Abstract
Unlike other chromatin remodelers, INO80 preferentially mobilizes hexasomes, which can form during transcription. Why INO80 prefers hexasomes over nucleosomes remains unclear. Here, we report structures of S. cerevisiae INO80 bound to a hexasome or a nucleosome. INO80 binds the two substrates in substantially different orientations. On a hexasome, INO80 places its ATPase subunit, Ino80, at superhelical location (SHL)-2, across from SHL-6/−7 as previously seen on nucleosomes. Our results suggest that INO80 action on hexasomes resembles action by other remodelers on nucleosomes, such that Ino80 is maximally active near SHL-2. The SHL-2 position also plays a critical role for nucleosome remodeling by INO80. Overall, the mechanistic adaptations used by INO80 for preferential hexasome sliding imply that sub-nucleosomal particles play considerable regulatory roles.
One-Sentence Summary
Structural studies reveal that INO80 binds hexasomes and nucleosomes in dramatically different orientations, providing mechanistic insight into how hexasomes are regulated.
In eukaryotes, central nuclear processes such as gene expression, DNA replication and DNA repair are coordinated with dynamic changes in chromatin states (1-3). ATP-dependent chromatin remodeling enzymes play essential roles in catalyzing such changes. These enzymes are broadly categorized into four major families: SWI/SNF, ISWI, CHD, and INO80 (4, 5). Each of them contains a core remodeling ATPase subunit and several auxiliary subunits that regulate the core ATPase. It has typically been presumed that the preferred substrate of these enzymes is a nucleosome, the smallest unit of chromatin, containing ~147 bp of DNA wrapped around an octamer of histone proteins (6). Consistent with this assumption, between them, these four classes slide the histone octamer, exchange histone variants, and transfer entire octamers (5, 7).
The INO80 complex has been shown to play roles in regulating transcription, DNA replication and DNA repair (8-11). How INO80’s biochemical activities relate to its diverse biological roles is not well understood. Unlike remodelers from other families, whose core ATPase subunits bind the nucleosome near superhelical location (SHL)-2, Ino80, the core ATPase subunit of the INO80 complex, binds nucleosomes near SHL-6/−7 (fig. S1A)(12-14). It has been speculated that this key difference in nucleosome engagement reflects a fundamentally different remodeling mechanism (15, 16). Indeed, we showed that the preferred substrate of the S. cerevisiae INO80 complex is not a nucleosome but a hexasome, which is a sub-nucleosomal particle that lacks a histone H2A-H2B dimer (17). Hexasomes are generated during transcription and may also be formed during DNA replication and repair (18-21). Further, INO80’s activity on nucleosomes is more dependent on flanking DNA length than on hexasomes (17, 22). These results suggested that INO80 has the versatility to act on hexasomes or nucleosomes based on the density of nucleosomes and hexasomes at a given locus. Yet, fundamental mechanistic questions remain. It is not clear how INO80 can act on both nucleosomes and hexasomes, which differ substantially in their structures. Additionally, why INO80 has different flanking DNA length dependencies on hexasomes versus nucleosomes is unclear.
Here, we report cryogenic-electron microscopy (cryo-EM) structures of endogenously purified S. cerevisiae INO80 bound to a hexasome and a nucleosome. We find that INO80 binds hexasomes and nucleosomes in opposite orientations, with Ino80 binding near SHL-2 on hexasomes and near −6/−7 on nucleosomes. The location of the Arp8 module suggests how flanking DNA length differentially regulates nucleosome and hexasome sliding. DNA gaps near SHL-2 inhibit sliding of both substrates by INO80. Together, our findings provide mechanistic insights into how INO80 slides both hexasomes and nucleosomes.
Structures of the INO80-hexasome and -nucleosome complexes
To visualize how INO80 binds to a hexasome or a nucleosome, we prepared hexasomes and nucleosomes on the same DNA templates containing the 147 bp 601 nucleosome positioning sequence with 80 bp of additional DNA as described previously (+80H and +80N, with definition explained in Fig. 1A, and fig. S1, A and B, and Supplementary Text) (17, 23, 24). Complexes were formed by mixing hexasomes or nucleosomes with endogenously purified S. cerevisiae INO80 without adding nucleotide (fig. S1, C to H).
