Highlights
-
•
There is lacking knowledge about how automated tools for lesion detection/segmentation in multiple sclerosis (MS) perform within a clinical setting and about how they might be integrated in clinical practice.
-
•
The value and economic impact of those tools on patient management is unclear.
-
•
The development of new tools for automated MS lesions detection/segmentation should include their integration in the clinical workflow.
Keywords: MRI, Multiple sclerosis, Systematic review, Lesion segmentation, Lesion detection
Abstract
Introduction: Over the past few years, the deep learning community has developed and validated a plethora of tools for lesion detection and segmentation in Multiple Sclerosis (MS). However, there is an important gap between validating models technically and clinically. To this end, a six-step framework necessary for the development, validation, and integration of quantitative tools in the clinic was recently proposed under the name of the Quantitative Neuroradiology Initiative (QNI).
Aims: Investigate to what extent automatic tools in MS fulfill the QNI framework necessary to integrate automated detection and segmentation into the clinical neuroradiology workflow.
Methods: Adopting the systematic Cochrane literature review methodology, we screened and summarised published scientific articles that perform automatic MS lesions detection and segmentation. We categorised the retrieved studies based on their degree of fulfillment of QNI’s six-steps, which include a tool’s technical assessment, clinical validation, and integration.
Results: We found 156 studies; 146/156 (94%) fullfilled the first QNI step, 155/156 (99%) the second, 8/156 (5%) the third, 3/156 (2%) the fourth, 5/156 (3%) the fifth and only one the sixth.
Conclusions: To date, little has been done to evaluate the clinical performance and the integration in the clinical workflow of available methods for MS lesion detection/segmentation. In addition, the socio-economic effects and the impact on patients’ management of such tools remain almost unexplored.
1. Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system, which affects almost 3 million people worldwide (Walton et al., 2020). MS is the most prevalent neurological disease among young adults, and it is associated with a progressive increase in disability, which can significantly affect an individual’s quality of life as well as impose a substantial economic burden on patients, their families and the entire society (Feinstein, 2004). MS mostly exhibits focal inflammatory and degenerative lesions, but also diffused brain and spinal cord damage, which ultimately results in permanent brain volume loss (Reich et al., 2018). Hence, assessing the impact of neuroinflammation and neurodegeneration in patients through the identification of adequate imaging biomarkers is fundamental.
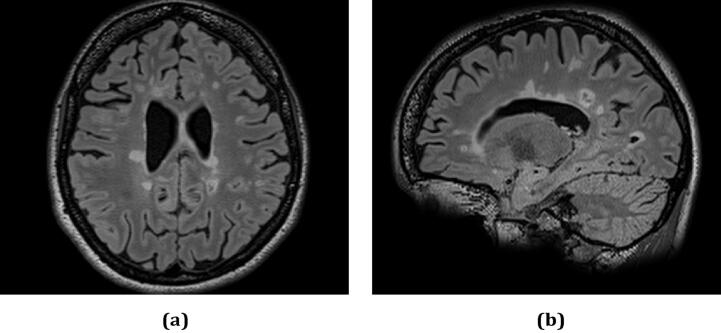
MS diagnosis requires the demonstration of dissemination in space (i.e., specific regions of the brain and spinal cord must be affected by areas of focal inflammation/damage, which are named plaques or lesions) and time (i.e., assessment of the increase in lesions’ number and volume over time). The information provided by Magnetic Resonance Imaging (MRI) can address both requirements and, therefore, it is essential for MS diagnosis (Thompson et al., 2018). Fig. 1 shows the appearance of MS lesions in brain MRI.
Fig. 1.
MS lesions in brain MRI axial (a) and sagittal (b) view. The figure comes from a dataset described in Lesjak et al. (2016).
The process of MS lesion detection and segmentation is usually performed manually by trained neuroradiologists and it is a time-consuming task and prone to errors (Egger et al., 2017). As a consequence, the development of automatic tools to support this procedure is urgently needed.
To date, several automated approaches have been proposed to support this key task, leading to a plethora of tools (reviewed in Llado et al., 2012; García-Lorenzo et al., 2012; Alrabai et al., 2022, Zeng et al., 2020, Ma et al., 2022, Diaz-Hurtado et al., 2022, Commowick et al., 2023) that are more or less mature towards clinical application and use. In the last 15 years, many international challenges, organised in the context of the Medical Image Computing and Computer Assisted Intervention (MICCAI) conference (Commowick et al., 2018, Kuijf et al., 2019, Styner et al., 2008, Commowick et al., 2021) and the International Symposium on Biomedical Imaging (ISBI) (Carass et al., 2017), provided benchmark datasets to promote a fair evaluation. In addition, the Shifts Challenge (Malinin et al., 2022) focused on the estimation of robustness and uncertainty of such methods.
To facilitate the adoption of automated image analysis tools in the practice of clinical neuroradiology, Goodkin et al. (2019) proposed a framework based on a sequence of six steps named Quantitative Neuroradiology Initiative (QNI). The six steps can be summarised in providing:
-
1.
the target clinical area and biomarkers;
-
2.
the structure of the automated method;
-
3.
a quantitative report;
-
4.
a technical and clinical validation;
-
5.
details about the integration into the clinical workflow;
-
6.
an in-use evaluation.
Although these requirements were originally applied to the radiological assessment of dementia, the framework was adopted later on to conduct systematic reviews on commercial volumetric MRI reporting tools in dementia (Pemberton et al., 2021) and MS (Mendelsohn et al., 2022). While in the present work we aim to present the state of the art of scientific literature, the mentioned reviews strictly focused on studies related to commercial devices.
Table 1 describes the requirements to fulfill the six QNI steps. The first and second steps include the identification of the target clinical area, the associated imaging biomarkers (lesional in the case of MS), the automated model’s structure, and reference datasets. A third phase consists of filing a visually informative quantitative report, to be integrated into the radiology report. The fourth step relates to the technical and pre-use clinical validation, which encapsulates a “credibility” and “accuracy” study. The former concept suggests a data quality check and a review of the technical performance of the method. The latter term refers to a blinded rating of a limited number of cases and an assessment of the clinical reporting process: radiologists’ accuracy and reporting efficiency should be examined, with and without the automated tool, in the closest possible environment to the usual radiology setting. The fifth step is the integration of tools into the clinical workflow, from data format compatibility to data protection and the joint visualisation of Digital Imaging and Communications in Medicine (DICOM) series and model output. The final phase describes an in-use pipeline evaluation for what concerns patient management and the socio-economic impact of the tool. Key concepts are the smoothness of the tool’s integration into a hospital’s radiology department, speed of diagnosis, cost in resources, productivity, general perception, and mid-term economic impact.
Table 1.
Description of QNI steps.
| QNI step | Requirements | |
|---|---|---|
| 1st | area of clinical need and corresponding imaging biomarkers (lesional for MS) | |
| 2nd | structure of automatic method, benchmark dataset for training/testing | |
| 3rd | quantitative (radiology) report with clinically and visually meaningful information | |
| 4th | technical and clinical validation (quality check, with and without tool reporting assessment) | |
| 5th | integration into the clinical workflow (data format compatibility and protection, input–output viewer) | |
| 6th | in-use evaluation (patient management, socio-economic impact) |
With these criteria, the proposed review analyses to what extent current literature of reporting automatic tools for detection and segmentation of MS lesions follows the QNI steps and, thus, considers the integration into the clinical routine.
2. Material and methods
In this review, we adopt the methodology described in the “Cochrane Handbook for Systematic Review of Interventions” (Lefebvre et al., 2022) to collect published articles till June 2023. To broaden and differentiate the screening pool, we targeted two databases, respectively medicine- and engineering-oriented: PubMed (https://pubmed.ncbi.nlm.nih.gov/) and IEEE (https://ieeexplore.ieee.org/Xplore/home.jsp). We adapted Cochrane’s threefold subdivision of screening keywords to our case, from “population, intervention and study design” to “population, task, and design of the tool”. While population refers to clinically confirmed MS patients, task and design describe what we expect as the automatic model’s first output and general characteristics.
By searching within both databases using Cochrane’s threefold subdivision of keywords, 770 studies were extracted (123 from IEEE and 647 from PubMed). Please note the word including a * is a “wild-card”, covering suffixes from a word stem, such as “automat*” stands for “automated” and “automatic”:
-
1.
“multiple sclerosis”;
-
2.
“segment*” OR “detect*”;
-
3.
“machine learning” OR “deep learning” OR “automat*” OR “digital tool”.
The above criteria were applied to all metadata, including title, abstract, and keywords.
2.1. Study inclusion criteria
Screened articles were included in the review when they met all the following inclusion criteria:
-
1.
original research published after 2011 in academic peer-reviewed journals or conferences in the English language;
-
2.
studies targeting fully automatic detection or segmentation of white matter non-enhanced (without contrast agents) lesions, as either a primary or a secondary objective;
-
3.
studies targeting brain MRI modalities;
-
4.
studies targeting clinical MS population alone or mixed with patients with a clinically isolated syndrome (CIS, a first symptomatic episode of potential MS);
-
5.
studies performing either technical, clinical, or in-use validation.
As a consequence, papers including a wider population than MS (in separate datasets), performing longitudinal or cross sectional evaluations, presenting a different primary goal or other lesion types (Rosa et al., 2022), have been reported in this review only if they also met the mentioned conditions. For each QNI framework’s step, the methodology of reviewed articles was discussed and evaluated as compliant or not compliant. It must be noted that failure to comply with some steps towards clinical use of those methods does not imply any superficiality in the methodology applied. It indicates, instead, that an article focuses primarily on other objectives.
In our work we distinguish among technical, clinical and in-use assessment as follows:
-
•
Technical validation: comparing results to manual segmentation and/or state-of-the-art segmentation software, and data quality checks.
-
•
Clinical validation: refers to any evaluation of the tool’s impact on clinical management, diagnostics, and reliability with respect to the reference annotated “ground truth”.
-
•
In-use evaluation: includes any study measuring how easily the tool can be integrated into reporting workflow, benefits for patients, general perception, and socio-economic effects of the tool.
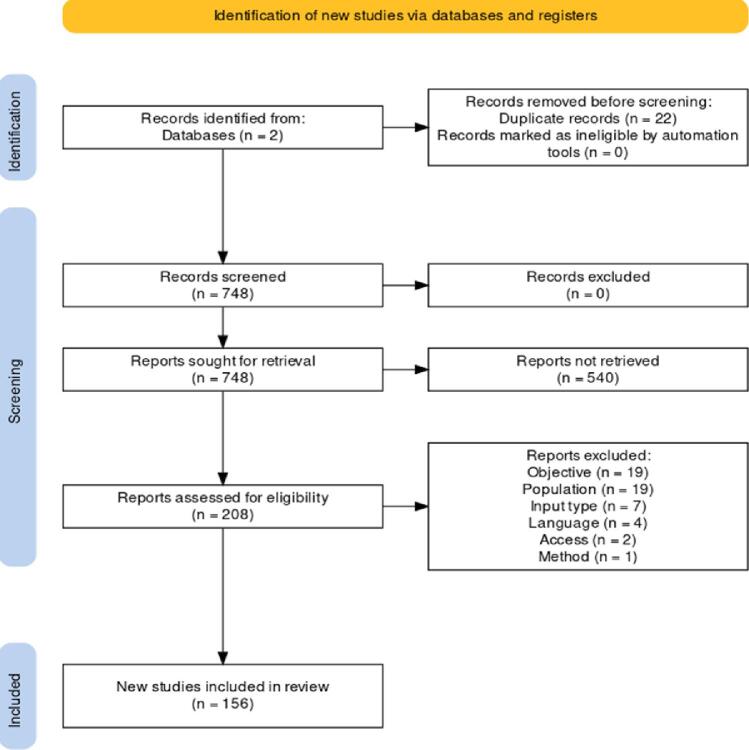
Merging results from the two databases, 22 records were excluded as duplicates leading to 748 studies to further review. Upon examination of the pool of abstracts, 562 occurrences were not retrieved as not compliant with the inclusion criteria. After carefully reviewing the full texts from the remaining 208 studies, 52 articles were further excluded due to their objective, population (e.g., dementia), input type (e.g., synthetic data), language, availability and method (only fully automatic methods were considered). The PRISMA flow diagram (Page et al., 2021) describing the procedure to select 156 studies to include in the review is reported in Fig. 2.
Fig. 2.
PRISMA flowchart applied during the screening process. The terms “objective”, “population”, “input type”, “language” refer to the inclusion criteria of Section 2.1. The term “access” refers to an exclusion due to the impossibility to access the full text of a paper. The term “method” refers to “not fully automatic methods”.
The search strategy was peer-reviewed by S.S., an experienced information specialist within our team. All data used in the review are available and can be accessed through PubMed and IEEE databases.
3. Results
Following the described methodology, 156 studies were identified (see the list of abbreviations in Table 2 and first columns of Table 3, Table 4, Table 5), which met all the inclusion criteria (Fig. 2).
Table 2.
List of abbreviations.
| Abbreviation | Meaning |
|---|---|
| FLAIR | fluid attenuated inversion recovery |
| MPRAGE | magnetisation-prepared rapid gradient echo |
| MP2RAGE | magnetisation-prepared 2 rapid gradient echo |
| PD-w | proton density weighted |
| T1-w | T1-weighted |
| T2-w | T2-weighted |
| DIR | double inversion recovery |
| PSIR | phase-sensitive inversion recovery |
| DP | diffusion perfusion |
| MNI | Montreal neurological institute |
| CLAHE | contrast limited adaptive histogram equalization |
| GM | gray matter |
| WM | white matter |
| CSF | cerebro spinal fluid |
| EPI | echo-planar imaging |
| CNN | convolutional neural network |
| FOV | field of view |
| SVM | support vector machine |
| FCNN | fully connected neural network |
| (A) VD | (absolute) volume difference |
| DSC | dice score |
| (L) TPR | (lesion) true positive rate |
| (L) FPR | (lesion) false positive rate |
| b.c. | bias field correction |
| reg. | registration |
| PPV | positive predicted value |
| TNR | true negative rate |
| FNR | false negative rate |
| IoU | intersection over union |
| AUC | area under the curve |
| NPV | negative predicted value |
| SI | similarity index |
| (p) AUC | partial area under the curve |
| HC | healthy controls |
| HD | Hausdorff distance |
| SD | surface distance |
Table 3.
