Abstract
Aims
Nuclear envelope integrity is essential for the compartmentalization of the nucleus and cytoplasm. Importantly, mutations in genes encoding nuclear envelope (NE) and associated proteins are the second highest cause of familial dilated cardiomyopathy. One such NE protein that causes cardiomyopathy in humans and affects mouse heart development is Lem2. However, its role in the heart remains poorly understood.
Methods and results
We generated mice in which Lem2 was specifically ablated either in embryonic cardiomyocytes (Lem2 cKO) or in adult cardiomyocytes (Lem2 iCKO) and carried out detailed physiological, tissue, and cellular analyses. High-resolution episcopic microscopy was used for three-dimensional reconstructions and detailed morphological analyses. RNA-sequencing and immunofluorescence identified altered pathways and cellular phenotypes, and cardiomyocytes were isolated to interrogate nuclear integrity in more detail. In addition, echocardiography provided a physiological assessment of Lem2 iCKO adult mice. We found that Lem2 was essential for cardiac development, and hearts from Lem2 cKO mice were morphologically and transcriptionally underdeveloped. Lem2 cKO hearts displayed high levels of DNA damage, nuclear rupture, and apoptosis. Crucially, we found that these defects were driven by muscle contraction as they were ameliorated by inhibiting myosin contraction and L-type calcium channels. Conversely, reducing Lem2 levels to ∼45% in adult cardiomyocytes did not lead to overt cardiac dysfunction up to 18 months of age.
Conclusions
Our data suggest that Lem2 is critical for integrity at the nascent NE in foetal hearts, and protects the nucleus from the mechanical forces of muscle contraction. In contrast, the adult heart is not detectably affected by partial Lem2 depletion, perhaps owing to a more established NE and increased adaptation to mechanical stress. Taken together, these data provide insights into mechanisms underlying cardiomyopathy in patients with mutations in Lem2 and cardio-laminopathies in general.
Keywords: Heart failure, Cardiac development, Nuclear Envelope, LINC complex, Laminopathy
Graphical Abstract
Graphical Abstract.
Time of primary review: 71 days
1. Introduction
The nuclear envelope (NE) in eukaryotic cells separates nuclear and cytoplasmic compartments. Underlying the NE is the nuclear lamina, which consists of Lamins A/C, B1, and B2.1 The nuclear lamina is thought to provide the nucleus with structural rigidity and plays roles in chromatin tethering and gene expression regulation.2,3 The NE is interspersed with the linker of nucleoskeleton and cytoskeleton (LINC) complexes that span the double membrane.4–6 In striated muscle, such as the heart, the LINC complex and its associated proteins have been shown to be essential for normal function.7–12 Associated with the LINC complex are the Lamina-associated peptide 2 (Lap2), Emerin, MAN1 (LEM)-domain-containing proteins, which also play critical roles in the heart. Indeed, mutations in Emerin, Lap2, and LEM-domain-containing 2 (Lem2) are associated with Emery-Dreifuss Muscular Dystrophy with cardiac conduction defects, dilated cardiomyopathy, and arrhythmogenic cardiomyopathy (AC), respectively.13–17 Specifically, a leucine 13 to arginine (L13R) mutation in Lem2 leads to AC and sudden death.15,16 Furthermore, a C-terminal mutation that changes serine 479 to phenylalanine (S479F) leads to the right bundle branch block and septal hypertrophy.17
Lem2 is reported to play roles in vitro and in lower organisms. These range from NE repair and resealing after mitosis, maintenance of nuclear shape, regulation of heterochromatin and gene expression, and regulation of mitogen-activated protein kinase (MAPK) signalling.18–26 In mice, global loss of Lem2 resulted in embryonic lethality between E10.5 and E11.5, with the overall body size and most tissues being substantially smaller in size.27 More specific developmental defects included impaired neurogenesis in neural tissues and thinner myocardial walls in the heart. However, the specific roles that Lem2 plays in the heart remain poorly understood.
To this end, we aimed to investigate whether Lem2 plays similar roles in vivo as those proposed in vitro and in lower organisms. Here, we found that Lem2 plays an essential role in heart development. Ablation of Lem2 in cardiomyocytes led to embryonic lethality, with mutant hearts showing marked signs of developmental delay. Transcriptomics revealed the enrichment of cell stress pathways and suppression of cardiac developmental pathways, which led us to investigate the cellular effects of Lem2 loss in cardiomyocytes. Lem2-null cardiomyocytes were highly apoptotic, concomitant with both increased DNA damage and micronuclei. The isolation of primary cardiomyocytes revealed aberrant nuclear morphology as well as high levels of nuclear rupture in Lem2-null cells, suggesting that Lem2 plays important roles in maintaining NE integrity. As these cells are highly contractile, we hypothesized that the inhibition of mechanical strain on the NE may rescue the nuclear phenotypes. Indeed, we found that inhibiting both myosin contraction and calcium channels attenuated nuclear defects. Conversely, increased activation of sarcomeric myosin exacerbated nuclear damage. Next, we explored roles of Lem2 in adult cardiomyocytes, which are highly adapted to withstand the mechanical load, as muscle contractile forces are larger compared with those found in embryonic hearts. In contrast to foetal cardiomyocytes, the ablation of Lem2 in adult cardiomyocytes did not affect the cardiac function or nuclear shape.
Our data suggest that Lem2 is critical for maintaining nuclear integrity in cardiac development in which the NE is in an immature state. Conversely, the presence of a more established nuclear lamina, NE, and LINC complex in adult cardiomyocytes is better able to adapt to both mechanical force and the absence of Lem2.
2. Methods
2.1. Animal studies
All procedures were performed in accordance with the Guidance on the Operation of the Animals (Scientific Procedures) Act, 1986 (UK Home Office). Mice carrying a knockout-first Lemd2tm1a allele were generated by blastocyst injection of targeted ES cells (#EPD0240_4_D10, EUCOMM) using standard techniques.28,29 Germ line transmission of Lemd2tm1a was confirmed by genotyping PCR analysis. Heterozygous Lemd2+/tm1a mice were bred to the FLPeR deleter strain,30 to remove the FRT-flanked knockout-first cassette, generating the Lemd2tm1c floxed allele in which exon2 was retained flanked by LoxP sites. For the Lem2 cKO mouse line, Lem2 floxed (Lem2f/f) were crossed with Xenopus laevis Myosin Light-Chain 2 (XMLC2)-Cre mice31 to generate Lem2f/+; XMLC2-Cre. Male Lem2f/+; XMLC2-Cre mice were crossed with female Lem2f/f mice to generate Lem2f/f; XMLC2-Cre (Lem2 cKO), Lem2f/+; XMLC2-Cre and Lem2f/f (controls). For the Lem2 cardiac Troponin T (cTnT)-Cre cKO mouse line, Lem2 floxed (Lem2f/f) were crossed with cTnT-Cre32 to generate Lem2f/+; cTnT-Cre. Male Lem2f/+; cTnT-Cre mice were crossed with female Lem2f/f mice to generate Lem2f/f; cTnT-Cre (Lem2 cTnT-Cre cKO), Lem2f/+; cTnT-Cre and Lem2f/f (controls). For the iCKO line, Lem2f/f mice were crossed with tamoxifen-inducible cardiac troponin T-Cre (Tnnt2MerCreMer/+) mice33 to generate Lem2f/+; Tnnt2MerCreMer/+ mice. Male Lem2f/f; Tnnt2MerCreMer/+ mice were crossed with Lem2f/f females to generate Lem2f/f; Tnnt2MerCreMer/+ (Lem2 iCKO) and Lem2f/f; Tnnt2+/+ (control). Tamoxifen dissolved in peanut oil was intraperitoneally injected for three consecutive days at a dose of 0.1 mg/g body weight, at 6 and 9 weeks of age for isolated cardiomyocyte and echocardiography studies, respectively. For the isolation of adult hearts, mice were sacrificed by cervical dislocation and exsanguination.
2.2. Echocardiography
Mice were anaesthetized with 0.5% isoflurane and underwent echocardiography using VisualSonics, SonoSite FUJIFILM, Vevo 3100 ultrasound system with a linear transducer 32–55 MHz as described previously.34,35 Measurements of heart rate, left ventricular internal dimension at the end of diastole and systole (LVIDd, LVIDs, respectively), end-diastolic inter-ventricular septal thickness (IVSd), and LV posterior wall thickness (LVPWd) were determined from the LV M-mode tracing. The percentage fractional shortening was used as an indicator of systolic cardiac function.
2.3. Cardiomyocyte isolations and analysis
Adult cardiomyocytes were isolated from mice under full anaesthesia with 5% isoflurane that was confirmed with reduced breathing rate and lack of toe-pinch response as described previously.36 Foetal hearts at E14.5 were isolated after cervical dislocation and exsanguination of pregnant dams and foetal cervical dislocation and decapitation. Foetal cardiomyocytes were isolated using HBSS supplemented with Trypsin (0.5 mg/mL) for 2.5 h with gentle rotation at 4°C. Hearts were then agitated at 150 rpm on an orbital shaker for 10 min at 37°C. Trypsin was inhibited with an equal volume of cold culture medium (3:1 mixture of DMEM:M199 media with 4.5 g/L glucose, 10% horse serum, 5% foetal calf serum, 10 mM HEPES, and penicillin/streptomycin). Hearts were then disaggregated gently with a 1 mL pipette, filtered with a 100 μm cell sieve, pelleted at 150 g for 5 min, and resuspended in a culture medium. Cells were then plated on collagen/laminin-coated glass. For experiments with 6 kPa PDMS hydrogels, PDMS was prepared and spin-coated onto coverslips as previously described.36 Fibronectin coating was used for comparison between glass and hydrogels. Adenoviral transduction of cGAS-mScarlet and/or BAF-GFP probes was carried out at four infectious particles per cell in each case, to identify nuclear ruptures. Cardiomyocytes were treated with 20 μM para-nitroblebbistatin, 5 μM verapamil, 1 μM Omecamtiv mecarbil, or DMSO (vehicle alone) for 4 h prior to fixation.
For two-dimensional (2D) nuclear shape analysis, the DAPI channel was thresholded by pixel intensity in ImageJ/FIJI, to highlight the full area of each nuclei, followed by measurement using inbuilt algorithms. For semi-quantitative analyses, abnormal nuclear shapes were defined as those showing a bleb/herniation or those showing a concavity in the surface. For three-dimensional (3D) shape analysis, z-stacks of images were imported into Volocity software (Perkin Elmer), followed by thresholding of the DAPI channel in 3D, and measurement using inbuilt algorithms.
2.4. Immunofluorescence
Hearts were fixed overnight in 4% PFA and cryoprotected using sucrose gradients followed by embedding in a sucrose/optimal cutting temperature mix as previously described.37 Tenmicrometres-thick sections were cut and stored at −80°C until use. After permeabilization with washing buffer (PBS with 0.2% TX-100), sections were incubated with the indicated antibodies (see Supplementary material online, Table S1) overnight in blocking buffer (PBS/2% normal donkey serum/3% BSA) in a humidified chamber at 4°C. Sections were rinsed in wash buffer and incubated at room temperature with the indicated fluorescently conjugated secondary antibodies and DAPI (1:1000) diluted in blocking buffer for 1 h. Slides were rinsed in wash buffer and mounted in mounting medium (Dako). Sections were imaged as described previously.38,39 Two hundred microlitres of 5-ethynyl-2′-deoxyuridine (EdU, molecular probes; 3 g/L) were injected intraperitoneally in pregnant females 2 h before embryo isolation.
