies. Genome Med. 2020;12(1):92. doi: 10.1186/s13073-020-00789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, Hirota K, Matsushita M, Furuta Y, Narazaki M, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68(11):2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 64.Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, Steere AC. Evidence of the immune relevance of Prevotella copri , a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017;69(5):964–975. doi: 10.1002/art.40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwon EJ, Ju JH. Impact of posttranslational modification in pathogenesis of rheumatoid arthritis: focusing on citrullination, carbamylation, and acetylation. Int J Mol Sci. 2021;22(19):22. doi: 10.3390/ijms221910576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seifert JA, Bemis EA, Ramsden K, Lowell C, Polinski K, Feser M, Fleischer C, Demoruelle MK, Buckner J, Gregersen PK, et al. Association of antibodies to Prevotella copri in anti–cyclic citrullinated peptide-positive individuals at risk of developing rheumatoid arthritis and in patients with early or established rheumatoid arthritis. Arthritis Rheumatol. 2023;75(4):507–516. doi: 10.1002/art.42370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lorenzo D, GianVincenzo Z, Carlo Luca R, Karan G, Jorge V, Roberto M, Parvizi J. Oral–gut microbiota and arthritis: Is there an evidence-based axis? J Clin Med. 2019;8(10):8. doi: 10.3390/jcm8101753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wells PM, Williams FMK, Matey-Hernandez ML, Menni C, Steves CJ. ‘RA and the microbiome: do host genetic factors provide the link? J Autoimmun. 2019;99:104–115. doi: 10.1016/j.jaut.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nii T, Maeda Y, Motooka D, Naito M, Matsumoto Y, Ogawa T, Oguro-Igashira E, Kishikawa T, Yamashita M, Koizumi S, et al. Genomic repertoires linked with pathogenic potency of arthritogenic Prevotella copri isolated from the gut of patients with rheumatoid arthritis. Ann Rheum Dis. 2023;82(5):621–629. doi: 10.1136/ard-2022-222881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhai J, Rong J, Li Q, Gu J. Immunogenetic study in Chinese population with ankylosing spondylitis: are there specific genes recently disclosed? Clin Dev Immunol. 2013;2013:419357. doi: 10.1155/2013/419357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, He Z, Zhong W, Fan Y, Zhang L, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017;18(1):142. doi: 10.1186/s13059-017-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou C, Zhao H, Xiao XY, Chen BD, Guo RJ, Wang Q, Chen H, Zhao L-D, Zhang C-C, Jiao Y-H, et al. Metagenomic profiling of the pro-inflammatory gut microbiota in ankylosing spondylitis. J Autoimmun. 2020;107:102360. doi: 10.1016/j.jaut.2019.102360. [DOI] [PubMed] [Google Scholar]
- 73.Fox TJ, Anastasopoulou C. Graves orbitopathy. StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC; 2022. [Google Scholar]
- 74.Kahaly GJ, Wüster C, Olivo PD, Diana T. High titers of thyrotropin receptor antibodies are associated with orbitopathy in patients with Graves disease. J Clin Endocrinol Metab. 2019;104(7):2561–2568. doi: 10.1210/jc.2018-02705. [DOI] [PubMed] [Google Scholar]