Studies’ information containing details on datasets, inputs, and architecture of the automatic algorithm, pre-processing steps, and evaluation metrics (part 1).
| Data | Inputs | Method | Pre-processing | Evaluation metrics | QNI steps fulfilled | ||
|---|---|---|---|---|---|---|---|
| (Yildirim and Dandil, 2021b) | MICCAI 2008, ISBI 2015 | FLAIR | Mask R-CNN | / | DSC, AVD, lesion-wise TPR and FPR | 1st, 2nd | |
| (Ghosal et al., 2020) | MICCAI 2016 | T1-w, MPRAGE, FLAIR, T1-w gadolinium and T2/DP contrast enhanced |
U-Net | denoising, intensity correction and skull-stripping | DSC accuracy, sensitivity, specificity | 1st, 2nd | |
| (Kats et al., 2019) | ISBI 2015 | T1-w, T2-w, PD-w and FLAIR |
U-Net | / | DSC, precision, recall | 1st, 2nd | |
| (Kumar et al., 2019) | MICCAI 2016 | T1-w, MPRAGE, FLAIR, T1-w gadolinium-enhanced and T2/DP contrast enhanced |
combination of Dense- and U-Net | remove null slices, z-score normalisation | DSC sensitivity, specificity, accuracy | 1st, 2nd | |
| (Vang et al., 2020) | ISBI 2015 | FLAIR | Mask R-CNN | skull stripping, b.c., z-score normalisation | DSC, precision, LTPR, LFPR, sensitivity | 1st, 2nd | |
| (Joshi and Sharma, 2022) | MICCAI 2008 and 30 extra MS | T1-w, T2-w and FLAIR | CNN and graph convolutional networks | skull stripping, b.c. | DSC | 1st, 2nd | |
| (Kolarik et al., 2021) | MICCAI 2016 | FLAIR | VGG-16 encoder, residual U-Net decoder | normalisation | DSC and recall | 1st, 2nd | |
| (Zhang et al., 2019) | ISBI 2015 and 15 extra simulated MS | T1-w MPRAGE, FLAIR | 2D fully convolutional densely connected network | ISBI: b.c., skull and dura stripping, 2nd b.c. and MNI reg.; extra: reg. to T1-w, skull stripping, b.c. |
DSC, precision, recall, LFPR, LTPR, VD | 1st, 2nd | |
| (Kamraoui et al., 2021) | ISBI 2015, MICCAI 2016, extra 43 subjects | T1-w and FLAIR | U-Net | ISBI: b.c., skull and dura stripping, b.c., MNI reg; MICCAI: denoising, reg. on FLAIR, skull stripping, b.c., MNI reg.; extra: denoising, MNI reg., skull stripping, b.c., denoising, normalisation |
DSC, precision, recall, LTPR, LFPR | 1st, 2nd | |
| (Chen et al., 2021) | ISBI 2015, extra 157 MS | FLAIR | local attention feature and graph attention clustering | ISBI: skull stripping, reg. and normalisation; extra: b.c., normalisation, resampling | DSC, TPR, LTPR, LFPR, absolute VD | 1st, 2nd | |
| (Billot et al., 2021) | MICCAI 2016, ISBI 2015 | T1-w and FLAIR | U-Net | ISBI: skull stripping | DSC and brain ROI | 1st, 2nd | |
| (Alijamaat et al., 2021) | MICCAI 2016 | MPRAGE, FLAIR, T1w gadolinium, PD-w | CNN with wavelet pooling | remove null slices, 0–1 normalisation | accuracy, TPR, DSC | 1st, 2nd | |
| (Basaran et al., 2022) | MSSEG-2 | FLAIR | nnU-Net | skull stripping, b.c., baseline and follow-up reg. | F1, DSC, volume of FP | 1st, 2nd | |
| (Hashemi et al., 2018) | MICCAI 2016, ISBI 2015 | MPRAGE, FLAIR, T1 contrast enhanced, PD-w, T2; MPRAGE, FLAIR, PD-w, T2 |
U-Net, FC Dense-Net | reg. | DSC, recall, F2, Jaccard index, LTPR, LFPR, VD | 1st, 2nd | |
| (Roura et al., 2015) | MICCAI 2008, extra 14 MS | T1-w and FLAIR | Statistical Parametric Mapping | skull stripping, denoising, b.c., MNI reg. | DSC, TPR, PPV | 1st, 2nd | |
| (Essa et al., 2020) | MICCAI 2008 | T2-w and FLAIR | R-CNN | MNI reg., skull stripping, b.c. | TPR, PPV, DSC, VD | 1st, 2nd | |
| (Zhang et al., 2021) | ISBI 2015, extra 176 MS | T1-w, FLAIR, T2-w, anatomical coordinates | encoder-decoder backbone, anatomical convolutional modules, region-based loss modules |
MNI reg. | DSC, precision, sensitivity, F1, LTPR, LFPR, Lesion F1 | 1st, 2nd | |
| (Sadeghibakhi et al., 2022) | ISBI 2015 | T1-w and FLAIR | ResNet | skull stripping, normalisation, MNI reg., CLAHE | DSC, LTPR, LFPR, absolute VD | 1st, 2nd | |
| (Brosch et al., 2016) | MICCAI 2008, ISBI 2015, extra 195 subjects | T1-w, T2-w, PD-w, FLAIR or T1-w, T2-w, PD-w | CNN with shortcuts | skull stripping, 0–1 normalisation, reg. | DSC, VD, LTPR, LFPR | 1st, 2nd | |
| (Ackaouy et al., 2020) | MICCAI 2016 | T1-w and FLAIR | CNN | denoising, skull stripping, b.c. | DSC and F1 | 1st, 2nd | |
| (Ashtari et al., 2022) | MSSEG-2 | FLAIR | U-Net | zero regions removal, z-score normalisation, resampling | DSC, HD, sensitivity, PPV, F1, number and volume of predicted lesions |
1st, 2nd | |
| (Fenneteau et al., 2021) | ISBI 2015 | FLAIR | U-Net | z-score normalisation | DSC, sensitivity, precision | 1st, 2nd | |
| (Sarica and Seker, 2022) | MSSEG-2 | FLAIR | U-Net | skull stripping, b.c., normalisation, baseline and follow-up fusion | DSC, F1, n. and volume of predictions, PPV, sensitivity, specificity, mean SD |
1st, 2nd | |
| (Aslani et al., 2019) | ISBI 2015, extra 37 MS | T1-w, T2-w, FLAIR | ResNet | skull stripping, MNI reg., normalisation | DSC, LTPR, LFPR, avg symmetric SD, HD, PPV, VD | 1st, 2nd | |
| (Valverde et al., 2017) | MICCAI 2008 | T1-w, T2-w, FLAIR | CNN | co-reg. | VD, TPR, FPR | 1st, 2nd | |
| (Filho, 2017) | MICCAI 2016, extra 17 subjects | T1-w and FLAIR | iterative contrast enhancement and logistic classification | MNI reg., WM-GM segmentation, b.c., denoising | sensitivity, specificity, DSC, volume similarity | 1st, 2nd | |
| (Guizard et al., 2015) | MICCAI 2008, extra 108 MS | T1-w, T2-w, FLAIR | rotation-invariant multi-contrast non-local mean segmentation | denoising, normalisation, MNI reg., skull stripping | DSC, TPR LTPR, PPV, LPPV, VD, FPR, symmetric SD | 1st, 2nd | |
| (Abdullah et al., 2012) | MICCAI 2008, extra 10 MS and synthetic data | T1-w, T2-w and FLAIR | SVM with textural features, position features, co-registered intensities, tissues priors and neighbouring blocks features |
MNI reg., normalisation | DSC, detected lesion load, TPR, PPV | 1st, 2nd | |
| (Geremia et al., 2011) | MICCAI 2008 | T1-w, T2-w and FLAIR | discriminative random decision forest | sub-sampling and cropping, b.c., normalisation, MNI reg., segmentation of WM, GM, CSF |
TNR, TPR, FPR, PPV, volume overlap, VD, symmetric SD | 1st, 2nd | |
| (Joshi and Sharma, 2021) | 50 MS | T1-w, T2-w and FLAIR | graph convolutional network and cnn autoencoder | b.c. | DSC, precision and loss | 1st, 2nd | |
| (Sepahvand et al., 2020) | multi-centric 886 MS | T1-w, T2-w, PD-w and FLAIR | U-Net | skull stripping, b.c., normalisation, MNI reg. | AUC, specificity and sensitivity | 1st, 2nd | |
| (Papadopoulos et al., 2022) | 30 MS | FLAIR | U-Net | cropping window on label mask, 0–1 normalisation | accuracy, IoU, DSC, precision, recall | 1st, 2nd | |
| (Nair et al., 2019) | multi-centric 1064 MS | T2-w, T1-w, PD-w, FLAIR | CNN | skull stripping, b.c., MNI reg. | ROC with TPR/FPR at voxel and lesion level | 1st, 2nd | |
| (Rosa et al., 2020) | 54 MS | MP2RAGE and FLAIR | U-Net | co-reg. | DSC, absolute VD, TPR, LTPR, FPR, LFPR, WML and CL detection rate |
1st, 2nd | |
| (Gabr et al., 2019) | multi-centric 1008 MS | T1-w, T2-w, PD-w and FLAIR | U-Net | denoising, skull stripping, b.c., normalisation | DSC, TPR, FPR, classification based on volume | 1st, 2nd | |
| (Hermann et al., 2021) | bi-centric 35 MS | MR fingerprinting EPI | U-Net | denoising, distortion correction | DSC and lesion detection rate | 1st, 2nd | |
| (Rakic et al., 2021) | multi-centric 159 MS | T1-w and FLAIR | U-Net with attention gate layers | MNI reg., skull stripping, z-score normalisation | lesion/voxel-wise DSC, confusion matrix, lesion load | 1st, 2nd | |
| (Krüger et al., 2021) | multi-centric 1809 MS | T1-w and FLAIR | CNN | strong artifacts exclusion, reg. on FLAIR | detection (1 voxel overlap), sensitivity, F1 lesion-wise, DSC voxel-wise | 1st, 2nd | |
| (Krishnan et al., 2021) | multi-centric 1574 MS | T1-w and FLAIR or T1-w post-contrast and FLAIR | 2.5D U-Net | b.c., MNI reg., skull stripping | PPV, sensitivity, absolute VD | 1st, 2nd | |
| (Elliott et al., 2013) | multi-centric 255 MS | T1-w, T2-w and FLAIR | Bayesian and random-forest based lesion-level classifier | skull stripping, b.c., normalisation | sensitivity, false detection rate | 1st, 2nd | |
| (Cabezas et al., 2014) | multi-centric 45 MS | T1-w, PD-w, T2-w, FLAIR, prob. maps of CSF, GM, WM, an outlier map, 800 region-based comparison meta-features |
Gentleboost classifier | skull stripping, b.c., denoising, normalisation, reg. | DSC | 1st, 2nd | |
| (Sajja et al., 2006) | 23 MS | T2-w and FLAIR | Parzen estimator with Gaussian kernel | removal of lesions close to the brain surface, FP/FN minimisation | similarity index, % of correct, over- and under-estimation | 1st, 2nd | |
| (Sweeney et al., 2013) | bi-centric 208 MS | T1-w, T2-w, PD-w and FLAIR | logistic regression with gaussian kernel | MNI reg., b.c., skull stripping, intensity thresholding, normalisation, multi-resolution smoothed volumes |
FPR, sensitivity and DSC | 1st, 2nd | |
| (Steenwijk et al., 2013) | 20 MS | T1-w and FLAIR | k-nearest neighbour | skull stripping, MNI reg., b.c. | DSC, sensitivity | 1st, 2nd | |
| (Schmidt et al., 2011) | 52 MS | T1-w and FLAIR | adaptive maximum a posteriori estimations and Markov random field | b.c., MNI reg., classification of CSF, WM, GM | DSC, correlation and regression of lesion volume | 1st, 2nd | |
| (Todea et al., 2023) | multi-centric 206 MS | MPRAGE and FLAIR at 2 timepoints | k-nearest neighbour | b.c., normalisation, reg. | sensitivity, specificity, accuracy, F1, PPV, NPV | 1st, 2nd | |
| (Gessert et al., 2020) | bi-centric 122 MS | FLAIR | LST toolbox vs U-Net and two path CNNs with attention-guided interaction modules |
z-score normalisation | LTPR, LFPR, DSC | 1st, 2nd | |
| (Hitziger et al., 2022) | MSSEG-2 | FLAIR | U-Net | MNI reg., crop FOV to area around brain, z-score normalisation | F1, recall, precision, DSC | 1st, 2nd | |
| (Mengin et al., 2022) | MSSEG-2, extra 17 MS | FLAIR | U-Net | z-score normalisation | DSC, sensitivity, PPV, F1 | 1st, 2nd | |
| (Krüger et al., 2020) | multi-centric 1791 MS | FLAIR | U-Net | / | LTPR, LFPR, DSC lesion-wise | 1st, 2nd |
Table 4.
