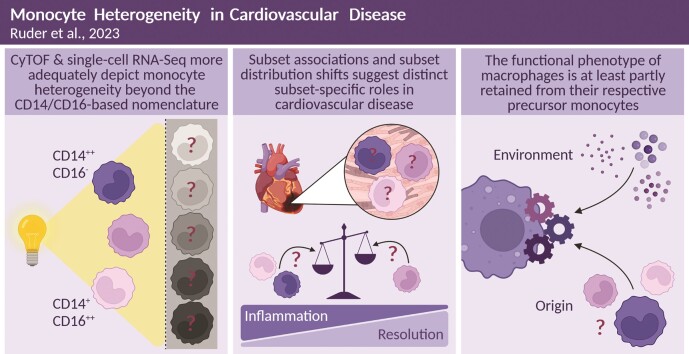
Abstract
Monocytes circulate the vasculature at steady state and are recruited to sites of inflammation where they differentiate into macrophages (MФ) to replenish tissue-resident MФ populations and engage in the development of cardiovascular disease (CVD). Monocytes display considerable heterogeneity, currently reflected by a nomenclature based on their expression of cluster of differentiation (CD) 14 and CD16, distinguishing CD14++CD16− classical (cMo), CD14++CD16+ intermediate (intMo) and CD14+CD16++ non-classical (ncMo) monocytes. Several reports point to shifted subset distributions in the context of CVD, with significant association of intMo numbers with atherosclerosis, myocardial infarction, and heart failure. However, clear indications of their causal involvement as well as their predictive value for CVD are lacking.
As recent high-parameter cytometry and single-cell RNA sequencing (scRNA-Seq) studies suggest an even higher degree of heterogeneity, better understanding of the functionalities of these subsets is pivotal. Considering their high heterogeneity, surprisingly little is known about functional differences between MФ originating from monocytes belonging to different subsets, and implications thereof for CVD pathogenesis. This paper provides an overview of recent findings on monocyte heterogeneity in the context of homeostasis and disease as well as functional differences between the subsets and their potential to differentiate into MФ, focusing on their role in vessels and the heart. The emerging paradigm of monocyte heterogeneity transcending the current tripartite subset division argues for an updated nomenclature and functional studies to substantiate marker-based subdivision and to clarify subset-specific implications for CVD.
Keywords: Monocytes, Cardiovascular, Heterogeneity, Inflammation, Atherosclerosis
Graphical Abstract
Graphical Abstract.
1. Introduction
With their high functional diversity, monocytes and monocyte-derived macrophages (MФ) play a key role in the pathogenesis and progression of cardiovascular disease (CVD) and atherosclerosis-related ischaemic heart disease and heart failure (HF).1
MФ are abundant in every tissue in the body, including heart and vessel wall, where they regulate tissue homeostasis and monitor trauma and infection.2,3 Recent evidence suggests that tissue-resident MФ pools have mixed ontological origins arising both from embryonic progenitors and blood monocytes.4 The phenotype of tissue-resident MФ is influenced by their microenvironment, resulting in high location-dependent heterogeneity.5,6 Monocytes are hematopoietic cells that not only serve as MФ precursors, complementing the tissue-resident MФ pools, but also play an important role in innate immunity. Upon inflammation, monocytes will locally extravasate towards the affected tissue where they differentiate into monocyte-derived MФ and exert MФ functions.7
In humans, monocytes circulating in blood can be divided into three subsets: CD14++CD16− ‘classical’ monocytes (cMo), CD14++CD16+ ‘intermediate’ monocytes (intMo), and CD14+CD16++ ‘non-classical’ monocytes (ncMo).8 Although many publications report functional differences between the three subsets, a delineating characterization of their functions to assign distinct functional phenotypes is lacking.9 Advances in the study of the monocyte subsets, especially recent high-parameter cytometry studies,10,11 have overhauled the prevailing view on monocyte heterogeneity and suggest that the current classification scheme may be inadequate to cover the functional diversity of monocytes. In addition, despite studies describing the potential of all three monocyte subsets to differentiate into MФ,12 it remains unclear whether they give rise to functionally distinct MФ. In this review, we will present an overview of current insights on monocyte heterogeneity and implications for monocyte taxonomy. We will address subset-specific monocyte functions in health and CVD, as well as subset correlation to CVD. Moreover, we will outline the current knowledge on the impact of the subset of the monocytic progenitor on MФ phenotype and activity in tissue, with particular focus on heart and vessel wall, and implications thereof for CVD progression.
2. Monocyte heterogeneity and consequences for monocyte taxonomy
First attempts to define monocyte heterogeneity were solely based on cell size and volume, distinguishing two major subpopulations referred to as large and small monocytes.13,14 These featured differences in antibody-dependent cellular cytotoxicity14 and chemotaxis towards zymosan-activated human serum,13 providing first evidence for functional differences between monocyte subpopulations. Flow cytometry and fluorescence-activated cell sorting (FACS) enabled a more granular analysis, leading to the identification and isolation of a monocyte subset smaller in size, characterized by low expression of cluster of differentiation (CD) 14, a lipopolysaccharide (LPS) receptor,15 and co-expression of CD16, an FcγIII receptor.16,17 These differences in expression of CD14 and CD16 serve as the basis for the nomenclature introduced in 2010, which is predominantly used today, classifying three human monocyte subsets: CD14++CD16− cMo, CD14++CD16+ intMo, and CD14+CD16++ ncMo.8 Accordingly, the terms cMo, intMo, and ncMo will be used to refer to the monocyte subsets in this review, and CD14/CD16 expression patterns will be included if subset definitions used in previously published studies deviate from the 2010 nomenclature.
In mice, two main monocyte subsets have been described: Ly6Chigh monocytes, which resemble CD14++CD16− cMo, and Ly6Clow monocytes, which represent the murine counterpart to CD14+CD16++ ncMo.18,19 Although considered counterparts, CD14/CD16 and Ly6C are not inert in the other species, respectively, and may modulate subset function differently. Moreover, the proportions of circulating monocyte subsets in mice and humans differ substantially (∼95% CD14++CD16− cMo vs. 50% Ly6Chigh),19 a note that should be considered for the interpretation of data from mouse studies (Figure 1). In addition, monocyte heterogeneity varies between mice of different sexes and strains, including the widely used BALB/c strain as well as C57Bl/6, the background strain of apolipoprotein E-knockout (ApoE−/−) and low-density lipoprotein receptor-knockout (LDLR−/−) mice, models commonly used in CVD research.20,21
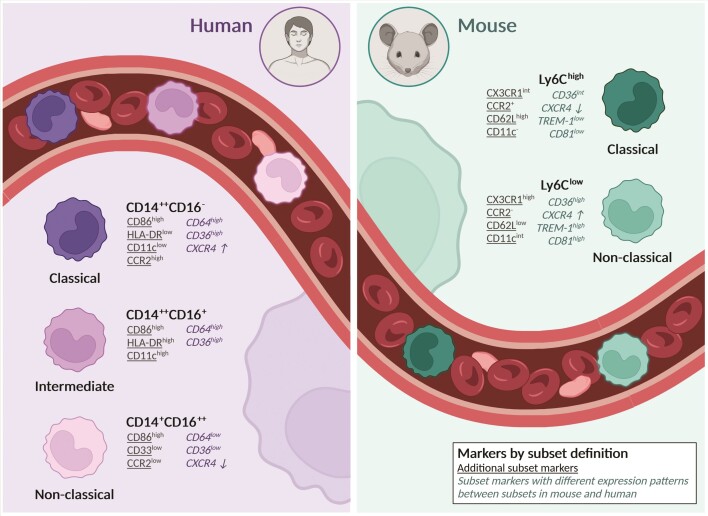
Figure 1.
Monocyte subset markers in human and mouse. Overview of markers by definition of the current monocyte subset nomenclature (bold) and additional markers to distinguish between monocyte subset in human and mouse (underlined). Although human classical CD14++CD16− and murine Ly6Chigh, likewise non-classical CD14+CD16++ and Ly6Clow, are considered counterparts, markers that identify a subset in one species are not necessarily transferable to the same subset in the other species. For example, while CD36 expression is lower on classical Ly6Chigh than in non-classical Ly6Clow monocytes, classical CD14++CD16− express high levels and non-classical CD14+CD16++ low levels of CD36. Markers with different expression patterns when comparing corresponding subsets in mouse and human are contained in this overview (cursive). Expression of TREM-1 and CD81 in intermediate CD14++CD16+ and non-classical CD14+CD16++ was not included as published studies did not distinguish between the two subsets. CD64 expression is similar between Ly6Chigh and Ly6Clow monocyte subsets and thus not mentioned for the murine subsets. CXCR4 expression levels are not clearly definable as high or low but show a higher (↑) or lower (↓) expression between subsets, indicated by arrows. All expression levels depicted refer to protein-based expression and were gathered from Ingersoll et al.19, Mildner et al.87, Thomas et al.32, Ong et al.11, and Mueller et al.138.
The CD14/CD16-based definition of monocyte subsets provided clear terminology for monocyte subsets but might not be sufficient for unambiguous identification. CD14 and CD16 expression depends on activation status and disease context,11,22 making a gating strategy relying solely on these markers insufficiently robust. More worrisome, natural killer (NK) cells, like ncMo, also express CD16 and are CD14-negative, potentially giving rise to contamination of ncMo by these CD56 (NCAM)dim cells. Inclusion of human leucocyte antigen (HLA)-DR in the FACS panel allows more selective discrimination of ncMo (Figure 2).23,24 Consequently, a separation of monocyte subsets solely based on CD14 and CD16 may be insufficient to unequivocally distinguish monocyte subsets.
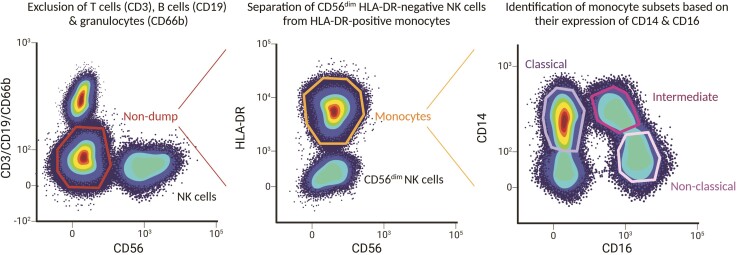
Figure 2.
Schematic gating strategy to discriminate human monocyte subsets. The plots depicted serve as an illustration of a gating strategy to identify human subset populations from peripheral blood mononuclear cells and to allow fluorescence-activated cell sorting at high purity. After gating of live and single cells, CD3-positive T cells, CD19-positive B cells, CD66b-positive granulocytes, and CD56high NK cells are excluded by dump gating. Next, monocytes are selected based on HLA-DR expression, and HLA-DR negative NK56dim cells which may not have been excluded in the previous gating step are separated from HLA-DR-positive monocytes. Finally, the three human monocyte subsets as defined by the current nomenclature are distinguished based on their expression of CD14 and CD16, with classical CD14++CD16−, intermediate CD14++CD16+, and non-classical CD14+ CD16++ monocytes. Although the ratio of the subsets, especially of non-classical and intermediate monocytes, varies greatly between donors, the proportion of classical monocytes consistently predominates over the two other subsets.
