Abstract
Background
High-grade gliomas (HGG) are aggressive brain tumors associated with short median patient survival and limited response to therapies, driving the need to develop tools to improve patient outcomes. Patient-derived xenograft (PDX) models, such as mouse PDX, have emerged as potential Avatar platforms for personalized oncology approaches, but the difficulty for some human grafts to grow successfully and the long time required for mice to develop tumors preclude their use for HGG.
Methods
We used a rapid and efficient ex-ovo chicken embryo chorioallantoic membrane (CAM) culture system to evaluate the efficacy of oncologic drug options for HGG patients.
Results
Implantation of fresh glioma tissue fragments from 59 of 60 patients, that include difficult-to-grow IDH-mutated samples, successfully established CAM tumor xenografts within 7 days, with a tumor take rate of 98.3%. These xenografts faithfully recapitulate the histological and molecular characteristics of the primary tumor, and the ability of individual fragments to form tumors was predictive of poor patient prognosis. Treatment of drug-sensitive or drug-resistant xenografts indicates that the CAM-glioma assay enables testing tumor sensitivity to temozolomide and carboplatin at doses consistent with those administered to patients. In a proof-of-concept study involving 14 HGG patients, we observed a correlation of 100% between the CAM xenograft response to temozolomide or carboplatin and the clinical response of patients.
Conclusion
The CAM-glioma model is a fast and reliable assay that has the potential to serve as a complementary model to drug discovery and a real-time Avatar platform to predict the best treatment for HGG patients.
Keywords: Avatar model, chick chorioallantoic membrane assay, glioblastoma, patient-derived xenograft model, personalized medicine
Key Points.
- Patient glioma tumor fragments undergo fast and robust expansion on CAM assay.
- Xenografts preserved the pathological characteristics of the parental tumor.
- CAM-glioma model has a high rate of prediction of patient response to treatments.
Importance of the Study.
Despite recent advances in the treatment of HGGs, the prognosis has not improved significantly over the past few decades. A major problem is the lack of rapid and reliable methods to develop new and more efficient drug therapies and to predict individual patient response. Here we demonstrate that the glioma CAM Avatar is a time-efficient model that recapitulates the important characteristics of glioma tumors. Furthermore, our proof-of-concept study shows that this model can accurately predict patient clinical outcomes in terms of progression-free and overall survival and, more importantly, chemoresistance/sensitivity of the tumor to patient’s drug regimen. These findings underline the potential of this model to be used for drug discovery and implementation of personalized medicine to improve survival of HGG patients.
High-grade gliomas (HGGs), which arise from neural stem cells or oligodendrocyte precursor cells in the brain, are malignant tumors that invade the surrounding brain tissue. They are traditionally classified based on histologic/genetic criteria and grade that includes oligodendroglioma IDH-mutant and 1p/19q codeleted (OLIG, CNS WHO grade 3), astrocytoma IDH-mutant (ASTRO, CNS WHO grade 3 or 4), and glioblastoma IDH-wild type (GBM, CNS WHO grade 4), which is the most aggressive form of the disease, accounting for the majority (60% to 70%) of all HGGs.1
All these malignant brain tumors are almost infallibly associated with recurrence and progression, regardless of the subtype. Aggressive therapeutic ionizing radiation in combination with alkylating chemotherapy, usually temozolomide (TMZ), is the mainstay of nonsurgical therapy for gliomas.2 Unfortunately, resistance to treatment almost inevitably occurs, leading to second-line interventions that include reirradiation, alkylating chemotherapy rechallenge, alternative chemotherapeutic approaches, and/or clinical trials. Despite these multimodal treatment regimens, the prognosis of patients with HGGs remains poor. For example, the median overall survival (OS) of patients with glioblastoma is only 15 months with a median progression-free survival (PFS) of 6.2–7.5 months.2,3 Advances in gene expression profiling and genomic sequencing have raised hope that cancer driver genes and mutations shared by a large number of tumors could be identified and targeted to improve tumor response and reduce unnecessary drug toxicity.4 However, precision oncology in gliomas has not proven successful because of the high degree of molecular and genetic heterogeneity between individual tumors and within tumors, and the lack of identification of clear gene targets.5 The genetic variability of HGGs, variable response to chemotherapeutic drugs, and poor clinical outcomes have encouraged the development of individualized patient models for drug discovery and personalized therapy.
There has been increasing interest in the use of patient-derived xenograft (PDX) models for basic and translational research in oncology.6 Traditional PDX models typically involves implanting patient tumor cells or tumor fragments into immunodeficient mice. Such models are a mainstay in preclinical and co-clinical drug assessment due to their capacity to maintain the principal histopathological and genetic characteristics of the original tumor better than cell lines or genetically engineered mice.7 Moreover, the sensitivity/resistance of PDXs to anticancer drugs was shown to correlate closely with clinical response in patients from whom they were derived.8 These favorable outcomes motivated the use of PDXs as personalized Avatars, where a patient and a derivative PDX are treated with the same therapeutic regimen.9,10 PDX models have been successfully established in most cancer types including neurological cancers such as medulloblastoma,11 ependymoma,12 and HGG.13 Despite their usefulness, traditional PDX models are associated with ethical and financial limitations. More importantly, there are strong limitations for their direct application in fast-progressing cancers such as HGGs. A period of time that can vary from 4 to 8 months is usually required for the establishment and amplification of sufficient tissues to assess drug sensitivity, so many patients are set to die before results become available.14 In addition, unless using a technically challenging orthotopic model of brain tumors, the engraftment rate of glioma fragments in mice is very low. This leads to clonal selection and replacement of the human tumor microenvironment by mouse components, during the multiple passages in mice necessary for tissue amplification, two factors that can alter drug responses.15,16 Thus, there is a critical need to develop alternative approaches for drug discovery and to drug sensitivity assessment in HGG patients.