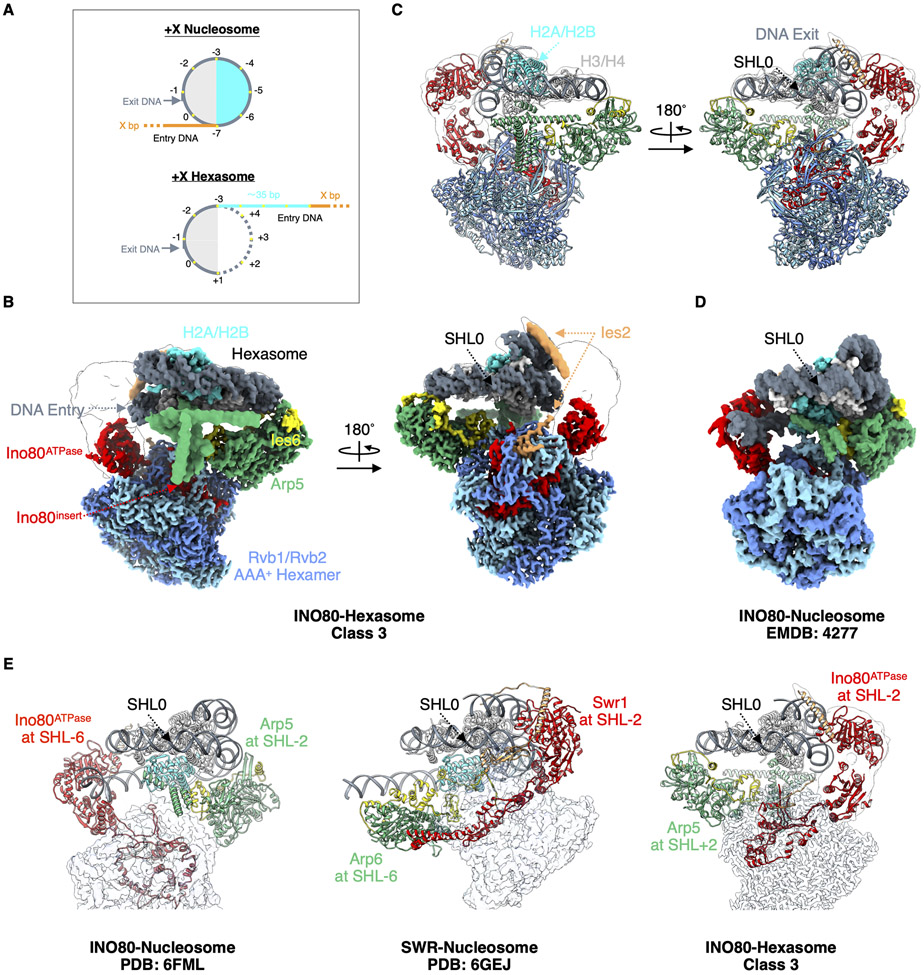
Fig. 1. Structure of the INO80-hexasome complex reveals large rotation.
(A) Cartoon illustration of a +X Nucleosome and a +X Hexasome. H2A-H2B dimer proximal to the flanking DNA (entry side dimer): cyan; H3-H4: light gray; 601 DNA: dark gray; flanking DNA: orange; additional free (unwrapped) DNA: cyan; super helical locations: yellow dots; DNA from the bottom gyre: dotted line. (B) Two different views of cryo-EM density map of the INO80-hexasome complex (class 3). (C) Atomic model of the INO80-hexasome complex (class 3), viewed in the same orientation as the map is viewed in (B). (D) Cryo-EM density map of Chaetomium thermophilum INO80-nucleosome complex (EMDB: 4277 (14)) displayed with its nucleosome dyad and H3-H4 tetramer aligned with that of the hexasome in the right panel of (B). Note that INO80 on a hexasome rotates ~180° from where it sits on a nucleosome when keeping the nucleosome/hexasome dyad and H3-H4 aligned. (E) Structural comparisons of INO80-nucleosome complex (left), SWR-nucleosome complex (middle) and INO80-hexasome complex (right), with nucleosome/hexasome dyad and H3-H4 aligned.
We determined cryo-EM structures of the INO80-hexasome complex in three different conformational snapshots (Fig. 1, B and C, and figs. S2 to S6). The overall shape of INO80 is similar within these structures and also to previously published structures of the nucleosome in complex with human (12) and Chaetomium thermophilum (14) INO80. We group subunits of the INO80 complex into four modules: Rvb module (Rvb1/Rvb2), Arp8 module (Arp8/Arp4/Actin/Ies4 and Taf14), Ino80 module (Ino80/Ies2) and Arp5 module (Arp5/Ies6). The Ino80 protein consists of three major regions: the N-terminal domain (NTD), the HSA region (Ino80HSA) and the ATPase domain (Ino80ATPase). Detailed descriptions of these modules in our structures are in the Supplementary Text.
While the INO80 architecture appears similar to that in the INO80-nucleosome structures, a major difference is that it is rotated ~180° on a hexasome compared to a nucleosome (Fig. 1, B to E). We identified two primary interactions between INO80 and the hexasome: Ino80ATPase binds the hexasome near SHL-3 (class 1), −2.5 (class 2) and −2 (class 3), and the Arp5/Ies6 module binds near SHL+1, +1.5 and +2 (fig. S6, A and B) respectively. Class 3 is the predominant INO80-hexasome class. All Ino80ATPase locations on hexasomes are different than on nucleosomes, which are near SHL-6 or SHL-7 (12-14). However, the Ino80 orientation on hexasomes is consistent with structures of other major chromatin remodelers on nucleosomes such as S. cerevisiae ISW1 (25-27), Chd1 (28-30), RSC (31-33), Snf2 (34) and in particular the SWR1 complex (35), which is from the same sub-family as the INO80 complex. In these structures the ATPase domains interact with nucleosomes near either SHL+2 or SHL-2 (Fig. 1E).