2.5. Fluorescence intensity analysis
For fluorescence intensity analysis of lamina, NE and LINC complex proteins, line profiles/scans were drawn in ImageJ/FIJI to bisect nuclei. Using the relevant fluorescent channel for each stain, peak intensities corresponding to the nuclear periphery were taken, and the cytoplasmic background signal was subtracted. For quantification of SM22a and CD31, images were thresholded in Fiji using either using Moments or Max Entropy for SM22a and CD31, respectively, and values divided by the total area.
2.6. High-resolution episcopic microscopy analysis
Samples were prepared as described previously.40 Brightness/contrast adjustments were made using Adobe Photoshop and images assembled using Horos software. Wall thickness measurements were taken at two points using sections generated in silico by eroding from the base of the heart: at the apex when both ventricles become visible and at the mid-point between the apical section and the top of the ventricles (examples shown in Figure 1D and Supplementary material online, Figure S2B). The measurements were then made relative to their respective axis section width and length, and mean values reported. Fractal analysis was carried out using the FIJI plugin FracLac, with box-counting dimension (DB) values used as a readout for the morphological complexity of the trabecular organization.41
Figure 1.
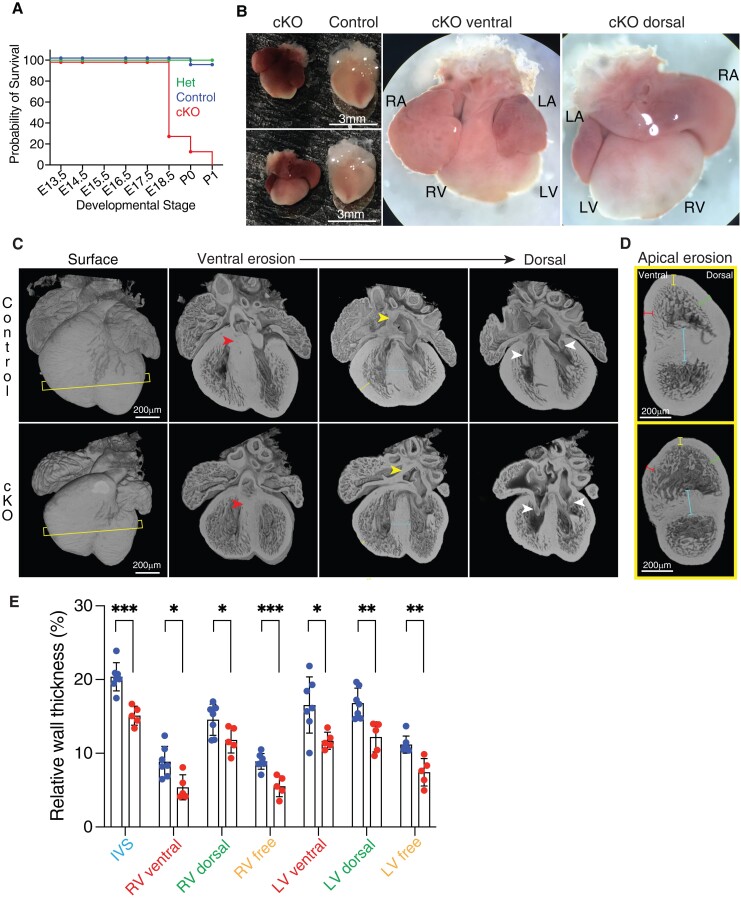
Lem2 is essential for normal heart development. (A) Kaplan–Meier survival curve during development from embryonic day 13.5 (E13.5) to postnatal day 1 (P1). Note the foetal demise of Lem2 cardiomyocyte-specific Lem2 knockout (cKO) mice (n = 36–98 mice at each developmental stage). (B) Wholemount images taken prior to birth at E18.5 of cKO and control mice. RA, LA, RV, and LV denote right and left atria, right and left ventricles, respectively. Note the enlarged atria with congested blood in cKO hearts. (C) HREM images of whole hearts taken at E16.5 with erosions from the ventral to dorsal surface. Yellow and white arrowheads denote atria septum (AS) and atrio-ventricular (AV) connections, respectively. Slice lines indicate sections imaged in (D). Yellow and blue arrows denote right ventricle freewalls and inter-ventricular septa (IVS), respectively. Note the normal formation of AS and AV connections, but the smaller hearts and underdeveloped wall thicknesses in cKO mice. (D and E) HREM images eroded from the heart apex showing a mid-ventricle view. Colour-coded lines represent wall thicknesses measured in (E). Blue lines, IVS; red lines, ventral walls; green lines, dorsal walls; yellow lines, freewalls. Note that the walls are all significantly thinner in the cKO compared with controls, indicating delayed cardiac development. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t-test; n = 5–7 mice per genotype. Graphs show mean ± SD.
2.7. Western blotting
Ventricles were lysed in 50 μL/mg tissue in a Precellys homogenizer and sonicated to shear DNA. Lysates were centrifuged at 13 900g at 4°C for 15 min, supernatants aspirated and loaded onto 4–12% NuPage Bis/Tris gels for separation before transfer onto a Nitrocellulose membrane (Biorad) at 4°C at a constant current of 350 mA in transfer buffer (25 mM Tris, 190 mM glycine, 20% methanol, pH 8.3) for 90 min. After blocking for 1 h in Tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% non-fat dry milk, membranes were incubated overnight at 4°C with the indicated primary antibody (see Supplementary material online, Table S1) in blocking buffer (TBS/0.1% Tween 20/2% non-fat dry milk). Blots were washed and incubated with fluorescent secondary antibodies for 1 h at room temperature and imaged with an infrared imaging system (ODYSSEY CLx; LI-COR Biosciences). Image Studio software (LI-COR Biosciences) was used for quantitative densitometry analysis to evaluate protein expression levels.
2.8. Histology
Hearts were isolated from age- and sex-matched littermates at 83 weeks of age, and washed in PBS before being fixed overnight in 10% formalin. Hearts were subsequently dehydrated in 70% ethanol, embedded in paraffin and cut into 7 μm sections. Sections were stained using H&E or picrosirius red, mounted, and imaged on an All-in-one fluorescence microscope (BZ-X700, Keyence).
2.9. Real-time polymerase chain reaction
Total RNA was extracted from mouse hearts using ReliaPrep™ RNA Tissue miniprep system (Promega). cDNA was synthesized using M-MLV Reverse Transcriptase (Promega). Primers for real-time polymerase chain reaction (RT-PCR) are listed in Supplementary material online, Table S2. RT-PCR reactions were performed using SYBR-Green (PCR biosystems) in a StepOnePlus Real-time PCR System (Applied Biosystems).
2.10. RNA-sequencing
RNA was isolated from whole hearts with their atria removed and its quality was assessed using a Bioanalyser (2100 Instrument, Agilent). High-quality RNA (RIN score > 8) was used for library preparation. One microgram of RNA was used to generate sequencing libraries using NEBNext Ultra II Directional RNA library Prep Kit for Illumina (NEB) according to the manufacturer’s recommendation. Purification steps were performed using the ‘ProNex Size-Selective Purification System’ (fragment cut-off size 100 bp). The index primers employed during the library preparation corresponded to the ‘NEBNext Multiplex Oligos for Illumina (Index Primers Set 1)’. Once the libraries were generated, their quality was evaluated on the Bioanalyzer. Those libraries whose electropherograms showed a narrow distribution with a peak size of ∼300 bp were sent for sequencing. Paired-end multiplexed sequencing of libraries to generate reads of 100 bp was performed on a HiSeq 1000 instrument with TruSeq SBS and PE Cluster v3 Kits (Illumina); 70–100 million reads per sample were obtained.
A minimum of four hearts per genotype were used for sequencing and analysis. Data will be made available in the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) database.
2.11. RNA-sequencing data analysis
The raw read counts (FastQ files) were trimmed and aligned (STAR) to the UCSC Mus musculus reference genome using Partek Flow software. The gene counts obtained were normalized (GSA) and differential expression analysis was performed by applying the DESeq2 method. For further analyses, P-values were corrected for multiplicity and the threshold used was false discovery rate (FDR) ≤0.1.
2.12. Functional annotation
Gene set enrichment analysis was performed using fgsea package,42 version 1.10.1 and molecular signatures database (MSigDB) version 6.243 on all genes that were expressed above background levels (readcounts > 600) with settings ‘minSize = 15’, ‘maxSize = 500’, and ‘nperm = 10 000’. P < 0.05 was considered significant.
2.13. Molecular biology
pTRIP-CMV-GFP-FLAG-cGAS E225A-D227A was a gift from Nicolas Manel (Addgene plasmid # 86674; http://n2t.net/addgene: 86674; RRID:Addgene_86674), and Lem2 were subcloned into mScarlet-1 and mEOS-C1, respectively, using XhoI restriction enzyme, then into pShuttle-CMV with EcoRV using the In-Fusion HD Cloning Plus System kit (Takara Bio, USA). These were linearized with Pme1 and cotransformed with pAdEasy-1 into BJ5183 cells. Restriction digest and DNA sequencing identified correct clones, which were transformed into JM109 cells. Purified recombinant plasmid DNA was digested with PacI and transfected into HEK293 cells with Lipofectamine (Invitrogen) according to the manufacturer’s instructions. Transfected cells were harvested and freeze-thawed, centrifuged at 4000 rpm for 10 min, and supernatant collected, which was used for subsequent rounds of infection and amplification.
2.14. Adenovirus purification and quantification
Adenovirus purification was performed according to the manufacturer’s instructions (Adeno-X Maxi Purification kit). Briefly, HEK293 cells were seeded in 12 wells of a 24-well plate at a density of 2.5 × 105 cells/mL (1 mL/well). The next day, 10-fold serial dilutions (from 10−2 to 10−7) of the purified viral samples were prepared using DMEM (10% FBS, 2% GPS) and 50 μL of every dilution was added dropwise to each well. Infected cells were incubated at 37°C and 5% CO2 and after 48 h, titration procedure was carried out by counting the number of fluorescent cells in five fields per well. The infectious units per millilitre were calculated using the following formula: [(average number of infected cells/field)×(total fields/well)]/[(amount of viral dilution in mL)×(dilution factor)]. BAF-GFP was purchased from VectorBuilder.