Studies’ information containing details on datasets, inputs, and architecture of the automatic algorithm, pre-processing steps, and evaluation metrics (part 2).
| Data | Inputs | Method | Pre-processing | Evaluation metrics | QNI steps fulfilled | ||
|---|---|---|---|---|---|---|---|
| (Schmidt et al., 2019) | bi-centric 60 MS | FLAIR | analysis of FLAIR intensities distribution | tissue classification, b.c. and co-reg. | DSC, FPR, TPR | 1st, 2nd | |
| (Combès et al., 2021) | multi-centric 54 MS | T1-w, T2-w, FLAIR | FCNN | orientation in RAS coordinates, skull stripping, baseline and follow-up reg., cropping, b.c., intensity histogram linear rescaling and Nyul standardisation |
n. of lesions and annotation and reporting time with and without tool, inter-rater variability |
1st, 2nd, 3rd, 4th | |
| (Jannat et al., 2021) | 30 MS and 100 controls | T1-w, T2-w and FLAIR | CNN | / | precision, recall, F1 | 1st, 2nd | |
| (Salem et al., 2020) | 60 MS | T1-w, T2-w, PD-w and FLAIR | FCNN | skull stripping, b.c., Nyul standardisation | TPF, FPF, DSC | 1st, 2nd | |
| (Zhang et al., 2022) | multi-centric 507 MS | FLAIR | 2D U-Net | normalisation and resampling | DSC, sensitivity, precision | 1st, 2nd | |
| (Rachmadi et al., 2019) | bi-centric 40 MS | FLAIR | irregularity maps generation | brain and CSF mask extraction, co-reg. and b.c. | DSC, PPV, spec, TPR, non-parametric Spearman’s correlation coefficient | 1st, 2nd | |
| (Chen et al., 2021) | ISBI 2015 | FLAIR, MPRAGE, T2-w, PD-w | U-Net | b.c., z-score normalisation | DSC, PPV, TPR, LFPR, LTPR | 1st, 2nd | |
| (Mehta et al., 2021) | multi-centric 1073 MS | T1-w, T2-w, PD-w and FLAIR | Bayesian U-Net and another U-Net | AUC with TPR and FDR | 1st, 2nd | ||
| (Opbroek et al., 2014) | multi-centric 70 MS | T1-w, T2-w and FLAIR | reduced SVM on 33 features | Nyul standardisation | relative AVD, average symmetric SD, TPR, FPR | 1st, 2nd | |
| (de Oliveira et al., 2022) | ISBI 2015, MICCAI 2016 | FLAIR | U-Net | anisotropic diffusion filter, normalisation, skull stripping, b.c. | DSC, accuracy, precision, sensitivity, specificity | 1st, 2nd | |
| (Yamamoto et al., 2022) | 28 MS | FLAIR | CNN | denoising | absolute VD, PPV, TPR, DSC, HD, F1 | 1st, 2nd | |
| (McKinley et al., 2019) | multi-centric 139 MS | T1-w, T2-w and FLAIR | FCNN | skull stripping, co-reg. | DSC | 1st, 2nd | |
| (Zhang et al., 2021b) | multi-centric 200 MS | T1-w, T2-w and FLAIR | U-Net | normalisation | DSC, LTPR, LPPV, lesion-wise F1 | 1st, 2nd | |
| (Abolvardi et al., 2019) | ISBI 2015 | FLAIR | U-Net | reg. | DSC | 1st, 2nd | |
| (Fenneteau et al., 2021a) | ISBI 2015, extra 30 MS | FLAIR | MPU-net | skull stripping, z-score normalisation | DSC | 1st, 2nd | |
| (de Oliveira et al., 2020) | ISBI 2015, extra 5 MS | T1-w and FLAIR | CNN | skull stripping, b.c. | volume of lesions | 1st, 2nd | |
| (Yildirim and Dandil, 2021a) | 38 MS | T2-w | mask R-CNN with ResNet101 as backbone | / | DSC, volume overlap error, LTPR, LFPR | 1st, 2nd | |
| (Tran et al., 2022) | 30 MS | T1-w and FLAIR | intensity-based | b.c. | WM hyperintensities volume agreement, DSC, FPR, TPR, F1 score | 1st, 2nd, 3rd | |
| (Cavedo et al., 2022) | 130 images multi-centric, different populations | FLAIR | intensity-based | / | WM hyperintensities volume, DSC, relative VD, absolute volume error | 1st, 2nd, 3rd | |
| (Brune et al., 2020) | 56 MS | MPRAGE and FLAIR | intensity-based | / | lesion count of tool vs neuroradiologists, single and multiple timepoints | 1st, 2nd, 3rd | |
| (Jain et al., 2017) | 22 MS | T1-w and FLAIR | maximum a posteriori model on image intensities of both time points | b.c., normalisation | DSC, F1, LTPR, LFPR, AVD | 1st, 2nd | |
| (Van Hecke et al., 2021) | batches of 10 and 25 MS, plus 87 subjects with CIS and MS |
T1-w and FLAIR | U-Net with attention gate layers | MNI reg., skull stripping, z-score normalisation | with vs without tool performances, surveys on patient’s perspective | 1st, 2nd, 3rd, 4th, 5th, 6th | |
| (Sousa et al., 2021) | ISBI 2015, MICCAI 2016, 33 MS bicentric | T1-w, FLAIR | CNN | skull stripping, b.c. | DSC, PPV, AVD | 1st, 2nd | |
| (Elsebely et al., 2021) | MICCAI 2008 | FLAIR | ensemble of SVMs and decision tree | contrast-brightness correction | DSC, accuracy, n. of TP/FP/FN, sensitivity, PPV | 1st, 2nd | |
| (Bhanumurthy and Anne, 2016) | MICCAI 2016 | FLAIR | modified histon based fast fuzzy C-means | normalisation | FPR, FNR, specificity, sensitivity, accuracy | 1st, 2nd | |
| (Ghodhbani et al., 2022) | MICCAI 2008, ISBI 2015 | T1-w, T2-w and FLAIR | U-Net | / | DSC | 1st, 2nd | |
| (Roy et al., 2013) | MICCAI 2008 | T1-w, T2-w and FLAIR | SVM | MNI reg., skull stripping, z-score normalisation | 1st, 2nd | ||
| (Chen et al., 2020) | MICCAI 2016, ISBI 2015 | PD-w, T1-w, T2-w and FLAIR | network with attention and graph convolution features | / | DSC, PPV, LFPR, LTPR, VD | 1st, 2nd | |
| (Tomas-Fernandez and Warfield, 2015) | synthetic and MICCAI 2008 | FLAIR | intensity-based using reference healthy population | b.c., denoising | PPV, LTPR, LFPR | 1st, 2nd | |
| (Ghribi et al., 2017) | MICCAI 2008, ISBI 2015 and 70 MS and HC | FLAIR | gaussian mixture model | b.c., skull stripping, denoising, z-score normalisation | DSC, PPV, LTPR, LFPR, SD, VD | 1st, 2nd | |
| (Salem et al., 2019) | 65 MS/CIS, synthetic and ISBI 2015 | T1-w and FLAIR | FCNN | b.c. | mean square error, structural similarity index, DSC, sensitivity, precision | 1st, 2nd | |
| (Hashemi et al., 2018) | MICCAI 2016, ISBI 2015 | MPRAGE, PD-w, FLAIR, T2-w | U-Net vs FC DenseNet | / | DSC, F2, sensitivity, precision, Jaccard index, PPV, LTPR, LFPR, VD | 1st, 2nd | |
| (Hou et al., 2019) | ISBI 2015 | FLAIR, t1-w, T2-w, PD-w | cross attention densely-connected network | / | DSC, Jaccard index, PPV, TPR, LFPR, LTPR, VD, SD | 1st, 2nd | |
| (Rondinella et al., 2023) | ISBI 2015 | FLAIR | U-Net with attention mechanism | skull stripping, normalisation and black images removal | DSC, sensitivity, specificity, extra fraction, IoU, PPV, NPV | 1st, 2nd | |
| (Weiss et al., 2013) | MICCAI 2008 | FLAIR | dictionary learning with sparsity constraint | skull stripping, normalisation | DSC, TPR, PPV | 1st, 2nd | |
| (Homayoun and Ebrahimpour-Komleh, 2017) | MICCAI 2008 | FLAIR | artificial neural network | skull stripping, b.c., denoising, normalisation | sensitivity, specificity, FPR, FNR, SI | 1st, 2nd | |
| (Andresen et al., 2022) | MS-SEG2 | FLAIR | CNN | skull stripping, 0–1 normalisation | F1, sensitivity, PPV (detection); DSC, SD, HD (segmentation) | 1st, 2nd | |
| (Valencia et al., 2022) | MS-SEG2 and 136 CIS | FLAIR and synthetic T1-w | FCNN | MNI reg., skull stripping, 0–1 normalisation | sensitivity, FDR, precision | 1st, 2nd | |
| (Salem et al., 2022) | MS-SEG2 | FLAIR | U-Net | skull stripping, Nyul normalisation, b.c. | F1, PPV, sensitivity, DSC | 1st, 2nd | |
| (Freire and Ferrari, 2016) | ISBI 2015 | FLAIR | intensity-based | denoising, b.c. | DSC, TPR, FPR, AVD | 1st, 2nd | |
| (Sarica et al., 2022) | ISBI 2015, MICCAI 2016 | T1-w, T2-w and FLAIR | residual U-Net | skull stripping, normalisation and zero padding (ISBI), denoising, skull stripping, b.c., normalisation (MICCAI) |
DSC, PPV, LTPR, LFPR, AVD | 1st, 2nd | |
| (Shahab et al., 2021) | ISBI 2015 | T1-w, T2-w and FLAIR | CNN | denoising, b.c., skull stripping, normalisation | DSC, Jaccard index, PPV, TPR, LFPR, LTPR, AVD | 1st, 2nd | |
| (Jog et al., 2015) | MICCAI 2008 and in-house 49 MS | T1-w, FLAIR, T2-w (MPRAGE for in-house) | decision trees | MNI reg., skull stripping, normalisation, b.c. | TPR, PPV, lesion volume | 1st, 2nd | |
| (Knight et al., 2018) | 96 MS multicentric, MICCAI 2016, ISBI 2015 | FLAIR | voxel-wise logistic regression | MNI reg., b.c., normalisation | SI, precision and recall | 1st, 2nd | |
| (Valcarcel et al., 2018) | 98 MS and ISBI 2015 | combinations of MPRAGE, PD-w, FLAIR, T2-w | local-level logistic regression | MNI reg., b.c., skull stripping, z-score normalisation | DSC, pAUC | 1st, 2nd | |
| (Gao et al., 2014) | MICCAI 2008 | T1-w, T2-w and FLAIR | energy minimisation and non-local means algorithm | b.c., reg. | DSC, specificity, FNR, VD | 1st, 2nd | |
| (Krishnan et al., 2023) | ISBI 2015, MICCAI 2016 and multicentric double blinded trial 798 + 714 + 416 MS |
T1-w, T2-w and FLAIR | U-Net | MNI reg., b.c., skull stripping | PPV, TPR, DSC on lesion volume (segmentation), LPPV, LTPR, LFPR on lesion count (detection), also AVD on ISBI |
||
| (Mechrez et al., 2016) | MICCAI 2008 and 38 MS | T1-w and FLAIR | intensity-based | b.c., skull stripping | VD, SSD, TPR, FPR | 1st, 2nd | |
| (Bouzidi et al., 2020) | 30 MS | T1-w, T2-w and FLAIR | otsu threshold and connected components filters | b.c., denoising | DSC, sensitivity, precision | 1st, 2nd | |
| (Bijar et al., 2012) | 20 MS | FLAIR | genetic algorithm and localised weighted filters | / | similarity criteria, overlap fraction, extra fraction | 1st, 2nd | |
| (Narayana et al., 2018) | multi-center, double-blinded, and randomized phase III clinical trial 1008 MS |
T1-w, T2-w, PD-w and FLAIR | CNN | skull stripping, b.c., normalisation | DSC | 1st, 2nd | |
| (Tripoliti et al., 2019) | / | / | / | / | 3rd, 5th | ||
| (Abhale et al., 2022) | 1000 MS from phase 3 multicentric trial | T2-w | FCNN | / | DSC | 1st, 2nd | |
| (Zangeneh and Yazdi, 2016) | 20 MS | FLAIR | gaussian mixture model and genetic algorithm | skull stripping | accuracy, number of FP, TP, FN, TN | 1st, 2nd |
Table 5.