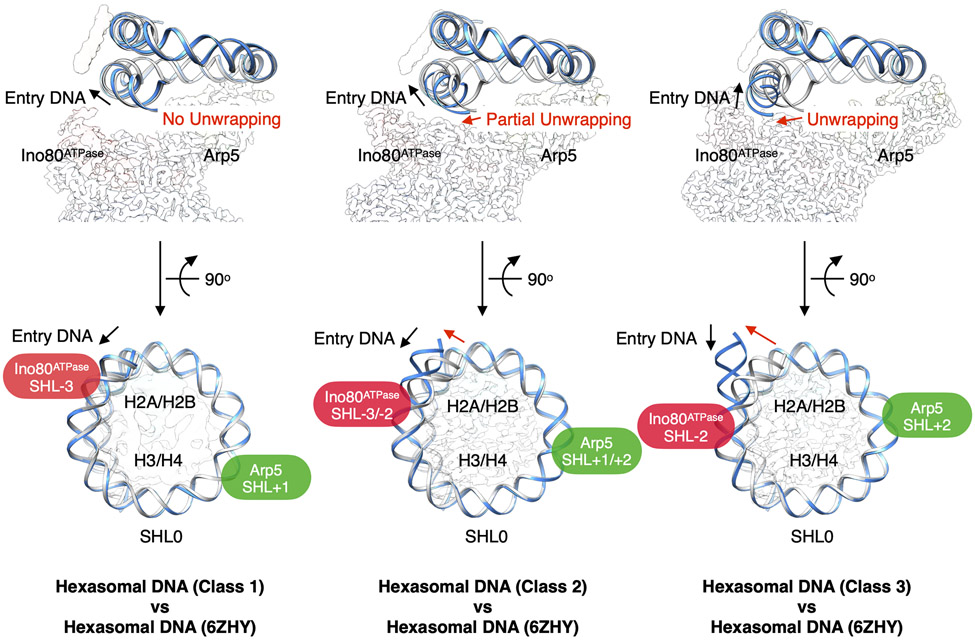
Loss of an H2A-H2B dimer in a hexasome causes an additional ~35 bp of DNA to unwrap from the histone core (free DNA) (Fig. 1A, and fig. S1B). Comparison of our hexasome structures with an unbound hexasome (PDB: 6ZHY, (36)) reveals different levels of further DNA unwrapping. In class 1, the hexasome is almost identical with the unbound hexasome, without detectable additional DNA unwrapping. The level of DNA unwrapping increases as the Ino80ATPase binding position changes from SHL-3 (class 1) to SHL-2 (class 3) (Fig. 2, and fig. S6C).
Fig. 2. Conformational snapshots of INO80-hexasome complexes.
Comparison of DNA from each INO80-hexasome class (blue) with DNA from an unbound hexasome (PDB: 6ZHY, gray), showing degree of DNA unwrapping (upper row) and binding locations of Ino80ATPase and Arp5 (bottom row).
For comparison, we also determined structures of S. cerevisiae INO80 bound to a nucleosome and captured two conformational snapshots (class 1 and 2) from the same dataset (figs. S7 to S9, Supplementary Text). Ino80ATPase in class 1 is located near SHL-7, similar to its location in the human INO80-nucleosome structure (12), while in class 2, it binds near SHL-6, similar to the C. thermophilum structure (14) (fig. S9, A and C). The Arp5/Ies6 module interacts with the nucleosome near SHL-3 and SHL-2 (fig. S9D), respectively. These observations are also consistent with previous findings showing that nucleosomal DNA between SHL-7 and −6 is protected by INO80 (13).
The SHL-2 position plays a critical role in nucleosome and hexasome sliding
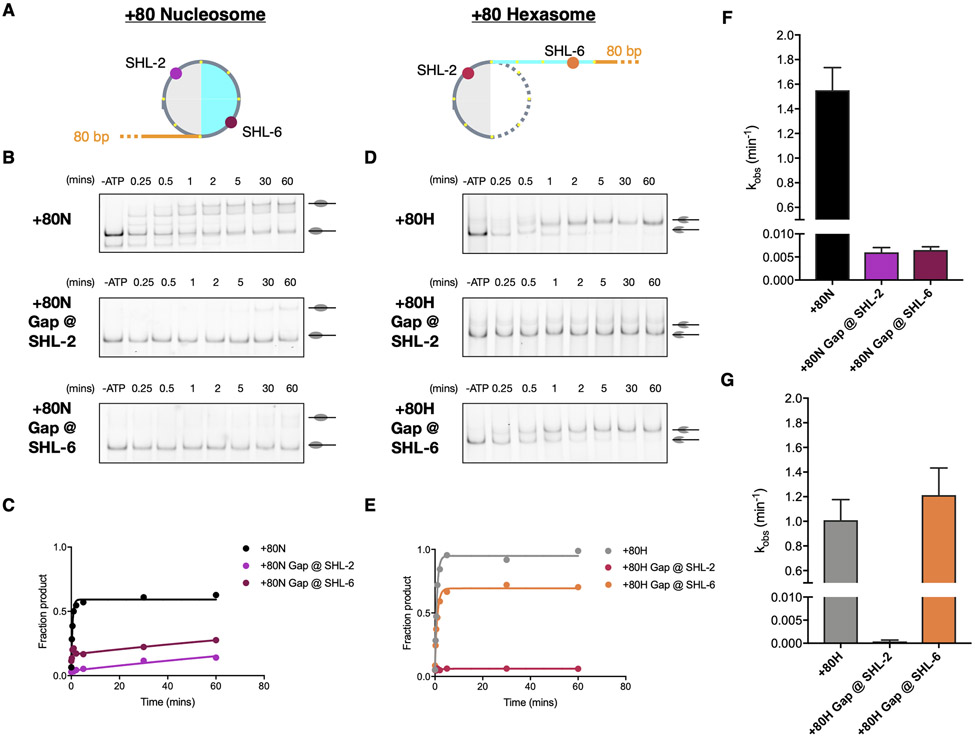
We observe that Ino80ATPase engages the hexasome predominantly near SHL −2. These results raise the possibility that Ino80ATPase acts near SHL-2 when sliding hexasomes. In contrast, consistent with prior findings (12, 14), we observe that Ino80ATPase engages the nucleosome near two positions, SHL-7 and −6. Also as previously proposed, our findings are consistent with the possibility that Ino80ATPase acts near SHL-6 when sliding nucleosomes (13). A commonly used assay to identify the DNA location from where the ATPase domain of a remodeler acts to translocate DNA is to place a single nucleotide gap at the proposed site of action and test if the gap inhibits DNA translocation (37-39). Therefore, to directly test the importance of the SHL-6 and SHL-2 locations, we assembled nucleosomes and hexasomes with single base gaps near SHL-2 or SHL-6 and measured INO80 activity using a gel-based sliding assay (Fig. 3A).