2.15. Pressure chamber
Hydrodynamic pressure stimulation was performed in a MechanoCulture TR stimulator, placed inside a tissue culture incubator. Briefly, adult cardiomyocytes were isolated and seeded on glass coverslips overnight. The stimulator was modified for low-pressure stimulation in the range of normal and high pressure and perfusion ports to supply presaturated cell culture medium. Cardiomyocytes were stimulated with a sinusoid profile (stretch: 0.5 s, duration: 1 s, hold: 0 s, recovery: 0.5 s) and alternating between set pressures of 16 kPa (peak load) and 2 kPa (pre-load) to reach a measured pressure profile of 120/15 mmHg for normal pressure stimulation and between 27 and 4 kPa for a measured pressure profile of 200/30 mmHg for high-pressure stimulation. Cyclic pressure stimulations were performed for 3 h as described previously.44 Cells were then fixed in 2% PFA/PBS for analysis of nuclear morphology.
2.16. Nanoindentation
To determine the stiffness of adult cardiomyocyte nuclei, nanoindentation was carried out using an Optics11 Chiaro nanoindenter attached to a Leica DMI-8 inverted microscope as described previously.45 Cardiomyocytes were stained with Hoechst to locate nuclei, and only nuclei at the nearest surface of the cardiomyocyte to the indenter were analysed. Measurements were taken above the nucleus with a 9 μm diameter spherical probe (2.1 µm contact radius at 0.5 µm indentation depth, corresponding to approximately half the nuclear radius, whereby an indentation at 0.5 µm depth measures primarily the nuclear vs. the cytoskeletal contribution to the stiffness).46 The contact point was identified from a contact point fit of data to 20% of the maximal load and was used to subsequently fit Young’s modulus using an indentation depth of 2 μm and a contact threshold of 0.005 arbitrary units. Approach and retraction speeds were set to 500 nm/s. The Hertzian contact model was used to fit the load-indentation data for the calculation of Young’s modulus.
2.17. Statistics
Data are presented as mean ± standard deviation (SD). We used a two-tailed Student’s t-test or two-way analysis of variance (ANOVA), with Sadik’s post hoc test for comparisons among groups as indicated. Analysis was performed using Microsoft Excel or GraphPad Prism software. P-values of <0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001).
3. Results
3.1. Lem2 is essential for normal heart development
To investigate the role of Lem2 in cardiac function, we generated cardiomyocyte-specific knockout mice using a Cre-LoxP approach by crossing Lem2f/f mice with cardiomyocyte-specific X. laevis myosin light-chain 2 (XMLC2V)-Cre transgenic mice,31 hereby designated as Lem2 cKO (see Supplementary material online, Figure S1A). Lem2 ablation was confirmed with western blotting (WB) and immunofluorescence analyses (Supplementary material online, Figure S1B–D). We found that the majority of Lem2 cKO mice did not survive past embryonic day 18.5 (E18.5) (Figure 1A and Supplementary material online, Figure S1E–G), indicating lethality in late foetal stages. Tissue harvesting at E18.5 revealed that the Lem2 cKO hearts were smaller with enlarged atria containing congested blood (Figure 1B), indicating abnormal cardiac function. To investigate this further, we performed high-resolution episcopic microscopy (HREM) for 3D organ reconstruction and morphological analyses.47 Surprisingly, we did not observe any obvious congenital defects involving abnormal chamber development, which may have explained the foetal demise in Lem2 cKO mice (Figure 1C and D and Supplementary material online, Figures S2A–C and Movies S1–S3). However, ventricle wall thicknesses were slightly reduced at E14.5 (see Supplementary material online, Figure S2D), which became more pronounced at E16.5 (Figure 1E), suggestive of developmental delay. To determine if there were changes to the vasculature and endothelial cell organization in Lem2 cKO, hearts were stained for SM22, CD31, respectively. However, no changes were observed between Lem2 cKO and their littermate controls (see Supplementary material online, Figure S1I–L). To investigate if there were alterations in trabecular organization, we performed fractal analyses on HREM data at E16.5 to quantify morphological complexity, but revealed no changes (see Supplementary material online, Figure S1M–O).
3.2. Lem2 ablation activates cell death and inhibits cardiac development pathways
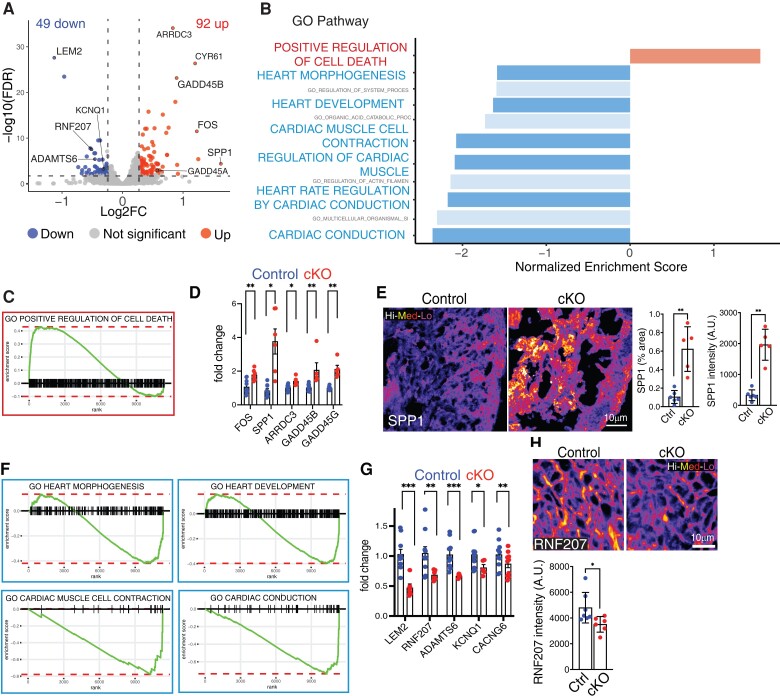
To further assess alterations in cardiac development, we performed RNA-sequencing analysis at E14.5 on control and Lem2 cKO whole hearts. Differential gene expression revealed slight but significant changes to gene expression, with 92 and 49 genes being significantly up- and down-regulated, respectively (Figure 2A). Gene ontology (GO) analysis revealed significant enrichment of the apoptotic cell death pathway and reduction of processes that are key for proper cardiac development, including those involved in morphogenesis, contraction, and conduction (Figure 2B, C, and F; Supplementary material online, Figure S2E and F). Up-regulated genes included Gadd45b and Gadd45g, which have been implicated in cell death,48,49 as well as Fos, Spp1, and Arrdc3 (Figure 2D and E). Down-regulated genes included Rnf207, Kcnq1, and Cacng6, which have been implicated in cardiac conduction and contraction as well as Adamts6 that regulates heart morphogenesis50–53 (Figure 2G and H). Importantly, we were able to validate these changes both at mRNA and protein levels (Figure 2E and H).
Figure 2.
Lem2 ablation activates cell death and inhibits cardiac development pathways. (A) Volcano plot showing differentially expressed genes between control and cardiomyocyte-specific Lem2 knockout (cKO) ventricles at embryonic day 14.5 (E14.5), from RNA-sequencing data (>1.2-fold change, FDR <0.01). Red and blue dots represent the 92 and 49 genes that were significantly up- and down-regulated, respectively. A selection of qPCR-validated genes is labelled. (B) GO pathway analysis was performed on differentially expressed genes. Selected terms are highlighted from the top 10 enriched GO-terms. (C) GO-term ranking plot shows an over-representation of genes involved in the positive regulation of cell death in cKO hearts. (D) RT–qPCR validation of genes identified from RNA-seq as being significantly up-regulated in cKO hearts, including cell death genes GADD45A and G. (E) Immunofluorescent validation of SPP1, showing increased levels in cKO hearts, further validating the RNA-seq data. (F) GO-term ranking plots for shows an under-representation of genes important for cardiac development in cKO hearts. (G) RT–qPCR validation of genes identified from RNA-seq as being significantly down-regulated in cKO hearts. Note that cardiac genes RNF207, KCNQ1, and CACNG6, as well as Lem2 and ADAMTS6 were significantly decreased. (H) Immunofluorescent validation of RNF207, showing decreased levels in cKO hearts, further validating the RNA-seq data. False colour (Fire LUT) used in (E) and (H). *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t-test; n = 4 mice per genotype for RNA-seq; n = 7–11 for qPCR validations; n = 5 for immunofluorescence validation. Graphs show mean ± SD.
Taken together, these data show that Lem2 is required for cell viability and indispensable for normal cardiac developmental gene expression.
3.3. Loss of Lem2 results in cardiomyocyte apoptosis, DNA damage accumulation, and micronuclei during cardiac development
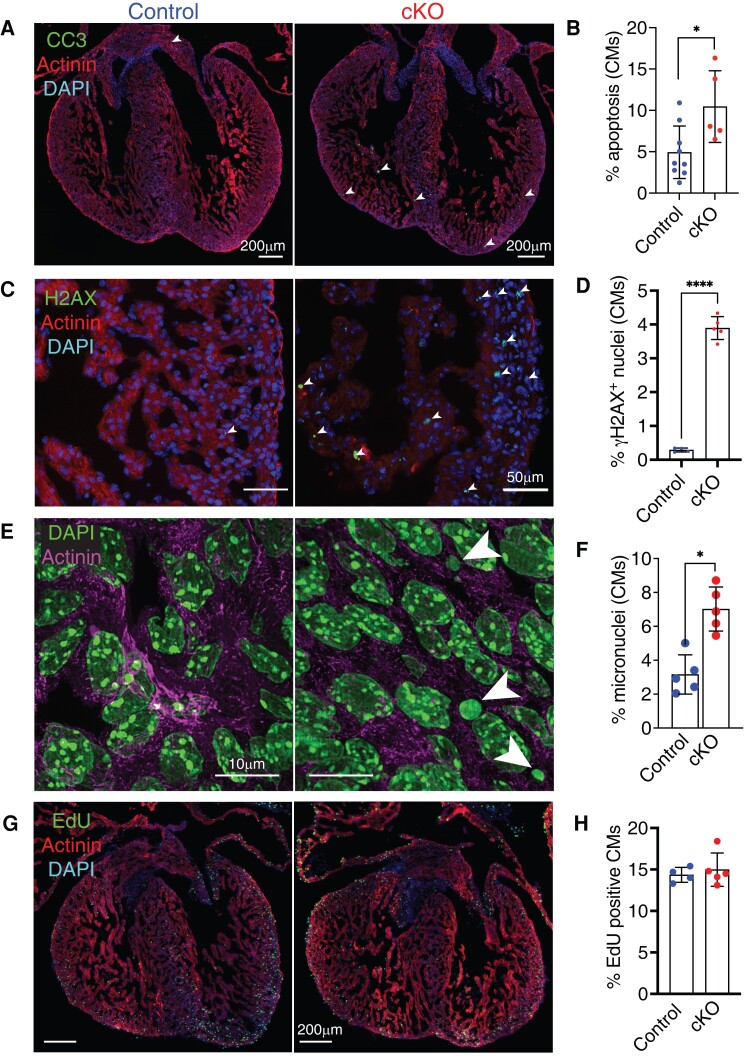
To test whether the up-regulated cell death pathway resulted in apoptosis, we labelled hearts for cleaved caspase 3, which confirmed high levels of apoptosis in Lem2 cKO hearts between E13.5 and E16.5 (Figure 3A and B and Supplementary material online, Figure S3A and B), consistent with findings in neural tubes of global Lem2 KO mice.27 Consistently, we routinely observed a ∼41% reduction in the number of cardiomyocytes isolated from E14.5 hearts from cKO compared with controls (mean ± SD: 94 500 ± 25 500 vs. 159 900 ± 34 700 cardiomyocytes, respectively) (data not shown).