Studies’ information containing details on datasets, inputs, and architecture of the automatic algorithm, pre-processing steps, and evaluation metrics (part 3).
| Data | Inputs | Method | Pre-processing | Evaluation metrics | QNI steps fulfilled | ||
|---|---|---|---|---|---|---|---|
| (Karpate et al., 2015) | 16 MS and 20 HC | MPRAGE, T2-w, FLAIR | Least squares probabilistic classification | b.c., denoising | precision, recall | 1st, 2nd | |
| (Mei et al., 2017) | 10 MS | FLAIR, T1-w (also with gadolinium) | self-organising maps (nerual network) | / | topographic and quantisation errors | 1st, 2nd | |
| (Zhang et al., 2018) | 69 MS | T1-w and FLAIR | generative adversarial network | b.c. on T1-w | DSC, recall, precision, F1 | 1st, 2nd | |
| (Dachraoui et al., 2020) | 30 MS | T1-w (also gadolinium), T2-w and FLAIR | Fuzzy C-Means clustering and geodesic models | skull stripping, denoising, contrast adjustment | number of TP, TN and precision | 1st, 2nd | |
| (Deshpande et al., 2015) | 14 MS | MPRAGE, PD-w, FLAIR | adaptive dictionary learning | b.c., skull stripping | PPV, sensitivity | 1st, 2nd | |
| (Harmouche et al., 2014) | 100 MS multicentric | T1-w, T2-w, PD-w, FLAIR (for half datasets) | Markov random fields | b.c., skull stripping, normalisation | DSC, sensitivity, PPR (ratio) | 1st, 2nd | |
| (Nass et al., 2022) | 30 MS | T1-w (also contrast enhanced), T2-w, FLAIR | Fuzzy C-Means | contrast adjustment | DSC | 1st, 2nd | |
| (Zhang et al., 2022) | 135 MS | FLAIR | 2D U-Net | normalisation | DSC, sensitivity, precision | 1st, 2nd | |
| (Ye et al., 2020) | 38 MS | diffusion basis spectrum imaging, T1-w and T2-w | FCNN | normalisation | number of predictions, AUC, sensitivity, specificity, F1 | 1st, 2nd | |
| (Thakur et al., 2022) | 200 MS per month for 10yrs | FLAIR | intensity subtraction between timepoints | b.c., skull stripping, resampling and reg. | number of clinical cases assessed with CAD, time per patient | 1st, 2nd, 3rd, 5th | |
| (Fartaria et al., 2019) | 25 MS bicentric | 7T MP2RAGE | partial volume estimation and topological constraints | skull stripping | % of detected lesions, FPR, AVD, F1 | 1st, 2nd | |
| (Hosseinipanah et al., 2019) | >80 MS | FLAIR | ensemble of SVMs | normalisation | DSC, JI, sensitivity, specificity, PPV | 1st, 2nd | |
| (Meier et al., 2017) | 29 MS + 13 MS and 15 HC | T1-w, T2-w and FLAIR | intensity-based with 2 thresholds for supra- and infra-tentorial | b.c., skull stripping, tissue segmentation, normalisation | sensitivity, specificity, DSC, Jaccard index, PPV, HD, TPR | 1st, 2nd | |
| (Jain et al., 2015) | 30 MS | T1-w and FLAIR | intensity-based | skull stripping on T1-w | DSC, AVD, total lesion VD, precision, sensitivity | 1st, 2nd | |
| (Bonanno et al., 2021) | 20 MS | FLAIR | Watershed-Clustering algorithm | denoising | accuracy, sesnitivity, specificity, AUC | 1st, 2nd | |
| (Huang et al., 2022) | 20 MS | FLAIR | V-Net | b.c., skull stripping, tissue segmentation | DSC, HD, AVD, TPR, F1 | 1st, 2nd | |
| (Narayana et al., 2019) | multicentric, double blinded, randomized trial 1008 MS | T1-w, T2-w, PD-w and FLAIR in combinations | U-Net | skull stripping, b.c., normalisation, denoising | DSC, FPR, TPR | 1st, 2nd | |
| (Arnold et al., 2022) | 33 MS bicentric | T1-w, T2-w, FLAIR | local-level logistic regression | b.c., normalisation | DSC, TPR, FDR | 1st, 2nd | |
| (Krüger et al., 2021) | 1809 MS multicentric | FLAIR or T1-w and FLAIR | CNN | strong artifacts removal | sensitivity, PPV, F1, DSC | 1st, 2nd | |
| (Fartaria et al., 2018) | 39 MS | FLAIR and MPRAGE | outlier rejection and region growing vs fuzzy clustering | skull stripping, b.c. | detection rate, FPR, DSC, lesion volume | 1st, 2nd | |
| (Karimian and Jafari, 2015) | 25 MS bicentric | T1-w, T2-w, FLAIR | Gaussian mixture model | skull stripping, normalisation | DSC, accuracy, specificity, sensitivity | 1st, 2nd | |
| (Sweeney et al., 2014) | 98 MS | T1-w, T2-w, FLAIR | intensity-based | MNI reg., skull stripping, b.c., normalisation | DSC, AUC, computational time | 1st, 2nd | |
| (Fartaria et al., 2015) | 39 MS | FLAIR, DIR, MPRAGE, MP2RAGE | k-nearest neighbour | reg. to MP2RAGE, skull stripping, b.c., normalisation | detection rate, sensitivity, specificity, accuracy, DSC | 1st, 2nd | |
| (Rosa et al., 2021) | 44 MS and 12 HC | MPRAGE to generate synthetic MP2RAGE | generative adversarial network | skull stripping, z-score normalisation | detection rate (WML and cortical), DSC, AVD | 1st, 2nd | |
| (Rovira et al., 2021) | 100 MS | PD-w, T2-w, MPRAGE, FLAIR | CNN | skull stripping, b.c., normalisation | number of new/enlarging lesions, per patient mean new/enlarging lesions | 1st, 2nd | |
| (Egger et al., 2016) | 50 MS | FLAIR | intensity-based | / | DSC, AVD, lesion count | 1st, 2nd | |
| (Cabezas et al., 2014) | 45 MS | FLAIR | intensity-based | atlas reg., skull stripping, b.c., denoising | TPF, FPF, DSC at voxel and lesion level | 1st, 2nd | |
| (Salem et al., 2017) | 60 MS/CIS | T1-w, T2-w, PD-w, FLAIR | image subtraction and logistic regression | b.c., skull stripping, Nyul normalisation | TPF, FPF, DSC | 1st, 2nd | |
| (Khotanlou and Afrasiabi, 2012) | 15 MS | T1-w, T2-w and FLAIR | SVM | denoising, morphological operations to exclude non-brain area | SI, overlap fraction, extra-fraction | 1st, 2nd | |
| (Valcarcel et al., 2018a) | 40 MS | T1-w, T2-w, FLAIR | local level logistic regression | skull stripping, b.c., normalisation | DSC, pAUC, root mean square, detection and outline errors | 1st, 2nd | |
| (Valcarcel et al., 2020) | 94 MS and 40 MS | T1-w, T2-w, FLAIR | intensity-based | MNI reg., skull stripping, normalisation | lesion volume bias, absolute volume error | 1st, 2nd | |
| (Dwyer et al., 2019) | multicentric 100 subjects, 192 MS/CIS, 15 MS, 125 MS and 76 HC | FLAIR | random forest classifier | b.c., normalisation | agreement with conventional T2-w lesion volume | 1st, 2nd | |
| (Battaglini et al., 2014) | multicentric, randomized, double-blind, placebo-controlled Phase II clinical trial 103 MS (randomly select 19 MS) |
T1-w, T2-w, PD-w | subtraction images between timepoints | skull stripping, normalisation | SI, lesion count | 1st, 2nd | |
| (Le et al., 2019) | 47 MS multicentric | FLAIR and T2-w combination (also T1-w depending on algorithm) |
comparison of three algorithms | b.c., skull stripping | lesion VD, DSC, sensitivity, symmetric SD | 1st, 2nd | |
| (Zhong et al., 2014) | 26 MS | FLAIR | high spatial frequency suppression | b.c., skull stripping, CSF sulcus and ventricle segmentation | SI, lesion volume | 1st, 2nd | |
| (Galimzianova et al., 2017) | 30 MS | FLAIR | Markov random fields | skull stripping, b.c. | DSC, total lesion load | 1st, 2nd | |
| (Schläger et al., 2022) | 74 MS multicentric | T1-w and FLAIR to generate synthetic DIR | generative adversarial network | b.c., normalisation | new lesions count (location based), disease activity assessment | 1st, 2nd | |
| (Subbanna et al., 2015) | 1195 MS | T1-w, T2-w, PD-w, FLAIR | two levels of Markov Random Fields | b.c., skull stripping, normalisation | sensitivity, PPV | 1st, 2nd | |
| (Galimzianova et al., 2015) | 30 MS | T1-w, T2-w and FLAIR | stratified mixture models | b.c. | DSC, Jeffrey’s divergence | 1st, 2nd | |
| (Spies et al., 2013) | 10 MS | MPRAGE | tissue segmentation, stereotactic normalisation and voxelwise stat analysis |
/ | DSC | 1st, 2nd | |
| (Ganiler et al., 2014) | 20 MS | T2-w, PD-w combinations | image subtraction | b.c., skull stripping, normalisation, WM masking | sensitivity, FDR, DSC | 1st, 2nd | |
| (Nguyen et al., 2018) | 30 MS | FLAIR | subtraction image | b.c., skull stripping, normalisation | sesnitivity, specificity, human review time | 1st, 2nd | |
| (Ong et al., 2012) | MICCAI 2008, 38 MS | T1-w, FLAIR | intensity-based | skull stripping, b.c. | lesion load, SI, Jaccard index, FPF, TPF | 1st, 2nd | |
| (Cerasa et al., 2011) | 11 MS | FLAIR | cellular neural network | skull stripping | DSC, total lesion load | 1st, 2nd | |
| (Kuwazuru et al., 2011) | 3 MS | T1-w, T2-w, FLAIR | SVM and artificial neural network | enhancement by subtraction of background | accuracy, SI, sensitivity, number of FP | 1st, 2nd | |
| (Bilello et al., 2013) | 88 MS | FLAIR | image subtraction | skull stripping, b.c. | with and without CAD: lesion count and location, PPV, NPV, sensitivity, specificity, efficiency, AUC, lesion-wise sensitivity, FPR and PPV, time spent, clinical reporting |
1st, 2nd, 3rd, 4th, 5th | |
| (Hindsholm et al., 2021) | 93 MS | FLAIR | 2D CNN | normalisation, cropping or zero padding | DSC, recall, F1, precision, qualitative assessment of output lesion masks | 1st, 2nd | |
| (Roy et al., 2015) | 10 MS | MPRAGE and FLAIR | patch based with temporal information from timepoints | normalisation | DSC, LTPR, LFPR, AVD | 1st, 2nd | |
| (Bouman et al., 2023) | 198 MS | DIR or PSIR generated artificially from T1-w, T2-w, PD-w or FLAIR |
U-Net-like | MNI reg., skull stripping, b.c. | lesions count, precision | 1st, 2nd | |
| (Sitter et al., 2017) | 69 MS + 1 CIS | 2D FLAIR and 3D T1-w | multiple models comparison | MNI reg., b.c. | SI, volumes of FP and FN | 1st, 2nd | |
| (Cabezas et al., 2016) | 36 MS/CIS | FLAIR | image subtraction | skull stripping, b.c., normalisation | DSC, FPR, TPR | 1st, 2nd |
3.1. Target population
In ten articles, MS patients were mixed with subjects presenting CIS (Salem et al., 2020, Jannat et al., 2021, Salem et al., 2019, Valencia et al., 2022, Salem et al., 2017, Dwyer et al., 2019, Sitter et al., 2017, Cabezas et al., 2016), or neuromyelitis optica spectrum disorders and cerebral small vessel disease (Zhang et al., 2022), or mild cognitive impairment, Alzheimer’s disease, Parkinson’s disease and frontotemporal dementia (Cavedo et al., 2022). The remaining studies targeted at least one dataset with only MS patients (see second columns of Table 3, Table 4, Table 5).
3.2. Magnetic resonance imaging
Fluid attenuated inversion recovery (FLAIR) was the most common MRI contrast used as input for the proposed automatic methods. It was used alone or in combination with a T1-weighted (T1-w) image, a T2-weighted (T2-w), a proton density weighted (PD-w) image, or with contrast enhancement (see third columns of Table 3, Table 4, Table 5). In six cases, the only input provided to the network were either T2-w images (Abhale et al., 2022, Yildirim and Dandil, 2021a), MPRAGE (Magnetisation-prepared rapid gradient echo) (Galimzianova et al., 2015, Spies et al., 2013), MP2RAGE (Magnetisation-prepared 2 rapid gradient echo) (Fartaria et al., 2019) or MR fingerprinting EPI (Echo-planar imaging) (Hermann et al., 2021). Less common contrasts, such as diffusion basis spectrum imaging (Ye et al., 2020), DIR (Fartaria et al., 2015, Schläger et al., 2022, Bouman et al., 2023) and PSIR (Bouman et al., 2023) were also adopted.
3.3. Datasets
The methods developed by 92 studies were (at least partially) based on datasets from international challenges: MICCAI 2008 (Styner et al., 2008), MICCAI 2016 (MS-SEG) (Commowick et al., 2018), MS-SEG2 (Commowick et al., 2021) and ISBI 2015 (Carass et al., 2017). Earlier works focused on relatively small cohorts due to the limited sample size provided in the challenges, such as 5 and 20 patients, respectively, in the training set of ISBI 2015 in Vang et al. (2020) and of MICCAI 2008 in Joshi and Sharma (2022).
A single case (Tripoliti et al., 2019) did not provide any reference dataset. The authors proposed the architecture of a tool for the estimation of MS progression, announcing a future proof of concept study with 30 patients for its validation. Since the target area was clearly determined, the first QNI step was considered satisfied.
The remaining 83 studies were based on data from large clinical trials, University hospitals or publicly available sources (see second columns of Table 3, Table 4, Table 5).
3.4. Automatic detection and segmentation
Many different automatic methods were developed for lesion detection and segmentation. Other studies used automatic methods, such as k-nearest neighbour (Fartaria et al., 2015, Todea et al., 2023, Steenwijk et al., 2013), Support Vector Machines (SVMs) (Abdullah et al., 2012, Opbroek et al., 2014, Elsebely et al., 2021, Roy et al., 2013, Hosseinipanah et al., 2019, Khotanlou and Afrasiabi, 2012, Kuwazuru et al., 2011), Markov random fields (Schmidt et al., 2011, Harmouche et al., 2014, Galimzianova et al., 2017, Subbanna et al., 2015), random forest (Geremia et al., 2011, Elliott et al., 2013, Dwyer et al., 2019), or ad hoc intensity-based algorithms (Tran et al., 2022, Cavedo et al., 2022, Brune et al., 2020, Tomas-Fernandez and Warfield, 2015, Freire and Ferrari, 2016, Mechrez et al., 2016, Meier et al., 2017, Jain et al., 2015, Sweeney et al., 2014, Egger et al., 2016, Cabezas et al., 2014, Valcarcel et al., 2020, Ong et al., 2012).