Fig. 3. Inhibition of DNA translocation at specific SHL sites influence nucleosome and hexasome sliding by INO80.
(A) Cartoon illustration of a +80 Nucleosome (left) and a +80 Hexasome (right) with approximate locations of site-specific single base gaps indicated. Colors are the same as in Fig. 1A. (B-C) Example gels and time courses of native gel-based remodeling assays of WT INO80 on +80 nucleosomes with no gap, gap near SHL-2, and gap near SHL-6. (D-E) Example gels and time courses of native gel-based remodeling assays of WT INO80 on +80 hexasomes with no gap, gap near SHL-2, and gap near SHL-6. (F-G) Average observed rate constants of INO80 sliding activity. kobs (min−1): +80N: 1.551 ± 0.1846; +80N Gap @ SHL-2: 0.005995 ± 0.001054; +80N Gap @ SHL-6: 0.006497 ± 0.0007117; +80H: 1.01 ± 0.1668; +80H Gap @ SHL-2: 0.000379 ± 0.0002849; +80H Gap @ SHL-6: 1.213 ± 0.2209. Data represent the mean ± SEM for three technical replicates performed under single-turnover conditions with saturating enzyme and ATP.
We found that a gap at SHL-6 inhibits INO80’s sliding activity on nucleosomes by ~ 200-fold but so did a gap at SHL-2 (Fig. 3, B to G). In contrast, a gap at SHL-6 did not inhibit INO80’s sliding activity on hexasomes, but a gap at SHL-2 slowed hexasomes sliding by ~ 2000-fold (Fig. 3, B to G). These results are consistent with Ino80ATPase acting near SHL-2 when sliding hexasomes and raise new questions about why both the SHL-2 and SHL-6 locations are critical for nucleosome sliding by INO80. We describe possible explanations in the Discussion.
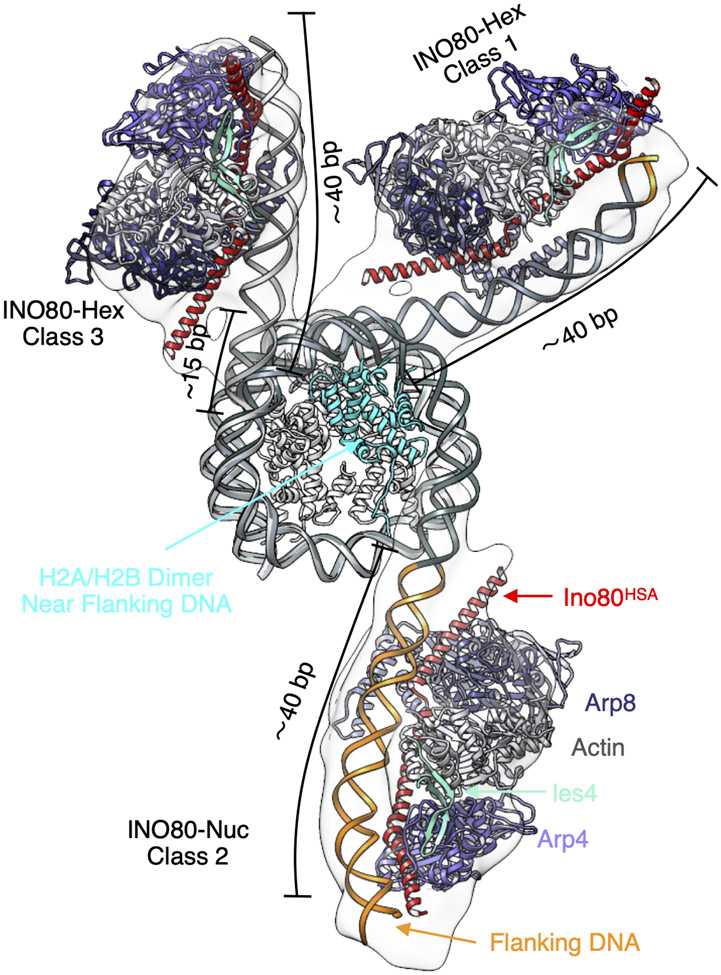
The role of the Arp8 module in flanking DNA length dependence
S. cerevisiae INO80 slides +40 nucleosomes ~100-fold slower than +80 nucleosomes (17, 22). However, sliding hexasomes is less flanking DNA dependent. Our structures suggest that the Arp8 module requires ~40 bp of DNA for appropriate engagement. In class 1 of the INO80-hexasome structure, Arp8 engages with the ~35 bp of DNA unwrapped from removal of the H2A-H2B dimer and an additional ~5 bp of flanking DNA. In class 3 of the INO80-hexasome structure, the Arp8 module engages entirely with ~40 bp of unwrapped DNA that now includes additional DNA unwrapped relative to the unbound hexasome (Fig. 4). In contrast, in class 2 of the INO80-nucleosome structure, the Arp8 module engages entirely with flanking DNA consistent with previous findings (40) (Fig. 4). Our structural data with hexasomes along with the previous data with nucleosomes suggest that 40 bp may be the minimum amount of DNA needed for the Arp8 module to bind and that proper Arp8 module engagement is essential for maximal remodeling activity (40).
Fig. 4. The Arp8 module engages different regions of DNA in nucleosomes versus hexasomes.