Figure 3.
Loss of Lem2 results in cardiomyocyte apoptosis, DNA damage accumulation, and micronuclei during cardiac development. (A–F) Foetal hearts at embryonic day 14.5 (E14.5) were stained with antibodies raised against cleaved caspase 3 (to mark apoptotic cells) (A); γH2aX (DNA double-strand breaks) (C); and sarcomeric α-actinin and DAPI (nuclei). Note the strong increase in cardiomyocytes (CMs) positive for apoptosis, γH2aX, and micronuclei (arrowheads) observed in cardiomyocyte-specific Lem2 knockout (cKO) hearts, quantified in (B, D, and F). (G and H) E14.5 hearts labelled with EdU to delineate cells undergoing DNA synthesis, quantified in cardiomyocytes (H). Note that no changes in EdU incorporation were observed. *P < 0.05, ****P < 0.0001, two-tailed t-test; n = 4–8 biological replicates per genotype, with 500–1000 cells quantified/replicate. Graphs show mean ± SD.
These findings were confirmed using an independent model in which Cre expression was driven by the cardiac troponin T (cTnT) promoter to generate Lem2f/f;cTnT−Cre/+ cKO hearts (see Supplementary material online, Figures S1H and S3F and G).32 Given that Lem2 is a regulator of MAPK signalling,27 we investigated these pathways and found that Lem2 cKO hearts displayed elevated ERK1/2, but not p38 activation levels (see Supplementary material online, Figure S3L and M). Lem2 has previously been implicated in NE resealing and repair;18–21 therefore, we reasoned that Lem2 ablation may lead to genomic damage and subsequent instability in cardiomyocytes. Indeed, Lem2 cKO hearts showed increased levels of γH2AX (which marks DNA double-strand breaks)54 and micronuclei at E14.5 and E16.5 (Figure 3C–F and Supplementary material online, Figure S3C–E).
Cardiac development is tightly regulated and requires a precise balance between proliferation and apoptosis. We used EdU and anti-phospho-histone H3 staining to mark cells undergoing DNA synthesis and mitosis, respectively. Importantly, proliferation levels were comparable between genotypes (Figure 3G and H and Supplementary material online, Figure S3H–K), suggesting that the observed phenotype of smaller hearts and thinner ventricular walls in Lem2 cKO mice is governed by increased apoptosis rather than reduced proliferation.
3.4. Cardiomyocyte nuclei lacking Lem2 are aberrantly shaped and more susceptible to mechanical rupture
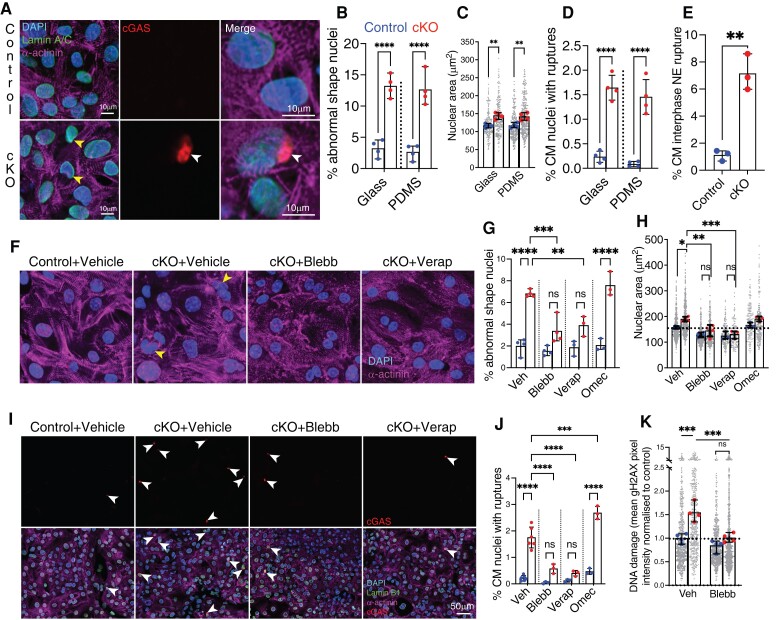
After observing increased apoptosis, DNA damage, and micronuclei in vivo, we next sought to understand the underlying mechanisms using cell biology that is amenable to drug treatments. We plated cardiomyocytes on soft, 6 kPa PDMS hydrogels to mimic in vivo heart stiffness.55–57 Strikingly, we observed an aberrant nuclear shape and enlarged nuclear area in E14.5 Lem2 cKO cardiomyocytes compared with control cells (Figure 4A–C).
Figure 4.
Foetal cardiomyocyte nuclei lacking Lem2 are aberrantly shaped and more susceptible to mechanical rupture and DNA damage. (A–D) Foetal cardiomyocytes from control and cardiomyocyte-specific Lem2 knockout (cKO) mice expressing a cGAS-mScarlet probe were isolated from E14.5 hearts and plated on soft, 6 kPa PDMS-coated coverslips to mimic stiffnesses encountered in vivo. Cells were stained for DAPI (nuclei), sarcomeric a-actinin (cardiomyocytes), and Lamin B1 (nuclear lamina). Note the aberrant nuclear shapes (yellow arrows in A), larger nuclear sizes, and increased nuclear rupture frequency (white arrow in A) observed in the cKO compared with control cardiomyocytes, quantified in (B–D). (E) NERDI events were detected in live cardiomyocytes over 16 h. Note the significant increase in NE rupture in cKO compared with control. (F–J) E14.5 cardiomyocytes from control and cKO mice expressing cGAS probe were plated on glass and treated with blebbistatin (Blebb) or verapamil (Verap) to inhibit muscle contraction or with Omecamtiv mecarbil (Omec) to stimulate contraction. Nuclear shapes and rupture were quantified in (F, G, and I). Note the striking attenuation of nuclear shape abnormalities and rupture events observed by inhibiting contraction. In contrast, stimulating muscle contraction increased nuclear rupture. (K) Lem2 cKO and control cardiomyocytes were treated with vehicle or blebbistatin (Blebb), fixed and stained for gH2AX, and mean intensities measured and normalized to control. Note the increased levels of DNA damage in cKO compared with control, which was attenuated by inhibiting contraction. Large data points show overall means from each experiment, small points show individual nuclear measurements within each experiment. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA; n = 3–5 experiments with 150–300 cells quantified/experiment. Graphs show mean ± SD.
Interestingly, others have shown associations between nuclear rupture, DNA damage, and cell death in migrating neurons depleted of Lamin B1.58 To assess NE integrity and observe if a similar process was occurring in Lem2 cKO cardiomyocytes, we transiently expressed catalytically inactive cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS) fused to an mScarlet fluorescent tag in Lem2 cKO cardiomyocytes to mark cytoplasmic chromatin exposure, hence nuclear rupture events.59,60 Notably, we saw a significantly increased incidence of nuclear rupture in Lem2 cKO cells compared with control, demonstrating an important role for Lem2 in maintaining nuclear integrity (Figure 4D). Importantly, similar observations were made when cells were plated on glass, which is much stiffer than hydrogel (Figure 4B–D), suggesting that the phenotype was independent of forces from the external environment. cGAS has previously been shown to associate with chromatin after NE breakdown during mitosis and aberrant NE resealing after mitosis,61–63 which may contribute to the levels of cGAS observed at the NE. To reveal whether cGAS accumulation at the NE was due to rupture during interphase (NERDI) or defective NE resealing after mitosis,64 we performed overnight live-cell imaging on cardiomyocytes expressing cGAS and tracked the incidence of cGAS accumulation at the NE in cardiomyocytes that specifically did not undergo mitosis. Importantly, we observed high levels of NERDI in Lem2 cKO compared with controls (∼8 vs. ∼1%, respectively) over 16 h (Figure 4E). Further evidence to suggest that cGAS accumulation at the NE was largely the result of NERDIs, we transduced cardiomyocytes with BAF-GFP as a marker for NERDI scars.64 Importantly, we observed significantly increased levels of BAF localization to the NE in Lem2 cKO compared with control, that was reminiscent of the cGAS data (see Supplementary material online, Figure S4A). Consistent with this, we observed a near 100% coincidence of BAF localization with cGAS to NE foci (see Supplementary material online, Figure S4B and C).
Next, we sought to understand the mechanisms underlying nuclear rupture. Given that cardiomyocyte nuclei are under constant mechanical load via the LINC complex, which transmits the force from the sarcomeres to the nucleoskeleton (see Supplementary material online, Movie S4), we hypothesized that inhibiting muscle contraction might attenuate nuclear shape defects and ruptures. To this end, we treated cardiomyocytes with para-nitroblebbistatin which inhibits myosin and arrests muscle contraction.65 This agent was able to fully attenuate nuclear shape defects and rupture (Figure 4F–J), suggesting that contractility is the driver of nuclear pathology in Lem2-null cardiomyocytes.
Given that para-nitroblebbistatin is a general inhibitor of the myosin Class II family, we hypothesized that these nuclear defects could potentially be driven by non-muscle contractility, i.e. myosin forces originating from the non-sarcomeric cytoskeleton (e.g. cortical actin network and/or actin stress fibres).66 Thus, we specifically inhibited muscle contraction via a different mechanism using verapamil, a clinically used drug which inhibits voltage-gated calcium channels to target excitation–contraction coupling. Similarly, verapamil was also able to fully rescue aberrant shapes and ruptures in cardiomyocyte nuclei (Figure 4F–J).67 To further test whether nuclear integrity was affected by muscle contraction, we used a cardiac myosin activator, Omecamtiv mecarbil, to stimulate muscle contraction. In agreement with our hypothesis, we observed increased nuclear rupture in Lem2 cKO nuclei after stimulating contraction (Figure 4G, H, and J).
Knowing that muscle contraction induced nuclear rupture, we hypothesized that it may also drive DNA damage, as observed in vivo (Figure 3). Indeed, increased DNA damage was apparent in cKO cardiomyocytes relative to controls, which was rescuable by inhibiting muscle contraction (Figure 4K). Together, these results show that in cardiomyocytes, Lem2 is essential for maintaining NE and genome integrity under mechanical load from muscle contractile forces.
To characterize potential changes in NE composition, we used detailed WB and immunofluorescence analyses. However, no detectable differences in the majority of NE and lamina proteins were observed between genotypes (see Supplementary material online, Figure S3L–N). These data suggest that the nuclear defects we observe are driven by the lack of Lem2, rather than changes to NE composition.