Recent high-parameter cytometry and single-cell RNA sequencing (scRNA-Seq) studies have revolutionized our view on monocyte heterogeneity and led to an improved resolution of the aforementioned monocyte subsets, especially of intMo and ncMo.10 Previously, 6-sulfo LacNAc (slan) expression had been shown to more reliably distinguish slan-negative CD14++CD16+ intMo from slan-positive CD14+CD16++ ncMo using FACS and massive analysis of complimentary DNA ends (MACE).22 To complicate matters, a high-resolution approach using cytometry by time-of-flight (CyTOF) to profile monocyte heterogeneity by Roussel et al.25 revealed that slan expression may also mark separate subsets within the ncMo population. Although slan-positive monocytes show similar cytokine production patterns26 and transcriptional profiles27 as ncMo, they seem to differ in their expression of CXCR6.28,29 As a study by Cros et al.18 failed to confirm the slan-based sub-differentiation within the ncMo, the jury is still out on the added value of slan as a monocyte subset marker. In parallel, Villani et al.30 aimed at improving monocyte subset identification using scRNA-Seq of human monocytes, which were FACS-sorted with a CD14/CD16-based gating strategy. Their study revealed four monocyte clusters of which two were located within the intMo subset. However, as one of the subsets within intMo was typified by established NK markers such as KLRC4, cathepsin W and Prf1, the true monocyte nature of this subset remains to be addressed. Conversely, Mildner et al.31 describe a newly identified Ly6Cint intMo subset in mice showing transcriptional as well as phenotypic overlap to both Ly6Chigh cMo and Ly6Clow ncMo. Similarly, Thomas et al.32 found human intMo to cluster with both cMo and ncMo using CyTOF. Addition of CD11c (integrin α-X), HLA-DR, CD36 (GP4, GP3B, SCARB3), and CCR2 (CD192) to their CD14/CD16-based gating strategy allowed clear separation of the three conventional monocyte subsets. Although the introduction of additional subset markers improved differentiation between intMo and ncMo, the latter still relies on prior subset identification using CD14 and CD16. However, as Ong et al.11 showed, a CD64 (FCGR1)-, CD86 (B7.2)-, CD33 (SIGLEC3)-, CCR2-, and HLA-DR-based marker panel performs equally well to reliably identify CD16− cMo, CD16+CD14high intMo, and CD16+CD14low ncMo, suggesting a CD14/CD16-independent separation is possible.
Next to enabling improved identification of the conventional three subsets, high-parameter cytometry and scRNA-Seq technology combined with high-dimensional analyses advanced the identification of new subsets within the three monocyte populations.11,25,30,32,33 In human, Hamers et al.29 identified eight monocyte subsets using CyTOF, with three subsets falling within ncMo and four within cMo. Merah-Mourah et al.,33 using 17-colour FACS, identified a set of large and a set of small monocytes that could be further divided by their expression of CD16, resulting in a total of six monocyte subsets. Large and small monocyte sets showed differences in the production of tumour necrosis factor (TNF) and interleukin (IL) −1β in response to toll-like receptor (TLR) agonists, as well as in their expression of adhesion molecules, suggesting distinct functions. However, the conventional tripartite differentiation of monocytes was underpinned by Dutertre et al.34 who applied a machine learning algorithm for subset discrimination in scRNA-Seq and CyTOF including 14 backbone lineage markers plus one of 332 variable markers each. These partly conflicting findings could be caused by minor differences in the source sample but most likely by differences in strategy of and settings for data analysis. Notably, the cMo and intMo populations identified by Dutertre et al.34 each embedded two marginally different subsets, such as CD35 (CR1)+/CD89 (FCAR)+ cMo and CD55 (DAF)+/Mac2+ cMo.
In conclusion, by both the use of new surface markers and the inclusion of additional markers to the conventional CD14- and CD16-based panel, greater heterogeneity within the human monocyte population than the current trinomial terminology suggests was revealed, and new monocyte subpopulations were identified. Although recent findings mostly agree on heterogeneity within the original cMo and ncMo subsets, a lack of a clear consensus is evident by differences in the number of subpopulations reported. This may be attributable to differences in number and combination of markers as well as (unsupervised) clustering methods used. The reproducibility and comparability may be further impacted by substantial differences in the identification and annotation of cell clusters. Whether or not these new subpopulations represent truly separate phenotypes or reflect the stochastic diversity within the mother populations remains to be established by lineage tracing and differentiation studies. It is not unlikely that some studies have considerably over-clustered this cell species. Moreover, standardized cluster annotation approaches such as the reference mapping included in Seurat v435 are needed to consistently define monocyte subpopulations.
In 2016, an update to the 2010 nomenclature was proposed, suggesting the numerical labels ‘Mon1’ for cMo, ‘Mon2’ for intMo, and ‘Mon3’ for ncMo.36 Despite resemblance to the M1/M2 terminology for MФ polarization states, these labels were not chosen to express a predisposition of MФ to adopt a M1 or M2 polarization state depending on the monocyte subset they originate from, but to simplify monocyte subset terminology.36 A numerical nomenclature does not only allow detachment from a marker-based subset definition, which may change with the discovery of additional or alternative subset markers, it could also facilitate the coherent naming of newly identified monocyte subsets. However, it appears that the ambiguity observed in literature to date may at least partly be attributed to an incomplete subset definition, exclusively based on surface marker expression, which should be complemented with distinctive functions and differentiation potential of monocyte subsets. Thus, a taxonomy putting newly identified monocyte subsets into context, as suggested by Bassler et al.,37 will still be necessary to adequately describe monocyte heterogeneity. Moreover, functional differences of established original and newly identified monocyte subsets remain to be assessed and related to their role as precursors of tissue-resident MФ and to pathogen and endogenous trauma responses.
3. Monocyte subset distribution in CVD
Monocytes are major players in the development of CVD, illustrated by the observation that the number of circulating monocytes has a predictive value for cardiovascular risk and mortality.38–40 However, most studies did not stratify for monocyte subsets. Moreover, the interpretation of findings reported in studies that did discriminate between monocyte subsets is complicated by differences in subset definition, in staining and gating protocols, as well as the number and combination of markers used to identify subsets. In a prospective study using flow cytometry to determine monocyte subset counts, Rogacev et al.41 showed that intMo were the only subset to independently predict CVD risk in elective coronary angiography patients in 3 years of follow-up. These findings are in contrast with a study by Berg et al., showing elevated cMo counts to predict cardiovascular events, defined as fatal or non-fatal myocardial infarction (MI), ischaemic stroke, or coronary heart disease-related death. The use of frozen leucocyte samples and the smaller study population (n = 700) in the study by Berg et al.42 compared to fresh samples in Rogacev et al.41 (n = 951) might account for the different observations between the two studies. Wildgruber et al.43 found increased intMo and decreased ncMo numbers in patients with progression of peripheral artery occlusive disease, the number of cMo remaining unchanged. Similarly, while CD106 (VCAM-1) expression on cMo was reported to be increased in coronary artery disease (CAD) patients, these patients also featured elevated intMo counts, possibly due to a shift from cMo to ncMo at levels that were predictive of adverse CVD outcome.44 Interestingly, CAD patients with elevated levels of lipoprotein (a) [Lp(a)], a pro-atherogenic lipoprotein and independent risk factor for CVD,45 had increased intMo compared to patients with normal Lp(a) levels, whereas ncMo and cMo proportions remained unchanged in both patient groups.46 As compared to cMo, telomere length of intMo was shorter, which might indicate senescence, although comparisons of telomere length [reviewed in Vaiserman and Krasnienkov47] between different cell types/subsets should be taken with caution. Moreover, intMo expression of chemokine receptors like CCR2, CCR5 (CD195), CCR7 (CD197), and CX3CR1 was higher than in the other subsets,48 suggesting they are more responsive to chemotactic cues in CVD.
Several studies also investigated correlations between monocyte subset counts and atherosclerotic plaque composition and/or stability, with inconsistent outcomes (Table 1). Interrogating the AtheroExpress plaque cohort, none of the monocyte subsets was found to be associated with critical plaque characteristics such as fat deposition, collagen deposition, calcification, intraplaque haemorrhage, intraplaque vessel, and smooth muscle cell density.49 However, in a cohort of 588 individuals that underwent coronary CT angiography for routine health check-up, intMo levels correlated with mixed and calcified plaque types, whereas cMo counts correlated with non-calcified plaques.50 In a smaller cohort (n = 51) in which asymptomatic coronary artery plaques were analysed by virtual histology based on intravascular ultrasound, intMo were significantly correlated with plaque vulnerability traits such as burden of fibrous components, necrotic core size, and calcification.51 These findings were confirmed by Yamamoto et al.52 in a small prospective optical coherence tomography and coronary angiography study of coronary artery patients. Although findings are not completely consistent, the overall picture emerging from these studies is that intMo numbers seem most predictive of CVD and strongest associated with plaque vulnerability. Whether intMo are causally involved in CVD or whether the CVD context leads to aberrant cMo-to-ncMo conversion or increased expression of intMo markers remains subject to further studies.
Table 1.
Reported shifts in monocyte subsets distributions in atherosclerosis
| Source | Cohort size | Outcome |
|---|---|---|
| Meeuwsen et al.,49 The Netherlands | 175 undergoing CEA (63.4% male) | No association of any monocyte subset with vulnerable plaque traits |
| Lo et al.,50 Taiwan | 588 undergoing general health check with coronary CT (68.7% male) | Correlation of intMo with mixed and calcified plaque types Correlation of cMo with non-calcified plaque type |
| Yoshida et al.,51 Japan | 51 asymptomatic CAD (75% male) | Correlation of intMo with vulnerable plaque traits |
| Yamamoto et al.,52 Japan | 50 CAD (80% male) | Counts of intMo associated with vulnerable plaque traits |
CEA, carotid endarterectomy; CAD, coronary artery disease; CVD, cardiovascular disease.
Plaque destabilization and rupture can eventually lead to an ischaemic event causing myocardial necrosis. Recruited monocytes and cardiac resident MФ are instrumental in the trauma repair response to infarct [reviewed in Frantz and Nahrendorf53]. According to Tsujioka et al.,54 CD14++CD16− cMo are the monocyte subset recruited first, peaking 2.6 days after infarct onset, while CD16-positive intMo and ncMo are recruited in a later phase to peak around day 5 after onset. This timely sequence might reflect the developmental trajectory of cMo to ncMo (as discussed below) after monocytosis in response to trauma. Interestingly, peak recruitment of cMo, but not CD16-positive intMo and ncMo, was inversely correlated with myocardial salvage and left ventricular ejection fraction 6 months after onset54 (Table 2).
Table 2.
