Abstract
We developed and evaluated a two-step PCR-based assay with universal primers and genus- or species-specific primers for the detection of the most prevalent bacterial etiologies of otitis media with effusion (OME) in children from Lebanese hospitals. These etiologies included Haemophilus, Streptococcus, and Moraxella (Branhamella) catarrhalis, which were detected in middle-ear effusion (MEE) samples taken from children with OME. A total of 47 MEE samples were aspirated from 36 patients during insertion of a tympanostomy tube performed particularly for OME. The duration of effusion in all patients was ≥2 months. DNA was extracted from MEE samples, and PCR was initially done with DNA extracts by using the universal primers RW01 and DG74, which flank an ∼370-bp fragment found in the 16S rRNA gene of all bacterial species. For the identification of specific bacteria, we used in three separate reaction mixtures the following genus- or species-specific primers: (i) a Haemophilus-specific probe (probe RDR125) as a primer along with DG74, (ii) a Streptococcus-specific primer (primer STR1; designed by us) along with DG74, and (iii) an M. catarrhalis-specific primer pair (primer pair MCA1-MCA2). Thirty-five MEE samples (74.5%) gave the expected 370-bp band, indicating the presence of bacterial DNA in the tested samples. Of the 35 PCR-positive samples tested, 33 (94.3%) were positive for Haemophilus, 3 (8.6%) were positive for Streptococcus, and 10 (28.6%) were positive for M. catarrhalis. Ten samples (28.6%) exhibited a mixed infection and were positive for both Haemophilus and M. catarrhalis. Culture was simultaneously performed for all 47 MEE samples. Ten of the 47 MEE samples (21.3%) exhibited bacterial growth. These 10 were PCR positive for bacterial DNA. The remaining 25 PCR-positive samples were negative by culture, thus showing about 53% discordance between PCR results and those of culture. The PCR assay proved to be more sensitive than culture, more rapid, less cumbersome, and more cost-effective than the available PCR-Southern hybridization-based assays.
Otitis media with effusion (OME) is a common disease of childhood characterized by the presence of fluid in the middle-ear cavity behind an intact tympanic membrane, also known as middle-ear effusion (MEE) (5). OME often follows an episode of acute otitis media (AOM) (12), which is a highly frequent childhood disease due to its association with upper respiratory tract viral infections (4). In OME, symptoms of acute disease like fever and ear pain are usually lacking, and the disease is often noticed when the affected child displays a delay in language acquisition or inattentiveness due to the conductive type of hearing loss caused by fluid accumulation in the middle-ear cavity. In OME, the effusion is produced by the inflamed mucosa lining the cavity, and according to the duration of persistence of this effusion, OME may be classified as acute when the effusion lasts for ≤3 weeks, as subacute when the effusion lasts between 3 weeks and 3 months, and as chronic when the effusion persists for ≥3 months (10).
In the past, OME was thought to be an entirely inflammatory process and the MEE produced was considered sterile. This belief held true until 1958, when Senturia et al. (11) were able to recover bacteria from cultures of MEE samples. Since then, several studies in various regions of the world have been conducted in order to reveal the bacteriology of OME, and almost all of these studies relied upon culturing of the MEE samples (1, 2, 15); in addition, PCR-based assays attempting to detect bacterial DNA in MEE samples were also recently developed (9, 14). However, most of the available PCR-based assays often use Southern hybridization, which is time-consuming, costly, and cumbersome due to the involvement of multiple procedures. According to the findings of culture and PCR-based assays, the most common bacteria found in MEE samples are Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella (Branhamella) catarrhalis (9, 16). This is in addition to other bacteria like Streptococcus pyogenes, Staphylococcus, Pseudomonas aeruginosa, and anaerobes like Prevotella and Porphyromonas (2).
The present work aimed at (i) the use and evaluation of a two-step PCR-based assay that initially uses the universal primers RW01 and DG74 to amplify a 370-bp sequence in the conserved 16S rRNA gene common to all bacteria to demonstrate the presence of bacteria and, in a second step, single genus- or species-specific primers along with DG74 for the specific identification of the bacteria involved in OME; (ii) evaluation of the utility of a probe (probe RDR125), developed by Greisen et al. (3), as a primer along with DG74 for the PCR detection of Haemophilus and the utility of primer STR1, developed by us, for the detection of Streptococcus when used with DG74; and (iii) determination of the bacterial etiology of OME in Lebanese patients by using the two-step PCR-based assay and culture, since no such studies have been conducted in Lebanon.