3.5. No overt defects in cardiac function are observed in Lem2 iCKO mice
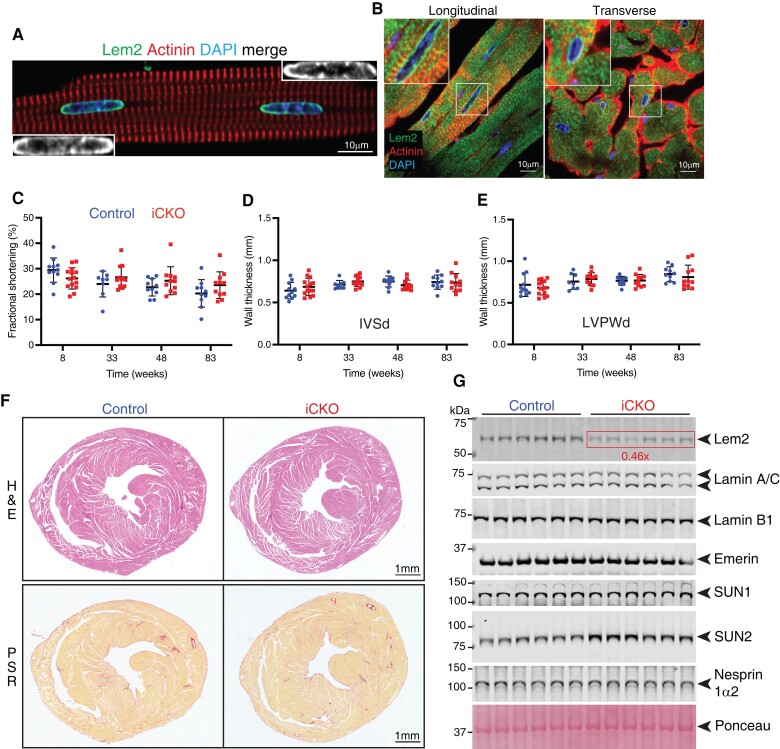
Having established that Lem2 is essential in foetal cardiomyocytes, we aimed to determine whether it plays a similar role in adults. After establishing expression of Lem2 in adult cardiomyocytes using isolated cells and heart sections (Figure 5A and B), we generated Lem2 inducible conditional knockout (iCKO) mice, using a tamoxifen-inducible cardiomyocyte-specific Cre mouse line Tnnt2MerCreMer/+.33
Figure 5.
Lem2 is expressed in adult cardiomyocytes, but is dispensable for maintaining cardiac function in adult mice. Adult cardiomyocytes (A) or longitudinal/transverse sections of adult heart (B) from 8-week-old control mice were stained using antibodies raised against Lem2 (green), sarcomeric a-actinin to denote cardiomyocytes (red) and DNA (blue). Note the discreet localization of Lem2 to the NE in cardiomyocytes. (C–E) Echocardiography was performed on Lem2f/f (control) and Lem2f/f; Cre/+ (cardiomyocyte-specific tamoxifen-inducible Lem2 knockout, iCKO) mice at 8 weeks of age, 1 week prior to tamoxifen injection, and subsequently followed up at 33, 48, and 83 weeks of age. Measured parameters were as follows: (C) fractional shortening; (D and E) inter-ventricular septum thickness during diastole and left ventricular posterior wall thickness during diastole (LVPWd). No differences were observed between control and iCKO. (F) Heart sections from 83-week-old mouse hearts stained with haemotoxylin and eosin (H&E) to delineate nuclei and muscle tissue, respectively (top panels); picrosirius red to delineate collagen in red (bottom panels). Note that there were no differences in morphology or fibrosis between genotypes. (G) WB were performed on protein extracted from whole hearts from 83-week-old mice. Note that as expected, Lem2 levels were reduced in iCKO compared with control, but Lamins A/C, B1, Emerin, SUN1, and Nesprin-1a2 levels were comparable between genotypes (quantifications shown in Supplementary material online, Figure S4G). n = 10–14 mice per genotype for echocardiography, n = 6 for WB, and n = 3 for immunohistochemistry. Graphs show mean ± SD.
To investigate whether the loss of Lem2 specifically affected adult stages of life, we assessed in vivo cardiac function in Lem2 iCKO mice. We performed echocardiography at 8 weeks of age, prior to Lem2 ablation at 9 weeks, followed by serial echocardiograms at 33, 48, and 83 weeks of age. Cardiac function and wall thicknesses of Lem2 iCKO mice were comparable to control littermates across all measurements and ages (Figure 5C–E and Supplementary material online, Figure S4D–F). Consistently, we observed no changes in expression levels of the foetal gene programme and pro-fibrotic markers by quantitative RT-PCR analysis (see Supplementary material online, Figure S4G), suggestive of no adverse remodelling or fibrosis. Complementary to the echocardiography and gene expression analyses, histological analysis revealed no evidence of morphological defects, changes in extracellular matrix deposition, nor changes in myocyte size (Figure 5F and Supplementary material online, Figure S4H). Furthermore, there were also no changes in heart weight/tibia length and heart weight/body weight ratios of Lem2 iCKO mice compared with control littermates (see Supplementary material online, Figure S4I).
These data suggest that once the adult mouse heart has fully developed, removal of Lem2 does not affect heart function up to 83 weeks of age.
3.6. The majority of NE protein levels are unchanged in Lem2 iCKO hearts and isolated cardiomyocytes
Given the lack of baseline phenotype in the Lem2 iCKO mice, we hypothesized that other NE proteins may compensate for Lem2, as shown previously in other systems.68,69 To test this, we performed WBs on whole hearts from control and Lem2 iCKO mice and probed them for a panel of NE and lamina proteins (Figure 5G). As expected, Lem2 levels were reduced to 46% of control levels in Lem2 iCKO. Other proteins were unchanged, except for the LINC complex protein SUN2, which was increased 1.5-fold compared with control (P < 0.01) (see Supplementary material online, Figure S4J).
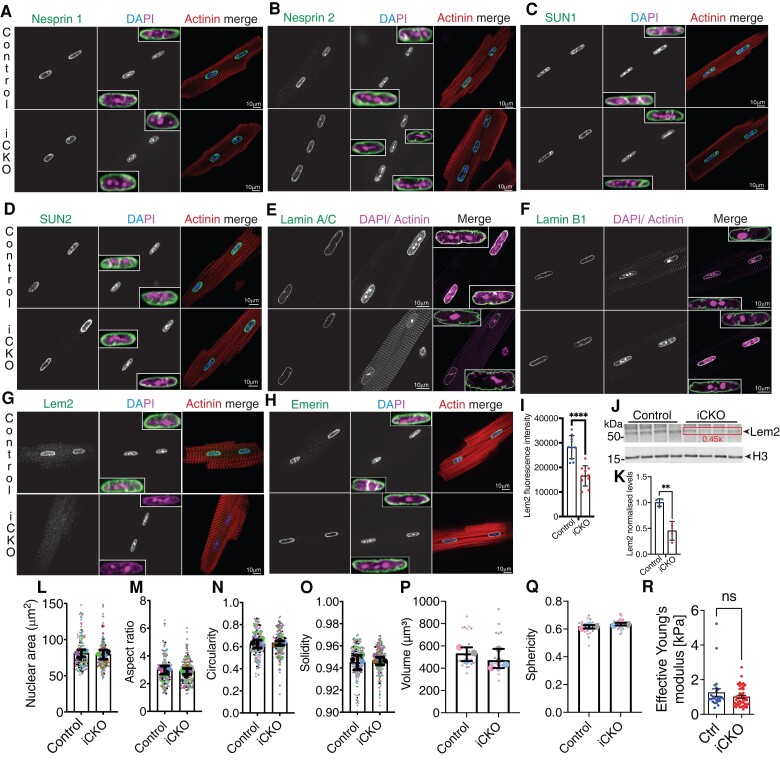
Immunofluorescence levels and localization of most NE proteins were unchanged, with the exception of Lem2 which was significantly reduced in iCKO cardiomyocytes, and a slight increase in SUN2 that was consistent with WBs (Figure 6A–K and Supplementary material online, FigureS5A–F). mRNA transcript levels of Sun2 (as well as other LINC complex components) were similar between genotypes (see Supplementary material online, Figure S5C), suggesting that the increase in SUN2 observed in Lem2 iCKO cardiomyocytes is likely regulated at the protein, rather than the gene expression level.
Figure 6.
Nuclear shape and NE composition are largely unaffected by loss of Lem2 in adult cardiomyocytes. (A–H) Adult cardiomyocytes isolated from 8- to 10-week-old control and cardiomyocyte-specific inducible Lem2 knockout (iCKO) mice were stained using antibodies raised against various NE components (green), sarcomeric a-actinin or phalloidin (red), and DNA (blue/magenta). (A and B) Nesprin 1 and 2, respectively (C and D), SUN1 and 2, respectively, (E and F) Lamins A/C and B1, respectively, (G) Lem2, (H) Emerin. (I) Quantification of Lem2 fluorescence intensity in adult cardiomycoytes. (J) WB were performed on protein extracted from isolated adult cardiomyocytes from 8- to 10-week-old mice using antibodies raised against Lem2 and Histone H3 (loading control), quantified in (K). 2D nuclear shape parameters were analysed: nuclear area (L), aspect ratio (M), circularity (N), solidity (O). 3D shape parameters: nuclear volume (P) and sphericity (Q). Insets show enlarged NE components merged with DAPI and maximum intensity projections of Lamin A/C. (L–Q) Large data points represent means from individual mice, and small points represent individual nuclear measurements within each mouse. (R) Nanoindentation was performed on living adult cardiomyocytes to measure nuclear stiffness between genotypes. Note that nuclear stiffness was comparable between ink and control. n = 3–5 mice/genotype. Graphs show mean ± SD.
Knowing that ERK1/2 activation was elevated in foetal Lem2 cKO hearts, we assessed MAPK pathways in adult iCKO cardiomyocytes, but found no differences in activation levels (see Supplementary material online, Figure S5E and G).
3.7. Nuclear integrity and stiffness are unaffected in Lem2 iCKO adult cardiomyocytes
Given that we observed alterations to nuclear shape in Lem2 cKO cardiomyocytes, we hypothesized that similar findings might be observed in adulthood. We carried out comprehensive 2D and 3D analyses of cardiomyocyte nuclei, but found no differences in shape parameters between genotypes (Figure 6L–Q). Interestingly, we observed invaginations of the NE/lamina in both controls and iCKO mice, but to similar extents in both genotypes (see Supplementary material online, Movies S5–S8).
Previous work from others has shown that mutations in Lem2’s interaction partners, Lamins A and C, affect nuclear stiffness, rendering nuclei more deformable to mechanical stress.3 To test whether this was occurring in Lem2 iCKO, we performed nanoindentation on adult cardiomyocytes from Lem2 iCKO and control mice. The calculated Young’s modulus revealed comparable nuclear stiffnesses between Lem2 iCKO and controls (1.01 vs. 1.26 kPa, respectively) (Figure 6R).
Together these data suggest that Lem2 is not required for normal nuclear morphology and mechanics in adult mouse cardiomyocytes.