Reported shifts in monocyte subsets distributions in myocardial infarction
| Source | Cohort size | Outcome |
|---|---|---|
| Shantsila et al.,62 UK | 245 STEMI (78% male) | Increased absolute number of intMo |
| Tapp et al.,55 UK | 50 STEMI, 40 CAD and 40 healthy controls (80–86% male) | Counts of circulating cMo and intMo increased on first day after STEMI intMo levels correlated with cytokine and troponin levels as well as recovery of left ventricular function |
| Zhou et al.,57 China | 100 STEMI, 60 CAD and 35 healthy controls (68–78% male) | Expansion of intMo predictive of adverse cardiovascular events within 2-year follow-up |
| Zeng et al.,58 China | 96 STEMI (78% male) | Persisting elevation of intMo after STEMI associated with risk for major adverse cardiovascular events within 2.5-year follow-up |
| Tsuijoka et al.,54 Japan | 36 AMI (75% male) | Peak levels of cMo inversely correlated with myocardial salvage and LVEF after infarct onset |
| Van der Laan et al.,56 The Netherlands | 28 AMI and 12 other causes (67% male) | Accumulation of cMo at border of infarct zone 12 h to 5 days after AMI, followed by infiltration of intMo/ncMo |
STEMI, ST-elevated myocardial infarction; AMI, acute myocardial infarction; CAD, coronary artery disease; LVEF, left ventricular ejection fraction.
As shown by Tapp et al.,55 circulating cMo and intMo counts were increased in the first day after ST-elevation myocardial infarction (STEMI), upon which intMo levels were significantly correlated with plasma cytokine and cardiac troponin levels, and recovery of left ventricular function, suggesting this subset to play a key role in cardiac healing. Histopathological examination of myocardial tissue collected from autopsy indicated cMo accumulation at the border of the infarct zone surrounding the necrotic area during the inflammatory phase at 12 h to 5 days after acute myocardial infarction (AMI). In the following proliferative phase at 5–14 days after AMI, infiltration of the infarct zone by CD16-positive monocytes was observed, though these were not further classified into CD14++CD16+ intMo or CD14+CD16++ ncMo.56 Apparently, apart from CD14++CD16− cMo, CD16-positive monocytes (probably ncMo) are also needed for proper AMI repair, likely because they serve different functions. As described above, the AMI-associated expansion of intMo was predictive of adverse cardiovascular events within 2 years after the primary event,57 suggesting a detrimental role of intMo in AMI repair or in atherosclerosis. According to Zeng et al.,58 particularly STEMI patients with persistent elevations of circulating intMo after infarct were at risk of a major adverse cardiovascular event during 2.5-year follow-up, suggesting that not only the subset profile but also subset dynamics upon infarct can impact the outcome of cardiovascular events. This may support the finding of Dutta et al.59 in mice where increased monocyte recruitment in the aftermath of an infarct was not limited to the infarct zone itself but also implicated distal sites of chronic atherosclerosis, resulting in recurrent events.
AMI-associated loss of cardiac function can develop into HF, the major cause of morbidity and mortality after AMI.60 HF patients had an increased percentage of CD14++CD16+ intMo compared to healthy subjects at increased cardiovascular risk, which correlated with disease severity,61 a finding that was confirmed by Shantsila et al.62 for absolute subset numbers (Table 2). Moreover, intMo had higher expression of the cell adhesion molecules ICAM-1 (CD54) and VCAM-1, of which the latter was associated with adverse clinical outcome for acute HF.63 In keeping with this, significant increases in intMo counts in (acute) HF were reported by Wrigley et al. (cross-sectional study)64 and by Elchinova et al.65 Interestingly, while the former also noted elevated cMo levels, the latter, larger study found reduced cMo levels. In this study, also the number of intMo (cells/µL) in HF patients was independently associated with all-cause death.65 Finally, a study by Amir et al.66 failed to reproduce these findings, reporting an unchanged intMo proportion in HF patients, but reduced percentages of cMo and increased percentages of ncMo (Table 3). Moreover, the percentage of ncMo, but not cMo or intMo, was inversely correlated with severe HF. Discrepancies between these study outcomes may be attributable to differences in the study populations’ risk and environmental factor profile. Indeed, the secretome of adipose tissue of obese HF patients was able to induce a shift towards ncMo differentiation, in contrast to that of non-obese HF patients.67 Oxysterol-mediated activation of NR4A1, a nuclear receptor critically involved in ncMo differentiation, could explain this finding.68,69 It remains unclear whether this obesity-associated ncMo increase is a protective response or merely a consequence of larger disease characteristics. MФ origin in the heart was also shown to change with ageing, resulting in increased monocyte recruitment and a shift towards M1-like cardiac MФ,70 suggesting increased susceptibility for and worse outcome of CVD in aged individuals. Thus, the increased presence of monocyte-derived MФ in the heart and arteries may have disease-relevant repercussions for MФ phenotype in these organs.
Table 3.
Reported shifts in monocyte subsets distributions in heart failure
| Source | Cohort size | Outcome |
|---|---|---|
| Barisione et al.,61 Italy | 30 congestive HF and 26 healthy controls (all male) | Increased percentage of intMo in HF correlating with disease severity |
| Wrigley et al.,64 UK | 51 acute HF, 42 stable HF, 44 CAD and 40 healthy controls (58–83% male) | Increased intMo count in acute HF compared to stable HF and CAD Increased intMo count in stable HF compared to CAD Elevated cMo levels in acute HF compared to stable HF, CAD, and healthy controls No difference in ncMo counts |
| Wrigley et al.,63 UK | 51 acute HF, 42 stable HF and 44 CAD (68–83% male) | Higher expression of ICAM-1 on intMo of acute HF compared to stable HF and CAD VCAM-1 expression on intMo of acute HF associated with death or hospitalization |
| Elchinova et al.,65 Spain | 400 HF (72.8% male) | Increased intMo count Absolute number of intMo per µL blood associated with all-cause death Reduced cMo levels |
| Amir et al.,66 Israel | 59 systolic HF and 29 controls without heart disease (53–76% male) | Reduced percentage of cMo in HF Increased percentage of ncMo in HF Percentage of ncMo inversely correlated with severe HF No difference in intMo percentage |
HF, heart failure; CAD, coronary artery disease.
Overall, most studies agree that monocyte subset distribution shifts in CVD, although cause and direction of this shift still need to be further investigated. A shift towards ncMo is also observed with ageing, which may reflect accelerated immune ageing and increased susceptibility to CVD. Several studies suggest an association of the intMo subset with CVD, atherosclerosis, AMI, and HF, although not all studies agree on their predictiveness of prognosis.
4. Functional diversity of monocyte subsets in health and disease
Today, monocytes are no longer viewed as mere precursors of MФ, as this dogma has been challenged by several studies showing clear functional specializations of cMo and ncMo subsets (Figure 3).71,72 The recently reported high heterogeneity of this lineage may have further eroded this view. Monocytes functionally adapt to inflammatory stimuli and carry out effector functions in innate immune responses to pathogens in circulation and in trauma monitoring of the endothelium.72–74 However, assigning functional phenotypes to monocyte subsets has been impeded by conflicting results, differences in methodology and readout, and functions being assigned to multiple subsets.75,76 The official CD14/CD16-based nomenclature intentionally avoids terms like ‘inflammatory’ and ‘anti-inflammatory’ to allow broader function-based categorization. Further investigation of functional differences between the subsets is still necessary,8,77 taking into account the potential heterogeneity of the monocyte subsets as suggested by the aforementioned single-cell studies.
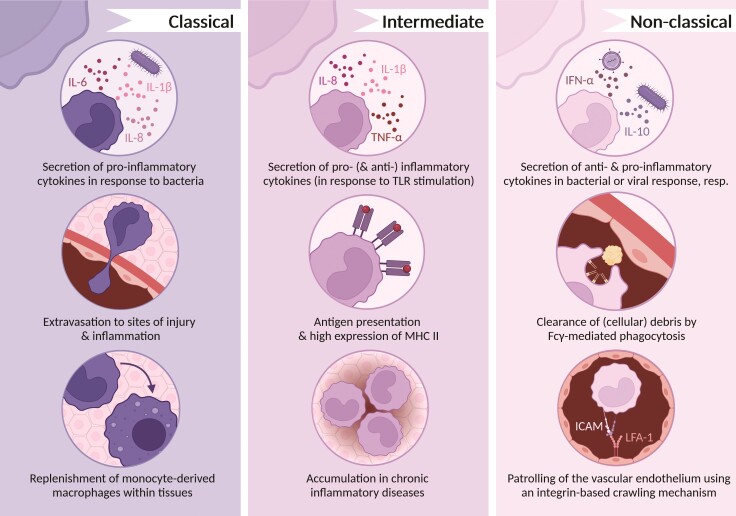
Figure 3.
Functions of classical, intermediate, and non-classical monocytes. Monocytes of different subsets exert different functions, illustrated in this figure. The current nomenclature purposefully avoids the terms ‘pro-inflammatory‘and ‘anti-inflammatory’ to describe monocyte subsets. Considering their functions, especially cytokine secretion by the different monocyte subsets, the importance of a nomenclature independent of single functions becomes apparent, as non-classical monocytes produce pro-inflammatory cytokines in response to viruses but are often termed ‘anti-inflammatory’.
CMo sense cues from sites of injury and inflammation are released into circulation via CCL2-CCR2 signalling, and extravasate into affected tissues.78–80 They are highly active in phagocytosis and production of reactive oxygen species (ROS).75,81 In response to bacterial signals, cMo secrete inflammatory cytokines such as IL-6, IL-8, and IL-1β.12,30 Although they are able to clear debris,79 this capacity is less developed than in ncMo,82 a difference that has been used for selective bead-based tracking of ncMo in vivo.83 Moreover, cMo were found to differentiate into intMo, which in turn differentiate into ncMo, following a sequential ontogeny scenario as shown by Patel et al.84 using a humanized mouse model, the MISTRG mouse.85 In humans, after endotoxin-induced depletion of blood monocytes, re-population followed a sequential pattern of cMo appearing first, followed by intMo and finally ncMo.84 In a smaller study (n = 12), Thaler et al.86 observe cMo to be the first subset to appear after LPS challenge and cMo counts to recover within 6 h. Moreover, they report a reduction in cMo counts and considerable shift towards intMo in the next 18 h, suggesting enhanced differentiation of cMo into intMo in response to inflammatory cues. The time needed for cell trafficking from bone marrow to circulation seems to be decreased under inflammatory conditions, which might be caused by the release of cMo retained in the bone marrow and spleen as an ‘emergency pool’.84 Most evidence on this emergency release and the contribution of bone marrow-derived monocytes was obtained from mouse experiments, where two main monocyte subsets are distinguished based on their expression of Ly6C.87 The circulating pool of Ly6Chigh cMo is rapidly replenished by egress of CXCR4high Ly6Chigh bone marrow monocytes,88 which may have alternative phenotypes and functional adaptations, similar to neutrophil-like Ly6Chigh cMo or segregated nucleus-containing atypical Ly6Clow ncMo [reviewed in Guilliams et al.89]. The control of this emergency release of monocytes is associated with microRNA (miR)-146a, leading to expansion and trafficking of Ly6Chigh cMo and resulting in an increased inflammatory response against bacteria [reviewed in Duroux-Richard et al.90]. Circulating Ly6Chigh cMo may also convert into Ly6Clow ncMo, as proposed by Yona et al.91 regarding the rapid appearance of BrdU-labelled Ly6Chigh cMo in circulation, whereas BrdU-spositive Ly6Clow ncMo became detectable after 5 days.