- 75.Shi TT, Hua L, Wang H, Xin Z. The potential link between gut microbiota and serum TRAb in Chinese patients with severe and active Graves’ orbitopathy. Int J Endocrinol. 2019;2019:9736968. doi: 10.1155/2019/9736968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leite AZ, Rodrigues NC, Gonzaga MI, Paiolo JCC, de Souza CA, Stefanutto NAV, Omori WP, Pinheiro DG, Brisotti JL, Matheucci Junior E, et al. Detection of increased plasma interleukin-6 levels and prevalence of Prevotella copri and Bacteroides vulgatus in the feces of type 2 diabetes patients. Front Immunol. 2017;8:1107. doi: 10.3389/fimmu.2017.01107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.d’Hennezel E, Abubucker S, Murphy LO, Cullen TW, Lozupone C. Total lipopolysaccharide from the human gut microbiome silences toll-like receptor signaling. mSystems. 2017;2(6). doi: 10.1128/mSystems.00046-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao M, Xu S, Cavagnaro MJ, Zhang W, Shi J. Quantitative analysis and visualization of the interaction between intestinal microbiota and type 1 diabetes in children based on multi-databases. Front Pediatr. 2021;9:752250. doi: 10.3389/fped.2021.752250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shilo S, Godneva A, Rachmiel M, Korem T, Bussi Y, Kolobkov D, Karady T, Bar N, Wolf BC, Glantz-Gashai Y, et al. The gut microbiome of adults with type 1 diabetes and its association with the host glycemic control. Diabetes Care. 2022;45(3):555–563. doi: 10.2337/dc21-1656. [DOI] [PubMed] [Google Scholar]
- 80.Ericsson AC, Akter S, Hanson MM, Busi SB, Parker TW, Schehr RJ, Hankins MA, Ahner CE, Davis JW, Franklin CL, et al. Differential susceptibility to colorectal cancer due to naturally occurring gut microbiota. Oncotarget. 2015;6(32):33689–33704. doi: 10.18632/oncotarget.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pianta A, Chiumento G, Ramsden K, Wang Q, Strle K, Arvikar S, Costello CE, Steere AC. Identification of novel, immunogenic HLA–DR-Presented Prevotella copri peptides in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021;73(12):2200–2205. doi: 10.1002/art.41807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yao Y, Ni H, Wang X, Xu Q, Zhang J, Jiang L, Wang B, Song S, Zhu X. A new biomarker of fecal bacteria for non-invasive diagnosis of colorectal cancer. Front Cell Infect Microbiol. 2021;11:744049. doi: 10.3389/fcimb.2021.744049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP, White BA. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Obuya S, Elkholy A, Avuthu N, Behring M, Bajpai P, Agarwal S, Kim H-G, El-Nikhely N, Akinyi P, Orwa J, et al. A signature of Prevotella copri and Faecalibacterium prausnitzii depletion, and a link with bacterial glutamate degradation in the Kenyan colorectal cancer patients. J Gastrointest Oncol. 2022;13(5):2282–2292. doi: 10.21037/jgo-22-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vitali F, Colucci R, Di Paola M, Pindo M, De Filippo C, Moretti S, Cavalieri D. Early melanoma invasivity correlates with gut fungal and bacterial profiles. Br J Dermatol. 2022;186(1):106–116. doi: 10.1111/bjd.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336–348. doi: 10.1016/j.ebiom.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Panda M, Rai AK, Rahman T, Das A, Das R, Sarma A, Kataki AC, Chattopadhyay I. Alterations of salivary microbial community associated with oropharyngeal and hypopharyngeal squamous cell carcinoma patients. Arch Microbiol. 2020;202(4):785–805. doi: 10.1007/s00203-019-01790-1. [DOI] [PubMed] [Google Scholar]
- 88.Liu Z, Li J, Liu H, Tang Y, Zhan Q, Lai W, Ao L, Meng X, Ren H, Xu D, et al. The intestinal microbiota associated with cardiac valve calcification differs from that of coronary artery disease. Atherosclerosis. 2019;284:121–128. doi: 10.1016/j.atherosclerosis.2018.11.038. [DOI] [PubMed] [Google Scholar]