3.8. Nuclear shape is maintained in response to elevated pressure in Lem2 iCKO adult cardiomyocytes
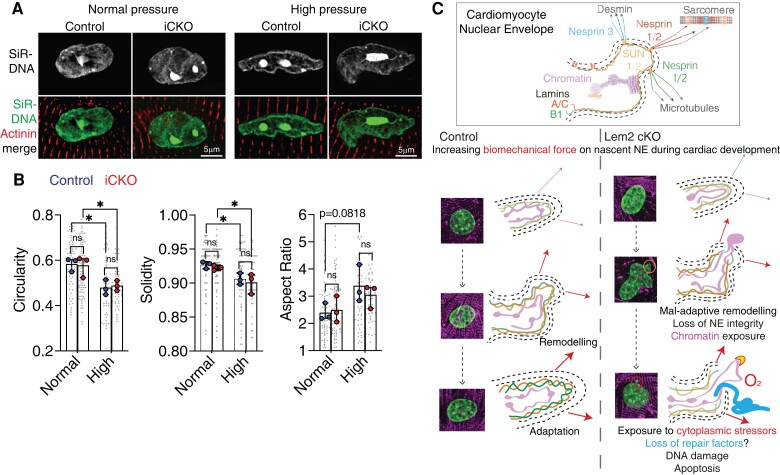
Given the lack of baseline phenotype in Lem2 iCKO cardiomyocytes, we next wanted to stress nuclei in living Lem2 iCKO cardiomyocytes. To perform this, we isolated adult cardiomyocytes from Lem2 iCKO or littermate controls and subjected them to hydrodynamic pressures that are analogous to those observed in either healthy, normal hearts or pressure-overloaded hearts in vivo using a pressure stimulator as shown previously70 (CellScale MechanoCulture TR, modified for a low-pressure range).44
We subjected adult cardiomyocytes from Lem2 iCKO and controls to oscillatory stimulation of normal (120/15 mmHg) or high pressure (200/30 mmHg), and nuclear shape analyses were carried out. Importantly, cardiomyocyte nuclei from both genotypes responded comparably to high pressure and became more irregularly shaped and less circular, as measured by nuclear circularity, solidity, and aspect ratios (Figure 7A). However, there were no observed changes in nuclear shape between Lem2 iCKO and control cells (Figure 7B).
Figure 7.
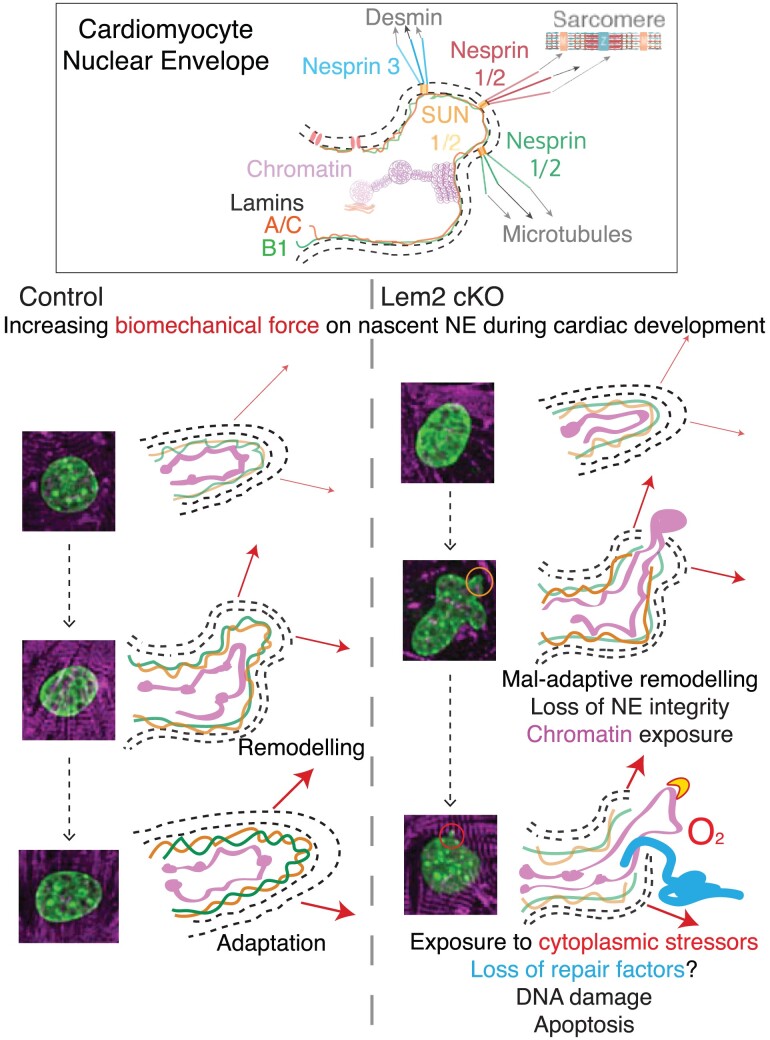
Nuclear shape is maintained in response to elevated pressure in Lem2 iCKO adult cardiomyocytes. (A) Adult cardiomyocytes were isolated from control and iCKO animals and subjected to either normal (120/15 mmHg) or high (200/30 mmHg) oscillatory isotopic pressure and stained for DNA (green) and alpha actinin (red). (B) 2D nuclear shape parameters were analysed: circularity, solidity, aspect ratio. Note the alterations to nuclear shape parameters caused by high pressure compared with normal pressure conditions, but no differences were observed between control and iCKO cells. (C) Working model: in control cardiomyocytes during foetal heart development, the nascent NE adapts and remodels to mitigate the effects of increases in biomechanical load. Conversely, in Lem2 KO cardiomyocytes, the NE undergoes maladaptive remodelling, which results in nuclear rupture, chromatin exposure, and subsequent DNA damage. These ultimately lead to cell death as observed in our cardiomyocyte-specific Lem2 knockout (cKO) model. Conversely, in adult cardiomyocytes that have adapted to withstanding the physical demands on the NE, removal of Lem2 apparently does not affect cardiac function or nuclear shape. Our data suggest that Lem2 is essential for foetal heart development and that removal of Lem2 in adult cardiomyocytes is well tolerated at baseline.
Together, these data suggest that Lem2 is not required to maintain nuclear shape under high pressure in adult cardiomyocytes. Having said this, we cannot exclude the possibility that the remaining Lem2 in Lem2 iCKO hearts is sufficient to maintain Lem2’s function in this role.
4. Discussion
Overall, our data demonstrate that Lem2 is critical for foetal heart development and survival. Lem2 cKO foetal hearts display elevated apoptosis and reduced expression of cardiac developmental genes. Additionally, Lem2-null foetal cardiomyocyte nuclei are highly susceptible to mechanically induced rupture, resulting in DNA damage, micronuclei, and apoptosis. Conversely, ablation of Lem2 in adult cardiomyocytes results in no detectable effects on the cardiac function or nuclear shape. Various factors may be able to explain the dichotomy of phenotypes observed between foetal and adult cardiomyocytes including:
Differing Lamin A/C levels between foetal and adult cardiomyocytes: Lamin A/C, which contributes to nuclear rigidity,71 is present at low levels in embryogenesis, gradually increasing as cells undergo differentiation and tissues become stiffer.72 One could therefore envisage that Lem2 plays a structural role in foetal myocytes prior to the establishment of a more robust, Lamin A/C-containing nuclear lamina. In support of this, Lem2 is expressed in E8.5 mouse embryos, prior to the onset of Lamin A/C expression at E12.27,73,74 In contrast to foetal cardiomyocytes, fully differentiated adult cardiomyocytes express higher levels of Lamin A/C, and are therefore better able to mitigate the effect of Lem2 loss. In addition, Lem2 ablation resulted in increased SUN2 levels in adulthood but not in foetal stages, which may be a further compensatory mechanism in fully developed cells. Interestingly, others have shown that SUN2 levels can be modulated in response to forces such as cell stretch and cytoskeletal stiffness,75,76 and it is possible that the loss of Lem2 results in mechanical adaptation at the NE via effects on SUN2.
Differentiation status of foetal compared with adult cardiomyocytes: roles of Lem2 in NE resealing after mitosis point to differing functions in dividing (foetal) vs. terminally differentiated (adult) cardiomyocytes.19,21,68 Our data cannot exclude the possibility that Lem2 plays a role in NE resealing after mitosis, since foetal cardiomyocytes are still dividing and there was evidence of micronuclei in Lem2 cKO hearts. However, we did not observe changes to DNA synthesis or mitosis in Lem2 cKO foetal hearts (Figure 3G and H and Supplementary material online, Figure S3F–I). In addition, our live-cell imaging experiments in culture suggested a high occurrence of rupture events in cardiomyocytes that did not undergo division. Furthermore, the relative subtlety of the cardiac size and morphology defects in foetal Lem2 cKO hearts suggest that Lem2 does not profoundly affect cardiomyocyte division, when compared with other well-established genes involved in this process.77 These data favour the hypothesis that the pathophysiological mechanism is likely through defective repair of the NE during interphase.
An intriguing finding of our study was that cardiomyocyte nuclear rupture may be a normal occurrence during cardiac development. In particular, these ruptures are largely the result of forces originating from muscle contraction, which require Lem2-orchestrated repair. We speculate this process may act as a quality control mechanism to ensure nuclear robustness prior to the significant haemodynamic load changes observed postnatally and into adolescence.55 Future studies to test this hypothesis would require ablation of Lem2 expression immediately prior to birth to reveal whether these stresses associated with postnatal heart development drive nuclear rupture and if Lem2 is similarly required for this process.
A limitation of our study was that we achieved around ∼55% knockdown of Lem2 protein in iCKO mice. Therefore, we cannot exclude the possibility that the remaining Lem2 was able to functionally compensate and mask a potential phenotype. Future studies to achieve greater Lem2 removal will be required to reveal whether there are overt changes in cardiac function after Lem2 ablation in adult cardiomyocytes. Alternatively, it would be interesting to perform transverse aortic constriction to uncover whether Lem2 plays an important role in the adult heart in response to pressure overload in vivo.
Interestingly, a variant in Lem2 that changes leucine 13 to arginine (L13R) is associated with AC and sudden cardiac death.15,16 However, limited patient data make it hard to discern the molecular mechanisms in play. For example, data from one patient suggest that the levels of Lem2 comparable to control samples, suggesting the L13R mutation could be gain-of-function. However, the autosomal-recessive nature of inheritance would suggest it is most likely loss-of-function. Nevertheless, transcriptomics data from our study showed down-regulation of cardiac conduction genes (Figure 2), which is in line with the arrhythmogenic phenotype observed in humans. Furthermore, other parallels between the human and mouse phenotype include thinning of myocardial walls and apparent cardiac dysfunction. Despite this, the Lem2 L13R variant in humans results in relatively late-onset AC, and death between the ages of 20 and 50, compared with the foetal lethality observed in Lem2 cKO mice. Therefore, further work using Lem2 L13R animal models will be important for understanding the underlying pathophysiological mechanisms.