Overlay of atomic models of the hexasome (class 1 and class 3) and the nucleosome (class 2) with the Arp8 module (PDB: 8A5O), aligned by the H3-H4 tetramer.
Altered interactions by the Arp5 module
To understand why Ino80 may not bind a nucleosome directly near SHL-2, we compared interactions made by Arp5/Ies6 in hexasomes versus nucleosomes (Supplementary Text). When INO80 binds to a hexasome, the Arp5/Ies6 regions used in the context of a nucleosome are repurposed for different interactions. Modeling the missing H2A-H2B dimer into our INO80-hexasome structure reveals steric clashes of the Arp5 module with the entry side proximal H2A-H2B dimer and with part of the DNA that wraps around the H2A-H2B dimer near SHL-2 (fig. S11). These clashes could be avoided if the H2A-H2B dimer is sufficiently dislodged. To test for this possibility, we inhibited dimer dislodgement by introducing a site-specific disulfide crosslink between the two H2A molecules (N38C) (41) or promoted dimer dislodgement by using an H2A mutant (R81A) that destabilizes the H2A-H2B/H3-H4 interface (42) (fig. S12, A, B and H). The disulfide crosslink did not inhibit nucleosome sliding while the H2A mutant did not promote nucleosome sliding (fig. S12, C to G), indicating that complete dimer dislodgement is not necessary for INO80-mediated nucleosome sliding. In the absence of dimer dislodgement, another way to avoid these clashes could be by substantial rearrangement of the Arp5 module together with subtle rearrangements of the H2A-H2B dimer (fig. S9E).
DISCUSSION
Implications of the INO80-hexasome structure for nucleosome sliding by INO80
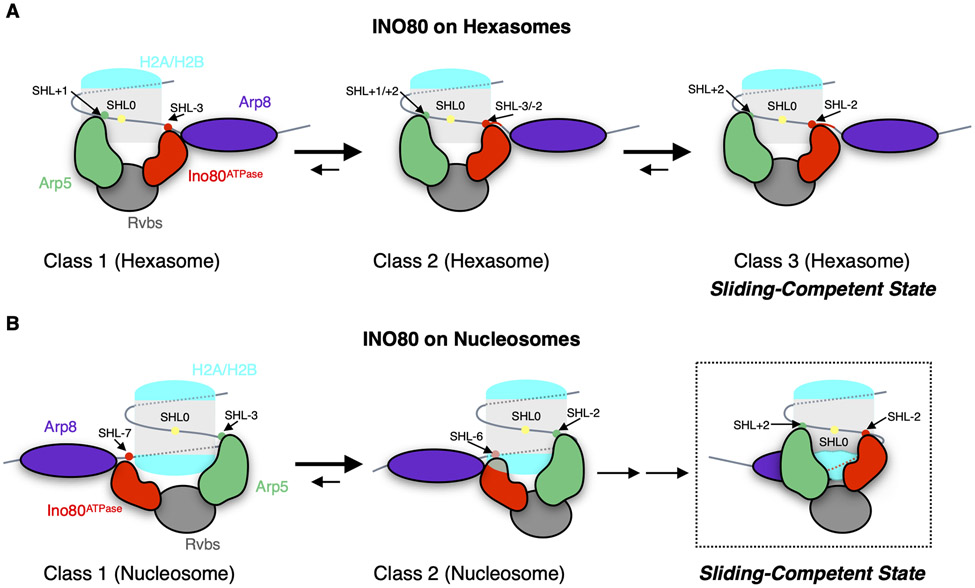
The major conformation of the INO80-hexasome complex (class 3) has Ino80ATPase near SHL-2 and approximately ~15bp of unwrapped DNA from the entry site in addition to the ~35bp of DNA that is unwrapped from removal of an H2A-H2B dimer. The placement of Ino80ATPase near SHL-2 is consistent with how the ATPase subunits of other remodelers bind the nucleosome. Together with our prior finding that hexasomes are remodeled faster than nucleosomes, these results suggest that the class 3 structure represents the sliding-competent conformation of INO80 on hexasomes (Fig. 5A, and fig. S13A). In contrast, the states of INO80 bound to a nucleosome have Ino80ATPase bound near either SHL-6 or −7 consistent with previous findings. These differences raise the question of whether the INO80-nucleosome structures represent sliding-competent conformations or whether a rearrangement of Ino80ATPase to SHL-2 is necessary to achieve efficient nucleosome sliding.
Fig. 5. Model of INO80-induced hexasome and nucleosome sliding.
(A) Hexasome sliding: the Ino80ATPase samples different positions between SHL-3 and SHL-2 but binds predominantly near SHL-2. The INO80 complex becomes sliding-competent when Ino80ATPase engages near SHL-2. (B) Nucleosome sliding: INO80 initially binds with Ino80ATPase at SHL-7 or −6. Upon ATP-hydrolysis, Ino80ATPase moves toward SHL-2 where INO80 becomes sliding-competent.