The chicken embryo chorioallantoic membrane (CAM) is a simple, highly vascularized extraembryonic membrane that functions as the respiratory organ of avian embryos. Due to the immunodeficient nature of the chick embryo and the abundant vascularization of the CAM, this in vivo model has proven to be very useful for the implantation of various types of cancer cell lines and patient-derived tumor cells or explants.17,18 The CAM-PDX model is well suited for rapid tumor engraftment and subsequent testing of therapeutic approaches. As such, the duration of tumor amplification and drug regimen screening is reduced to 7 days. In addition, several studies have suggested the potential of this assay to assess the chemosensitivity of brain tumors.19–21 An ethical advantage of the CAM assay is that the CAM is not innervated, and experiments are terminated before the establishment of pain perception in the embryo.22 All these features indicate that the CAM model has the potential to become a rapid and efficient precision medicine platform for HGGs.
In this study, we report the development of a personalized CAM avatar model that retains crucial characteristics of the original tumor specimen and reliably predicts chemotherapeutic drug sensitivity for high-grade gliomas.
Materials and Methods
Collection of Patient Tumor Specimens and Tissue Preparation
Patient glioma tissues were collected between the years 2016 and 2020, according to a protocol approved by the research ethics committee of the Centre hospitalier universitaire de Sherbrooke (ID#11-088). Written informed consent was obtained from all patients. Pathological diagnosis was established by a neuropathologist using the histologic and genetic criteria described by the 2021 World Health Organization (WHO) classification of brain tumors.23,24 Fresh tumor specimens were collected and prepared as detailed in Supplementary Methods.
Glioma Cell Lines
The glioblastoma cell lines U-87 MG and LN-18 were obtained from the American Type Culture Collection and cultured as detailed in Supplementary Methods.
Establishment of the CAM Xenograft Model
Fertilized eggs from white leghorn chickens were obtained from the Couvoir Boire et Frères Inc. (Wickham, QC, Canada). The ex-ovo culture was performed in a standard incubator at 37°C as previously described.25 Between embryonic day 8 and 10, fragments of freshly resected tumor specimens or suspensions of glioma cell lines were implanted on CAM as detailed in Supplementary Methods. On day 16, chick embryos were euthanized by decapitation. Vascularized tumor masses with no visible signs of necrosis were considered successfully engrafted and were removed from the CAM. Xenograft volumes were calculated using the formula (Dd2/2) and tissues were either snap-frozen in liquid nitrogen and stored at −80°C for DNA extraction or fixed in formalin for 24 hours and embedded into paraffin for histopathological analysis or recut into 1–2 mm diameter fragments for reimplantation.
Histology and Immunostaining
Hematoxylin and eosin (HE) and immunohistochemistry (IHC) staining of original tumors and CAM xenografts was performed according to the standard HE and avidin-biotin immunoperoxidase complex technique respectively as detailed in Supplementary Methods.
Genomic Analysis
DNA was extracted from frozen original tissues and CAM tumors at the Rnomics Platform at the Université de Sherbrooke (Qc, Canada). Whole-exome sequencing was performed at the Mayo Clinic (Rochester, MN) using the SureSelect capture kit (Agilent). Chicken sequencing reads were removed using Xenome prior to mutation calling.
CAM Xenograft Drug Sensitivity Assays and Clinical Evaluation
Two days after the implantation of tumor fragments or glioma cell lines, drugs were injected into the CAM vasculature at the following concentrations: 8 mg/kg of carboplatin (Centre hospitalier universitaire de l’Université de Sherbrooke, QC, Canada) or 4 mg/kg of TMZ (Sigma, cat#T2577). On day 16, chick embryos were euthanized, and xenograft volumes calculated. Drug treatments were considered effective when they resulted in at least a 30% decrease (P < .05) in the volume of the lesions compared with the control xenograft group. Patients were followed monthly (for carboplatin treatment) or every 1–4 months (for TMZ treatment) by magnetic resonance imaging scan. According to the Revised Assessment in Neuro Oncology (RANO) criteria,26 patient response was classified as either complete response (CR), partial response (PR), or stable disease (SD), which are generally considered therapeutically beneficial in a clinical setting, or progressive disease (PD).
Statistical Analysis
The GraphPad software (version 9.3.1) was used for statistical analysis. Significance was assessed by an unpaired Student’s t-test (Mann-Whitney), a one-way ANOVA (Kruskal-Wallis), or a Gehan-Breslow-Wilcoxon test as indicated in figure legends. A P value smaller than .05 was considered significant.
Results
Establishment of a High-efficiency Glioma CAM-PDX Model that Retains the Histopathological and Molecular Characteristics of the Original Tumor
To develop the glioma CAM-PDX model, we used an ex-ovo chicken embryo culture system to allow broad access to the CAM for external manipulations, including tumor implantation and drug treatments. Surgically resected oligodendroglioma, astrocytoma, and glioblastoma tissues were obtained from a total of 60 individual patients undergoing treatment at the Centre hospitalier universitaire de Sherbrooke between April 2016 and December 2020. The demographic and clinical characteristics of these patients are summarized in Supplementary Table 2, and the detailed clinicopathological characteristics of each patient are shown in Supplementary Table 3. The median age in this cohort is 53 years with a 2:1 male to female ratio. We also observed a prevalence of HGGs (CNS WHO grade 4) (68.3%) as opposed to lower grade tumors (CNS WHO grade 2–3) (31.7%). Furthermore, primary and recurrent tumors are present in similar proportions (55% and 45%, respectively).
To preserve the morphological characteristics of the original tumor as accurately as possible, fresh tumor explants were used.27 The tissue was cut into fragments of less than 5 mm3 (between 1 and 2 mm in diameter) (Figure 1A), which were implanted on embryonic development day (EDD) 9 or 10 and cultured on CAMs for an additional 6 or 7 days as illustrated in Figure 1B. Initial studies indicated that these culture conditions resulted in optimal growth (Figures 1C, D) and greater uptake (Figures 1E, F) of the xenografted fragments. Using this optimized model, 59 out of 60 glioma specimens established xenograft tumors on CAM, corresponding to a success rate of 98.3% (Figure 1G). We also observed an uptake of 74.4% for individual tumor fragments of the specimen (Figure 1G). An example of successfully implanted CAM-glioma tumor is shown in Figure 1H. HE staining of xenografts reveal a network of capillaries filled with nucleated chicken erythrocytes28 as shown in Figure 1I. There was no association between tumor type (primary or recurrent), IDH status (wild type or mutated) or the cell type/grade (oligodendroglioma, astrocytoma, or glioblastoma) and the extent of glioma fragment engraftment (Figure 1J, K and L).