The conversion to Ly6Clow ncMo was found to be dependent on CCR2,91 which is expressed exclusively by Ly6Chigh cMo.18 The mechanism underlying the conversion of Ly6Chigh cMo to Ly6Clow ncMo was elucidated by Thomas et al.69 who studied the regulation of the Nr4a1 gene, which encodes transcription factor Nur77. In line with previous results, Ly6Clow ncMo showed higher expression of Nr4a1 than Ly6Chigh cMo.68,69 Nr4a1 knockout led to partial depletion of the Ly6Clow ncMo pool, indicating its crucial role for the generation of Ly6Clow monocytes.68 Thomas et al.69 found Nr4a1 to be regulated by the interaction of Klf2 with the Nr4a1 super-enhancer domain E2. In line with this finding, Patel et al.84 addressed the kinetics of human monocyte subsets by isotopic 6,6-2H2-glucose labelling in healthy volunteers, showing turnover rates of 1 day for cMo, 4.3 days for intMo, and 7.4 days for ncMo, corresponding to the observations in mice. These data could be partly confirmed by Tak et al.93 (n = 14) who also observed consecutive isotopic labelling of the three monocyte subsets, pointing to a linear differentiation model. Only few cMo were differentiating into intMo, while almost all intMo were on their way to become ncMo, suggesting that the first step is rate limiting. Blood residence times for cMo and ncMo were ∼2.5 days; however, intMo were seen to have much shorter blood residence time in this study (<1 day). Additional studies will be needed to fully clarify this. Taken together, under steady-state conditions, cMo can either differentiate into MФ to replace tissue-resident MФ or develop into ncMo with involvement of Nr4a1.
In contrast to cMo, ncMo produce anti-inflammatory cytokines like IL-10 in response to bacterial stimuli.18 However, this does not render ncMo the anti-inflammatory counterpart of cMo, as they produce high levels of inflammatory cytokines in a TLR7-mediated response to viruses and nucleic acids.30,80,90 NcMo were shown to initiate recruitment and activation of other innate immune cells, such as NK cells and neutrophils through TNF-α-induced upregulation of E-selectin on endothelial cells.89,93 The main function of ncMo seems to be patrolling of the vascular endothelium and removal of cell debris via Fcγ-mediated phagocytosis.79,80,89 Their patrolling behaviour rests on a crawling mechanism slower than the rolling adhesion preceding cMo extravasation and is dependent on LFA-1/ICAM interaction.75,80,89 NcMo showed high expression of genes associated with adhesion and cytoskeleton arrangement, which may serve to facilitate motility for patrolling.92,94 As reported by Chimen et al.,93 pooled ncMo and intMo transmigrate through unstimulated endothelial cell monolayers faster in vitro, although fewer ncMo/intMo adhere compared to cMo. Moreover, the former did not re-enter the blood stream, whereas cMo did, in line with findings on the regress of Ly6Chigh cMo but not Ly6Clow ncMo to the bone marrow in mice.92,95 Apparently, ncMo are either not equipped to respond to signals for reverse transmigration or more firmly retained in the subendothelial space.93 NcMo are also believed to play a role in the resolution of inflammation, as they were shown to differentiate into wound-healing MФ83 and have increased expression of miR-150 and miR-21, although substantial and tissue-specific evidence is still missing.80,83,90 It has been reported that CD16-positive monocytes have higher antigen presentation capacity96 as well as decreased lipid accumulation and migration towards complement component C5a,97 but a sufficient delimitation between ncMo and intMo is still outstanding. Whether ncMo produce ROS is still unclear, as studies have shown both high levels of ROS production at baseline11 as well as low ROS levels in response to IgG-opsonized bovine serum albumin.18 Altogether, ncMo patrol the vasculature at steady state and contribute to the maintenance of vascular integrity.
Compared to ncMo and cMo, little is known about the functions of intMo in both homeostasis and inflammation, partly due to the low and variable numbers of this subset in circulation. Moreover, due to their low abundance, intMo were often examined after pooling with ncMo, complicating the interpretation of findings. The question whether intMo merely represent an intermediate step in the conversion of cMo to ncMo or a separate population exerting a distinct functional role is legitimate. However, several studies do report an increase of intMo in the context of chronic inflammatory diseases such as CVD. For example, the Prospective Halle Monocyte Study showed a shift from cMo to intMo in patients with CAD.98 IntMo express high levels of major histocompatibility complex (MHC) II and have high antigen presentation capacity.79,99 Like cMo, they show phagocytic activity and basal production of ROS.75,94 IntMo were found to produce both pro- and anti-inflammatory cytokines upon TLR stimulation.79 Cros et al.18 report highest secretion of TNF-α and IL-1β by intMo compared to cMo and cMo. Moreover, they found intMo to secrete IL-8, although at lower levels than cMo. In contrast, Wong et al.99 report ncMo to secrete the highest levels of TNF-α and IL-1β and similar levels of IL-8 secreted by all subsets. Thorough functional mapping of intMo and its differentiation kinetics is needed to clarify their role in homeostasis and disease, and more specifically, to address whether the observed association with disease reflects adverse functions or is a bystander effect of a failure to mature into ncMo.
In conclusion, ncMo and cMo exert specialized functions in innate host defence to pathogens and injury, with prominent viral response and injury monitoring activity for the former and pro-inflammatory activity for the latter. Although the functions exerted by intMo are less clear, they seem to be associated with inflammatory diseases. The sequential development of monocyte subsets seems to follow the course of inflammation and its resolution. Additional studies, in particular functional assays, are needed to complement the already existing knowledge from transcriptional studies. Generally, further research considering monocyte heterogeneity beyond the conventional nomenclature is needed to understand the role of monocyte subpopulations in homeostasis and inflammation.
5. Role of monocytes as MФ precursors in health and disease
The foundation for our current model of the mononuclear phagocyte system was laid by a concept proposed by van Furth et al.7, postulating that monocytes arising from (bone marrow) myeloid precursors extravasate to differentiate into MФ and repopulate the tissue-resident MФ pool. However, over the last years, lineage tracing studies in mice have revealed that tissue-resident MФ originate from the yolk sac and foetal liver and have self-renewal capacity, ensuring homeostatic MФ levels in the tissue.100 The tissue-resident MФ pools in the skin (Langerhans cells) and brain (microglia) are maintained by self-renewal throughout life without contribution of bone marrow-derived monocytes.101–103 Yet, the MФ pools in heart,104,105 dermis,106 and gut107–109 were shown to contain both embryo- and bone marrow-derived MФ in steady state, with a ratio that gradually shifts towards the latter during life. Although it was long unclear whether this also held for humans, Bajpai et al.110 were able to unravel the contribution of bone marrow-derived monocytes to the tissue MФ pool in the human heart with gender mismatched heart transplantation. Their data revealed that, similar to mice, maintenance of the heart-resident MФ pool partly relies on the self-renewal capacity of donor heart MФ and partly on the influx of recipient bone marrow-derived MФ. Moreover, Bigley et al.111 showed that in subjects with autosomal dominant and sporadic monocytopenia, Langerhans cells and skin-resident MФ levels were unaffected and preserved. This suggests that not only in mice but also in human, tissue-resident MФ pools are gradually replaced by monocyte influx in some organs like heart, but not all. Which of the monocyte subsets is responsible for replenishment of the cardiac and vascular MФ pool at steady state is an outstanding question. Under pathogen-induced or sterile inflammatory conditions, the pool of tissue-resident MФ will be partly depleted during a first-response wave, and tissue compartments will open for monocytes to enter, which re-populate the tissue and assist in defence and repair.112 After MI, circulating monocytes are recruited to the heart via CCL2 where they differentiate into MФ [reviewed in Ma et al.70]. In atherosclerosis-susceptible apolipoprotein E-deficient mice, combined inhibition of CCR2, CCR5, and CXC3R1 led to an almost complete absence of lesions and was found to be more effective in reducing atherosclerosis than individual inhibition of these chemokine receptors.113 Considering their differential expression on monocyte subsets, with higher CX3CR1 expression levels on ncMo in both mice and humans,114 these findings indicate different pathways of infiltration and subsequent MФ accumulation.113 Indeed, in human atherosclerosis, more cMo were found to enter lesions than ncMo, although ncMo were shown to bind and become loaded with oxidised low-density lipoprotein (LDL),115 suggesting differential roles of the monocyte subsets in atherosclerosis [reviewed in Gautier et al.116].
Recently, the MФ niche concept has been coined [reviewed in Guilliams et al.117], in which the niche ensures maintenance of the tissue-resident MФ pool by the production of trophic factors that induce proliferation of tissue MФ. MФ consume these factors and self-regulate the proliferation rate in this tissue niche. MФ death will result in increased availability of trophic factors, thereby inducing MФ proliferation. In parallel, MФ death, especially in secondary necrosis or necroptosis, will induce chemokine release, leading to the recruitment of circulatory monocytes. In health, the former route will prevail (proliferation being faster than recruitment), but in disease, the latter pathway may dominate, (temporarily) tipping the balance in tissue towards monocyte-derived MФ.
CMo exit the circulation at steady state and differentiate into MФ in tissues that require monocyte influx to replace tissue-resident MФ and fill available niches, as discussed above. CMo that are not recruited and remain in circulation develop into ncMo, following a sequential ontogeny scenario.84,89,118 Transcriptome profiling of cMo and ncMo revealed several differentially expressed genes that hint towards subset-specific functions.119 CMo were found to express more CD11b (integrin α-M), CCR2, and TLR4 (CD284), suggesting an important role in bacterial infection and inflammation. Lack of CCR2, which prevents cMo stromal egress, leads to a reduction of the tissue MФ pool in the intestine and dermis compartments, known to be replenished by monocytes in steady state,100,107,120,121 suggesting that cMo are the main source contributing to the tissue MФ pool. In the same study, ncMo were seen to express low levels of CD11b and CCR2, but high levels of CX3CR1, LFA-1 (CD11a), HLA-DR, and genes necessary for cytoskeletal dynamics.119 This confirms earlier work by Geissmann et al.73 and others showing that ncMo patrol the vessels to maintain vessel integrity and can be viewed as luminal end-stage differentiated ‘MФ’.91,122,123 Whether or not ncMo are able to differentiate into full-fledged tissue MФ remains subject to debate. However, blocking LFA-1 or ICAM-1 resulted in a reduced patrolling phenotype and concomitantly increased circulatory ncMo numbers,124 suggesting that the ncMo pool may partly reside in the sub-endothelium. Moreover, as shown by Schyns et al.,125 a subset of lung interstitial MФ was derived from intravascular ncMo in steady state, collectively suggesting that also ncMo are able to populate tissue during steady state, although in this setting, they appear to be vessel-confined as well.