MATERIALS AND METHODS
Case definition.
MEE samples were aspirated from children undergoing myringotomy with or without the insertion of a tympanostomy tube for OME. For all patients considered, the effusion (MEE) had persisted for at least 2 months prior to the date of operation; i.e., all cases of OME were of the subacute (MEE duration, 3 weeks to 3 months) and chronic (MEE duration, 3 months) types. All patients had been on antibiotic therapy for at least 2 weeks prior to the operation date.
Source of MEE samples.
A total of 47 MEE samples were collected between September 1996 and May 1997 from 36 Lebanese children undergoing tympanostomy-tube placement for OME. The ages of the children ranged from 2 to 10 years. MEE samples were provided by three medical centers in Beirut, Lebanon.
Sample aspiration and culture.
The surgeon aspirated MEE samples from the OME patients during tympanostomy-tube insertion. Each MEE sample was cultured on chocolate agar, and the culture was incubated in 5% CO2 at 37°C overnight. Positive cultures were further identified by Gram staining and standard identification assays.
DNA extraction.
DNA was extracted from MEE samples by the method of Loutit and Tompkins (7), with some modifications. The method uses 800 μg of proteinase K per ml and 5% Triton X-100.
PCR.
PCR was done with DNA extracts first by using the universal primers RW01 (5′-AAC TGG AGG AAG GTG GGG AT-3′) and DG74 (5′-AGG AGG TGA TCC AAC CGC A-3′) which flank an ∼370-bp region in the 16S rRNA gene. MEE samples showing the 370-bp amplicon with the universal primers were further assessed by using genus- or species-specific primers for the identification of Haemophilus, Streptococcus, and M. catarrhalis. For Haemophilus, the primer pair used was RDR125 (5′-GG AGT GGG TTG TAC CAG AAG TAG AT 3′) and DG74; the RDR125 primer initially used as a probe by Greisen et al. (3) hybridizes with an internal sequence within the 370-bp fragment and was used with DG74 as a primer to flank a 124-bp region. The data for Haemophilus were further confirmed by using a second pair of specific primers, F1 (5′-AAC TTT TGG CGG TTA CTC TG 3′) and R1 (5′-CTA ACA CTG CAC GAC GGT TT-3′), designed by Ueyama et al. (14). Primers F1 and R1 flank a 351-bp fragment within the P6-encoding gene (outer membrane protein-encoding gene) of Haemophilus. The results obtained with the primer-probe combination were compared with those obtained with the F1-R1 primer pair, and the degree of concordance between the two results was recorded. The specificities of all these primers were assessed by testing them with DNA from other genera. For Streptococcus, DG74 was used together with primer STR1, whose sequence was designed by us (5′-AGT CGG TGA GGT AAC CGT AAG-3′). Primer STR1 lies within the 370-bp sequence and flanks, along with DG74, a region of 105 bp. The specificity of primer STR1 was assessed by using it along with DG74 to amplify DNA from other genera. The primers used for M. catarrhalis were those designed by Post et al. (9), and the expected band size is about 140 bp. These primers are MCAT1 (5′-TTG GCT TGT GCT AAA ATA TC-3′) and MCAT2 (5′-GTC ATC GCT ATC ATT CAC CT-3′). They were also used to amplify DNA from other genera to assess their specificities.
PCR amplification was carried in 100-μl reaction mixtures each containing 56.5 μl of sterile distilled water, 16 μl of deoxyribonucleoside triphosphate (0.2 mM), 10 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2), 1 μl of each one of the primer pairs to be used (0.3 μg/μl), 0.5 μl of Taq DNA polymerase (5 U/μl), and 15 μl of the DNA extract. Amplification was carried in a minicycler (MJ Research, Watertown, Mass.), and the PCR programs used were optimized for each primer set. The PCR programs for all primer pairs except F1 and R1 consisted of a three-step cycle repeated 34 times: denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. This was followed by a final extension at 72°C for 10 min. For the F1-R1 primer pair, the same PCR conditions were used, except that a primer annealing temperature of 60°C rather than one of 55°C was used. The amplicons were separated on 1.5% SeaKem agarose (FMC BioProducts, Rockland, Maine) gels stained with ethidium bromide at a final concentration of 0.5 μg/ml. The gels were then visualized on a UV-light transilluminator (HaakeBuchler, Saddle Brook, N.J.) and photographed with Polaroid type 667 film (Polaroid Ltd., St. Albans, Hertford, United Kingdom).