4.1. Working model of Lem2 function in cardiomyocytes
In control cardiomyocytes during foetal heart development, the nascent NE adapts and remodels to mitigate the effects of increases in biomechanical load (Figure 7C). Conversely, in Lem2 cKO cardiomyocytes, the NE undergoes maladaptive remodelling, which results in gene expression changes, nuclear rupture, chromatin exposure, and subsequent DNA damage. These lead to increased cardiomyocyte cell death, which ultimately results in the underdevelopment of the heart as observed in our Lem2 cKO model. Conversely, in adult cardiomyocytes, which harbour nuclei that are stiffer and more resistant to mechanical load, removal of Lem2 apparently does not affect cardiac function or nuclear integrity. Our data suggest that Lem2 is essential for foetal heart development and that depletion of Lem2 in adult cardiomyocytes is well tolerated at baseline.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
J.A.R., L.G., and M.J.S.: conceptualization; J.A.R., E.B., N.A.-V., C.B., D.H., F.P., T.M., E.M., N.C., O.T., T.I., A.M.S., and M.J.S.: methodology; J.A.R., E.B., N.A.-V., C.A.O., C.B., E.M., P.S., F.P., T.M., N.C., and M.J.S.: investigation; J.A.R., N.A.-V., and M.J.S.: visualization; M.J.S.: funding acquisition, project administration, and supervision; J.A.R. and M.J.S.: writing—original draft; J.A.R., L.G., A.M.S., and M.J.S: writing—review and editing.
Supplementary Material
Acknowledgements
We thank the British Heart Foundation for funding, the Wohl Centre for Cellular Imaging for their support, and Camille Charoy for technical assistance.
Translational perspective.
Cardiac laminopathies, caused by mutations in proteins associated with the nuclear envelope (NE), are the second highest cause of inherited dilated cardiomyopathy. However, the underlying pathological mechanisms are poorly understood, and the molecular players involved in nuclear integrity remain obscure. Mutations in the NE protein Lem2 lead to human cardiomyopathy, but its roles in the heart remain unknown.
Here, we demonstrate that Lem2 protects the NE against muscle contraction that causes catastrophic nuclear ruptures. Moreover, our data provide mechanistic insight into pathophysiological mechanisms that underlie Lem2 cardiomyopathy and cardiac laminopathies in general.
Contributor Information
Jacob A Ross, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Nathaly Arcos-Villacis, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Edmund Battey, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK; Centre of Human and Applied Physiological Sciences, School of Basic and Medical Biosciences, Faculty of Life Sciences and Medicine, King’s College London, London SE1 1UL, UK.
Cornelis Boogerd, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW), University Medical Center Utrecht, Utrecht 3584 CT, The Netherlands.
Constanza Avalos Orellana, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Emilie Marhuenda, Division of Bioengineering, School of Engineering and Materials Science, Queen Mary University of London, London E1 4NS, UK.
Pamela Swiatlowska, Division of Bioengineering, School of Engineering and Materials Science, Queen Mary University of London, London E1 4NS, UK.
Didier Hodzic, Department of Developmental Biology, Washington University School of Medicine, 660S. Euclid Avenue, St Louis, MO 63110, USA.
Fabrice Prin, Crick Advanced Light Microscopy Facility, The Francis Crick Institute, London NW1 1AT, UK.
Tim Mohun, Crick Advanced Light Microscopy Facility, The Francis Crick Institute, London NW1 1AT, UK.
Norman Catibog, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Olga Tapia, Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA; Research Group on Foods, Nutritional Biochemistry and Health, Universidad Europea del Atlántico, Santander 39011, Spain; Department of Basic Medical Sciences, Institute of Biomedical Technologies, University of La Laguna, Tenerife 38200, Spain.
Larry Gerace, Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Thomas Iskratsch, Division of Bioengineering, School of Engineering and Materials Science, Queen Mary University of London, London E1 4NS, UK.
Ajay M Shah, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Matthew J Stroud, British Heart Foundation Centre of Research Excellence, School of Cardiovascular & Metabolic Medicine & Sciences, Faculty of Life Sciences & Medicine, King’s College London, James Black Centre, 125 Coldharbour Lane, London SE5 9NU, UK.
Funding
This work was supported by British Heart Foundation fellowship FS/17/57/32934 (J.A.R., E.B., N.A.-V., and M.J.S.); British Heart Foundation grant RE/18/2/34213 (J.A.R., E.B., N.C., A.M.S., and M.J.S.); British Heart Foundation Chair CH/1999001/11735 (A.M.S.); British Heart Foundation grant (PG/20/6/34835) (T.I.); Medical Research Council of the UK (MR/S023593/1) (E.B. and M.J.S.); Biotechnology and Biological Sciences Research Council of the UK (BB/S001123/1) (T.I.); and National Institutes of Health GM028521 (L.G.).
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author. RNA-seq data are available on the GEO database with accession number GSE217693.
References
- 1. Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol 1978;79:546–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med 2009;13:1059–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004;113:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ross JA, Stroud MJ. THE NUCLEUS: mechanosensing in cardiac disease. Int J Biochem Cell Biol 2021;137:106035. [DOI] [PubMed] [Google Scholar]
- 5. Stroud MJ. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiomyopathy. Biophys Rev 2018;10:1033–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 2006;172:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stroud MJ, Feng W, Zhang J, Veevers J, Fang X, Gerace L, Chen J. Nesprin 1alpha2 is essential for mouse postnatal viability and nuclear positioning in skeletal muscle. J Cell Biol 2017;216:1915–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banerjee I, Zhang J, Moore-Morris T, Pfeiffer E, Buchholz KS, Liu A, Ouyang K, Stroud MJ, Gerace L, Evans SM, McCulloch A, Chen J. Targeted ablation of nesprin 1 and nesprin 2 from murine myocardium results in cardiomyopathy, altered nuclear morphology and inhibition of the biomechanical gene response. PLoS Genet 2014;10:e1004114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stroud MJ, Banerjee I, Veevers J, Chen J. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ Res 2014;114:538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, Pytel P, McNally EM. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet 2009;18:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chai RJ, Werner H, Li PY, Lee YL, Nyein KT, Solovei I, Luu TDA, Sharma B, Navasankari R, Maric M, Sim LYE, Loh YJ, Aliwarga E, Cheong JWL, Chojnowski A, Autio MI, Haiyang Y, Boon Tan KK, Keng CT, Ng SL, Chew WL, Ferenczi M, Burke B, Foo RSY, Stewart CL. Disrupting the LINC complex by AAV mediated gene transduction prevents progression of Lamin induced cardiomyopathy. Nat Commun 2021;12:4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stewart RM, Rodriguez EC, King MC. Ablation of SUN2-containing LINC complexes drives cardiac hypertrophy without interstitial fibrosis. Mol Biol Cell 2019;30:1664–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 1994;8:323–327. [DOI] [PubMed] [Google Scholar]
- 14. Taylor MR, Slavov D, Gajewski A, Vlcek S, Ku L, Fain PR, Carniel E, Di Lenarda A, Sinagra G, Boucek MM, Cavanaugh J, Graw SL, Ruegg P, Feiger J, Zhu X, Ferguson DA, Bristow MR, Gotzmann J, Foisner R, Mestroni L. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum Mutat 2005;26:566–574. [DOI] [PubMed] [Google Scholar]
- 15. Boone PM, Yuan B, Gu S, Ma Z, Gambin T, Gonzaga-Jauregui C, Jain M, Murdock TJ, White JJ, Jhangiani SN, Walker K, Wang Q, Muzny DM, Gibbs RA, Hejtmancik JF, Lupski JR, Posey JE, Lewis RA. Hutterite-type cataract maps to chromosome 6p21.32-p21.31, cosegregates with a homozygous mutation in LEMD2, and is associated with sudden cardiac death. Mol Genet Genomic Med 2016;4:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abdelfatah N, Chen R, Duff HJ, Seifer CM, Buffo I, Huculak C, Clarke S, Clegg R, Jassal DS, Gordon PMK, Ober C; Care4Rare Canada Consortium; Frosk P, Gerull B. Characterization of a unique form of arrhythmic cardiomyopathy caused by recessive mutation in LEMD2. JACC Basic Transl Sci 2019;4:204–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marbach F, Rustad CF, Riess A, Dukic D, Hsieh TC, Jobani I, Prescott T, Bevot A, Erger F, Houge G, Redfors M, Altmueller J, Stokowy T, Gilissen C, Kubisch C, Scarano E, Mazzanti L, Fiskerstrand T, Krawitz PM, Lessel D, Netzer C. The discovery of a LEMD2-associated nuclear envelopathy with early progeroid appearance suggests advanced applications for AI-driven facial phenotyping. Am J Hum Genet 2019;104:749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Halfmann CT, Sears RM, Katiyar A, Busselman BW, Aman LK, Zhang Q, O'Bryan CS, Angelini TE, Lele TP, Roux KJ. Repair of nuclear ruptures requires barrier-to-autointegration factor. J Cell Biol 2019;218:2136–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu M, LaJoie D, Chen OS, von Appen A, Ladinsky MS, Redd MJ, Nikolova L, Bjorkman PJ, Sundquist WI, Ullman KS, Frost A. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc Natl Acad Sci USA 2017;114:E2166–E2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vietri M, Schultz SW, Bellanger A, Jones CM, Petersen LI, Raiborg C, Skarpen E, Pedurupillay CRJ, Kjos I, Kip E, Timmer R, Jain A, Collas P, Knorr RL, Grellscheid SN, Kusumaatmaja H, Brech A, Micci F, Stenmark H, Campsteijn C. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat Cell Biol 2020;22:856–867. [DOI] [PubMed] [Google Scholar]
- 21. von Appen A, LaJoie D, Johnson IE, Trnka MJ, Pick SM, Burlingame AL, Ullman KS, Frost A. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 2020;582:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ulbert S, Antonin W, Platani M, Mattaj IW. The inner nuclear membrane protein Lem2 is critical for normal nuclear envelope morphology. FEBS Lett 2006;580:6435–6441. [DOI] [PubMed] [Google Scholar]
- 23. Barrales RR, Forn M, Georgescu PR, Sarkadi Z, Braun S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev 2016;30:133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morales-Martinez A, Dobrzynska A, Askjaer P. Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J Cell Sci 2015;128:1090–1096. [DOI] [PubMed] [Google Scholar]
- 25. Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 2012;150:934–947. [DOI] [PubMed] [Google Scholar]
- 26. Gatta AT, Olmos Y, Stoten CL, Chen Q, Rosenthal PB, Carlton JG. CDK1 controls CHMP7-dependent nuclear envelope reformation. Elife 2021;10:e59999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tapia O, Fong LG, Huber MD, Young SG, Gerace L. Nuclear envelope protein Lem2 is required for mouse development and regulates MAP and AKT kinases. PLoS One 2015;10:e0116196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011;474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC; Sanger Institute Mouse Genetics Project; Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 2013;154:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 2000;28:106–110. [PubMed] [Google Scholar]
- 31. Breckenridge R, Kotecha S, Towers N, Bennett M, Mohun T. Pan-myocardial expression of Cre recombinase throughout mouse development. Genesis 2007;45:135–144. [DOI] [PubMed] [Google Scholar]
- 32. Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BLM. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev 2003;17:2362–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan J, Sultana N, Zhang L, Park DS, Shekhar A, Hu J, Bu L, Cai CL. Generation of a tamoxifen inducible Tnnt2MerCreMer knock-in mouse model for cardiac studies. Genesis 2015;53:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stroud MJ, Fang X, Zhang J, Guimaraes-Camboa N, Veevers J, Dalton ND, Gu Y, Bradford WH, Peterson KL, Evans SM, Gerace L, Chen J. Luma is not essential for murine cardiac development and function. Cardiovasc Res 2018;114:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z, Stroud MJ, Zhang J, Fang X, Ouyang K, Kimura K, Mu Y, Dalton ND, Gu Y, Bradford WH, Peterson KL, Cheng H, Zhou X, Chen J. Normalization of Naxos plakoglobin levels restores cardiac function in mice. J Clin Invest 2015;125:1708–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ackers-Johnson M, Li PY, Holmes AP, O'Brien SM, Pavlovic D, Foo RS. A simplified, Langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ Res 2016;119:909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stroud MJ, Fang X, Veevers J, Chen J. Generation and analysis of striated muscle selective LINC complex protein mutant mice. Methods Mol Biol 2018;1840:251–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stroud MJ, Kammerer RA, Ballestrem C. Characterization of G2L3 (GAS2-like 3), a new microtubule- and actin-binding protein related to spectraplakins. J Biol Chem 2011;286:24987–24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stroud MJ, Nazgiewicz A, McKenzie EA, Wang Y, Kammerer RA, Ballestrem C. GAS2-like proteins mediate communication between microtubules and actin through interactions with end-binding proteins. J Cell Sci 2014;127:2672–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Geyer SH, Maurer-Gesek B, Reissig LF, Weninger WJ. High-resolution episcopic microscopy (HREM) - simple and robust protocols for processing and visualizing organic materials. J Vis Exp 2017;125:e56071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Captur G, Wilson R, Bennett MF, Luxan G, Nasis A, de la Pompa JL, Moon JC, Mohun TJ. Morphogenesis of myocardial trabeculae in the mouse embryo. J Anat 2016;229:314–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2021; doi:10.1101/060012.