6. Impact of monocyte subset origin on MФ phenotype or function
MФ display immense phenotypic diversity37,126 with subsets displaying specialized organ- and condition-specific functions127,128 and were previously described to differentially affect CVD.129,130 As discussed earlier, this heterogeneity results from the plastic adaptation to their microenvironment, and also to differences in origin (embryonic vs. monocyte-derived). Hence, the functional specialization of the MФ pool in heart and arteries is likely to be impacted by monocyte heterogeneity and (age/disease-dependent shifts therein).131 This legitimates the question whether cardiac and arterial MФ partly retain the functional phenotype of the monocyte they were derived from.89
Colony-stimulating factors (CSF) such as macrophage (M-) CSF and granulocyte-macrophage (GM-) CSF, which control monocyte survival and differentiation, may act differentially on cMo, intMo, and ncMo. GM-CSF receptor CD116 (CSF2R) was found to be expressed on all monocyte subsets, with highest expression in cMo.132 In contrast, expression of M-CSF receptor CD115 (CSF1R) was high on ncMo,133 and blocking M-CSF receptor signalling reduced ncMo but not cMo numbers.134
In vivo support for monocyte subset-specific MФ differentiation was provided by Menezes et al.,128 showing that MHCII− Ly6Chigh cMo differentiate into iNOS-positive MФ upon microbial challenge, a process regulated by the myeloid lineage commitment factor PU.1. As Olingy et al.83 demonstrated, Ly6Clow ncMo preferentially develop into CD206 (MRC1)-positive wound-healing MФ in peri-implant tissue as compared to Ly6Chigh cMo. These observations underpin the notion that ncMo are primarily involved in tissue repair and suggest that the phenotype of monocyte-derived MФ mirrors their precursors’ phenotype to some extent. Both studies support the concept that MФ phenotypes are determined by their monocyte progenitor.37
These murine finding however do not necessarily translate directly to human monocyte-to-macrophage differentiation, where the picture is less clear. A study by Goudot et al.127 showed that only CD14-positive cMo are capable of differentiating into bona fide MФ, while CD16-positive monocytes had low survival in culture and did not contribute greatly. While committed to differentiate into MФ, cMo need additional cues for dendritic cell differentiation, a process controlled by the aryl hydrocarbon receptor.127 However, other groups have succeeded in differentiating MФ from CD16-positive intMo and ncMo.12,135 For instance, Frankenberger et al.135 reported both CD16-negative cMo and CD16-positive intMo/ncMo to develop into MФ after differentiation with M-CSF. Interestingly, cMo-derived MФ (cMo-MФ) showed higher expression of CD14, CD163 (M130, SCARI1), and versican compared to MФ from CD16-positive monocytes, a difference already present at the monocyte stage. Moreover, M-CSF-matured MФ derived from CD16-positive monocytes showed higher phagocytic activity, similar to their precursor. Again, ncMo and intMo were pooled to CD16-positive monocytes, so intMo may be accountable for the observed effects. In contrast, Boyette et al.12 report an inherent commitment of ncMo to become MФ even in the absence of growth factors, as they displayed signs of MФ differentiation already after 2 days in culture. In fact, all monocyte subsets developed into MФ and secreted increasing amounts of MФ-associated cytokines with progressing differentiation and increasing phagocytic activity along the way. The highest phagocytic activity was observed in cMo-MФ in comparison to intMo-MФ and ncMo-MФ.12 This seems opposite to the findings of Frankenberger et al., although it should be noted that the platforms (antibody-opsonized Escherichia coli bacteria vs. polystyrene beads) and consequently scavenger receptors and mechanism involved were different. Collectively, findings on subset-dependent functional differences of monocyte-derived MФ are conflicting, and a clear consensus is lacking. The general notion is that MФ phenotype reflects the phenotypic characteristics of the precursor population at least to some extent, and suggests that the response of monocyte-derived MФ to environmental stimuli may indeed be influenced by their origin. This may be especially relevant in the context of inflammation, in which monocyte-derived MФ may respond differently to inflammatory signals than tissue-resident MФ.131
7. Conclusions and perspectives
Monocytes are a heterogeneous population that allows classification into subgroups based on characteristics like size, marker expression, or function, although the current terminology distinguishing three monocyte subsets is based on CD14 and CD16 expression. The main functions of cMo are phagocytosis and production of ROS in response to bacterial stimuli, and maturation into ncMo in the absence of inflammation. The ncMo subset responds to viral challenge with the secretion of pro-inflammatory cytokines and patrols the vasculature at steady state. The intMo subset seems to be associated strongest with CVD, as intMo were found to be predictive of CVD and secondary cardiovascular events and increase in acute HF.
Due to the low abundance of intMo and ncMo compared to cMo, they have commonly been studied as a pool of CD16-positive monocytes. This complicates the interpretation of the results and does not allow to assign specific functions to these two subsets. Especially the functional phenotype of intMo lacks a distinct characterization to define its role in inflammation and steady state. Thus, more studies focusing on functional differences between intMo and ncMo are needed. The capacity of human monocytes of different subsets to extravasate and their fate after they have left the circulation are still unknown. Although all monocyte subsets have been shown to be able to differentiate into MФ in vitro, it is not clear if and to what extent subsets contribute to the tissue-resident MФ pool in heart and vasculature. Considering the patrolling behaviour of ncMo, it is still an open question whether ncMo can be considered circulating MФ with specialized sensing functions as such, bypassing their differentiation to bona fide tissue MФ. In general, it is not clear to what extent MФ function is influenced by environmental factors of the niche they have been recruited to or the retention of at least part of the functional phenotype of their precursor monocyte. Thus, differences between the phenotype of monocyte-derived and tissue-resident MФ might be preserved.
The classification of monocytes into subgroups has become more sophisticated, thanks to emerging technologies like high-parameter cytometry and scRNA-Seq, and transcends a taxonomy defined by merely two surface markers. Future studies, especially employing novel methods such as multiplex staining and multi-omics approaches that combine transcriptomics and surface marker information, will further expand our understanding of monocyte heterogeneity.136,137 The reappraisal of monocyte heterogeneity demonstrates the importance of unequivocal subset definition and demands an updated terminology to account for the heterogeneity of the monocyte population. Moreover, subset-specific intervention studies are needed to assess functional diversity of monocyte subsets and monocyte subset-derived MФ to dissect their individual role in CVD.
Authors’ contributions
A.V.R. and S.M.W.W. wrote and revised the manuscript. L.T., E.A.L.B., and P.G. revised the manuscript.
Acknowledgements
All figures were created with BioRender.com.
Contributor Information
Adele V Ruder, Department of Pathology, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Center (MUMC+), P. Debyelaan 25, 6229 HX Maastricht, The Netherlands.
Suzan M W Wetzels, Department of Pathology, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Center (MUMC+), P. Debyelaan 25, 6229 HX Maastricht, The Netherlands.
Lieve Temmerman, Department of Pathology, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Center (MUMC+), P. Debyelaan 25, 6229 HX Maastricht, The Netherlands.
Erik A L Biessen, Department of Pathology, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Center (MUMC+), P. Debyelaan 25, 6229 HX Maastricht, The Netherlands; Institute for Molecular Cardiovascular Research, RWTH Aachen University, Pauwelsstraße 30, 52074 Aachen, Germany.
Pieter Goossens, Department of Pathology, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Center (MUMC+), P. Debyelaan 25, 6229 HX Maastricht, The Netherlands.
Funding
This work was supported by the Dutch Heart Foundation (Dekker grant 2020T042 to P.G.); European Research Area Network on Cardiovascular Diseases (ERA-CVD JTC2017 "AtheroMacHete" to E.A.L.B. and P.G.); Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO, transatlantic collaboration grant FAPESP "DNAMoving" to E.A.L.B. and S. M.W.W. and STW 13568 "Barcoding the Obese" to E.A.L.B.).
Data availability
Data availability is not applicable to this review as no datasets were generated or analysed.