RESULTS
Our data have shown that of the 47 MEE samples tested, 35 (74.5%) gave a band of ∼370 bp in tests with universal primers, and of these, 33 (94%) were positive for Haemophilus in tests with the genus-specific primer RDR125 and the universal primer DG74. These samples exhibited the expected 124-bp band. No DNA from other genera was amplified by these primers. Three (8.5%) samples were positive for Streptococcus in tests with the genus-specific primer STR1 and the universal primer DG74 and gave the expected 105-bp band. No DNA from other genera was amplified with these primers. Ten (28.5%) samples were positive for M. catarrhalis in tests with the species-specific primers MCAT1 and MCAT2 and gave the expected 140-bp band. No DNA from other genera was amplified. None of the MEE samples that were positive for Haemophilus in tests with genus-specific primer RDR125 and universal primer DG74 were negative for this organism in tests with the genus-specific F1-R1 primer pair which amplified the expected 351-bp segment, thus showing a 100% concordance between the results obtained by PCR with Haemophilus-specific primer RDR125 and universal primer DG74 and Haemophilus-specific primer pair F1-R1. Figures 1 and 2 show representative amplicons.
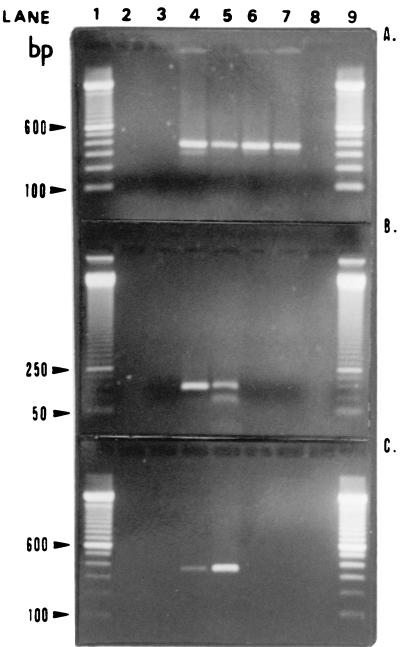
FIG. 1.
Agarose gels containing representative amplicons. (A) Universal primers RW01 and DG74. Lanes 1 and 9, 100-bp ladder; lane 2, negative control for the extraction process; lane 3, negative control for PCR; lane 4, positive control for PCR (Escherichia coli ATCC 25922 DNA); lanes 5 to 7, three PCR-positive MEE samples, respectively (370-bp band); lane 8, a PCR-negative MEE sample. (B) Haemophilus-specific primer-probe pair RDR125 and DG74. Lanes 1 and 9, 50-bp ladder; lane 2, empty; lane 3, negative control for PCR; lane 4, positive control for PCR (H. influenzae ATCC 49766 DNA); lane 5, PCR-positive MEE sample (124-bp band); lanes 6 and 7, two PCR-negative MEE samples, respectively; lane 8, empty. (C) Haemophilus-specific F1-R1 primer pair. Lanes 1 and 9, 100-bp ladder; lane 2, empty; lane 3, negative control for PCR; lane 4, positive control for PCR (H. influenzae ATCC 49766 DNA); lane 5, PCR-positive MEE sample (351-bp band); lanes 6 and 7, two PCR-negative MEE samples, respectively; lane 8, empty.