Previous crosslinking studies have shown that detachment of nucleosomal DNA from H2A-H2B close to the entry site occurs during INO80 remodeling (13). Our data show that progressively more DNA is unwrapped as Ino80ATPase binds closer to SHL-2 on hexasomes (Fig. 2, and fig. S6C). Together these results suggest that DNA unwrapping is coupled to Ino80ATPase accessing its most sliding-competent state. Foot-printing studies have shown that while binding of INO80 to nucleosomes mainly protects nucleosomal DNA from SHL-5 to SHL-6 and near SHL-3, there is modest but detectable protection near SHL-2 (13). Nicks and gaps between SHL-7 and SHL −2 have been shown to inhibit nucleosome sliding to different extents (13, 43). Here we show that site-specific gaps near SHL-2 or SHL-6 substantially inhibit INO80’s sliding of nucleosomes (by ~200 fold). DNA gaps are commonly used to identify the site of action of the ATPase domain of remodelers (37-39). We therefore speculate that INO80 initially binds the nucleosome with Ino80ATPase near SHL-6/−7, and this is followed by an ATP-dependent rotation around the nucleosome to position Ino80ATPase near SHL-2 from where Ino80ATPase then translocates nucleosomal DNA (Fig. 5B, and fig. S14A). A gap at SHL-6 would then inhibit ATP-dependent movement of Ino80ATPase on the nucleosome while the gap at SHL-2 would inhibit translocation of nucleosomal DNA by INO80 relative to the histone octamer (fig. S14). Single-molecule FRET studies have identified an ATP-dependent pause phase prior to ATP-dependent nucleosome sliding (22). The pause could represent the reorientation of Ino80ATPase from SHL-6/−7 towards SHL-2 and add a step that slows remodeling of nucleosomes compared to hexasomes. Simply placing the INO80 complex as is on nucleosomes with the Ino80ATPase near SHL-2 results in steric clashes of the Arp5 module with the nucleosome (fig. S11). While partial H2A-H2B dimer dislodgment, as previously proposed, could avoid such clashes (17), our biochemical data here indicate that dimer dislodgement is not essential for nucleosome sliding by INO80 (fig. S12). Thus more structural studies are needed to understand how INO80 might rotate around a nucleosome.
Alternatively, a gap near SHL-2 may affect the action of the Arp5 module. For such a scenario we speculate that Ino80ATPase translocates DNA near SHL-6 and effective translocation also requires action of the Arp5 module near SHL-2 as previously proposed (12, 14). A gap at SHL-6 would then inhibit translocation of nucleosomal DNA by Ino80ATPase and a gap at SHL-2 would inhibit productive engagement of the Arp5 module (fig. S15).
Clearly distinguishing between these two models will require substantial additional structural analysis of INO80 remodeling intermediates on nucleosomes.
Implications for hexasome sliding by INO80
Our structures provide a view into how INO80 engages a hexasome. In the predominant INO80-hexasome structure, Ino80ATPase binds near SHL-2. A site-specific gap at SHL-2 substantially inhibits INO80’s sliding of hexasomes (~2000 fold) while a gap near SHL-6 does not have a major effect. We therefore hypothesize that Ino80ATPase bound at SHL-2 on a hexasome represents the active structure. Compared to the subtle changes at SHL-2 observed when other remodelers bind nucleosomes (16), the 15 bp of unwrapped DNA (up to SHL-2.5) in class 3 substantially loosens histone DNA interactions and thus may allow more ready translocation from SHL-2. We further propose that the new contacts made by the Arp5/Ies6 module with the exposed H3-H4 surface provide an anchor allowing the Ino80 motor to efficiently pump DNA through the hexasome. These findings also explain the differential effects of the Arp5/Ies6 module on hexasome versus nucleosome sliding (17). The location of the Arp8 module is also different on hexasomes than on nucleosomes. On nucleosomes the Arp8 module binds ~ 40 bp entirely on the flanking DNA (Fig. 4). In the most prevalent INO80-hexasome state (class 3), the Arp8 module is bound entirely to the unwrapped DNA, substantially reducing the need to bind flanking DNA (Fig. 4). These different binding modes of the Arp8 module could explain why hexasome sliding by INO80 is less dependent on flanking DNA length compared to nucleosome sliding.
Supplementary Material
Acknowledgements
Cryo-EM facility at UCSF is managed by Dr. David Bulkley and Mr. Glenn Gilbert. Computation at Cheng laboratory is supported by Mr. Matthiew Harrington and Dr. Junrui Li. We thank Dr. Lvqin Zheng for advice on model building, Dr. Zanlin Yu for providing GO grids, Julia Tretyakova for expressing and purifying histones, Upneet Kaur for providing INO80 WT enzyme, and members of Narlikar and Cheng laboratories for helpful discussions. We thank Dr. Carl Wu and Dr. Anand Ranjan for sharing unpublished data showing inhibition of INO80 activity on nucleosomes containing a gap near SHL-2. We also thank Dr. Sebastian Diendl and Dr. Anton Sabantsev for detailed advice on generating DNAs with site-specific base gaps.
Funding:
This work is supported by grants from National Institute of Health (R35GM140847 to Y.C., R35GM127020 to G.J.N., F31GM136187 to M.A.G., F31GM142271 to E.N.M., F32GM137463 to U.S.C) and by an American Cancer Society—Roaring Fork Valley Research Fund Postdoctoral Fellowship (PF-18-155-01-DMC to L.J.H.). Equipment at UCSF cryo-EM facility was partially supported by National Institutes of Health (NIH) grants (S10OD020054, S10OD021741, and S10OD025881). Y.C. is an Investigator of Howard Hughes Medical Institute.
Footnotes
Competing interests: Y.C. is scientific advisory board member of ShuiMu BioSciences Ltd.