- 43. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Swiatlowska P, Sit B, Feng Z, Marhuenda E, Xanthis I, Zingaro S, Ward M, Zhou X, Xiao Q, Shanahan C, Jones GE, Yu CH, Iskratsch T. Pressure and stiffness sensing together regulate vascular smooth muscle cell phenotype switching. Sci Adv 2022;8:eabm3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Battey E, Ross JA, Hoang A, Wilson DGS, Han Y, Levy Y, Pollock RD, Kalakoutis M, Pugh JN, Close GL, Ellison-Hughes GM, Lazarus NR, Iskratsch T, Harridge SDR, Ochala J, Stroud MJ. Myonuclear alterations associated with exercise are independent of age in humans. J Physiol 2023. [DOI] [PubMed] [Google Scholar]
- 46. Guerrero CR, Garcia PD, Garcia R. Subsurface imaging of cell organelles by force microscopy. ACS Nano 2019;13:9629–9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN; Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center; Jackson L; Infrastructure Nationale Phenomin ICdlS; MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bucan M, Murray SA. High-throughput discovery of novel developmental phenotypes. Nature 2016;537:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim MY, Seo EJ, Lee DH, Kim EJ, Kim HS, Cho HY, Chung EY, Lee SH, Baik EJ, Moon CH, Jung YS. Gadd45beta is a novel mediator of cardiomyocyte apoptosis induced by ischaemia/hypoxia. Cardiovasc Res 2010;87:119–126. [DOI] [PubMed] [Google Scholar]
- 49. Lucas A, Mialet-Perez J, Daviaud D, Parini A, Marber MS, Sicard P. Gadd45gamma regulates cardiomyocyte death and post-myocardial infarction left ventricular remodelling. Cardiovasc Res 2015;108:254–267. [DOI] [PubMed] [Google Scholar]
- 50. Roder K, Werdich AA, Li W, Liu M, Kim TY, Organ-Darling LE, Moshal KS, Hwang JM, Lu Y, Choi BR, MacRae CA, Koren G. RING finger protein RNF207, a novel regulator of cardiac excitation. J Biol Chem 2014;289:33730–33740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prins BP, Mead TJ, Brody JA, Sveinbjornsson G, Ntalla I, Bihlmeyer NA, van den Berg M, Bork-Jensen J, Cappellani S, Van Duijvenboden S, Klena NT, Gabriel GC, Liu X, Gulec C, Grarup N, Haessler J, Hall LM, Iorio A, Isaacs A, Li-Gao R, Lin H, Liu CT, Lyytikainen LP, Marten J, Mei H, Muller-Nurasyid M, Orini M, Padmanabhan S, Radmanesh F, Ramirez J, Robino A, Schwartz M, van Setten J, Smith AV, Verweij N, Warren HR, Weiss S, Alonso A, Arnar DO, Bots ML, de Boer RA, Dominiczak AF, Eijgelsheim M, Ellinor PT, Guo X, Felix SB, Harris TB, Hayward C, Heckbert SR, Huang PL, Jukema JW, Kahonen M, Kors JA, Lambiase PD, Launer LJ, Li M, Linneberg A, Nelson CP, Pedersen O, Perez M, Peters A, Polasek O, Psaty BM, Raitakari OT, Rice KM, Rotter JI, Sinner MF, Soliman EZ, Spector TD, Strauch K, Thorsteinsdottir U, Tinker A, Trompet S, Uitterlinden A, Vaartjes I, van der Meer P, Volker U, Volzke H, Waldenberger M, Wilson JG, Xie Z, Asselbergs FW, Dorr M, van Duijn CM, Gasparini P, Gudbjartsson DF, Gudnason V, Hansen T, Kaab S, Kanters JK, Kooperberg C, Lehtimaki T, Lin HJ, Lubitz SA, Mook-Kanamori DO, Conti FJ, Newton-Cheh CH, Rosand J, Rudan I, Samani NJ, Sinagra G, Smith BH, Holm H, Stricker BH, Ulivi S, Sotoodehnia N, Apte SS, van der Harst P, Stefansson K, Munroe PB, Arking DE, Lo CW, Jamshidi Y. Exome-chip meta-analysis identifies novel loci associated with cardiac conduction, including ADAMTS6. Genome Biol 2018;19:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet 1996;12:17–23. [DOI] [PubMed] [Google Scholar]
- 53. Burgess DL, Gefrides LA, Foreman PJ, Noebels JL. A cluster of three novel Ca2+ channel gamma subunit genes on chromosome 19q13.4: evolution and expression profile of the gamma subunit gene family. Genomics 2001;71:339–350. [DOI] [PubMed] [Google Scholar]
- 54. Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 55. Jacot JG, Martin JC, Hunt DL. Mechanobiology of cardiomyocyte development. J Biomech 2010;43:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pandey P, Hawkes W, Hu J, Megone WV, Gautrot J, Anilkumar N, Zhang M, Hirvonen L, Cox S, Ehler E, Hone J, Sheetz M, Iskratsch T. Cardiomyocytes sense matrix rigidity through a combination of muscle and non-muscle myosin contractions. Dev Cell 2018;44:326–336.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ward M, Iskratsch T. Mix and (mis-)match - the mechanosensing machinery in the changing environment of the developing, healthy adult and diseased heart. Biochim Biophys Acta Mol Cell Res 2020;1867:118436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen NY, Yang Y, Weston TA, Belling JN, Heizer P, Tu Y, Kim P, Edillo L, Jonas SJ, Weiss PS, Fong LG, Young SG. An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc Natl Acad Sci USA 2019;116:25870–25879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Earle AJ, Kirby TJ, Fedorchak GR, Isermann P, Patel J, Iruvanti S, Moore SA, Bonne G, Wallrath LL, Lammerding J. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat Mater 2020;19:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, Piel M. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016;352:359–362. [DOI] [PubMed] [Google Scholar]
- 61. Guey B, Wischnewski M, Decout A, Makasheva K, Kaynak M, Sakar MS, Fierz B, Ablasser A. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 2020;369:823–828. [DOI] [PubMed] [Google Scholar]
- 62. Zhong L, Hu MM, Bian LJ, Liu Y, Chen Q, Shu HB. Phosphorylation of cGAS by CDK1 impairs self-DNA sensing in mitosis. Cell Discov 2020;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li T, Huang T, Du M, Chen X, Du F, Ren J, Chen ZJ. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science 2021;371:eabc5386.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wallis SS, Ventimiglia LN, Otigbah E, Infante E, Cuesta-Geijo MA, Kidiyoor GR, Carbajal MA, Fleck RA, Foiani M, Garcia-Manyes S, Martin-Serrano J, Agromayor M. The ESCRT machinery counteracts Nesprin-2G-mediated mechanical forces during nuclear envelope repair. Dev Cell 2021;56:3192–3202.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kepiro M, Varkuti BH, Vegner L, Voros G, Hegyi G, Varga M, Malnasi-Csizmadia A. para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor. Angew Chem Int Ed Engl 2014;53:8211–8215. [DOI] [PubMed] [Google Scholar]
- 66. Fenix AM, Neininger AC, Taneja N, Hyde K, Visetsouk MR, Garde RJ, Liu B, Nixon BR, Manalo AE, Becker JR, Crawley SW, Bader DM, Tyska MJ, Liu Q, Gutzman JH, Burnette DT. Muscle-specific stress fibers give rise to sarcomeres in cardiomyocytes. Elife 2018;7:e42144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bell D, McDermott BJ. Inhibition by verapamil and diltiazem of agonist-stimulated contractile responses in mammalian ventricular cardiomyocytes. J Mol Cell Cardiol 1995;27:1977–1987. [DOI] [PubMed] [Google Scholar]
- 68. Barkan R, Zahand AJ, Sharabi K, Lamm AT, Feinstein N, Haithcock E, Wilson KL, Liu J, Gruenbaum Y. Ce-emerin and LEM-2: essential roles in Caenorhabditis elegans development, muscle function, and mitosis. Mol Biol Cell 2012;23:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol 2009;29:5718–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hershberger KA, Abraham DM, Martin AS, Mao L, Liu J, Gu HB, Locasale JW, Hirschey MD. Sirtuin 5 is required for mouse survival in response to cardiac pressure overload. J Biol Chem 2017;292:19767–19781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F, Speicher DW, Discher DE. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013;341:1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, Joffe B. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013;152:584–598. [DOI] [PubMed] [Google Scholar]
- 73. Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 1989;105:365–378. [DOI] [PubMed] [Google Scholar]
- 74. Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell 1987;51:383–392. [DOI] [PubMed] [Google Scholar]
- 75. Gilbert HTJ, Mallikarjun V, Dobre O, Jackson MR, Pedley R, Gilmore AP, Richardson SM, Swift J. Nuclear decoupling is part of a rapid protein-level cellular response to high-intensity mechanical loading. Nat Commun 2019;10:4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mu X, Tseng C, Hambright WS, Matre P, Lin CY, Chanda P, Chen W, Gu J, Ravuri S, Cui Y, Zhong L, Cooke JP, Niedernhofer LJ, Robbins PD, Huard J. Cytoskeleton stiffness regulates cellular senescence and innate immune response in Hutchinson-Gilford Progeria Syndrome. Aging Cell 2020;19:e13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol 1996;271:H2183–H2189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author. RNA-seq data are available on the GEO database with accession number GSE217693.