Figure 1.
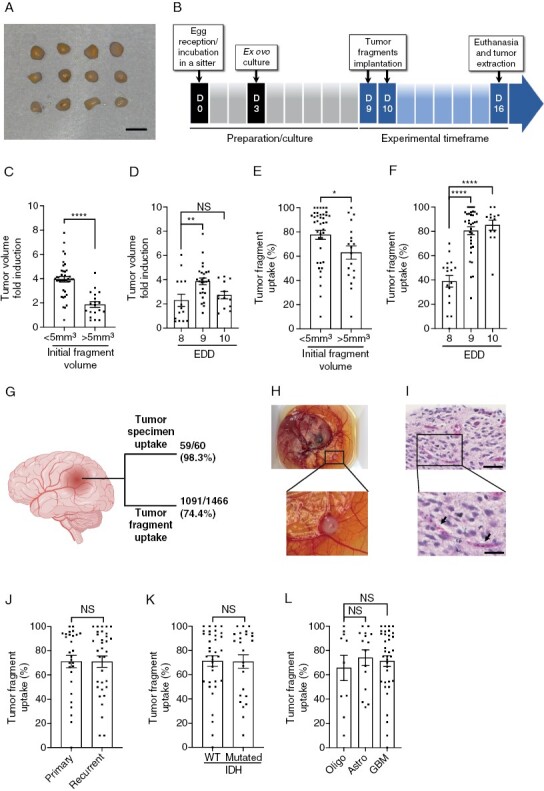
Optimization of the glioma CAM-PDX model. (A) Representative images of the tumor fragments used for implantation. Scale bar = 2mm. (B) Timeline of the procedure. (C and D) Xenograft volume 6 or 7 days post-implantation based on initial fragment size (C) and embryonic development day (EDD) of implantation (D). (E and F) Tumor uptake as a function of initial fragment size (E) and EDD of implantation (F). (G) Tumor specimen and tumor fragment uptake on CAM. Created with BioRender.com (H) Representative image and (I) representative H&E staining of a CAM-glioma xenograft. Arrows show chicken blood vessels. Scale bar = 50 μm. Zoom in scale bar = 25 μm. (J–L) Tumor take rate according to primary or recurrent tumors (J), IDH status (K), or cell types/grades (L) of gliomas. Oligo: oligodendroglioma, Astro: astrocytoma, GBM: glioblastoma. Values are expressed as mean ± SEM. *P < .05, **P < .01, ****P < .0001, (C,E,J,K) Mann-Whitney test, (D,F,L) Kruskal-Wallis test.
The ability of the CAM to support the growth of glioma tissue fragments was evaluated by tumor volume measurements before and after 7 days of implantation. Xenografts were found to undergo robust expansion on the CAM, with fold induction between 1.5 and 7.8 of the mean volume of the developed tumors (Supplementary Figure S1A). A significant increase in fold induction was observed for recurrent tumor compared to primary tumors (Supplementary Figure S1B). In contrast, no significant difference was observed according to IDH status (Supplementary Figure S1C), or for the 3 different grades/types of gliomas implanted (Supplementary Figure S1D). In addition, patient-derived tumor fragments that successfully engrafted onto CAM could be serially passaged across multiple recipient eggs for up to 3 amplification cycles (Supplementary Figure S1E). Together, these results demonstrate that the CAM-based xenograft model is a robust approach for the amplification of patient glioma tumors.
We next sought to determine whether the tumors grown on CAM retained some important characteristics of the original tumor. Histopathologic examination by H&E staining demonstrated morphologic features characteristic of HGGs, including necrosis and pleomorphic astrocytic cells with marked nuclear atypia and mitosis in xenografted tumors, similar to those observed in the original tumors (Figure 2A). In some cases, the presence of gemistocytic cells, multinucleated giant cells, and cells with abundant vacuoles were also observed in both tissue types (Supplementary Figure S2). Specific staining for GFAP and vimentin, two important markers of malignant gliomas,29,30 and two neuronal markers synaptophysin and neuro-specific enolase (NSE) showed similar expression between the original and implanted tumor tissues. In addition, the relative abundance of stem-like cells, identified by CD44 and nestin, and actively proliferating cells, identified by Ki67 was also comparable between the two sample types (Figure 2A).
Figure 2.
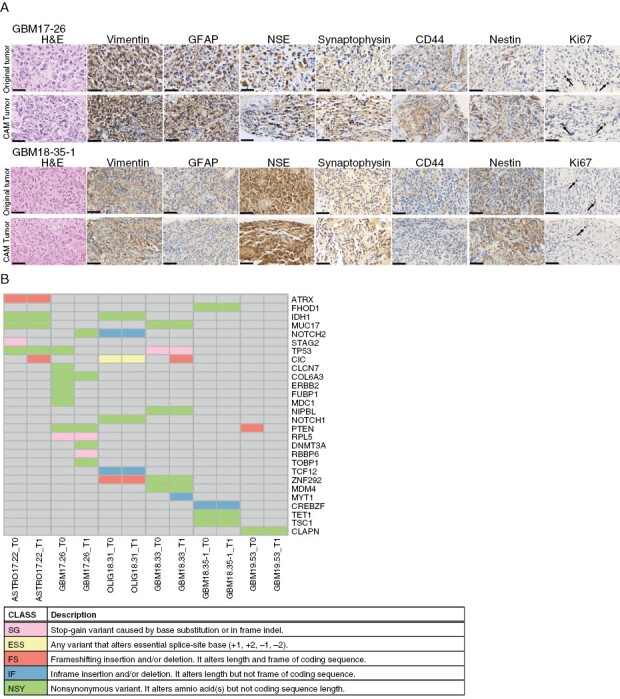
Histopathological and genetic characteristics of original and xenografted tumors. (A) Representative images of H&E, vimentin, GFAP, CD44, nestin, synaptophysin, NSE, and Ki67 staining of the original and xenografted tumors; arrows show Ki67 positive cells; N = 5. Scale bar = 50 μm. (B) Mutations in 28 glioblastoma-associated genes identified by whole-exome sequencing of parental (T0) and xenografted (T1) tumors; N = 6. ASTRO: astrocytoma, GBM: glioblastoma, OLIG: oligodendroglioma.