Data and materials availability: For the core INO80 of the INO80-Hexasome complex (class 1, class 2 and class 3) and the core INO80 of the INO80-Nucleosome complex (class 1 and class 2), the coordinates are deposited in the Protein Data Bank with the accession codes 8ETS, 8ETU, 8ETW, 8EU9, and 8EUF; the cryo-EM density maps are deposited in the Electron Microscopy Data Bank (EMDB) with the accession codes EMD-28597, EMD-28599, EMD-28601, EMD-28609, and EMD-28613. For the hexasome of the INO80-Hexasome complex (class 1, class 2 and class 3) and the nucleosome of the INO80-Nucleosome complex (class 1 and class 2), the coordinates are deposited in the Protein Data Bank with the accession codes 8ETT, 8ETV, 8EU2, 8EUE, and 8EUJ; the cryo-EM density maps are deposited in the Electron Microscopy Data Bank (EMDB) with the accession codes EMD-28598, EMD-28600, EMD-28602, EMD-28612, and EMD-28614.
References and Notes
- 1.Bar-Ziv R, Voichek Y, Barkai N, Chromatin dynamics during DNA replication. Genome Res 26, 1245–1256 (2016). doi: 10.1101/gr.201244.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrenhofer-Murray AE, Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem 271, 2335–2349 (2004). doi: 10.1111/j.1432-1033.2004.04162.x [DOI] [PubMed] [Google Scholar]
- 3.Hubner MR, Spector DL, Chromatin dynamics. Annu Rev Biophys 39, 471–489 (2010). doi: 10.1146/annurev.biophys.093008.131348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clapier CR, Cairns BR, The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273–304 (2009). doi: 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 5.Zhou CY, Johnson SL, Gamarra NI, Narlikar GJ, Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu Rev Biophys 45, 153–181 (2016). doi: 10.1146/annurev-biophys-051013-022819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). doi: 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 7.Clapier CR, Iwasa J, Cairns BR, Peterson CL, Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 18, 407–422 (2017). doi: 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao Y, Shen X, INO80 subfamily of chromatin remodeling complexes. Mutat Res 618, 18–29 (2007). doi: 10.1016/j.mrfmmm.2006.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrison AJ, Shen X, Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10, 373–384 (2009). doi: 10.1038/nrm2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poli J, Gasser SM, Papamichos-Chronakis M, The INO80 remodeller in transcription, replication and repair. Philos Trans R Soc Lond B Biol Sci 372, (2017). doi: 10.1098/rstb.2016.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen X, Mizuguchi G, Hamiche A, Wu C, A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544 (2000). doi: 10.1038/35020123 [DOI] [PubMed] [Google Scholar]
- 12.Ayala R et al. , Structure and regulation of the human INO80-nucleosome complex. Nature 556, 391–395 (2018). doi: 10.1038/s41586-018-0021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brahma S et al. , INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun 8, 15616 (2017). doi: 10.1038/ncomms15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eustermann S et al. , Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 (2018). doi: 10.1038/s41586-018-0029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markert J, Luger K, Nucleosomes Meet Their Remodeler Match. Trends Biochem Sci 46, 41–50 (2021). doi: 10.1016/j.tibs.2020.08.010 [DOI] [PubMed] [Google Scholar]
- 16.Yan L, Chen Z, A Unifying Mechanism of DNA Translocation Underlying Chromatin Remodeling. Trends Biochem Sci 45, 217–227 (2020). doi: 10.1016/j.tibs.2019.09.002 [DOI] [PubMed] [Google Scholar]
- 17.Hsieh LJ et al. , A hexasome is the preferred substrate for the INO80 chromatin remodeling complex, allowing versatility of function. Mol Cell 82, 2098–2112 e2094 (2022). doi: 10.1016/j.molcel.2022.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S, Mechanisms of Nucleosome Dynamics In Vivo. Cold Spring Harb Perspect Med 6, (2016). doi: 10.1101/cshperspect.a026666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kireeva ML et al. , Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 9, 541–552 (2002). doi: 10.1016/s1097-2765(02)00472-0 [DOI] [PubMed] [Google Scholar]
- 20.Kulaeva OI, Hsieh FK, Studitsky VM, RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci U S A 107, 11325–11330 (2010). doi: 10.1073/pnas.1001148107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran S, Ahmad K, Henikoff S, Transcription and Remodeling Produce Asymmetrically Unwrapped Nucleosomal Intermediates. Mol Cell 68, 1038–1053 e1034 (2017). doi: 10.1016/j.molcel.2017.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou CY et al. , The Yeast INO80 Complex Operates as a Tunable DNA Length-Sensitive Switch to Regulate Nucleosome Sliding. Mol Cell 69, 677–688 e679 (2018). doi: 10.1016/j.molcel.2018.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levendosky RF, Bowman GD, Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. Elife 8, (2019). doi: 10.7554/eLife.45472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levendosky RF, Sabantsev A, Deindl S, Bowman GD, The Chd1 chromatin remodeler shifts hexasomes unidirectionally. Elife 5, (2016). doi: 10.7554/eLife.21356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armache JP et al. , Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. Elife 8, (2019). doi: 10.7554/eLife.46057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chittori S, Hong J, Bai Y, Subramaniam S, Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res 47, 9400–9409 (2019). doi: 10.1093/nar/gkz670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan L, Wu H, Li X, Gao N, Chen Z, Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol 26, 258–266 (2019). doi: 10.1038/s41594-019-0199-9 [DOI] [PubMed] [Google Scholar]
- 28.Farnung L, Vos SM, Wigge C, Cramer P, Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017). doi: 10.1038/nature24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nodelman IM et al. , Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state. Nat Struct Mol Biol 29, 121–129 (2022). doi: 10.1038/s41594-021-00719-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundaramoorthy R et al. , Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. Elife 7, (2018). doi: 10.7554/eLife.35720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel AB et al. , Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. Elife 8, (2019). doi: 10.7554/eLife.54449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner FR et al. , Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 579, 448–451 (2020). doi: 10.1038/s41586-020-2088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Y et al. , Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019). doi: 10.1126/science.aay0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Li M, Xia X, Li X, Chen Z, Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445 (2017). doi: 10.1038/nature22036 [DOI] [PubMed] [Google Scholar]