The high-level category of deep neural networks was predominant, where convolutional neural networks (CNNs) as U-Nets were most represented (see fourth column of Table 3, Table 4, Table 5). Basaran et al. (2022) adopted nnU-Net (Isensee et al., 2021), a method that automatically configures pre-processing steps, architecture, training and post-processing to better adapt to dataset properties and available hardware.
In Tripoliti et al. (2019) no details were disclosed about their automatic method and, as mentioned in Section 3.3, the reference dataset was not described. As a consequence, this conference paper did not fulfill the second QNI step.
Longitudinal methods (i.e., assessing changes in lesions' number and volume across two or more time points) adopt different approaches compared to cross-sectional methods (i.e., those using images acquired at a single time point). In fact, the evaluation of follow-up scans presents challenges, such as the one related to image registration—if patient positioning is not consistent—, and the one concerning the required pre-processing steps to account for variations in image acquisition between scans. Moreover, new lesions in follow-up scans are usually small and there is currently no threshold defining a significant lesion enlargement. To overcome these challenges, different approaches have been proposed to date such as the one proposed by Salem et al. (2022)—using a cascade of two FCNN’s to refine possible misclassifications—or the one suggested by Sepahvand et al. (2020), where an attention mechanism based on image subtraction between two timepoints was applied to help a U-Net differentiating between anatomical and artifactual change.
3.5. Data quality check and pre-processing
Data quality check, if mentioned, consisted of the removal of null slices (Ghosal et al., 2020, Kumar et al., 2019, Alijamaat et al., 2021, Rondinella et al., 2023), control of the scanning protocol and a thorough visual inspection (Schmidt et al., 2019). In Cavedo et al. (2022), before computing MRI analysis, a quality check of MRI parameters is performed to verify that the parameters align with those recommended. An image quality assessment was also explored in Valencia et al. (2022), through the median absolute error and the structural similarity index. Other metrics, such as lesion conspicuity, SNR (signal to noise ratio), contrast to noise ratio, and variance of the Laplacian were selected in Arnold et al. (2022). Narayana et al. (2018) used the automated pipeline validated in Narayana et al. (2013) to check headers and the SNR of DICOM images.
A more careful approach was developed in Rakic et al. (2021), dealing with T1-w and FLAIR modalities of 159 MS patients from multiple centers and scanners. In order to preserve robustness and minimize data bias, the authors followed a carefully designed protocol: the stratification of training, validation, and test set was obtained in a way to equally represent all data characteristics, such as screening site, scanner model, magnetic field strength, scan quality, slice thickness.
In Todea et al. (2023), two experts performed an image quality assessment (SNR, artifacts, contrast, good registration between time points) and a longitudinal analysis was evaluated on the whole dataset and on images with the same quality score. The same concept was applied to images obtained with a 1.5T and 3T scanner. On the other hand, in Combès et al. (2021), data with lower quality were intentionally not excluded from the study to mimic a real-world scenario.
Most studies include the following data pre-processing steps: bias field inhomogeneities correction, intensity normalization, skull stripping, denoising, resampling, and co-registration in the case of multiple input modalities.
3.6. Quantitative reports
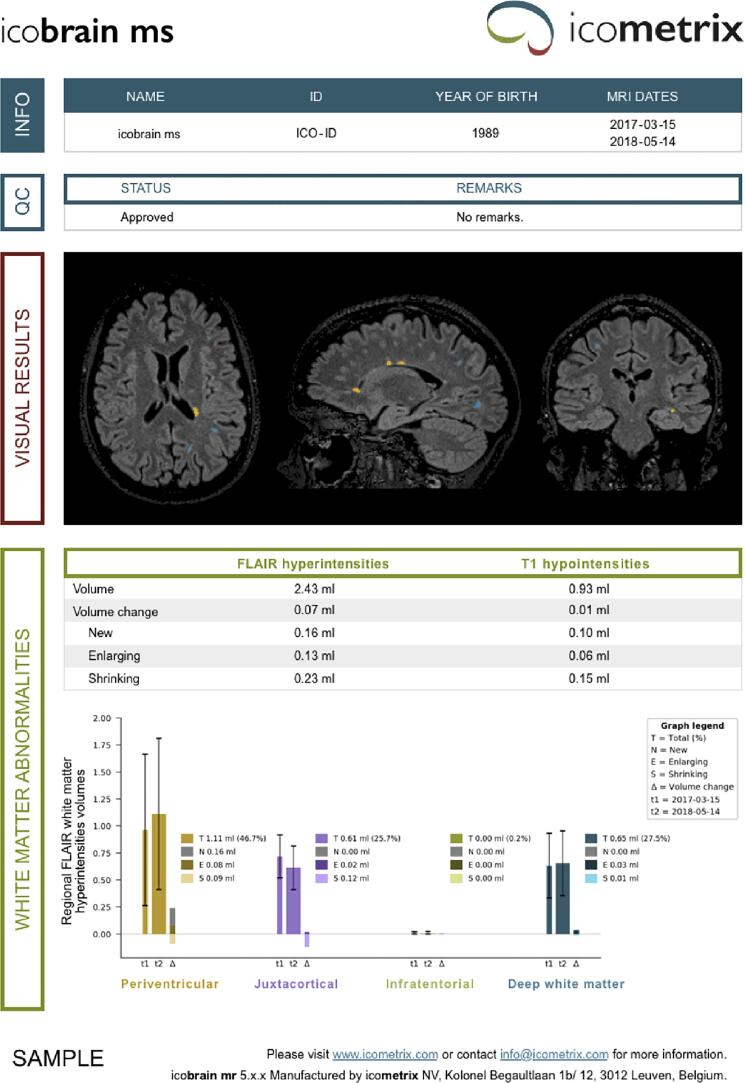
The results presented in 148 studies did not provide radiologists with a summary report. Combès et al., 2021, Brune et al., 2020, Tripoliti et al., 2019, Thakur et al., 2022, Van Hecke et al., 2021 explicitly explored the use of the developed tool to assist radiologists in generating a quantitative report, providing information such as the number, the volume and the location of lesions. Two examples are reported in Fig. 3.
Fig. 3.
Report example published in Van Hecke et al. (2021) (a) and Brune et al. (2020) (b).
In Cavedo et al. (2022), the authors presented a report with detection scores and the overlay of predictions on original images, while Yildirim and Dandil, 2021a generated a similar documentation in a web-based user interface tested by two radiologists.
In Bilello et al. (2013), the generated report contained new (or enlarging) and resolved (or improving) lesions detected, their specific location and the cerebral hemisphere involved.
3.7. Technical validation
The commonly explored technical evaluation metrics were those required to participate in the international contests (Maier-Hein et al., 2022):
-
1.
Overlap-based metrics, such as Dice score coefficient (DSC), sensitivity (recall), specificity, precision, accuracy, lesion-wise true positive rate (TPR) and false positive rate (FPR), the absolute volume difference between ground truth and predicted segmentation;
-
2.
Surface-based metrics, such as the average symmetric surface distance.
The lesion annotation through consensus was improved in the latest challenges: the available ground truth (GT) masks are more reliable in terms of inter-observer variability, providing higher quality GT to train and evaluate the models. An exhaustive list of adopted metrics is reported in the sixth columns of Table 3, Table 4, Table 5.
The latest reviews (Diaz-Hurtado et al., 2022; Commowick et al., 2023) report satisfactory and already close to human rater performances for many detection/segmentation automatic methods. However, as also mentioned in Commowick et al. (2023), there are currently little data related to the integration and use of those methods in clinical routine, especially in relation to the quantification of the uncertainty of their predictions in clinical practice.
3.8. Clinical validation
Combès et al. (2021) proposed a pre-use validation of their tool involving clinicians. The authors assessed the impact of the segmentation tool on experts’ performances as follows: three experts were asked to annotate a point near each lesion’s center (for 48 patients) with and without the help of the automatic tool (referred to as phases one and two). The number of marked lesions and time spent during the procedure were recorded in both cases. All experts were exhorted to conduct this experiment in situations similar to clinical practice. In particular, they were explicitly instructed to spend a reading time comparable to that of clinical routine. A few days prior to the first phase, each expert followed a short training session to get acquainted with the tool.
This experiment was evaluated through several metrics and compared between the two phases, such as the number of detected lesions (by each rater and overall), the average patient-wise number of lesions detected by experts (compared between phases using a paired t-test), or the pooled inter-expert standard deviation associated to the number of detected lesions.
In addition, the impact on routine clinical practice was assessed on six patients, with and without the tool (the two phases were two weeks apart): the experts measured the time needed from loading and reading MRI in hospital Picture Archiving and Communication Systems (PACS) to generating a radiology report. Patients were categorized in the report as showing “no activity”, “1 lesion” or “>1 lesion” with respect to baseline. Time spent to perform radiological readings for each of the three experts and each of the two settings were summarized, and the mean times elapsed in the two settings were tested for equality using a paired t-test.
A post-experiment interview was conducted to ask experts whether they were satisfied with the tool’s level of information and performance.
In Van Hecke et al. (2021), lesion segmentations were compared with the assessment of two raters, one experienced radiologist and one assistant neurologist. The experiment consisted of marking and counting MS lesions on images from 10 patients. The two raters independently assessed all images, which were shuffled and presented first as original scans, then with automatic lesion annotations. The reporting time was recorded, and the agreement between the counts reported by the two raters with and without the tool was analysed. Moreover, a similar procedure was followed to test if the help of automatic reports might change radiological findings when assessing follow-up scans.
In Bilello et al. (2013), two neuroradiologists generated a clinical report without assistance from the CAD software. Independently, the same scans were assessed by another neuroradiologist using only the software output. In both cases, new, enlarging, resolved and improving detected lesions were compared, as well as the specified lesion location. The duration of the software-assisted pipeline was also recorded for each scan, not including the image processing time.
Yildirim and Dandil, 2021a reported having their pipeline tested by two radiologists and evaluated as an auxiliary tool for diagnosis and decision support in terms of ease of use, practicality, working speed, and automatic detection. Since no details on the modality of these tests were disclosed in the article, the fourth QNI step can not be considered fulfilled.
Similarly, Hindsholm et al. (2021) only presented a qualitative assessment of output masks by radiologists. Hence, their clinical validation does not comply with the QNI framework.
Technologists involved by Thakur et al. (2022) reported the time for manual intervention to execute the tool and the time to assess and generate a report for a single patient. However, they used these findings to compare two versions of the same software instead of evaluating advantages with respect to a manual assessment. For this reason this article did not fulfill the fourth QNI step.
3.9. Integration into clinical workflow
In Bilello et al. (2013), the DICOM series of all the paired examinations were available in PACS to be exported and used as inputs to the automated method. Similarly, in Tripoliti et al. (2019) the user can retrieve imaging data either from the PACS or the local disk of the computer where the automatic software is installed.
In Combès et al. (2021), once stored in the local clinical PACS, MR images were pseudonymized and securely transferred into a processing hosting (certified health data hosting provider), and new lesions were automatically segmented. Then, the processed images and corresponding segmentation maps were transferred back to PACS, which could be visualized in a dedicated web MRI viewer (using DICOM format).
Van Hecke et al. (2021) developed a platform including a web portal for healthcare professionals, volumetric brain reports, and the integration with hospitals’ PACS and electronic medical record systems.
In Thakur et al. (2022), the automated software was integrated and routinely used in clinical practice since April 2012. The images were stored in PACS and converted from DICOM to NIfTI (Neuroimaging informatics technology initiative) for processing. The authors mentioned their method needs MRI scans to be acquired at the same institution.
The integration of the tool into the clinical workflow was only partially investigated in Yildirim and Dandil, 2021a, including data compatibility and the visualisation of segmented lesions overlayed with the input image. Yet, the integration of their web-based system with a hospital electronic information system, such as PACS, was not considered. Thus, the fifth QNI step was not satisfied.
3.10. In-use validation
Van Hecke et al. (2021) presented and tested a care management system, including a patient mobile phone application (available on Android and iOS) and a website. A first patient’s perspective survey was conducted to understand patients’ attitude towards the app, different possible features, and their level of interest in using such application. A second survey collected information such as patients’ propensity to view MRI images on their own, or if they would be interested in knowing whether there were any changes in follow-ups (such as new lesions or brain volume loss).
3.11. QNI steps fulfillment
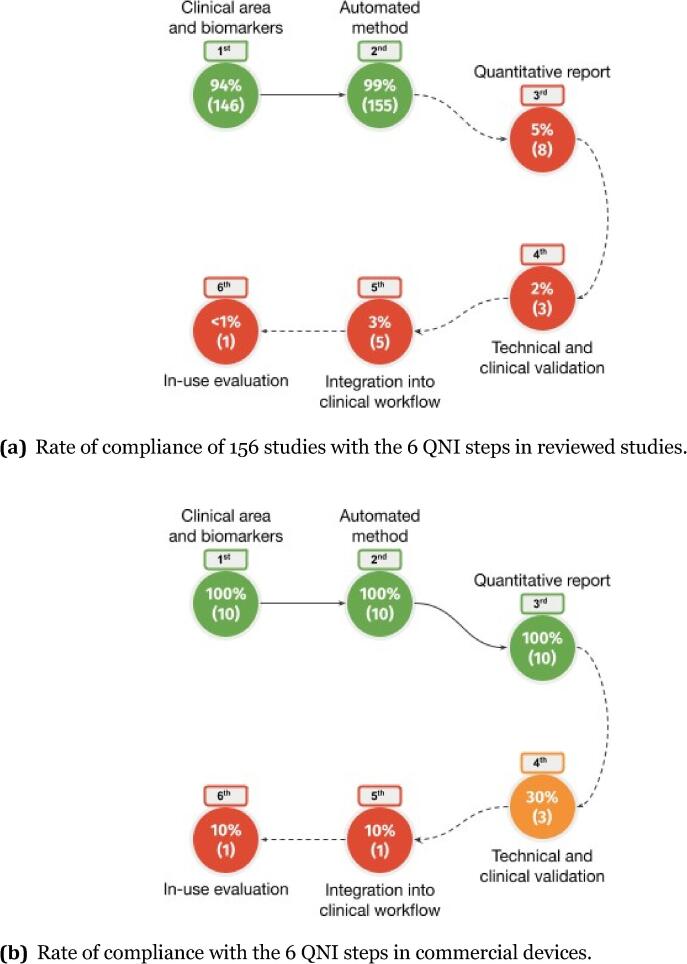
Based on the findings presented in 156 studies, 146 comply with the first QNI step, while 155 fulfill the second. The third step is considered by eight works, three studies fully investigate the fourth and five the fifth. Only a single article explores the last QNI step. An overview of the fulfillment of QNI steps in the screened literature is presented in the road map of Fig. 4a. A similar road map can be generated from data related to 10 commercial devices screened by Mendelsohn et al. (2022), reported in Fig. 4b. A summary of the fulfilled steps is reported in the last columns of Table 3, Table 4, Table 5.