To determine whether CAM-amplified tumors maintain genomic alterations found in patient tissues, we performed whole-exome sequencing of 6 HGG tumors and matched early-passage (P1) CAM xenografts. Analysis of genomic alterations in 4,927 genes indicated a high percentage (95.4%–97.7%) of identity between the parental and the xenograft tumors (Figure 2B and Supplementary Table 4), with the exception of patient GBM17-26, who also displayed the highest rate of total mutations (Supplementary Table 5). As expected, commonly mutated genes were found in glioma-related pathways, such as IDH1, TP53, and PTEN, suggesting that cancer-driven mutations are well conserved in the CAM model. These results indicate that, in the CAM model, glioma xenografts retain many of their original histological and genomic characteristics.
The CAM-glioma PDX Model Enables the Assessment of Sensitivity/Resistance to TMZ and Carboplatin
We first identified drug doses that distinguish sensitive from resistant gliomas by treating CAM xenografts derived from drug-sensitive U-87 MG or drug-resistant LN-18 human glioblastoma cell lines31–33 with escalating doses of TMZ or carboplatin. TMZ was chosen as it is part of the standard treatment for newly diagnosed HGGs34 and carboplatin as it is the most frequently used second-line treatment at the CHUS because of clinicians’ expertise in intra-arterial (intra-carotid) infusion of the compound that enhances its efficacy.35,36 Maximum growth inhibition of U-87 MG-derived tumor xenografts was observed after IV injection of 4 mg/kg TMZ or 8 mg/kg carboplatin (Figure 3A, B), which is comparable to the usual doses given to HGG patients2,35 (Supplementary Table 6). In contrast, and as expected, drug-resistant LN18-derived xenografts, failed to respond to TMZ or carboplatin (Figure 3C, D). Using these optimized drug doses, we next evaluated whether patient-derived glioma xenografts exhibit differential drug responses. Tumor fragments from 9 HGG patients were implanted on CAM and the developed tumors treated with TMZ or carboplatin. A differential drug response was observed in these xenografts (Figure 3E-G and Supplementary Figure 3). For example, while GBM20-65 xenografts responded to both TMZ and carboplatin (Figure 3E), GBM18-35-1 responded only to carboplatin (Figure 3F) whereas ASTRO19-48 was insensitive to both treatments (Figure 3G). Drug-induced inhibitory effects were associated with higher levels of caspase-3 activation (Figure 3H, I). These results suggest that the CAM-PDX model is appropriate for assessing sensitivity/resistance to chemotherapeutic drugs commonly used to treat HGGs at doses corresponding to the ones administered to patients.
Figure 3.
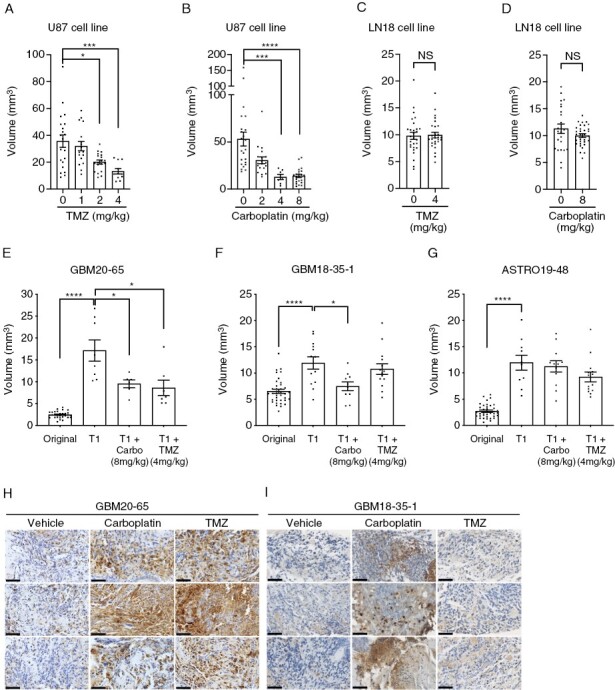
Effect of drug treatment on glioma xenografts grown on CAM. (A–D) Tumor volume of xenografts derived from U-87 MG or LN18 cell lines treated with different doses of (A and C) temozolomide (TMZ) (N = 3) or (B and D) carboplatin (Carbo) (N = 3). (E–G) Tumor volume of original glioma fragments (Original) or CAM xenografts (T1) treated with vehicle, carboplatin or TMZ for patient GBM20-65 (E), GBM18-35-1 (F), and ASTRO19-48 (G) and (H,I) representative images of cleaved caspase 3 staining for three separate xenografts per group (n = 3–7). GBM: glioblastoma, ASTRO: astrocytoma. Scale bar = 50 μm. Bars = mean ± SEM. *P < .05, **P < .01, ***P < .001, ****P < .0001, (A and B) Kruskall-Wallis test, (C–G) Mann-Whitney test.