References
- 1. Oh ES, Na M, Rogers CJ. The association between monocyte subsets and cardiometabolic disorders/cardiovascular disease: a systematic review and meta-analysis. Front Cardiovasc Med 2021;8:640124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeBerge M, Shah SJ, Wilsbacher L, Thorp EB. Macrophages in heart failure with reduced versus preserved ejection fraction. Trends Mol Med 2019;25:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watanabe S, Alexander M, Misharin AV, Budinger GRS. The role of macrophages in the resolution of inflammation. Journal of Clinical Investigation 2019;129:2619–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity 2014;41:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014;159:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014;159:1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ 1972;46:845–852. [PMC free article] [PubMed] [Google Scholar]
- 8. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJM, Liu Y-J, MacPherson G, Randolph GJ, Scherberich J, Schmitz J, Shortman K, Sozzani S, Strobl H, Zembala M, Austyn JM, Lutz MB. Nomenclature of monocytes and dendritic cells in blood. Blood 2010;116:e74–e80. [DOI] [PubMed] [Google Scholar]
- 9. Cormican S, Griffin MD. Human monocyte subset distinctions and function: insights from gene expression analysis. Front Immunol 2020;11:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pn L, Niewold TB. By CyTOF: heterogeneity of human monocytes. Arterioscler Thromb Vasc Biol 2017;37:1423–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ong S-M, Teng K, Newell E, Chen H, Chen J, Loy T, Yeo T-W, Fink K, Wong S-C. A novel, five-marker alternative to CD16–CD14 gating to identify the three human monocyte subsets. Front Immunol 2019;10:1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boyette LB, Macedo C, Hadi K, Elinoff BD, Walters JT, Ramaswami B, Chalasani G, Taboas JM, Lakkis FG, Metes DM. Phenotype, function, and differentiation potential of human monocyte subsets. PLOS ONE 2017;12:e0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arenson EB Jr, Epstein MB, Seeger RC. Volumetric and functional heterogeneity of human monocytes. Journal of Clinical Investigation 1980;65:613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Norris DA, Morris RM, Sanderson RJ, Kohler PF. Isolation of functional subsets of human peripheral blood monocytes. J Immunol 1979;123:166–172. [PubMed] [Google Scholar]
- 15. Devitt A, Moffatt OD, Raykundalia C, Capra JD, Simmons DL, Gregory CD. Human CD14 mediates recognition and phagocytosis of apoptotic cells. Nature 1998;392:505–509. [DOI] [PubMed] [Google Scholar]
- 16. Kruger M, Coorevits L, De Wit TP, Casteels-Van Daele M, Van De Winkel JG, Ceuppens JL. Granulocyte-macrophage colony-stimulating factor antagonizes the transforming growth factor-beta-induced expression of fc gamma RIII (CD16) on human monocytes. Immunology 1996;87:162–167. [PMC free article] [PubMed] [Google Scholar]
- 17. Passlick B, Flieger D, Ziegler-Heitbrock L. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 1989;74:2527–2534. [PubMed] [Google Scholar]
- 18. Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, Puel A, Biswas SK, Moshous D, Picard C, Jais JP, D'Cruz D, Casanova JL, Trouillet C, Geissmann F. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 2010;33:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin M, Tacke F, Habenicht AJ, Ziegler-Heitbrock L, Randolph GJ. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 2010;115:e10–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elderman M, van Beek A, Brandsma E, de Haan B, Savelkoul H, de Vos P, Faas M. Sex impacts Th1 cells, Tregs, and DCs in both intestinal and systemic immunity in a mouse strain and location-dependent manner. Biol Sex Differ 2016;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petkova SB, Yuan R, Tsaih S-W, Schott W, Roopenian DC, Paigen B. Genetic influence on immune phenotype revealed strain-specific variations in peripheral blood lineages. Physiol Genomics 2008;34:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hofer TP, Zawada AM, Frankenberger M, Skokann K, Satzl AA, Gesierich W, Schuberth M, Levin J, Danek A, Rotter B, Heine GH, Ziegler-Heitbrock L. slan-defined subsets of CD16-positive monocytes: impact of granulomatous inflammation and M-CSF receptor mutation. Blood 2015;126:2601–2610. [DOI] [PubMed] [Google Scholar]
- 23. Abeles D, McPhail MJ, Sowter D, Antoniades CG, Vergis N, Vijay GKM, Xystrakis E, Khamri W, Shawcross DL, Ma Y, Wendon JA, Vergani D. CD14, CD16 and HLA-DR reliably identifies human monocytes and their subsets in the context of pathologically reduced HLA-DR expression by CD14hi/CD16neg monocytes: expansion of CD14hi/CD16pos and contraction of CD14lo/CD16pos monocytes in acute liver failure. Cytometry Part A 2012;81A:823–834. [DOI] [PubMed] [Google Scholar]
- 24. Autissier P, Soulas C, Burdo TH, Williams KC. Evaluation of a 12-color flow cytometry panel to study lymphocyte, monocyte, and dendritic cell subsets in humans. Cytometry Part A 2010;77A:410–419. [DOI] [PubMed] [Google Scholar]
- 25. Roussel M, Ferrell PB Jr, Greenplate AR, Lhomme F, Le Gallou S, Diggins KE, Johnson DB, Irish JM. Mass cytometry deep phenotyping of human mononuclear phagocytes and myeloid-derived suppressor cells from human blood and bone marrow. J Leukoc Biol 2017;102:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hofer TP, van de Loosdrecht AA, Stahl-Hennig C, Cassatella MA, Ziegler-Heitbrock L. 6-Sulfo LacNAc (slan) as a marker for non-classical monocytes. Front Immunol 2019;10:2052–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Leeuwen-Kerkhoff N, Lundberg K, Westers TM, Kordasti S, Bontkes HJ, de Gruijl TD, Lindstedt M, van de Loosdrecht AA. Transcriptional profiling reveals functional dichotomy between human slan+ non-classical monocytes and myeloid dendritic cells. J Leukoc Biol 2017;102:1055–1068. [DOI] [PubMed] [Google Scholar]
- 28. Ahmad F, Döbel T, Schmitz M, Schäkel K. Current concepts on 6-sulfo LacNAc expressing monocytes (slanMo). Front Immunol 2019;10:948–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamers AAJ, Dinh HQ, Thomas GD, Marcovecchio P, Blatchley A, Nakao CS, Kim C, McSkimming C, Taylor AM, Nguyen AT, McNamara CA, Hedrick CC. Human monocyte heterogeneity as revealed by high-dimensional mass cytometry. Arterioscler Thromb Vasc Biol 2019;39:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356:eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mildner A, Schönheit J, Giladi A, David E, Lara-Astiaso D, Lorenzo-Vivas E, Paul F, Chappell-Maor L, Priller J, Leutz A, Amit I, Jung S. Genomic characterization of murine monocytes reveals C/EBPβ transcription factor dependence of Ly6C− cells. Immunity 2017;46:849–862.e7. [DOI] [PubMed] [Google Scholar]
- 32. Thomas GD, Hamers AAJ, Nakao C, Marcovecchio P, Taylor AM, McSkimming C, Nguyen AT, McNamara CA, Hedrick CC. Human blood monocyte subsets: a new gating strategy defined using cell surface markers identified by mass cytometry. Arterioscler Thromb Vasc Biol 2017;37:1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Merah-Mourah F, Cohen SO, Charron D, Mooney N, Haziot A. Identification of novel human monocyte subsets and evidence for phenotypic groups defined by interindividual variations of expression of adhesion molecules. Sci Rep 2020;10:4397–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dutertre CA, Becht E, Irac SE, Khalilnezhad A, Narang V, Khalilnezhad S, Ng PY, van den Hoogen LL, Leong JY, Lee B, Chevrier M, Zhang XM, Yong PJA, Koh G, Lum J, Howland SW, Mok E, Chen J, Larbi A, Tan HKK, Lim TKH, Karagianni P, Tzioufas AG, Malleret B, Brody J, Albani S, van Roon J, Radstake T, Newell EW, Ginhoux F. Single-Cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity 2019;51:573–589.e8. [DOI] [PubMed] [Google Scholar]
- 35. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weber C, Shantsila E, Hristov M, Caligiuri G, Guzik T, Heine GH, Hoefer IE, Monaco C, Peter K, Rainger E, Siegbahn A, Steffens S, Wojta J, Lip GY. Role and analysis of monocyte subsets in cardiovascular disease. Joint consensus document of the European Society of Cardiology (ESC) working groups “atherosclerosis & vascular biology” and “thrombosis”. Thromb Haemost 2016;116:626–637. [DOI] [PubMed] [Google Scholar]
- 37. Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC, Schultze JL. The myeloid cell compartment—cell by cell. Annu Rev Immunol 2019;37:269–293. [DOI] [PubMed] [Google Scholar]
- 38. Choi SH, Kim JH, Lim S, Lim JY, Kim KW, Park KS, Jang HC. Monocyte count as a predictor of cardiovascular mortality in older Korean people. Age Ageing 2017;46:433–438. [DOI] [PubMed] [Google Scholar]
- 39. Kim JH, Lee YJ, Park B. Higher monocyte count with normal white blood cell count is positively associated with 10-year cardiovascular disease risk determined by Framingham risk score among community-dwelling Korean individuals. Medicine (Baltimore) 2019;98:e15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waterhouse DF, Cahill RA, Sheehan F, McCreery C. Prediction of calculated future cardiovascular disease by monocyte count in an asymptomatic population. Vasc Health Risk Manag 2008;4:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogacev KS, Cremers B, Zawada AM, Seiler S, Binder N, Ege P, Grosse-Dunker G, Heisel I, Hornof F, Jeken J, Rebling NM, Ulrich C, Scheller B, Bohm M, Fliser D, Heine GH. CD14++CD16 + monocytes independently predict cardiovascular events: a cohort study of 951 patients referred for elective coronary angiography. J Am Coll Cardiol 2012;60:1512–1520. [DOI] [PubMed] [Google Scholar]
- 42. Berg KE, Ljungcrantz I, Andersson L, Bryngelsson C, Hedblad B, Fredrikson GN, Nilsson J, Bjorkbacka H. Elevated CD14++CD16- monocytes predict cardiovascular events. Circ Cardiovasc Genet 2012;5:122–131. [DOI] [PubMed] [Google Scholar]
- 43. Wildgruber M, Aschenbrenner T, Wendorff H, Czubba M, Glinzer A, Haller B, Schiemann M, Zimmermann A, Berger H, Eckstein HH, Meier R, Wohlgemuth WA, Libby P, Zernecke A. The “Intermediate” CD14(++)CD16(+) monocyte subset increases in severe peripheral artery disease in humans. Sci Rep 2016;6:39483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cappellari R, D'Anna M, Bonora BM, Rigato M, Cignarella A, Avogaro A, Fadini GP. Shift of monocyte subsets along their continuum predicts cardiovascular outcomes. Atherosclerosis 2017;266:95–102. [DOI] [PubMed] [Google Scholar]
- 45. Nordestgaard BG, Chapman MJ, Ray K, Boren J, Andreotti F, Watts GF, Ginsberg H, Amarenco P, Catapano A, Descamps OS, Fisher E, Kovanen PT, Kuivenhoven JA, Lesnik P, Masana L, Reiner Z, Taskinen MR, Tokgozoglu L, Tybjaerg-Hansen A. European Atherosclerosis Society consensus P. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur Heart J 2010;31:2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krychtiuk KA, Kastl SP, Hofbauer SL, Wonnerth A, Goliasch G, Ozsvar-Kozma M, Katsaros KM, Maurer G, Huber K, Dostal E, Binder CJ, Pfaffenberger S, Oravec S, Wojta J, Speidl WS. Monocyte subset distribution in patients with stable atherosclerosis and elevated levels of lipoprotein(a). J Clin Lipidol 2015;9:533–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vaiserman A, Krasnienkov D. Telomere length as a marker of biological age: state-of-the-art, open issues, and future perspectives. Front Genet 2021;11:630186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Merino A, Buendia P, Martin-Malo A, Aljama P, Ramirez R, Carracedo J. Senescent CD14 + CD16 + monocytes exhibit proinflammatory and proatherosclerotic activity. J Immunol 2011;186:1809–1815. [DOI] [PubMed] [Google Scholar]