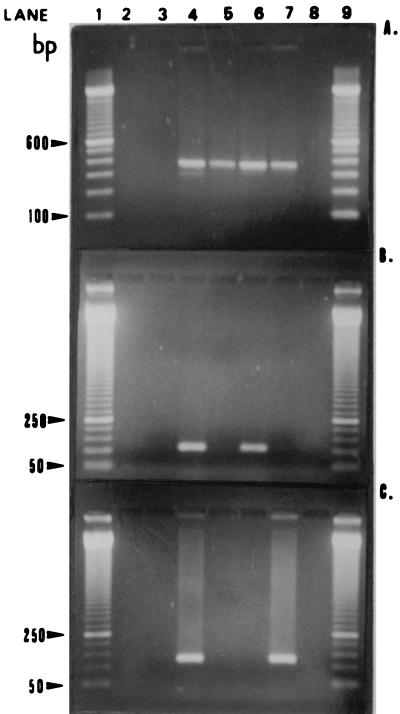
FIG. 2.
Agarose gels containing representative amplicons. (A) Universal primers RW01 and DG74. Lanes 1 and 9, 100-bp ladder; lane 2, negative control for the extraction process; lane 3, negative control for PCR; lane 4, positive control for PCR (E. coli ATCC 25922 DNA); lanes 5 to 7, three PCR-positive MEE samples, respectively (370-bp band); lane 8, PCR-negative MEE sample. (B) Streptococcus-specific STR1 and DG74 primer pair. Lanes 1 and 9, 50-bp ladder; lane 2, empty; lane 3, negative control for PCR; lane 4, positive control for PCR (S. pneumoniae ATCC 49619 DNA); lanes 5 and 7, two PCR-negative MEE samples, respectively; lane 6, PCR-positive MEE sample (105-bp band); lane 8, empty. (C) M. catarrhalis-specific MCAT1 and MCAT2 primer pair. Lanes 1 and 9, 50-bp ladder; lane 2, empty; lane 3, negative control for PCR; lane 4, positive control for PCR (M. catarrhalis DNA); lanes 5 and 6, two PCR-negative MEE samples, respectively; lane 7, a PCR-positive MEE sample (140-bp band); lane 8, empty.
Only 10 of the 47 MEE samples were culture positive, with 9 samples yielding H. influenzae and 2 samples yielding M. catarrhalis after performing the appropriate biochemical tests. Twelve of 33 (36.4%) samples from OME patients had mixed infections: 1 (3%) sample was positive for all 3 bacteria, 9 (27.3%) samples were positive for Haemophilus and M. catarrhalis, and 2 (6%) samples were positive for Haemophilus and Streptococcus. Few cases of OME were due to bacteria other than the three tested for (PCR positive with universal primers but PCR negative with specific primers), while others were not due to bacterial infection (PCR negative with universal primers) but were possibly due to viral infections or other causes. Table 1 presents the correlation between the culture and the PCR data.
TABLE 1.
Results of culture and PCR for the 47 MEE samples
| Bacterium or bacteria detected | No. of samples positive by the following test/no. of samples tested (%)
|
|
|---|---|---|
| Culture | PCR | |
| Bacteria | 10/47 (21.3) | 35/47 (74.5) |
| Haemophilus | 9/10 (90) | 33/35 (94.3) |
| M. catarrhalis | 2/10 (20) | 10/35 (28.6) |
| Streptococcus | 0/10 (0) | 3/35 (8.6) |
| Other bacteria | 0/10 (0) | 2/35 (5.7) |
| Mixed | ||
| Haemophilus, M. catarrhalis, and Streptococcus | 0/10 (0) | 1/33 (3.0) |
| Haemophilus and M. catarrhalis | 1/10 (10) | 9/33 (27.3) |
| Haemophilus and Streptococcus | 0/10 (0) | 2/33 (6.1) |
| Streptococcus and M. catarrhalis | 0/10 (0) | 0/33 (0) |
| Total mixed | 1/10 (10) | 12/33 (36.4) |
DISCUSSION
In the present study, we used a two-step PCR-based assay in order to detect Haemophilus, M. catarrhalis, and Streptococcus in MEE samples aspirated from Lebanese children with OME. The efficiency of the assay for the detection of bacteria was evaluated by comparing the PCR results to those of culture. In addition we evaluated the use of a Haemophilus-specific probe as a primer in an attempt to detect this organism by PCR alone without resorting to Southern hybridization.