CAM-Glioma PDX as a Predicting Model for Clinical Outcomes and Patient Response to Anticancer Drugs
We next evaluated the concordance between tumorigenicity in the CAM-PDX model and patient clinical outcome. Kaplan-Meier analysis of PFS curves indicates that recurrence tends to occur later in patients with low tumor fragment uptake on CAM compared to those with a high uptake (Figure 4A), whereas a significant association was observed between CAM-PDX engraftment and overall patient survival (Figure 4B). These results indicate that higher CAM-PDX tumorigenicity is associated with poorer prognosis in glioma patients.
Figure 4.
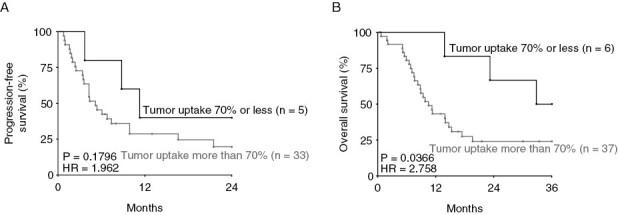
Prognostic value of the ability for glioma specimens to form xenografts in the CAM model. Kaplan-Meier analysis of (A) progression-free and (B) OS in glioma patients with high > 70% (n = 33–37) and low < 70% (n = 5–6) tumor fragment uptake on CAM, a cut-off chosen for maximal discrimination between high- and low-grade patients. HR: hazard ratio (logrank), Gehan-Breslow-Wilcoxon test.
To determine whether the CAM-PDX model can be used as an Avatar model to predict clinical responses in patients with glioma, we retrospectively evaluated treatment response in 15 HGG patients and their corresponding CAM-PDX models that were treated with optimized doses of TMZ, carboplatin, or vehicle alone. Treatments were considered effective in CAM when they resulted in a >30% and significant inhibition of tumor volume, according to the internationally standardized clinical Response Evaluation Criteria In Solid Tumors.37 These criteria were adapted to the CAM-PDX model by using the control group as a reference. Patient response to treatment was assessed according to the RANO criteria,26 which is the current standard for assessing response to treatment for HGGs. Patient response was classified as either CR, PR, or SD, which are generally considered therapeutically beneficial in a clinical setting, or PD. Table 1 summarizes the chemosensitivity profiles of these 14 glioma patients who generated 15 matched CAM-PDX models, 2 of which were derived from the same patient. Overall, the positive and negative predictive value was 100% (5/5 and 9/9 samples, respectively). As an example, the lack of effect of a drug on CAM xenograft growth corresponded to tumor progression in the patient treated with the same drug (Figure 5A, B). In contrast, stabilization of the patient’s pathology was observed in cases where the drug significantly inhibited tumor growth in CAM-PDX (Figure 5A, C). It should be noted that we could not clearly define the radiographic response for the ASTRO19-48 patient, in keeping with the finding that response assessment in astrocytoma IDH-mutant, CNS WHO grade 2–3 could be problematic using the current modified RANO criteria.38
Table 1.
Comparison of Patient and Xenografts Chemosensitivity Profiles
| Identification Number | Diagnosis | Tumor Type | IDH Status | Treatment | Patient Clinical Response | PDX Inhibitory Response |
|---|---|---|---|---|---|---|
| GBM18-33 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Recurrent | Wild Type | Carboplatin | PD | − |
| GBM18-35-1 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Primary | Wild Type | Temozolomide | PD | − |
| GBM18-35-2 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Recurrent | Wild Type | Carboplatin | PD | − |
| GBM18-38 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Recurrent | Wild Type | Carboplatin | PD | − |
| GBM18-39 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Recurrent | Wild Type | Carboplatin | PD | − |
| GBM19-42 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Primary | Wild Type | Temozolomide | PR | + |
| ASTRO19-43 | Astrocytoma, IDH-mutant, CNS WHO grade 4 | Recurrent | Mutated | Carboplatin | PD | − |
| OLIG19-45 | Oligodentroglioma, IDH-mutant, CNS WHO grade 3 | Recurrent | Mutated | Carboplatin | PD | − |
| ASTRO19-47 | Astrocytoma, IDH-mutant, CNS WHO grade 4 | Recurrent | Mutated | Temozolomide | SD | + |
| ASTRO19-48 | Astrocytoma, IDH-mutant, CNS WHO grade 3 | Recurrent | Mutated | Carboplatin | ? | − |
| OLIG19-50 | Oligodendroglioma, IDH-mutant, 1p/19q codeleted, CNS WHO grade 3 | Primary | Mutated | Temozolomide | CR | + |
| GBM20-60 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Primary | Wild Type | Temozolomide | PD | − |
| ASTRO20-63 | Astrocytoma, IDH-mutant, CNS WHO grade 4 | Recurrent | Mutated | Carboplatin | SD | + |
| GBM20-66 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Recurrent | Wild Type | Carboplatin | PD | − |
| GBM20-68 | Glioblastoma, IDH-wild type, CNS WHO grade 4 | Primary | Wild Type | Temozolomide | CR | + |
Abbreviations: GBM, glioblastoma; ASTRO, astrocytoma; OLIG, oligodendroglioma; PD, progressive disease; SD, stable disease; PR, partial response; CR, complete response.
Figure 5.