- 49. Meeuwsen JAL, de Vries JJ, van Duijvenvoorde A, van der Velden S, van der Laan SW, van Koeverden ID, van de Weg SM, de Borst GJ, de Winther MPJ, Kuiper J, Pasterkamp G, Hoefer IE, de Jager SCA, Queen of Hearts Consortium . Circulating CD14(+)CD16(-) classical monocytes do not associate with a vulnerable plaque phenotype, and do not predict secondary events in severe atherosclerotic patients. J Mol Cell Cardiol 2019;127:260–269. [DOI] [PubMed] [Google Scholar]
- 50. Lo SC, Lee WJ, Chen CY, Lee BC. Intermediate CD14++CD16 + monocyte predicts severe coronary stenosis and extensive plaque involvement in asymptomatic individuals. Int J Cardiovasc Imaging 2017;33:1223–1236. [DOI] [PubMed] [Google Scholar]
- 51. Yoshida N, Yamamoto H, Shinke T, Otake H, Kuroda M, Terashita D, Takahashi H, Sakaguchi K, Hirota Y, Emoto T, Amin HZ, Mizoguchi T, Hayashi T, Sasaki N, Yamashita T, Ogawa W, Hirata KI. Impact of CD14++CD16 + monocytes on plaque vulnerability in diabetic and non-diabetic patients with asymptomatic coronary artery disease: a cross-sectional study. Cardiovasc Diabetol 2017;16:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamamoto H, Yoshida N, Shinke T, Otake H, Kuroda M, Sakaguchi K, Hirota Y, Toba T, Takahashi H, Terashita D, Uzu K, Tahara N, Shinkura Y, Kuroda K, Nagasawa Y, Nagano Y, Tsukiyama Y, Yanaka KI, Emoto T, Sasaki N, Yamashita T, Ogawa W, Hirata KI. Impact of CD14(++)CD16(+) monocytes on coronary plaque vulnerability assessed by optical coherence tomography in coronary artery disease patients. Atherosclerosis 2018;269:245–251. [DOI] [PubMed] [Google Scholar]
- 53. Frantz S, Nahrendorf M. Cardiac macrophages and their role in ischaemic heart disease. Cardiovasc Res 2014;102:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsujioka H, Imanishi T, Ikejima H, Kuroi A, Takarada S, Tanimoto T, Kitabata H, Okochi K, Arita Y, Ishibashi K, Komukai K, Kataiwa H, Nakamura N, Hirata K, Tanaka A, Akasaka T. Impact of heterogeneity of human peripheral blood monocyte subsets on myocardial salvage in patients with primary acute myocardial infarction. J Am Coll Cardiol 2009;54:130–138. [DOI] [PubMed] [Google Scholar]
- 55. Tapp LD, Shantsila E, Wrigley BJ, Pamukcu B, Lip GY. The CD14++CD16 + monocyte subset and monocyte-platelet interactions in patients with ST-elevation myocardial infarction. J Thromb Haemost 2012;10:1231–1241. [DOI] [PubMed] [Google Scholar]
- 56. van der Laan AM, Ter Horst EN, Delewi R, Begieneman MP, Krijnen PA, Hirsch A, Lavaei M, Nahrendorf M, Horrevoets AJ, Niessen HW, Piek JJ. Monocyte subset accumulation in the human heart following acute myocardial infarction and the role of the spleen as monocyte reservoir. Eur Heart J 2014;35:376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou X, Liu XL, Ji WJ, Liu JX, Guo ZZ, Ren D, Ma YQ, Zeng S, Xu ZW, Li HX, Wang PP, Zhang Z, Li YM, Benefield BC, Zawada AM, Thorp EB, Lee DC, Heine GH. The kinetics of circulating monocyte subsets and monocyte-platelet aggregates in the acute phase of ST-elevation myocardial infarction: associations with 2-year cardiovascular events. Medicine (Baltimore) 2016;95:e3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zeng S, Yan LF, Luo YW, Liu XL, Liu JX, Guo ZZ, Xu ZW, Li YM, Ji WJ, Zhou X. Trajectories of circulating monocyte subsets after ST-elevation myocardial infarction during hospitalization: latent class growth modeling for high-risk patient identification. J Cardiovasc Transl Res 2018;11:22–32. [DOI] [PubMed] [Google Scholar]
- 59. Dutta P, Courties G, Wei Y, Leuschner F, Gorbatov R, Robbins CS, Iwamoto Y, Thompson B, Carlson AL, Heidt T, Majmudar MD, Lasitschka F, Etzrodt M, Waterman P, Waring MT, Chicoine AT, van der Laan AM, Niessen HW, Piek JJ, Rubin BB, Butany J, Stone JR, Katus HA, Murphy SA, Morrow DA, Sabatine MS, Vinegoni C, Moskowitz MA, Pittet MJ, Libby P, Lin CP, Swirski FK, Weissleder R, Nahrendorf M. Myocardial infarction accelerates atherosclerosis. Nature 2012;487:325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cahill TJ, Kharbanda RK. Heart failure after myocardial infarction in the era of primary percutaneous coronary intervention: mechanisms, incidence and identification of patients at risk. World J Cardiol 2017;9:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barisione C, Garibaldi S, Ghigliotti G, Fabbi P, Altieri P, Casale MC, Spallarossa P, Bertero G, Balbi M, Corsiglia L, Brunelli C. CD14CD16 Monocyte subset levels in heart failure patients. Dis Markers 2010;28:115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shantsila E, Ghattas A, Griffiths HR, Lip GYH. Mon2 predicts poor outcome in ST-elevation myocardial infarction. J Intern Med 2019;285:301–316. [DOI] [PubMed] [Google Scholar]
- 63. Wrigley BJ, Shantsila E, Tapp LD, Lip GY. Increased expression of cell adhesion molecule receptors on monocyte subsets in ischaemic heart failure. Thromb Haemost 2013;110:92–100. [DOI] [PubMed] [Google Scholar]
- 64. Wrigley BJ, Shantsila E, Tapp LD, Lip GY. CD14++CD16 + monocytes in patients with acute ischaemic heart failure. Eur J Clin Invest 2013;43:121–130. [DOI] [PubMed] [Google Scholar]
- 65. Elchinova E, Teubel I, Roura S, Fernandez MA, Lupon J, Galvez-Monton C, de Antonio M, Moliner P, Domingo M, Zamora E, Nunez J, Cediel G, Bayes-Genis A. Circulating monocyte subsets and heart failure prognosis. PLoS One 2018;13:e0204074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Amir O, Spivak I, Lavi I, Rahat MA. Changes in the monocytic subsets CD14(dim)CD16(+) and CD14(++)CD16(-) in chronic systolic heart failure patients. Mediators Inflamm 2012;2012:616384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eiras S, Varela-Roman A, Andrade MC, Castro A, Gonzalez-Ferreiro R, Vinuela JE, Fernandez-Trasancos A, Carreira MC, Alvarez E, Casanueva FF, Gonzalez-Juanatey JR. Non classical monocytes levels, increased by subcutaneous fat-secretome, are associated with less rehospitalization after heart failure admission. J Cardiovasc Transl Res 2017;10:16–26. [DOI] [PubMed] [Google Scholar]
- 68. Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, Geissmann F, Hedrick CC. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C− monocytes. Nat Immunol 2011;12:778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thomas Graham D, Hanna Richard N, Vasudevan Neelakatan T, Hamers Anouk A, Romanoski Casey E, McArdle S, Ross Kevin D, Blatchley A, Yoakum D, Hamilton Bruce A, Mikulski Z, Jain Mukesh K, Glass Christopher K, Hedrick Catherine C. Deleting an Nr4a1 super-enhancer subdomain ablates Ly6Clow monocytes while preserving macrophage gene function. Immunity 2016;45:975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ma Y, Mouton AJ, Lindsey ML. Cardiac macrophage biology in the steady-state heart, the aging heart, and following myocardial infarction. Transl Res 2018;191:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol 2017;17:349–362. [DOI] [PubMed] [Google Scholar]
- 72. Wacleche VS, Tremblay CL, Routy J-P, Ancuta P. The biology of monocytes and dendritic cells: contribution to HIV pathogenesis. Viruses 2018;10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science 2010;327:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014;14:392–404. [DOI] [PubMed] [Google Scholar]
- 75. Sampath P, Moideen K, Ranganathan UD, Bethunaickan R. Monocyte subsets: phenotypes and function in Tuberculosis infection. Front Immunol 2018;9:1726–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wong KL, Yeap WH, Tai JJY, Ong SM, Dang TM, Wong SC. The three human monocyte subsets: implications for health and disease. Immunol Res 2012;53:41–57. [DOI] [PubMed] [Google Scholar]
- 77. van de Veerdonk FL, Netea MG. Diversity: a hallmark of monocyte society. Immunity 2010;33:289–291. [DOI] [PubMed] [Google Scholar]
- 78. França CN, Izar MCO, Hortêncio MNS, do Amaral JB, Ferreira CES, Tuleta ID, Fonseca FAH. Monocyte subtypes and the CCR2 chemokine receptor in cardiovascular disease. Clin Sci 2017;131:1215–1224. [DOI] [PubMed] [Google Scholar]
- 79. Kapellos TS, Bonaguro L, Gemünd I, Reusch N, Saglam A, Hinkley ER, Schultze JL. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front Immunol 2019;10:2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Thomas G, Tacke R, Hedrick CC, Hanna RN. Nonclassical patrolling monocyte function in the vasculature. Arterioscler Thromb Vasc Biol 2015;35:1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sprangers S, de Vries TJ, Everts V. Monocyte heterogeneity: consequences for monocyte-derived immune cells. J Immunol Res 2016;2016:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Carlin Leo M, Stamatiades Efstathios G, Auffray C, Hanna Richard N, Glover L, Vizcay-Barrena G, Hedrick Catherine C, Cook HT, Diebold S, Geissmann F. Nr4a1-dependent Ly6Clow monocytes monitor endothelial cells and orchestrate their disposal. Cell 2013;153:362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Olingy CE, San Emeterio CL, Ogle ME, Krieger JR, Bruce AC, Pfau DD, Jordan BT, Peirce SM, Botchwey EA. Non-classical monocytes are biased progenitors of wound healing macrophages during soft tissue injury. Sci Rep 2017;7:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, Bigley V, Flavell RA, Gilroy DW, Asquith B, Macallan D, Yona S. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. Journal of Experimental Medicine 2017;214:1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK, Manz MG, Flavell RA. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 2014;32:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thaler B, Hohensinner PJ, Krychtiuk KA, Matzneller P, Koller L, Brekalo M, Maurer G, Huber K, Zeitlinger M, Jilma B, Wojta J, Speidl WS. Differential in vivo activation of monocyte subsets during low-grade inflammation through experimental endotoxemia in humans. Sci Rep 2016;6:30162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mildner A, Marinkovic G, Jung S, Gordon S. Murine monocytes: origins, subsets, fates, and functions. Microbiol Spectr 2016;4:4.5.01. [DOI] [PubMed] [Google Scholar]
- 88. Chong SZ, Evrard M, Devi S, Chen J, Lim JY, See P, Zhang Y, Adrover JM, Lee B, Tan L, Li JL, Liong KH, Phua C, Balachander A, Boey A, Liebl D, Tan SM, Chan JK, Balabanian K, Harris JE, Bianchini M, Weber C, Duchene J, Lum J, Poidinger M, Chen Q, Renia L, Wang CI, Larbi A, Randolph GJ, Weninger W, Looney MR, Krummel MF, Biswas SK, Ginhoux F, Hidalgo A, Bachelerie F, Ng LG. CXCR4 Identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J Exp Med 2016;213:2293–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guilliams M, Mildner A, Yona S. Developmental and functional heterogeneity of monocytes. Immunity 2018;49:595–613. [DOI] [PubMed] [Google Scholar]