Fig. 4.
Rate of compliance with the 6 QNI steps in reviewed studies (a) and commercial devices (b).
4. Discussion
The present systematic review exposes a considerable gap between methods’ development and the introduction of those methods into clinical practice. There are many possible cause for this gap.
A first explanation could be the difficulty to implement clinical trials: complying with clinical regulations and addressing ethical issues might result in an undesirable delay of the investigation. Participants’ insufficient knowledge about trial methods and the complexity of study protocols might also jeopardise patients’ recruitment process. The lack of trained medical personnel could represent a problem, when designing a clinical trial and even in the case of an internal clinical validation. All the above reasons are not specific to MS, meaning they could apply to many other neurological and non-neurological disorders.
Clinical integration presents, as well, some significant hurdles. To be applied in clinical practice, lesion segmentation methods should not only be integrated in the clinical workflow (i.e., be integrated in clinical PACS systems; be readily applicable to MR data that have not been preprocessed and sometimes acquired in different scanners, or with different image quality despite a consistent acquisition protocol, etc.) but also provide means to evaluate their outcome’s uncertainty and errors. Ad-hoc integration designs need to be developed considering the current clinical neuroradiological workflow as well as evaluating the reliability of those methods in a clinical routine setting, and the related clinicians’ trust in using them as clinical decision support tools. To help cover these aspects, an automatic tool could be conceived within a quality management framework for medical devices. The handling of possible failures, risk monitoring and data storage would also be addressed by following such guidelines. Data storage, management and sharing systems, such as KHEOPS (https://kheops.online/) or Flywheel (https://flywheel.io/), could be a way to deal with PACS and anonymise imaging data acquired at hospitals. Moreover, the use of a docker to execute software in an isolated and reproducible environment could help towards clinical integration. As to the real advantage of using automated methods in clinical routine, these should be carefully evaluated on site by providing means to assess errors and eventually also correct them for future evaluations, as for example could be done with uncertainty estimations/explainable AI and user-friendly interactive interfaces.
Along with this, the trade off between the economic costs of a clinical implementation and MS incidence may play an important role. In this sense, addressing medium to long-term effects (last QNI step) of the tool would be helpful. Studies should provide documentation such as:
-
1.
periodical reports on how easily the tool could be integrated and feedback from users
-
2.
the speed of diagnosis and failure rate, compared to pre-use cases
-
3.
the amount of required resources, productivity, patient perception, and economic impact.
On the other hand, if a tool is not clinically adopted, its efficacy and perception could be part of the reasons. An extremely wide range of solutions with respect to the methods characteristics, inputs, and processing steps is already available and discussed in reviews, such as (Llado et al., 2012; García-Lorenzo et al., 2012; Alrabai et al., 2022; Zeng et al., 2020; Ma et al., 2022; Diaz-Hurtado et al., 2022; Commowick et al., 2023). What is actually lacking is a validation that demonstrates the advantages of automatic methods with respect to the standard procedure.
Furthermore, many of the reviewed studies have been performed on data from international challenges, which were to some extent curated and, thus, did not reflect current “real-world” clinical scenarios. Feedback from radiologists and neurologists on clinical data could help methods explore and mitigate potential implementation biases (Vokinger et al., 2021; Varoquaux and Cheplygina, 2022). At the same time, this could change the way the tool is perceived in the clinical environment.
An additional reason may be that latest methods struggle to adapt to the heterogeneity of data acquired in clinical settings. Some recent works attempted at addressing the challenge of the use of images acquired with different contrast mechanisms and in scanners produced by different vendors and with different field strengths (Cerri et al., 2021; Billot et al., 2021). The issue represented by the different spatial resolution of clinical images, leading to variable partial volume effect during resampling, still requires ad hoc solutions and additional validation with on-site data. Also, an ad hoc integration of a method into a single institutional PACS may not generalise well in the case of a multicentric study.
Another possible motivation for the existing gap between development and clinical integration of methods could be the lack of national and international initiatives to promote their translation into clinical practice. In the current situation there is still a pronounced imbalance in favour of challenges supporting technical evaluations. Similar initiatives related to clinical validation and integration would certainly represent a boost in the implementation of solutions for MS lesion segmentation. Research focused on the integration of those methods into the clinical workflow as well as on the evaluation of their performance in a clinical routine setting might substantially help promoting their adoption and use by both neuroradiologists and neurologists.
Moreover, reducing the gap between the methods’ development and clinical translation might be highly beneficial also to improve the robustness and minimise the implementation bias of software solutions for MS lesion detection/segmentation. Ultimately, also patients would benefit from a more efficient and trustworthy process supporting disease diagnosis and monitoring of treatment effects.
5. Conclusions
We systematically reviewed automatic MS lesion detection and segmentation tools to assess their maturity towards clinical integration. Using the six steps of the QNI framework, we examined these quantitative tools’ development, validation, and integration level in the clinical workflow. In this review, we focused on the required development towards clinical application of MS lesion segmentation methods, and showed that—to date—there is no consistent evidence of tools’ integration into the clinical workflow. Our work demonstrates, therefore, that there is an important gap that needs to be filled by future research in this field. In addition, the socio-economic effects and the impact on patients’ management of those tools have yet to be studied.
Acknowledgements
This work was supported by the Hasler Foundation with the project MSxplain number 21042.
Data availability
Data will be made available on request.
References
- Abdullah B., Younis A., John N. Multi-Sectional Views Textural Based SVM for MS Lesion Segmentation in Multi-Channels MRIs. Open Biomed. Eng. J. 2012;6:56–72. doi: 10.2174/1874230001206010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhale P., Lashkare A., Deshpande A. Early Stage Detection of Multiple Sclerosis using FCNN. In 2022 10th International Conference on Emerging. Trends in Engineering and Technology - Signal and Information Processing. 2022;(ICETET-SIP-22),:01–04. doi: 10.1109/ICETET-SIP-2254415.2022.9791566. [DOI] [Google Scholar]
- Abolvardi, A., Hamey, L., and Ho-Shon, K. (2019). Registration Based Data Augmentation for Multiple Sclerosis Lesion Segmentation. In 2019 Digital Image Computing: Techniques and Applications (DICTA), pages 1–5. doi:10.1109/DICTA47822.2019.8946022.
- Ackaouy A., Courty N., Vallee E., Commowick O., Barillot C., Galassi F. Unsupervised Domain Adaptation With Optimal Transport in Multi-Site Segmentation of Multiple Sclerosis Lesions From MRI Data. Front. Comput. Neurosci. 2020;14:19. doi: 10.3389/fncom.2020.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alijamaat A., Nikravanshalmani A., Bayat P. Multiple sclerosis lesion segmentation from brain MRI using U-Net based on wavelet pooling. Int. J. Comput. Assist. Radiol. Surg. 2021;16 doi: 10.1007/s11548-021-02327-y. [DOI] [PubMed] [Google Scholar]
- Alrabai, A., Echtioui, A., and Hamida, A. (2022). Multiple sclerosis segmentation using deep learning models: Comparative study. In 2022 6th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP). doi:10.1109/ATSIP55956.2022.9805983.
- Andresen J., Uzunova H., Ehrhardt J., Kepp T., Handels H. Image registration and appearance adaptation in non-correspondent image regions for new ms lesions detection. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.981523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold T., Tu D., Okar S., Nair G., By S., Kawatra K., Robert-Fitzgerald T., Desiderio L., Schindler M., Shinohara R., Reich D., Stein J. Sensitivity of portable low-field magnetic resonance imaging for multiple sclerosis lesions. NeuroImage: Clinical. 2022;35 doi: 10.1016/j.nicl.2022.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashtari P., Barile B., Huffel S., Sappey-Marinier D. New multiple sclerosis lesion segmentation and detection using pre-activation U-Net. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.975862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslani S., Dayan M., Storelli L., Filippi M., Murino V., Rocca M., Sona D. Multi-branch convolutional neural network for multiple sclerosis lesion segmentation. NeuroImage. 2019;196 doi: 10.1016/j.neuroimage.2019.03.068. [DOI] [PubMed] [Google Scholar]
- Basaran B., Matthews P., Bai W. New lesion segmentation for multiple sclerosis brain images with imaging and lesion-aware augmentation. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.1007453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglini M., Rossi F., Grove R., Stromillo M., Whitcher B., Matthews P., De Stefano N. Automated identification of brain new lesions in multiple sclerosis using subtraction images. J. Magn. Reson. Imaging. 2014;39 doi: 10.1002/jMRI.24293. [DOI] [PubMed] [Google Scholar]
- Bhanumurthy Y., Anne K. An automated MRI segmentation framework for brains with tumors and multiple sclerosis lesions. In 2016 International Conference on Computation of Power. Energy Information and Commuincation (ICCPEIC) 2016:231–236. doi: 10.1109/ICCPEIC.2016.7557201. [DOI] [Google Scholar]
- Bijar A., Khayati R., Benavent A. Fuzzy based segmentation of multiple sclerosis lesions in magnetic resonance brain images. Proceedings - IEEE Symposium on Computer-Based Medical Systems. 2012 doi: 10.1109/CBMS.2012.6266317. [DOI] [Google Scholar]
- Bilello M., Arkuszewski M., Nucifora P., Nasrallah I., Melhem E., Cirillo L., Krejza J. Multiple sclerosis: Identification of temporal changes in brain lesions with computer-assisted detection software. Neuroradiol. J. 2013;26:143–150. doi: 10.1177/197140091302600202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot B., Cerri S., Van Leemput K., Dalca A., Iglesias J. Joint Segmentation Of Multiple Sclerosis Lesions And Brain Anatomy In MRI Scans Of Any Contrast And Resolution With CNNs. IEEE International Symposium on Biomedical Imaging. 2021;2021:1971–1974. doi: 10.1109/ISBI48211.2021.9434127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno L., Mammone N., De Salvo S., Bramanti A., Rifici C., Sessa E., Bramanti P., Marino S., Ciurleo R. Multiple sclerosis lesions detection by a hybrid watershed-clustering algorithm. Clin. Imaging. 2021;72:162–167. doi: 10.1016/j.clinimag.2020.11.006. [DOI] [PubMed] [Google Scholar]
- Bouman P., Noteboom S., Santos F., Beck E., Bliault G., Castellaro M., Calabrese M., Chard D., Eichinger P., Filippi M., Inglese M., Lapucci C., Marciniak A., Moraal B., Morales Pinzón A., Mühlau M., Preziosa P., Reich D., Rocca M., Steenwijk M. Multicenter evaluation of ai-generated dir and psir for cortical and juxtacortical multiple sclerosis lesion detection. Radiology. 2023;307 doi: 10.1148/radiol.221425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzidi, D., Fahmi, G., Taouil, K., and Fakhfakh, A. (2020). BrainSeg3D to Detect Multiple Sclerosis Lesions Using Magnetic Resonance Imaging. In 2020 IEEE International Conference on Design & Test of Integrated Micro & Nano-Systems (DTS), pages 1–6. doi:10.1109/DTS48731.2020.9196053.