Seventy-four percent of the MEE samples tested were PCR positive in tests with the universal primers. In addition, all except two of these samples were PCR positive in tests with genus- or species-specific primers. Only 21% of the samples yielded bacterial growth. This denotes that there is a 53% discordance between the PCR and the culture data. This observation correlates with the findings of Post et al. (9), who found that about 48% of the MEE samples tested were PCR positive for H. influenzae, M. catarrhalis, or S. pneumoniae but culture negative for these organisms. This implies that PCR is more efficient than culture in detecting bacteria in MEE samples. However, since PCR amplifies DNA from viable or dead bacteria, residual DNA from a previous episode of AOM may have been detected as well. However, a study conducted by Post et al. (8) with chinchilla models has shown that the bacterial DNA in MEE samples disintegrates within 2 days following bacterial death. This finding suggests that the bacterial DNA detected by PCR in our MEE samples may be originating from viable bacteria involved in disease and not residual DNA persisting from a previous episode of AOM unless the bacteria died only 2 days before the date of operation and sample aspiration. The fact that these bacteria, if they were viable, were not recovered by culture may be accounted for by the inherent limited sensitivity of culture assays in general and/or by the low bacterial count (low numbers of CFU) found in MEE samples. The presence of low bacterial counts on the order of 104 CFU/ml and less in MEE samples aspirated from OME patients has already been documented (13). Such low bacterial counts may in turn be due to the bactericidal effect of the antibiotics that are usually administered to patients prior to operation and to the fact that OME, unlike AOM, is due to a low-grade subclinical infection rather than an active one. According to Klimek et al. (6), amoxicillin administered to patients with chronic OME reaches levels of 6.2 μg/ml in MEE samples. Such high levels are bactericidal enough to lower the number of viable CFU in MEE samples, especially given the fact that the antibiotics were administered to the patients whom we considered for at least 10 days.
The two-step PCR-based assay which uses in step 1 universal primers for the detection of bacterial DNA and in step 2 genus- or species-specific primers for the detection of specific bacterial DNA offered a better approach for the identification of bacteria than existing PCR-Southern hybridization combinations. This is due to the fact that if bacterial DNA is present in a given MEE sample, then the first step would detect this DNA, while the second step would specifically define its identity within a shorter period of time (12 h) than PCR-Southern hybridization (2 days). This approach provided a rapid, cost-effective, and less cumbersome procedure for the detection of bacteria at the genus or species level.
For the specific PCR detection of Haemophilus in MEE samples, a sequence originally used as a Haemophilus-specific probe by Greisen et al. (3) was used as a Haemophilus-specific primer in the two-step PCR-based assay. This probe (RDR125) was used along with one of the universal primers (DG74) to amplify a 124-bp fragment within the 370-bp sequence of Haemophilus. The primer was shown to be genus specific since it did not amplify the DNA of other genera. The high prevalence (94%) of Haemophilus in our samples necessitated confirmatory testing by using Haemophilus-specific primer pair F1-R1; these primers flank a 351-bp region in the P6-encoding gene of Haemophilus. All samples were positive. This 100% concordance in the PCR results obtained with the combination of RDR125 and DG74 and those obtained with the F1-R1 primer pair shows the utility of RDR125 as a primer for the identification of Haemophilus. Streptococcus-specific primer STR1, which we designed, was used with DG74 to amplify a 105-bp fragment. The primer’s sequence was found to be unique for the genus since DNA from closely related genera as well as the genera tested in this study was not amplified when the primer was used along with DG74. Finally, the specific primers used by Post et al. (9) for the detection of M. catarrhalis in the two-step PCR generated a 140-bp amplicon. The specificities of these primers were also assessed by testing them with the DNA of a number of other genera.
In conclusion, the two-step PCR-based assay proved to be more sensitive than culture. In addition, it appeared to be more rapid, cost-efficient, and less cumbersome than available PCR-Southern hybridization-based assays. The use of a Haemophilus-specific probe as a primer proved to be efficient and allowed us to avoid cumbersome blotting and hybridization procedures. The data generated by the two-step PCR indicate that Haemophilus is the most prevalent bacterium among the bacteria for which we tested, followed by M. catarrhalis and Streptococcus. Mixed infection was also observed. Future work attempting to detect these three bacteria as well as other bacteria in a larger, statistically significant number of MEE samples is needed in order to determine the prevalence of the bacterial etiology of OME in Lebanon.