- 35.Willhoft O et al. , Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362, (2018). doi: 10.1126/science.aat7716 [DOI] [PubMed] [Google Scholar]
- 36.Lehmann LC et al. , Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch. Cell Rep 33, 108529 (2020). doi: 10.1016/j.celrep.2020.108529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha A, Wittmeyer J, Cairns BR, Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol 12, 747–755 (2005). doi: 10.1038/nsmb973 [DOI] [PubMed] [Google Scholar]
- 38.Schwanbeck R, Xiao H, Wu C, Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem 279, 39933–39941 (2004). doi: 10.1074/jbc.M406060200 [DOI] [PubMed] [Google Scholar]
- 39.Zofall M, Persinger J, Kassabov SR, Bartholomew B, Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol 13, 339–346 (2006). doi: 10.1038/nsmb1071 [DOI] [PubMed] [Google Scholar]
- 40.Kunert F et al. , Structural mechanism of extranucleosomal DNA readout by the INO80 complex. Sci Adv 8, eadd3189 (2022). doi: 10.1126/sciadv.add3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frouws TD, Barth PD, Richmond TJ, Site-Specific Disulfide Crosslinked Nucleosomes with Enhanced Stability. J Mol Biol 430, 45–57 (2018). doi: 10.1016/j.jmb.2017.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehmann K et al. , Effects of charge-modifying mutations in histone H2A alpha3-domain on nucleosome stability assessed by single-pair FRET and MD simulations. Sci Rep 7, 13303 (2017). doi: 10.1038/s41598-017-13416-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller-Planitz F, Klinker H, Becker PB, Nucleosome sliding mechanisms: new twists in a looped history. Nat Struct Mol Biol 20, 1026–1032 (2013). doi: 10.1038/nsmb.2648 [DOI] [PubMed] [Google Scholar]
- 44.Shen X, Preparation and analysis of the INO80 complex. Methods Enzymol 377, 401–412 (2004). doi: 10.1016/S0076-6879(03)77026-8 [DOI] [PubMed] [Google Scholar]
- 45.Dyer PN et al. , Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375, 23–44 (2004). doi: 10.1016/s0076-6879(03)75002-2 [DOI] [PubMed] [Google Scholar]
- 46.Luger K, Rechsteiner TJ, Richmond TJ, Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119, 1–16 (1999). doi: 10.1385/1-59259-681-9:1 [DOI] [PubMed] [Google Scholar]
- 47.Deindl S et al. , ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 152, 442–452 (2013). doi: 10.1016/j.cell.2012.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gamarra N, Narlikar GJ, Histone dynamics play a critical role in SNF2h-mediated nucleosome sliding. Nat Struct Mol Biol 28, 548–551 (2021). doi: 10.1038/s41594-021-00620-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gkikopoulos T et al. , A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333, 1758–1760 (2011). doi: 10.1126/science.1206097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhee HS, Bataille AR, Zhang L, Pugh BF, Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 159, 1377–1388 (2014). doi: 10.1016/j.cell.2014.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palovcak E et al. , A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J Struct Biol 204, 80–84 (2018). doi: 10.1016/j.jsb.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang F et al. , Amino and PEG-amino graphene oxide grids enrich and protect samples for high-resolution single particle cryo-electron microscopy. J Struct Biol 209, 107437 (2020). doi: 10.1016/j.jsb.2019.107437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohi M, Li Y, Cheng Y, Walz T, Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online 6, 23–34 (2004). doi: 10.1251/bpo70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology 152, 36–51 (2005). doi: 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 55.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 57.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). doi: 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 59.Grant T, Rohou A, Grigorieff N, cisTEM, user-friendly software for single-particle image processing. Elife 7, (2018). doi: 10.7554/eLife.35383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez-Garcia R et al. , DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874 (2021). doi: 10.1038/s42003-021-02399-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosenthal PB, Henderson R, Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745 (2003). doi: 10.1016/j.jmb.2003.07.013 [DOI] [PubMed] [Google Scholar]
- 62.Dang S et al. , Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017). doi: 10.1038/nature25024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jumper J et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 65.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367 (2012). doi: 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ, Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol 319, 1097–1113 (2002). doi: 10.1016/S0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- 67.Farnung L, Ochmann M, Garg G, Vos SM, Cramer P, Structure of a backtracked hexasomal intermediate of nucleosome transcription. Mol Cell 82, 3126–3134 e3127 (2022). doi: 10.1016/j.molcel.2022.06.027 [DOI] [PubMed] [Google Scholar]
- 68.Mayanagi K et al. , Structural visualization of key steps in nucleosome reorganization by human FACT. Sci Rep 9, 10183 (2019). doi: 10.1038/s41598-019-46617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brahma S, Ngubo M, Paul S, Udugama M, Bartholomew B, The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling. Nat Commun 9, 3309 (2018). doi: 10.1038/s41467-018-05710-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knoll KR et al. , The nuclear actin-containing Arp8 module is a linker DNA sensor driving INO80 chromatin remodeling. Nat Struct Mol Biol 25, 823–832 (2018). doi: 10.1038/s41594-018-0115-8 [DOI] [PubMed] [Google Scholar]
- 71.Oberbeckmann E et al. , Ruler elements in chromatin remodelers set nucleosome array spacing and phasing. Nat Commun 12, 3232 (2021). doi: 10.1038/s41467-021-23015-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.