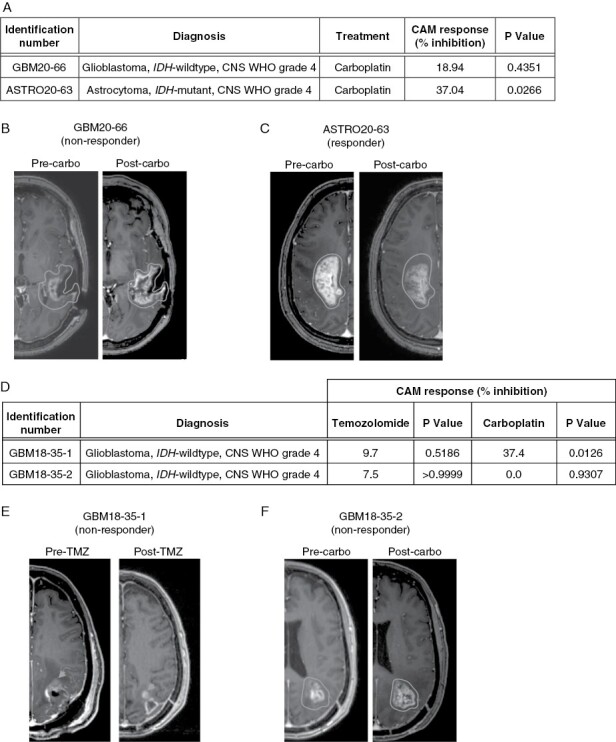
CAM-glioma and clinical response to chemotherapeutics. (A) Xenograft response to chemotherapeutic drugs for a non-responder (GBM20-66) and a responder (ASTRO20-63) PDX model. (B and C) Associated pre-and post-treatment MRI images for (B) GBM20-66 and (C) ASTRO20-63 patients. (D) Xenograft response to chemotherapeutic agents for two PDX models established from a single patient. (E and F) Pre- and post-treatment MRI images for the patient associated with (E) GBM18-35-1 and (F) GBM18-35-2 samples. Mann-Whitney test.
In addition, in one patient with primary glioblastoma IDH-wild type, CNS WHO grade 4, two sequential PDX models could be generated allowing longitudinal prediction of drug response (GBM18-35-1, GBM18-35-2). For the first CAM-PDX model, which was established after initial tumor resection, drug treatment of xenografted tumors showed significative tumor volume inhibition with carboplatin, whereas TMZ had no significant effect (Figure 5D). Clinically, the patient was treated with TMZ (as included in the STUPP protocol). Consistently with the lack of xenograft response to this drug, TMZ had no therapeutic benefit and the tumor progressed to a point of requiring furth 5E). In the second CAM-PDX model generated following this second surgery, carboplatin had no effect on xenograft volume (Figure 5D), which again was associated with a lack of clinical response of the patient to this drug (Figure 5F). Taken together, these results further support the idea that drug responses in CAM-PDX closely mimic patient clinical outcomes. More so, they highlight the potential importance of choosing a personalized treatment in first-line therapy.
Discussion
Despite recent advances in human glioma therapy, the prognosis has not improved significantly over the last several decades. One important issue is the lack of rapid and reliable methods for developing new and more efficient drug therapies and for predicting individual patient response. Here we demonstrate that the glioma CAM Avatar is a time-efficient model that faithfully recapitulates the histological and genetic characteristics of glioma tumors. Furthermore, our proof-of-concept study shows that this model can accurately predict patient clinical outcomes in terms of progression-free and OS and, more importantly, chemoresistance/sensitivity of the tumor to TMZ and carboplatin.
For this study, fresh tissue fragments from 60 glioma patients were implanted onto CAMs and 59 CAM-PDX models were successfully established. The engraftment rate was 100% for astrocytomas and oligodendrogliomas and 97.1% for glioblastomas for an average of 98.3%. This engraftment rate in the CAM-PDX model was higher than those observed in heterotopic mouse PDX glioma models, which ranged from 38% to 69%,13,39 but more comparable to, yet higher than, the orthotopic model of brain tumors, which showed a 76%–90% success rate.40,41 Of note, all 25 IDH-mutated samples successfully engrafted in the CAM model. This finding is of particular interest because IDH mutations are present in a high proportion of gliomas and a long-standing challenges in the development of glioma PDX models is the almost complete failure of IDH1-mutated specimens to engraft when transplanted heterotopically or orthotopically into mice.13,42,43 Consequently, it has been very difficult to establish appropriate models for preclinical study and drug testing for this molecular subtype of glioma. In fact, several small molecule inhibitors targeting the mutated form of IDH1 have been generated and are in various phases of development, although very few have reached phase III clinical trials. The CAM-PDX model, which allows the engraftment of tumors from patients with different types of gliomas regardless of their IDH status, would therefore be well suited for the evaluation of new therapeutic strategies designed to target the different genetic types of gliomas.
The mouse PDX model is known to preserve a high degree of concordance in histological features and genomic variation between primary tumors and corresponding xenografts and is therefore viewed as the most reliable for tumor biological research and is the gold standard in preclinical studies.40,44 Our histological study indicates that CAM xenografts also retain the cellular and morphologic heterogeneity as well as features typical of HGGs. For instance, features of the original tissue such as pleomorphic astrocytic cells or cells expressing stem-like and glial markers also populated the matching CAM-PDX. Furthermore, analysis of 6 whole-exome sequencing data showed that 91% of mutations, including the IDH status, were shared between original tissue samples and their CAM-amplified xenografts. The few differences between the original tumor and xenografts could be attributed to regional intratumoral heterogeneity, a hallmark of GBM.4,45 This may be particularly true for the PDX GBM17-26 sample, which has the lowest percentage of gene mutation retention in xenografts (3 out of 8) and the highest tumor mutation burden (Supplementary Table 4). In fact, this sample was identified as a giant cell glioblastoma, a rare variant of glioblastoma IDH-wild type characterized by frequent TP53 mutations, a marker associated with GBM subclonal heterogeneity.46,47 These results highlight that, while CAM xenografts generally retain key features of the parental tumor, further studies will be needed to define whether this GBM variant should be included in an eventual glioma CAM-PDX precision medicine platform.
Preclinical models are essential tools to study cancer development and drug screening. However, currently used models, such as organoids, cell line-derived xenografts, or patient-derived xenografts in zebrafish or murine models, all have certain limitations. For example, generating a full cohort of PDX mice for in vivo drug screening can take 4–8 months. Therefore, when used in a clinical setting, tumor progression or even death will often occur before screening results are available. Even though the zebrafish model is much more rapid, the required incubation temperature of 32–34°C and the normally used 48 h incubation time for drug testing may hamper the detection of some types of drug, leading to an underestimation of drug effects.48 In addition, cell line-derived xenografts are limited by poor predictability of drug sensitivity because they do not accurately reflect the genetic and biological heterogeneity of tumors,49 while organoids lack a method to generate large numbers of brain organoids with minimal inter-organoid variations. The glioma CAM-PDX model established herein can overcome some of these limitations. Xenografts grown on CAMs reproduce histological and genetic features of the original tissue while preserving its complex heterogeneity. Furthermore, the short experimental window of the CAM assay, which can be considered a limitation for long-term experiments, has proven to be an advantage because tumors can form on the CAM and respond to drug treatments in only 7 days, making this model particularly time efficient.