- Brosch T., Tang L., Yoo Y., Li D., Traboulsee A., Tam R. Deep 3D Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation. IEEE Trans. Med. Imaging. 2016;35:1. doi: 10.1109/TMI.2016.2528821. [DOI] [PubMed] [Google Scholar]
- Brune S., Høgestøl E., Cengija V., Berg-Hansen P., Sowa P., Nygaard G., Harbo H., Beyer M. LesionQuant for Assessment of MRI in Multiple Sclerosis–A Promising Supplement to the Visual Scan Inspection. Front. Neurol. 2020;11 doi: 10.3389/fneur.2020.546744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas M., Corral J., Oliver A., Diez Y., Tintorè M., Auger C., Montalban X., Llado M., Pareto D., Rovira A. Improved automatic detection of new t2 lesions in multiple sclerosis using deformation fields. Am. J. Neuroradiol. 2016;37 doi: 10.3174/ajnr.A4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas M., Oliver A., Roura E., Freixenet J., Vilanova J.C., Ramió-Torrentà L., Rovira A., Llado X. Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding. Comput. Methods Programs Biomed. 2014;115 doi: 10.1016/j.cmpb.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Cabezas M., Oliver A., Valverde S., Beltran B., Freixenet J., Vilanova J.C., Ramio-Torrenta L., Rovira A., Llado X. BOOST: A supervised approach for multiple sclerosis lesion segmentation. J. Neurosci. Methods. 2014;237:108–117. doi: 10.1016/j.jneumeth.2014.08.024. [DOI] [PubMed] [Google Scholar]
- Carass A., Roy S., Jog A., Cuzzocreo J.L., Magrath E., Gherman A., Button J., Nguyen J., Prados F., Sudre C.H., Jorge Cardoso M., Cawley N., Ciccarelli O., Wheeler-Kingshott C.A., Ourselin S., Catanese L., Deshpande H., Maurel P., Commowick O., Barillot C., Tomas-Fernandez X., Warfield S.K., Vaidya S., Chunduru A., Muthuganapathy R., Krishnamurthi G., Jesson A., Arbel T., Maier O., Handels H., Iheme L.O., Unay D., Jain S., Sima D.M., Smeets D., Ghafoorian M., Platel B., Birenbaum A., Greenspan H., Bazin P.-L., Calabresi P.A., Crainiceanu C.M., Ellingsen L.M., Reich D.S., Prince J.L., Pham D.L. Longitudinal multiple sclerosis lesion segmentation: Resource and challenge. NeuroImage. 2017;148:77–102. doi: 10.1016/j.neuroimage.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavedo E., Tran P., Thoprakarn U., Martini J.-B., Movschin A., Delmaire C., Gariel F., Heidelberg D., Pyatigorskaya N., Ströer S., Krolak-Salmon P., Cotton F., Santos C., Dormont D. Validation of an automatic tool for the rapid measurement of brain atrophy and white matter hyperintensity: QyScore. Eur. Radiol. 2022;32 doi: 10.1007/s00330-021-08385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerasa A., Bilotta E., Augimeri A., Cherubini A., Pantano P., Zito G., Lanza P., Valentino P., Gioia M., Quattrone A. A cellular neural network methodology for the automated segmentation of multiple sclerosis lesions. J. Neurosci. Methods. 2011;203:193–199. doi: 10.1016/j.jneumeth.2011.08.047. [DOI] [PubMed] [Google Scholar]
- Cerri S., Puonti O., Meier D., Wuerfel J., Mühlau M., Siebner H., Van Leemput K. A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. NeuroImage. 2021;225:117471. doi: 10.1016/j.neuroimage.2020.117471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Ru J., Zhou Y., Rekik I., Pan Z., Lin Y., Lu B., Shi J. MTANS: Multi-Scale Mean Teacher Combined Adversarial Network with Shape-Aware Embedding for Semi-Supervised Brain Lesion Segmentation. NeuroImage. 2021;244 doi: 10.1016/j.neuroimage.2021.118568. [DOI] [PubMed] [Google Scholar]
- Chen Z., Wang X., Huang J., Lu J., Zheng J. Deep Attention and Graphical Neural Network for Multiple Sclerosis Lesion Segmentation From MR Imaging Sequences. IEEE J. Biomed. Health Inform. 2021 doi: 10.1109/JBHI.2021.3109119. PP:1–1. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Wang, X., and Zheng, J. (2020). Hybrid Feature Network Driven by Attention and Graph Features for Multiple Sclerosis Lesion Segmentation from MR Images. In 2020 16th International Conference on Control, Automation, Robotics and Vision (ICARCV), pages 678–683. doi:10.1109/ICARCV50220.2020.9305404.
- Combès B., Kerbrat A., Pasquier G., Commowick O., Bon B., Galassi F., L’Hostis P., Graoui N., Chouteau R., Cordonnier E., Edan G., Ferré J.-C. A Clinically-Compatible Workflow for Computer-Aided Assessment of Brain Disease Activity in Multiple Sclerosis Patients. Front. Med. 2021;8 doi: 10.3389/fmed.2021.740248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commowick, O., Cervenansky, F., Cotton, F., and Dojat, M. (2021). MSSEG-2 challenge proceedings: Multiple sclerosis new lesions segmentation challenge using a data management and processing infrastructure. In MICCAI 2021-24th International Conference on Medical Image Computing and Computer Assisted Intervention, page 126.
- Commowick O., Combès B., Cervenansky F., Dojat M. Editorial: Automatic methods for multiple sclerosis new lesions detection and segmentation. Front. Neurosci. 2023;17 doi: 10.3389/fnins.2023.1176625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commowick O., Istace A., Kain M., Laurent B., Leray F., Simon M., Pop S., Girard P., Ameli R., Ferré J.-C., Kerbrat A., Tourdias T., Cervenansky F., Glatard T., Beaumont J., Doyle S., Forbes F., Knight J., Khademi A., Barillot C. Objective Evaluation of Multiple Sclerosis Lesion Segmentation using a Data Management and Processing Infrastructure. Sci. Rep. 2018;8:13650–13666. doi: 10.1038/s41598-018-31911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachraoui C., Labidi S., Mouelhi A. Computerized image segmentation of multiple sclerosis lesions using fuzzy level set model. International Conference on Advanced Technologies for Signal and Image Processing (ATSIP) 2020 doi: 10.1109/ATSIP49331.2020.9231765. [DOI] [Google Scholar]
- de Oliveira M., Santinelli F., Silva M., Coronetti F., Barbieri F., Lisboa-Filho P., Santos J., Cardoso J. In Conference: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2020. Quantification of Brain Lesions in Multiple Sclerosis Patients using Segmentation by Convolutional Neural Networks; pp. 2045–2048. [DOI] [Google Scholar]
- de Oliveira M., Silva M., Coronetti F., Santos J., Cardoso J., Lisboa-Filho P. Lesion Volume Quantification Using Two Convolutional Neural Networks in MRIs of Multiple Sclerosis Patients. Diagnostics. 2022;12:230. doi: 10.3390/diagnostics12020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande H., Maurel P., Barillot C. Adaptive dictionary learning for competitive classification of multiple sclerosis lesions. International Symposium on Biomedical Imaging, New York, USA. 2015;2015 doi: 10.1109/ISBI.2015.7163834. [DOI] [PubMed] [Google Scholar]
- Diaz-Hurtado M., Martínez-Heras E., Solana E., Casas-Roma J., Llufriu S., Kanber B., Prados F. Recent advances in the longitudinal segmentation of multiple sclerosis lesions on magnetic resonance imaging: a review. Neuroradiology. 2022;64 doi: 10.1007/s00234-022-03019-3. [DOI] [PubMed] [Google Scholar]
- Dwyer M., Bergsland N., Ramasamy D., Weinstock-Guttman B., Barnett M., Wang C., Tomic D., Silva D., Zivadinov R. Salient Central Lesion Volume: A Standardized Novel Fully Automated Proxy for Brain FLAIR Lesion Volume in Multiple Sclerosis. J. Neuroimaging. 2019;29 doi: 10.1111/jon.12650. [DOI] [PubMed] [Google Scholar]
- Egger C., Opfer R., Wang C., Kepp T., Sormani M.P., Spies L., Barnett M., Schippling S. Clinical; NeuroImage: 2016. MRI flair lesion segmentation in multiple sclerosis: Does automated segmentation hold up with manual annotation? p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger C., Opfer R., Wang C., Kepp T., Sormani M.P., Spies L., Barnett M., Schippling S. MRI FLAIR lesion segmentation in multiple sclerosis: Does automated segmentation hold up with manual annotation? NeuroImage Clinical. 2017;13:264–270. doi: 10.1016/j.nicl.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C., Collins L., Arnold D., Arbel T. Temporally Consistent Probabilistic Detection of New Multiple Sclerosis Lesions in Brain MRI. IEEE Trans. Med. Imaging. 2013;32 doi: 10.1109/TMI.2013.2258403. [DOI] [PubMed] [Google Scholar]
- Elsebely R., Hassan Yousef A., Salem A., Abdullah B. Automatic Segmentation of Multiple Sclerosis Lesions in Brain MR Images Using Ensemble Machine Learning. In 2021 International Mobile. Intelligent, and Ubiquitous Computing Conference (MIUCC) 2021:28–33. doi: 10.1109/MIUCC52538.2021.9447657. [DOI] [Google Scholar]
- Essa E., Al-Desouky D., Hussein S., Rashad M. Neuro-fuzzy patch-wiseR-CNN for multiple sclerosis segmentation. Med. Biolog. Eng. Comput. 2020;58 doi: 10.1007/s11517-020-02225-6. [DOI] [PubMed] [Google Scholar]
- Fartaria M., Bonnier G., Roche A., Kober T., Meuli R., Rotzinger D., Frackowiak R., Schluep M., Du Pasquier R., Thiran J.-P., Krueger G., Bach Cuadra M., Granziera C. Automated detection of white matter and cortical lesions in early stages of multiple sclerosis. J. Magn. Resonance Imaging: JMRI. 2015;43 doi: 10.1002/jMRI.25095. [DOI] [PubMed] [Google Scholar]
- Fartaria M., Sati P., Todea A., Radue E.-W., Rahmanzadeh R., O’Brien K., Reich D., Bach Cuadra M., Kober T., Granziera C. Automated detection and segmentation of multiple sclerosis lesions using ultra-high-field mp2rage. Investig. Radiol. 2019 doi: 10.1097/RLI.0000000000000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fartaria M., Todea A., Kober T., O’brien K., Krueger G., Meuli R., Granziera C., Roche A., Bach Cuadra M. Partial volume-aware assessment of multiple sclerosis lesions. NeuroImage: Clinical. 2018;18:245–253. doi: 10.1016/j.nicl.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein A. The Neuropsychiatry of Multiple Sclerosis. Can. J. Psychiatry. 2004;49(3):157–163. doi: 10.1177/070674370404900302. [DOI] [PubMed] [Google Scholar]
- Fenneteau A., Bourdon P., Helbert D., Fernandez-Maloigne C., Habas C., Guillevin R. Investigating efficient CNN architecture for multiple sclerosis lesion segmentation. J. Med. Imaging. 2021;8 doi: 10.1117/1.JMI.8.1.014504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire P., Ferrari R. Automatic iterative segmentation of multiple sclerosis lesions using Student’s t-mixture models and probabilistic anatomical atlases in FLAIR images. Comput. Biol. Med. 2016;73 doi: 10.1016/j.compbiomed.2016.03.025. [DOI] [PubMed] [Google Scholar]
- Gabr R., Coronado I., Robinson M., Sujit S., Datta S., Sun X., Allen W., Lublin F., Wolinsky J., Narayana P. Brain and lesion segmentation in multiple sclerosis using fully convolutional neural networks: A large-scale study. Multiple Sclerosis J. 2019;26 doi: 10.1177/1352458519856843. 135245851985684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimzianova A., Lesjak Z., Rubin D., Likar B., Pernus F., Spiclin Z. Locally adaptive magnetic resonance intensity models for unsupervised segmentation of multiple sclerosis lesions. J. Med. Imaging. 2017;5:1. doi: 10.1117/1.JMI.5.1.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimzianova A., Pernuš F., Likar B., Spiclin Z. Stratified mixture modeling for segmentation of white-matter lesions in brain MR images. NeuroImage. 2015;124 doi: 10.1016/j.neuroimage.2015.09.047. [DOI] [PubMed] [Google Scholar]
- Ganiler O., Oliver A., Diez Y., Freixenet J., Vilanova J.C., Beltran B., Ramio-Torrenta L., Rovira A., Llado X. A subtraction pipeline for automatic detection of new appearing multiple sclerosis lesions in longitudinal studies. Neuroradiology. 2014;56 doi: 10.1007/s00234-014-1343-1. [DOI] [PubMed] [Google Scholar]
- Gao J., Li C., Feng C., Xie M., Yin Y., Davatzikos C. Non-locally regularized segmentation of multiple sclerosis lesion from multi-channel MRI data. Magn. Resonance Imaging. 2014;32 doi: 10.1016/j.MRI.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Lorenzo D., Francis S., Narayanan S., Arnold D., Collins L. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Medical image analysis. 2012;17 doi: 10.1016/j.media.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Geremia E., Clatz O., Menze B., Konukoglu E., Criminisi A., Ayache N. Spatial Decision Forests for MS Lesion Segmentation in Multi-Channel Magnetic Resonance Images. NeuroImage. 2011;57:378–390. doi: 10.1016/j.neuroimage.2011.03.080. [DOI] [PubMed] [Google Scholar]
- Gessert N., Krüger J., Opfer R., Ostwaldt A.-C., Manogaran P., Kitzler H.H., Schippling S., Schlaefer A. Multiple sclerosis lesion activity segmentation with attention-guided two-path CNNs. Comput. Med. Imaging Graph. 2020;84 doi: 10.1016/j.compmedimag.2020.101772. [DOI] [PubMed] [Google Scholar]
- Fenneteau A., Bourdon P., Helbert D., Fernandez-Maloigne C., Habas C., Guillevin R. In 2021 Sixth International Conference on Advances in Biomedical Engineering (ICABME) 2021. CNN for multiple sclerosis lesion segmentation: How many patients for a fully supervised method? pp. 30–33. [DOI] [Google Scholar]
- Ghodhbani G., Sahnoun M., Fathi K., Patrick S. In 2022 6th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP) 2022. U-NET Architecture for automatic MS lesions segmentation using MR images. [DOI] [Google Scholar]
- Ghosal P., Prasad P., Nandi D. A Light Weighted Deep Learning Framework for Multiple Sclerosis Lesion Segmentation. International Conference on Image Information Processing (ICIIP) 2020 doi: 10.1109/ICIIP47207.2019.8985674. [DOI] [Google Scholar]
- Ghribi O., Sellami L., Slima M., Hamida A., Mhiri C., Mahfoudh K. An advanced MRI multi-modalities segmentation methodology dedicated to multiple sclerosis lesions exploration and differentiation. IEEE Trans. Nanobioscience. 2017:pp. doi: 10.1109/TNB.2017.2763246. [DOI] [PubMed] [Google Scholar]
- Goodkin O., Pemberton H., Vos S., Prados F., Sudre C., Moggridge J., Cardoso M.J., Ourselin S., Bisdas S., White M., Yousry T., Thornton J., Barkhof F. The quantitative neuroradiology initiative framework: application to dementia. Br. J. Radiol. 2019;92:20190365. doi: 10.1259/bjr.20190365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizard N., Coupé P., Fonov V., Phd J., Arnold D., Collins L. Rotation-invariant multi-contrast non-local means for MS lesion segmentation. NeuroImage: Clinical. 2015;46 doi: 10.1016/j.nicl.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmouche R., Subbanna N., Collins L., Arnold D., Arbel T. Probabilistic multiple sclerosis lesion classification based on modeling regional intensity variability and local neighborhood information. IEEE Trans. Bio-medical Eng. 2014;62 doi: 10.1109/TBME.2014.2385635. [DOI] [PubMed] [Google Scholar]
- Hashemi S.R., Salehi S.S., Erdogmus D., Prabhu S., Warfield S., Gholipour A. Asymmetric Loss Functions and Deep Densely Connected Networks for Highly Imbalanced Medical Image Segmentation: Application to Multiple Sclerosis Lesion Detection. IEEE Access. 2018;7:1721–1735. doi: 10.1109/ACCESS.2018.2886371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi S.R., Salehi S.S., Erdogmus D., Prabhu S., Warfield S., Gholipour A. Asymmetric loss functions and deep densely connected networks for highly imbalanced medical image segmentation: Application to multiple sclerosis lesion detection. IEEE Access. 2018;7:1721–1735. doi: 10.1109/ACCESS.2018.2886371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann I., Golla A., Martínez-Heras E., Schmidt R., Solana E., Llufriu S., Gass A., Schad L., Zöllner F. Lesion probability mapping in MS patients using a regression network on MR fingerprinting. BMC Med. Imaging. 2021;21 doi: 10.1186/s12880-021-00636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindsholm A., Cramer S., Simonsen H., Frederiksen J., Andersen F., Højgaard L., Ladefoged C., Lindberg U. Assessment of artificial intelligence automatic multiple sclerosis lesion delineation tool for clinical use. Clin. Neuroradiol. 2021;32 doi: 10.1007/s00062-021-01089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitziger S., Ling W., Fritz T., D’Albis T., Lemke A., Grilo J. Triplanar U-Net with lesion-wise voting for the segmentation of new lesions on longitudinal MRI studies. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.964250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayoun H., Ebrahimpour-Komleh H. Neural network-based learning kernel for automatic segmentation of multiple sclerosis lesions on magnetic resonance images. J. Biomed. Phys. Eng. 2017;7 [PMC free article] [PubMed] [Google Scholar]
- Hosseinipanah S., Zamani A., Emadi F., Hamtaeipourshirazi F. Multiple sclerosis lesions segmentation in magnetic resonance imaging using ensemble support vector machine (esvm) J. Biomed. Phys. Eng. 2019;9 doi: 10.31661/jbpe.v0i0.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, B., Kang, G., Xu, X., and Hu, C. (2019). Cross attention densely connected networks for multiple sclerosis lesion segmentation. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 2356–2361. doi:10.1109/BIBM47256.2019.8983149.