ACKNOWLEDGMENTS
We thank the Lebanese National Council for Scientific Research for financial support and the Hotel Dieu de France and Makassed hospitals for provision of some MEE samples.
REFERENCES
- 1.Bluestone C D, Stephenson J S, Martin L M. Ten-year review of otitis media pathogens. Pediatr Infect Dis J. 1992;11:S7. doi: 10.1097/00006454-199208001-00002. [DOI] [PubMed] [Google Scholar]
- 2.Brook I, Yocum P, Shah K, et al. Aerobic and anaerobic bacteriological features of serous otitis media in children. Am J Otolaryngol. 1983;4:389–392. doi: 10.1016/s0196-0709(83)80044-1. [DOI] [PubMed] [Google Scholar]
- 3.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoekelman R A. Infectious illness during the first year of life. Pediatrics. 1977;59:119. [PubMed] [Google Scholar]
- 5.Klein, J. O., M. Tos, B. Hussl, R. F. Naunton, M. Ohyama, and P. B. Van Cauwenberge. 1989. Definition and classification. Ann. Otol. Rhinol. Laryngol. 98(Suppl. 139):10. [PubMed]
- 6.Klimek J J, Nightingale C, Lehman W B, et al. Comparisons of concentrations of amoxicillin and ampicillin in serum and middle-ear fluids of children with chronic otitis media. J Infect Dis. 1977;135:999. doi: 10.1093/infdis/135.6.999. [DOI] [PubMed] [Google Scholar]
- 7.Loutit J S, Tompkins L S. PCR detection of Legionella pneumophila and L. dumoffii in water. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: American Society for Microbiology; 1993. pp. 261–263. [Google Scholar]
- 8.Post J C, Aul J J, White G J, et al. PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent, viable bacteria in the chinchilla model of otitis media. Am J Otolaryngol. 1996;17:106–111. doi: 10.1016/s0196-0709(96)90005-8. [DOI] [PubMed] [Google Scholar]
- 9.Post J C, Preston R A, Aul J J, Pettigrew M L, White J R, Anderson K W, Wadowsky R M, Reagan D R, Walker E S, Kingsley L A, Magit A E, Ehrlich G D. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA. 1995;273:1598–1604. [PubMed] [Google Scholar]
- 10.Senturia, B. H., C. D. Bluestone, J. O. Klein, D. J. Lim, and J. L. Paradise. 1980. Report of the Ad Hoc Committee on Definition and Classification of Otitis Media and Otitis Media with Effusion. Ann. Otol. Rhinol. Laryngol. 89(Suppl. 68):3.
- 11.Senturia B H, Gessart C F, Carr C D, Baumann E S. Studies concerned with tubotympanitis. Ann Otol Rhinol Laryngol. 1958;67:440–467. doi: 10.1177/000348945806700213. [DOI] [PubMed] [Google Scholar]
- 12.Shurin, P. A., S. I. Pelton, A. Donner, and J. O. Klein. 1997. Persistence of middle-ear effusion after acute otitis media in children. 300:1121–1123. [DOI] [PubMed]
- 13.Stenfors L E, Raisanen S. Quantitative analysis of the bacterial findings in otitis media. J Laryngol Otol. 1990;104:749–757. doi: 10.1017/s0022215100113842. [DOI] [PubMed] [Google Scholar]
- 14.Ueyama T, Kurono Y, Shirabe K, Takeshita M, Mogi G. High incidence of Haemophilus influenzae in nasopharyngeal secretions and middle ear effusions as detected by PCR. J Clin Microbiol. 1995;33:1835–1838. doi: 10.1128/jcm.33.7.1835-1838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Cauwenberge P B. Definition and character of acute and secretory otitis media. Adv Otorhinolaryngol. 1988;40:38–46. doi: 10.1159/000415671. [DOI] [PubMed] [Google Scholar]
- 16.Watson P, Voss L, Barber C, Aickin R, Bremner D, Lennon D. The microbiology of Chronic Otitis Media with Effusion in a group of Auckland children. N Z Med J. 1996;109:182–184. [PubMed] [Google Scholar]