An additional benefit of the CAM-glioma model is the fact that the large majority of the specimens can be successfully engrafted in the first round of implantation, regardless of the glioma subtype or variant. As such, no additional propagation is required for drug response testing. This is particularly important as murine-based PDX models will eventually lose human stromal components during the multiple in vivo passages needed to amplify enough material for drug testing, so a model requiring a low number of passages is ideal for maintaining the histological integrity of the primary tumor.39,50 In addition, clonal selection and evolution can occur during serial passaging, resulting in potential divergence in drug response.15
In our proof-of-concept study, the response of glioma PDX xenografts to chemotherapy was predictive of the patients’ clinical response to these drugs in all cases tested. This overall predictive value of 100% compares favorably with values, ranging from 67% to 100%, that have been reported in literature for different types of cancer implanted in mouse models, such as breast, ovarian, and pancreatic.51,52 In addition, results from 2 sequential CAM-PDX models established from one patient whose tumor did not respond to any treatments in the clinic indicate that carboplatin treatment might have been a better choice than TMZ used as a first-line treatment for this patient. Even though further evaluation of the correlation between patient responses and corresponding PDX in a larger prospective setting will be required, these data suggest that the glioma CAM-PDX model has a high potential to become a valuable platform for modeling clinical response in patients. This possibility is of great interest as CAM test results can be available early enough (in as little as 1–3 weeks) to guide clinicians in choosing the best treatment for patients afflicted by these rapidly progressing types of brain cancer.
Supplementary Material
Acknowledgments
We deeply thank Maxime Richer for sharing his expertise in glioma pathology. We also thank Gabriel Charest for the preparation of glioma tissue samples.
Contributor Information
Martine Charbonneau, Department of Immunology and Cell Biology, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Kelly Harper, Department of Immunology and Cell Biology, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Karine Brochu-Gaudreau, Department of Immunology and Cell Biology, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Alexis Perreault, Department of Immunology and Cell Biology, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Laurent-Olivier Roy, Department of Surgery, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Fabrice Lucien, Department of Urology, Mayo Clinic, Rochester, MN, USA.
Shulan Tian, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
David Fortin, Department of Surgery, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Claire M Dubois, Department of Immunology and Cell Biology, Faculty of Medicine and Health Sciences, Université de Sherbrooke, Sherbrooke, Québec, QC J1H 5N4, Canada.
Funding
This research was supported by the Centre de Recherche du CHUS (CRCHUS) (PAFI grant), Quebec Consortium for Industrial Research and Innovation (MEDTEQ+) (16E CAM-Avatar gliomas) and Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN-2016-03928). CMD is a member of FRSQ-funded CRCHUS.
Conflict of interest statement The authors declare that there is no conflict of interest.
Authorship statement
Conception and design: M.C., D.F., and C.M.D. Performing experiments: M.C., K.H., K.B.G., A.P., and L.O.R. Genomic analysis: F.L. and S.T. Data acquisition and analysis: M.C., D.F., and C.M.D. Drafting the manuscript: M.C., L.O.R., and C.M.D. All authors reviewed and approved the manuscript.
References
- 1. Stupp R, Brada M, van den Bent MJ, Tonn J-C, Pentheroudakis G, ESMO Guidelines working group. high-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 3):iiiiii9393–iii101. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Taillibert S, Kanner AA, et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314(23):2535–2543. [DOI] [PubMed] [Google Scholar]
- 4. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Staedtke V, Dzaye OD, Holdhoff M.. Actionable molecular biomarkers in primary brain tumors. Trends Cancer. 2016;2(7):338–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams JA. Using PDX for preclinical cancer drug discovery: the evolving field. J Clin Med. 2018;7(3):E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeRose YS, Wang G, Lin Y-C, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17(11):1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hidalgo M, Bruckheimer E, Rajeshkumar NV, et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther. 2011;10(8):1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garralda E, Paz K, López-Casas PP, et al. Integrated next-generation sequencing and avatar mouse models for personalized cancer treatment. Clin Cancer Res. 2014;20(9):2476–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stebbing J, Paz K, Schwartz GK, et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120(13):2006–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shu Q, Wong KK, Su JM, et al. Direct orthotopic transplantation of fresh surgical specimen preserves CD133+ tumor cells in clinically relevant mouse models of medulloblastoma and glioma. Stem Cells. 2008;26(6):1414–1424. [DOI] [PubMed] [Google Scholar]
- 12. Yu L, Baxter PA, Voicu H, et al. A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro Oncol. 2010;12(6):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeng W, Tang Z, Li Y, et al. Patient-derived xenografts of different grade gliomas retain the heterogeneous histological and genetic features of human gliomas. Cancer Cell Int. 2020;20:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Green S, Dam MS, Svendsen MN.. Mouse avatars of human cancers: the temporality of translation in precision oncology. Hist Philos Life Sci. 2021;43(1):27. [DOI] [PubMed] [Google Scholar]
- 15. Ben-David U, Ha G, Tseng Y-Y, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49(11):1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol. 2020;13(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeBord LC, Pathak RR, Villaneuva M, et al. The chick chorioallantoic membrane (CAM) as a versatile patient-derived xenograft (PDX) platform for precision medicine and preclinical research. Am J Cancer Res. 2018;8(8):1642–1660. [PMC free article] [PubMed] [Google Scholar]