- 89.Martín-Núñez GM, Cornejo-Pareja I, Roca-Rodríguez MDM, Clemente-Postigo M, Cardona F, Fernández-García JC, Moreno-Indias I, Tinahones FJ. H. pylori eradication treatment causes alterations in the gut microbiota and blood lipid levels. Front Med. 2020;7:417. doi: 10.3389/fmed.2020.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang L, Wang T, Wu Q, Dong X, Shen F, Liu D, Qin X, Yan L, Wan Q. Analysis of microbiota in elderly patients with acute cerebral infarction. PeerJ. 2019;7:e6928. doi: 10.7717/peerj.6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J, Drastich P, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. 2017;23(25):4548–4558. doi: 10.3748/wjg.v23.i25.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang B, Yuan G, Wu J, Wu Q, Li L, Jiang P. Prevotella copri ameliorates cholestasis and liver fibrosis in primary sclerosing cholangitis by enhancing the FXR signalling pathway. Biochim Biophys Acta Mol Basis Dis. 2022;1868(3):166320. doi: 10.1016/j.bbadis.2021.166320. [DOI] [PubMed] [Google Scholar]
- 93.Aly AM, Adel A, El-Gendy AO, Essam TM, Aziz RK. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016;8(1):42. doi: 10.1186/s13099-016-0124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moran-Ramos S, Cerqueda-García D, López-Contreras B, Larrieta-Carrasco E, Villamil-Ramírez H, Molina-Cruz S, Torres N, Sánchez‐Tapia M, Hernández‐Pando R, Aguilar‐Salinas C, et al. A metagenomic study identifies a Prevotella copri enriched microbial profile associated with non-alcoholic steatohepatitis in subjects with obesity. J Gastroenterol Hepatol. 2023;38(5):791–799. doi: 10.1111/jgh.16147. [DOI] [PubMed] [Google Scholar]
- 95.Liu P, Jiang Y, Gu S, Xue Y, Yang H, Li Y, Wang Y, Yan C, Jia P, Lin X, et al. Metagenome-wide association study of gut microbiome revealed potential microbial marker set for diagnosis of pediatric myasthenia gravis. BMC Med. 2021;19(1):159. doi: 10.1186/s12916-021-02034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Becattini S, Littmann ER, Carter RA, Kim SG, Morjaria SM, Ling L, Gyaltshen Y, Fontana E, Taur Y, Leiner IM, et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med. 2017;214(7):1973–1989. doi: 10.1084/jem.20170495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rolhion N, Chassaing B, Nahori MA, de Bodt J, Moura A, Lecuit M, Dussurget O, Bérard M, Marzorati M, Fehlner-Peach H, et al. A Listeria monocytogenes bacteriocin can target the commensal Prevotella copri and modulate intestinal infection. Cell Host & Microbe. 2019;26(5):691–701.e5. doi: 10.1016/j.chom.2019.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wright DP, Rosendale DI, Robertson AM. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett. 2000;190(1):73–79. doi: 10.1111/j.1574-6968.2000.tb09265.x. [DOI] [PubMed] [Google Scholar]
- 99.Leo S, Lazarevic V, Gaïa N, Estellat C, Girard M, Matheron S, Armand-Lefèvre L, Andremont A, Schrenzel J, Ruppé E, et al. The intestinal microbiota predisposes to traveler’s diarrhea and to the carriage of multidrug-resistant Enterobacteriaceae after traveling to tropical regions. Gut Microbes. 2019;10(5):631–641. doi: 10.1080/19490976.2018.1564431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gilchrist CA, Petri SE, Schneider BN, Reichman DJ, Jiang N, Begum S, Watanabe K, Jansen CS, Elliott KP, Burgess SL, et al. Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. J Infect Dis. 2016;213(10):1579–1585. doi: 10.1093/infdis/jiv772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ngobeni R, Samie A, Moonah S, Watanabe K, Petri WA Jr., Gilchrist C. Entamoeba species in South Africa: Correlations with the host microbiome, parasite burdens, and first description of Entamoeba bangladeshi outside of Asia. J Infect Dis. 2017;216(12):1592–1600. doi: 10.1093/infdis/jix535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pop M, Paulson JN, Chakraborty S, Astrovskaya I, Lindsay BR, Li S, Bravo HC, Harro C, Parkhill J, Walker AW, et al. Individual-specific changes in the human gut microbiota after challenge with enterotoxigenic Escherichia coli and subsequent ciprofloxacin treatment. BMC Genom. 