- 90. Duroux-Richard I, Robin M, Peillex C, Apparailly F. MicroRNAs: fine tuners of monocyte heterogeneity. Front Immunol 2019;10:2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, Perlman H, Malissen B, Zelzer E, Jung S. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013;38:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tak T, Drylewicz J, Conemans L, de Boer RJ, Koenderman L, Borghans JAM, Tesselaar K. Circulatory and maturation kinetics of human monocyte subsets in vivo. Blood 2017;130:1474–1477. [DOI] [PubMed] [Google Scholar]
- 93. Chimen M, Yates CM, McGettrick HM, Ward LSC, Harrison MJ, Apta B, Dib LH, Imhof BA, Harrison P, Nash GB, Rainger GE. Monocyte subsets coregulate inflammatory responses by integrated signaling through TNF and IL-6 at the endothelial cell interface. J Immunol 2017;198:2834–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zawada AM, Rogacev KS, Rotter B, Winter P, Marell R-R, Fliser D, Heine GH. SuperSAGE evidence for CD14++CD16 + monocytes as a third monocyte subset. Blood 2011;118:e50–e61. [DOI] [PubMed] [Google Scholar]
- 95. Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, Margalit R, Kalchenko V, Geissmann F, Jung S. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med 2007;204:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ziegler-Heitbrock L. The CD14+ CD16 + blood monocytes: their role in infection and inflammation. J Leukoc Biol 2007;81:584–592. [DOI] [PubMed] [Google Scholar]
- 97. Jackson WD, Weinrich TW, Woollard KJ. Very-low and low-density lipoproteins induce neutral lipid accumulation and impair migration in monocyte subsets. Sci Rep 2016;6:20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Höpfner F, Jacob M, Ulrich C, Russ M, Simm A, Silber RE, Girndt M, Noutsias M, Werdan K, Schlitt A. Subgroups of monocytes predict cardiovascular events in patients with coronary heart disease. The PHAMOS trial (Prospective Halle Monocytes Study). Hellenic J Cardiol 2019;60:311–321. [DOI] [PubMed] [Google Scholar]
- 99. Wong KL, Tai JJ-Y, Wong W-C, Han H, Sem X, Yeap W-H, Kourilsky P, Wong S-C. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 2011;118:e16–e31. [DOI] [PubMed] [Google Scholar]
- 100. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 2016;44:439–449. [DOI] [PubMed] [Google Scholar]
- 101. Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 2007;10:1538–1543. [DOI] [PubMed] [Google Scholar]
- 102. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010;330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, Weissman IL, Cyster JG, Engleman EG. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol 2002;3:1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, Brija T, Gautier EL, Ivanov S, Satpathy AT, Schilling JD, Schwendener R, Sergin I, Razani B, Forsberg EC, Yokoyama WM, Unanue ER, Colonna M, Randolph GJ, Mann DL. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014;40:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, Pinto AR, Klapproth K, Henri S, Malissen B, Rodewald HR, Rosenthal NA, Bajenoff M, Prinz M, Jung S, Sieweke MH. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med 2014;211:2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, Dalod M, Malissen B, Henri S. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 2013;39:925–938. [DOI] [PubMed] [Google Scholar]
- 107. Bain CC, Bravo-Blas A, Scott CL, Perdiguero EG, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 2014;15:929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. De Schepper S, Verheijden S, Aguilera-Lizarraga J, Viola MF, Boesmans W, Stakenborg N, Voytyuk I, Schmidt I, Boeckx B, Dierckx de Casterle I, Baekelandt V, Gonzalez Dominguez E, Mack M, Depoortere I, De Strooper B, Sprangers B, Himmelreich U, Soenen S, Guilliams M, Vanden Berghe P, Jones E, Lambrechts D, Boeckxstaens G. Self-Maintaining gut macrophages are essential for intestinal homeostasis. Cell 2018;175:400–415.e13. [DOI] [PubMed] [Google Scholar]
- 109. Shaw TN, Houston SA, Wemyss K, Bridgeman HM, Barbera TA, Zangerle-Murray T, Strangward P, Ridley AJL, Wang P, Tamoutounour S, Allen JE, Konkel JE, Grainger JR. Tissue-resident macrophages in the intestine are long lived and defined by tim-4 and CD4 expression. J Exp Med 2018;215:1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bajpai G, Schneider C, Wong N, Bredemeyer A, Hulsmans M, Nahrendorf M, Epelman S, Kreisel D, Liu Y, Itoh A, Shankar TS, Selzman CH, Drakos SG, Lavine KJ. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med 2018;24:1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bigley V, Haniffa M, Doulatov S, Wang XN, Dickinson R, McGovern N, Jardine L, Pagan S, Dimmick I, Chua I, Wallis J, Lordan J, Morgan C, Kumararatne DS, Doffinger R, van der Burg M, van Dongen J, Cant A, Dick JE, Hambleton S, Collin M. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med 2011;208:227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Teh YC, Ding JL, Ng LG, Chong SZ. Capturing the fantastic voyage of monocytes through time and space. Front Immunol 2019;10:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Combadière C, Potteaux S, Rodero M, Simon T, Pezard A, Esposito B, Merval R, Proudfoot A, Tedgui A, Mallat Z. Combined inhibition of CCL2, CX3CR1, and CCR5 abrogates Ly6Chi and Ly6Clo monocytosis and almost abolishes atherosclerosis in hypercholesterolemic mice. Circulation 2008;117:1649–1657. [DOI] [PubMed] [Google Scholar]
- 114. Tacke F, Alvarez D, Kaplan TJ, Jakubzick C, Spanbroek R, Llodra J, Garin A, Liu J, Mack M, van Rooijen N, Lira SA, Habenicht AJ, Randolph GJ. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. Journal of Clinical Investigation 2007;117:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mosig S, Rennert K, Krause S, Kzhyshkowska J, Neunübel K, Heller R, Funke H. Different functions of monocyte subsets in familial hypercholesterolemia: potential function of CD14 + CD16 + monocytes in detoxification of oxidized LDL. FASEB J 2009;23:866–874. [DOI] [PubMed] [Google Scholar]
- 116. Gautier EL, Jakubzick C, Randolph GJ. Regulation of the migration and survival of monocyte subsets by chemokine receptors and its relevance to atherosclerosis. Arterioscler Thromb Vasc Biol 2009;29:1412–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Guilliams M, Thierry GR, Bonnardel J, Bajenoff M. Establishment and maintenance of the macrophage niche. Immunity 2020;52:434–451. [DOI] [PubMed] [Google Scholar]
- 118. Poller WC, Nahrendorf M, Swirski FK. Hematopoiesis and cardiovascular disease. Circ Res 2020;126:1061–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Anbazhagan K, Duroux-Richard I, Jorgensen C, Apparailly F. Transcriptomic network support distinct roles of classical and non-classical monocytes in human. Int Rev Immunol 2014;33:470–489. [DOI] [PubMed] [Google Scholar]
- 120. Platt AM, Bain CC, Bordon Y, Sester DP, Mowat AM. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J Immunol 2010;184:6843–6854. [DOI] [PubMed] [Google Scholar]
- 121. Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, Shakhar G, Halpern Z, Jung S. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 2012;37:1076–1090. [DOI] [PubMed] [Google Scholar]
- 122. Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol 2009;27:669–692. [DOI] [PubMed] [Google Scholar]
- 123. Williams JW, Randolph GJ, Zinselmeyer BH. A polecat’s view of patrolling monocytes. Circ Res 2017;120:1699–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Buscher K, Marcovecchio P, Hedrick CC, Ley K. Patrolling mechanics of non-classical monocytes in vascular inflammation. Front Cardiovasc Med 2017;4:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schyns J, Bai Q, Ruscitti C, Radermecker C, De Schepper S, Chakarov S, Farnir F, Pirottin D, Ginhoux F, Boeckxstaens G, Bureau F, Marichal T. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat Commun 2019;10:3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Goudot C, Coillard A, Villani AC, Gueguen P, Cros A, Sarkizova S, Tang-Huau TL, Bohec M, Baulande S, Hacohen N, Amigorena S, Segura E. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 2017;47:582–596.e6. [DOI] [PubMed] [Google Scholar]
- 128. Menezes S, Melandri D, Anselmi G, Perchet T, Loschko J, Dubrot J, Patel R, Gautier EL, Hugues S, Longhi MP, Henry JY, Quezada SA, Lauvau G, Lennon-Duménil A-M, Gutiérrez-Martínez E, Bessis A, Gomez-Perdiguero E, Jacome-Galarza CE, Garner H, Geissmann F, Golub R, Nussenzweig MC, Guermonprez P. The heterogeneity of Ly6Chi monocytes controls their differentiation into iNOS+ macrophages or monocyte-derived dendritic cells. Immunity 2016;45:1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nagenborg J, Goossens P, Biessen EAL, Donners M. Heterogeneity of atherosclerotic plaque macrophage origin, phenotype and functions: implications for treatment. Eur J Pharmacol 2017;816:14–24. [DOI] [PubMed] [Google Scholar]
- 130. Stremmel C, Stark K, Schulz C. Heterogeneity of macrophages in atherosclerosis. Thromb Haemost 2019;119:1237–1246. [DOI] [PubMed] [Google Scholar]
- 131. Bonnardel J, Guilliams M. Developmental control of macrophage function. Curr Opin Immunol 2018;50:64–74. [DOI] [PubMed] [Google Scholar]
- 132. Evren E, Ringqvist E, Tripathi KP, Sleiers N, Rives IC, Alisjahbana A, Gao Y, Sarhan D, Halle T, Sorini C, Lepzien R, Marquardt N, Michaëlsson J, Smed-Sörensen A, Botling J, Karlsson MCI, Villablanca EJ, Willinger T. Distinct developmental pathways from blood monocytes generate human lung macrophage diversity. Immunity 2021;54:259–275.e7. [DOI] [PubMed] [Google Scholar]
- 133. Ożańska A, Szymczak D, Rybka J. Pattern of human monocyte subpopulations in health and disease. Scand J Immunol 2020;92:e12883. [DOI] [PubMed] [Google Scholar]
- 134. Kerkhofs D, van Hagen BT, Milanova IV, Schell KJ, van Essen H, Wijnands E, Goossens P, Blankesteijn WM, Unger T, Prickaerts J, Biessen EA, van Oostenbrugge RJ, Foulquier S. Pharmacological depletion of microglia and perivascular macrophages prevents vascular cognitive impairment in Ang II-induced hypertension. Theranostics 2020;10:9512–9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Frankenberger M, Hofer TPJ, Marei A, Dayyani F, Schewe S, Strasser C, Aldraihim A, Stanzel F, Lang R, Hoffmann R, Costa OP, Buch T, Ziegler-Heitbrock L. Transcript profiling of CD16-positive monocytes reveals a unique molecular fingerprint. Eur J Immunol 2012;42:957–974. [DOI] [PubMed] [Google Scholar]
- 136. Lawlor N, Nehar-Belaid D, Grassmann JDS, Stoeckius M, Smibert P, Stitzel ML, Pascual V, Banchereau J, Williams A, Ucar D. Single cell analysis of blood mononuclear cells stimulated through either LPS or anti-CD3 and anti-CD28. Front Immunol 2021;12:636720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rogers BM, Smith L, Dezso Z, Shi X, DiGiammarino E, Nguyen D, Sethuraman S, Zheng P, Choi D, Zhang D, Nguyen A, McGuire K, Liu W, Chung N, Chao DT, Ye S, Starbeck-Miller GR. VISTA Is an activating receptor in human monocytes. J Exp Med 2021;218:e20201601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Mueller KAL, Hanna DB, Ehinger E, Xue X, Baas L, Gawaz MP, Geisler T, Anastos K, Cohen MH, Gange SJ, Heath SL, Lazar JM, Liu C, Mack WJ, Ofotokun I, Tien PC, Hodis HN, Landay AL, Kaplan RC, Ley K. Loss of CXCR4 on non-classical monocytes in participants of the Women's Interagency HIV Study (WIHS) with subclinical atherosclerosis. Cardiovasc Res 2019;115:1029–1040. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability is not applicable to this review as no datasets were generated or analysed.