- 18. Chu P-Y, Koh AP-F, Antony J, Huang RY-J.. Applications of the chick chorioallantoic membrane as an alternative model for cancer studies. Cells Tissues Organs. 2022;211(2):222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pruss M, Dwucet A, Tanriover M, et al. Dual metabolic reprogramming by ONC201/TIC10 and 2-Deoxyglucose induces energy depletion and synergistic anti-cancer activity in glioblastoma. Br J Cancer. 2020;122(8):1146–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Power EA, Fernandez-Torres J, Zhang L, et al. Chorioallantoic membrane (CAM) assay to study treatment effects in diffuse intrinsic pontine glioma. PLoS One. 2022;17(2):e0263822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shoin K, Yamashita J, Enkaku F, et al. Chick embryo assay as chemosensitivity test for malignant glioma. Jpn J Cancer Res. 1991;82(10):1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ribatti D. The chick embryo chorioallantoic membrane (CAM). A multifaceted experimental model. Mech Dev. 2016;141:70–77. [DOI] [PubMed] [Google Scholar]
- 23. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Torp SH, Solheim O, Skjulsvik AJ.. The WHO 2021 Classification of Central Nervous System tumours: a practical update on what neurosurgeons need to know-a minireview. Acta Neurochir. 2022;164(9):2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leong HS, Steinmetz NF, Ablack A, et al. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nat Protoc. 2010;5(8):1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 27. Resau JH, Sakamoto K, Cottrell JR, Hudson EA, Meltzer SJ.. Explant organ culture: a review. Cytotechnology. 1991;7(3):137–149. [DOI] [PubMed] [Google Scholar]
- 28. Virtanen I, Kurkinen M, Lehto VP.. Nucleus-anchoring cytoskeleton in chicken red blood cells. Cell Biol Int Rep. 1979;3(2):157–162. [DOI] [PubMed] [Google Scholar]
- 29. Bleau A-M, Howard BM, Taylor LA, et al. New strategy for the analysis of phenotypic marker antigens in brain tumor-derived neurospheres in mice and humans. Neurosurg Focus. 2008;24(3–4):E28. [DOI] [PubMed] [Google Scholar]
- 30. Rutka JT, Ivanchuk S, Mondal S, et al. Co-expression of nestin and vimentin intermediate filaments in invasive human astrocytoma cells. Int J Dev Neurosci. 1999;17(5–6):503–515. [DOI] [PubMed] [Google Scholar]
- 31. Hermisson M, Klumpp A, Wick W, et al. O6-methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96(3):766–776. [DOI] [PubMed] [Google Scholar]
- 32. Happold C, Roth P, Wick W, et al. Distinct molecular mechanisms of acquired resistance to temozolomide in glioblastoma cells. J Neurochem. 2012;122(2):444–455. [DOI] [PubMed] [Google Scholar]
- 33. Huo J-F, Chen XB.. Long noncoding RNA growth arrest-specific 5 facilitates glioma cell sensitivity to cisplatin by suppressing excessive autophagy in an mTOR-dependent manner. J Cell Biochem. 2019;120(4):6127–6136. [DOI] [PubMed] [Google Scholar]
- 34. Fisher JP, Adamson DC.. Current FDA-approved therapies for high-grade malignant gliomas. Biomedicines. 2021;9(3):324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fortin D, Morin P-A, Belzile F, Mathieu D, Paré F-M.. Intra-arterial carboplatin as a salvage strategy in the treatment of recurrent glioblastoma multiforme. J Neurooncol. 2014;119(2):397–403. [DOI] [PubMed] [Google Scholar]
- 36. Fortin D, Desjardins A, Benko A, Niyonsega T, Boudrias M.. Enhanced chemotherapy delivery by intraarterial infusion and blood-brain barrier disruption in malignant brain tumors: the Sherbrooke experience. Cancer. 2005;103(12):2606–2615. [DOI] [PubMed] [Google Scholar]
- 37. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 38. Kazda T, Hardie JG, Pafundi DH, et al. Evaluation of RANO response criteria compared to clinician evaluation in WHO grade III anaplastic astrocytoma: implications for clinical trial reporting and patterns of failure. J Neurooncol. 2015;122(1):197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ji X, Chen S, Guo Y, et al. Establishment and evaluation of four different types of patient-derived xenograft models. Cancer Cell Int. 2017;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joo KM, Kim J, Jin J, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3(1):260–273. [DOI] [PubMed] [Google Scholar]
- 41. Fei XF, Zhang QB, Dong J, et al. Development of clinically relevant orthotopic xenograft mouse model of metastatic lung cancer and glioblastoma through surgical tumor tissues injection with trocar. J Exp Clin Cancer Res. 2010;29(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piaskowski S, Bienkowski M, Stoczynska-Fidelus E, et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer. 2011;104(6):968–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. William D, Mullins CS, Schneider B, et al. Optimized creation of glioblastoma patient derived xenografts for use in preclinical studies. J Transl Med. 2017;15(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4(9):998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Friedmann-Morvinski D. Glioblastoma heterogeneity and cancer cell plasticity. Crit Rev Oncog. 2014;19(5):327–336. [DOI] [PubMed] [Google Scholar]
- 46. Peraud A, Watanabe K, Schwechheimer K, et al. Genetic profile of the giant cell glioblastoma. Lab Invest. 1999;79(2):123–129. [PubMed] [Google Scholar]
- 47. Kim H, Zheng S, Amini SS, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cabezas-Sáinz P, Pensado-López A, Sáinz B, Sánchez L.. Modeling cancer using zebrafish xenografts: drawbacks for mimicking the human microenvironment. Cells. 2020;9(9): E1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gillet J-P, Calcagno AM, Varma S, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108(46):18708–18713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cassidy JW, Caldas C, Bruna A.. Maintaining tumor heterogeneity in patient-derived tumor xenografts. Cancer Res. 2015;75(15):2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Izumchenko E, Paz K, Ciznadija D, et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann Oncol. 2017;28(10):2595–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brown KM, Xue A, Julovi SM, et al. Using patient-derived xenograft models of colorectal liver metastases to predict chemosensitivity. J Surg Res. 2018;227:158–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