2016;17(1):440. doi: 10.1186/s12864-016-2777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kieser S, Sarker SA, Sakwinska O, Foata F, Sultana S, Khan Z, Islam S, Porta N, Combremont S, Betrisey B, et al. Bangladeshi children with acute diarrhoea show faecal microbiomes with increased Streptococcus abundance, irrespective of diarrhoea aetiology. Environ Microbiol. 2018;20(6):2256–2269. doi: 10.1111/1462-2920.14274. [DOI] [PubMed] [Google Scholar]
- 104.Kaur US, Shet A, Rajnala N, Gopalan BP, Moar P, D H, Singh BP, Chaturvedi R, Tandon R. High abundance of genus Prevotella in the gut of perinatally HIV-infected children is associated with IP-10 levels despite therapy. Sci Rep. 2018;8(1):17679. doi: 10.1038/s41598-018-35877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rizkallah MR, Saad R, Aziz RK. The human microbiome project, personalized medicine and the birth of pharmacomicrobiomics. Curr Pharmacogenomics Person Med. 2010;8(3):182–193. doi: 10.2174/187569210792246326. [DOI] [Google Scholar]
- 106.Aziz RK. Interview with Prof. Ramy K. Aziz, Cairo University. The dawn of pharmacomicrobiomics. Omics: J Integrative Biol. 2018;22(4):295–297. doi: 10.1089/omi.2018.0041. [DOI] [PubMed] [Google Scholar]
- 107.Yu C, Zhou B, Xia X, Chen S, Deng Y, Wang Y, Wu L, Tian Y, Zhao B, Xu H, et al. Prevotella copri is associated with carboplatin-induced gut toxicity. Cell Death Disease. 2019;10(10):714. doi: 10.1038/s41419-019-1963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun G, Liu H, Shi X, Tan P, Tang W, Chen X, Sun G, Yang W, Kong X, Zheng Z, et al. Treatment of patients with cancer using PD‑1/PD‑L1 antibodies: Adverse effects and management strategies (Review). Int J Oncol. 2022;60(6):60. doi: 10.3892/ijo.2022.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S, et al. The diversity of gut microbiome is associated with favorable responses to anti–programmed death 1 immunotherapy in Chinese patients with NSCLC. J Thorac Oncol. 2019;14(8):1378–1389. doi: 10.1016/j.jtho.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 110.Tsai CY, Lu HC, Chou YH, Liu PY, Chen HY, Huang MC, Lin C-H, Tsai C-N. Gut microbial signatures for glycemic responses of GLP-1 receptor agonists in type 2 diabetic patients: A pilot study. Front Endocrinol (Lausanne). 2021;12:814770. doi: 10.3389/fendo.2021.814770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tang Q, Jin G, Wang G, Liu T, Liu X, Wang B, Cao H. Current sampling methods for gut microbiota: A call for more precise devices. Front Cell Infect Microbiol. 2020;10:151. doi: 10.3389/fcimb.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348–1365. doi: 10.1038/s41587-021-01108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ardui S, Ameur A, Vermeesch JR, Hestand MS. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018;46(5):2159–2168. doi: 10.1093/nar/gky066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lo Presti A, Del Chierico F, Altomare A, Zorzi F, Monteleone G, Putignani L, Angeletti S, Cicala M, Guarino MPL, Ciccozzi M, et al. Phylogenetic analysis of Prevotella copri from fecal and mucosal microbiota of IBS and IBD patients. Therap Adv Gastroenterol. 2023;16:17562848221136328. doi: 10.1177/17562848221136328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yeoh YK, Sun Y, Ip LYT, Wang L, Chan FKL, Miao Y, Ng SC. Prevotella species in the human gut is primarily comprised of Prevotella copri. Prevotella Stercorea and related lineages. Sci Rep. 2022;12(1):9055. doi: 10.1038/s41598-022-12721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