- Huang F., Xia P., Vardhanabhuti V., Hui S., Lau G., Mak H., Cao P. Semisupervised white matter hyperintensities segmentation on MRI. Human Brain Mapp. 2022;44 doi: 10.1002/hbm.26109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Ribbens A., Sima D., Huffel S., Maes F., Smeets D. International MICCAI Workshop on Medical Computer Vision Bayesian and grAphical Models for Biomedical Imaging. 2017. Unsupervised Framework for Consistent Longitudinal MS Lesion Segmentation; pp. 208–219. [DOI] [Google Scholar]
- Isensee F., Jaeger P., Kohl S., Petersen J., Maier-Hein K. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods. 2021;18:1–9. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- Jain S., Sima D., Ribbens A., Cambron M., Maertens A., Van Hecke W., De Mey J., Barkhof F., Steenwijk M., Daams M., Maes F., Huffel S., Vrenken H., Smeets D. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage: Clinical. 2015;32 doi: 10.1016/j.nicl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannat S.A., Hoque T., Supti N.A., Alam M.A. 2021 Asian Conference on Innovation in Technology (ASIANCON) 2021. Detection of Multiple Sclerosis using Deep Learning; pp. 1–8. [DOI] [Google Scholar]
- Jog A., Carass A., Pham D., Prince J. Proceedings of SPIE–the. Vol. 9413. International Society for Optical Engineering; 2015. Multi-output decision trees for lesion segmentation in multiple sclerosis; p. 94131C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, A. and Sharma, K. (2021). Hybrid Topology of Graph Convolution and Autoencoder Deep Network For Multiple Sclerosis Lesion Segmentation. In 2021 International Conference on Artificial Intelligence and Smart Systems (ICAIS), pages 1529–1534. doi:10.1109/ICAIS50930.2021.9395914.
- Joshi, A. and Sharma, K. (2022). Multi-Modal Lesion Segmentation Using Deep Convolution Graph-Based Network. In 2022 IEEE Delhi Section Conference (DELCON), pages 1–6. doi:10.1109/DELCON54057.2022.9752917.
- Kamraoui R., Ta V.-T., Tourdias T., Mansencal B., Phd J., Coupé P. DeepLesionBrain: Towards a broader deep-learning generalization for multiple sclerosis lesion segmentation. Med. Image Anal. 2021;76 doi: 10.1016/j.media.2021.102312. [DOI] [PubMed] [Google Scholar]
- Karimian A., Jafari S. A new method to segment the multiple sclerosis lesions on brain magnetic resonance images. J. Med. Signals Sens. 2015;5:238–244. doi: 10.4103/2228-7477.168653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpate, Y., Commowick, O., and Barillot, C. (2015). Probabilistic one class learning for automatic detection of multiple sclerosis lesions. In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI 2015), pages 486–489. doi:10.1109/ISBI.2015.7163917.
- Kats, E., Goldberger, J., and Greenspan, H. (2019). Soft Labeling by Distilling Anatomical Knowledge for Improved MS Lesion Segmentation. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI), pages 1563–1566. doi:10.1109/ISBI.2019.8759518.
- Khotanlou H., Afrasiabi M. Feature selection in order to extract multiple sclerosis lesions automatically in 3d brain magnetic resonance images using combination of support vector machine and genetic algorithm. J. Med. Signals Sensors. 2012;2:211–218. doi: 10.4103/2228-7477.110333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J., Taylor G., Khademi A. Voxel-wise logistic regression and leave-one-source-out cross validation for white matter hyperintensity segmentation. Magn. Reson. Imaging. 2018;54 doi: 10.1016/j.MRI.2018.06.009. [DOI] [PubMed] [Google Scholar]
- Kolarik, M., Burget, R., Travieso, C., and Kocica, J. (2021). Planar 3D Transfer Learning for End to End Unimodal MRI Unbalanced Data Segmentation. In 2020 25th International Conference on Pattern Recognition (ICPR), pages 6051–6058. doi:10.1109/ICPR48806.2021.9412150.
- Krishnan A., Song Z., Clayton D., Gaetano L., Jia X., Crespigny A., Bengtsson T. Joint MRI T1 Unenhancing and Contrast-enhancing Multiple Sclerosis Lesion Segmentation with Deep Learning in OPERA Trials. Radiology. 2021;302 doi: 10.1148/radiol.211528. [DOI] [PubMed] [Google Scholar]
- Krishnan A., Song Z., Clayton D., Jia X., Crespigny A. Multi-arm u-net with dense input and skip connectivity for t2 lesion segmentation in clinical trials of multiple sclerosis. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-31207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J., Opfer R., Gessert N., Ostwaldt A.-C., Manogaran P., Kitzler H.H., Schlaefer A., Schippling S. Fully automated longitudinal segmentation of new or enlarged multiple sclerosis lesions using 3D convolutional neural networks. NeuroImage: Clinical. 2020;28 doi: 10.1016/j.nicl.2020.102445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J., Ostwaldt A.-C., Spies L., Geisler B., Schlaefer A., Kitzler H., Schippling S., Opfer R. Infratentorial lesions in multiple sclerosis patients: intra- and inter-rater variability in comparison to a fully automated segmentation using 3D convolutional neural networks. Eur. Radiol. 2021;32 doi: 10.1007/s00330-021-08329-3. [DOI] [PubMed] [Google Scholar]
- Krüger J., Ostwaldt A.-C., Spies L., Geisler B., Schlaefer A., Kitzler H., Schippling S., Opfer R. Infratentorial lesions in multiple sclerosis patients: intra- and inter-rater variability in comparison to a fully automated segmentation using 3d convolutional neural networks. Eur. Radiol. 2021;32 doi: 10.1007/s00330-021-08329-3. [DOI] [PubMed] [Google Scholar]
- Kuijf H.J., Biesbroek J.M., De Bresser J., Heinen R., Andermatt S., Bento M., Berseth M., Belyaev M., Cardoso M.J., Casamitjana A., Collins D.L., Dadar M., Georgiou A., Ghafoorian M., Jin D., Khademi A., Knight J., Li H., Lladó X., Luna M., Mahmood Q., McKinley R., Mehrtash A., Ourselin S., Park B.-Y., Park H., Park S.H., Pezold S., Puybareau E., Rittner L., Sudre C.H., Valverde S., Vilaplana V., Wiest R., Xu Y., Xu Z., Zeng G., Zhang J., Zheng G., Chen C., van der Flier W., Barkhof F., Viergever M.A., Biessels G.J. Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge. IEEE Trans. Med. Imaging. 2019;38(11):2556–2568. doi: 10.1109/TMI.2019.2905770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Murthy, O., S., Ghosal, P., Mukherjee, A., and Nandi, D. (2019). A Dense U-Net Architecture for Multiple Sclerosis Lesion Segmentation. In 2019 IEEE Region 10 Conference (TENCON). doi:10.1109/TENCON.2019.8929615.
- Kuwazuru J., Arimura H., Kakeda S., Yamamoto D., Magome T., Yamashita Y., Ohki M., Toyofuku F., Korogi Y. Automated detection of multiple sclerosis candidate regions in mr images: False-positive removal with use of an ann-controlled level-set method. Radiol. Phys. Technol. 2011;5:105–113. doi: 10.1007/s12194-011-0141-2. [DOI] [PubMed] [Google Scholar]
- La Rosa F., Abdulkadir A., Fartaria M., Rahmanzadeh R., Lu P.-J., Galbusera R., Barakovic M., Thiran J.-P., Granziera C., Cuadra M. Multiple sclerosis cortical and WM lesion segmentation at 3T MRI: a deep learning method based on FLAIR and MP2RAGE. NeuroImage: Clinical. 2020;27 doi: 10.1016/j.nicl.2020.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa F., Wynen M., Al-Louzi O., Beck E., Hülnhagen T., Maggi P., Thiran J.-P., Kober T., Shinohara R., Sati P., Reich D., Granziera C., Absinta M., Bach Cuadra M. Cortical lesions, central vein sign, and paramagnetic rim lesions in multiple sclerosis: Emerging machine learning techniques and future avenues. NeuroImage: Clinical. 2022;36 doi: 10.1016/j.nicl.2022.103205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa F., Yu T., Barquero G., Thiran J.-P., Granziera C., Bach Cuadra M. Mprage to mp2rage uni translation via generative adversarial network improves the automatic tissue and lesion segmentation in multiple sclerosis patients. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104297. [DOI] [PubMed] [Google Scholar]
- Le M., Tang L., Hernandez Torres E., Jarrett M., Brosch T., Metz L., Li D., Traboulsee A., Tam R., Rauscher A., Wiggermann V. Clinical; NeuroImage: 2019. FLAIR2 improves LesionTOADS automatic segmentation of multiple sclerosis lesions in non-homogenized, multi-center, 2D clinical magnetic resonance images; p. 101918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C., Glanville J., Briscoe S., Featherstone R., Littlewood A., Marshall C., Metzendorf M.-I., Noel-Storr A., Paynter R., Rader T., Thomas J., Wieland L. Cochrane Handbook for Systematic Reviews of Interventions. Cochrane. 2022 Available from URL:www.training.cochrane.org/handbook. [Google Scholar]
- Lesjak Z., Pernus F., Likar B., Spiclin Z. Validation of white-matter lesion change detection methods on a novel publicly available MRI image database. Neuroinformatics. 2016;14 doi: 10.1007/s12021-016-9301-1. [DOI] [PubMed] [Google Scholar]
- Llado X., Oliver A., Cabezas M., Freixenet J., Vilanova J.C., Quiles A., Valls L., Ramió-Torrentà L., Rovira A. Segmentation of multiple sclerosis lesions in brain MRI: A review of automated approaches. Inf. Sci. 2012;186:164–185. doi: 10.1016/j.ins.2011.10.011. [DOI] [Google Scholar]
- Ma Y., Zhang C., Cabezas M., Song Y., Tang Z., Liu D., Cai W., Barnett M., Wang C. Multiple sclerosis lesion analysis in brain magnetic resonance images: Techniques and clinical applications. IEEE J. Biomed. Health Inform. 2022 doi: 10.1109/JBHI.2022.3151741. PP:1–1. [DOI] [PubMed] [Google Scholar]
- Maier-Hein, L., Reinke, A., Godau, P., Tizabi, M.D., Büttner, F., Christodoulou, E., Glocker, B., Isensee, F., Kleesiek, J., Kozubek, M., Reyes, M., Riegler, M.A., Wiesenfarth, M., Kavur, E., Sudre, C.H., Baumgartner, M., Eisenmann, M., Heckmann-Nötzel, D., Rädsch, A.T., Acion, L., Antonelli, M., Arbel, T., Bakas, S., Benis, A., Blaschko, M., Cardoso, M.J., Cheplygina, V., Cimini, B.A., Collins, G.S., Farahani, K., Ferrer, L., Galdran, A., van Ginneken, B., Haase, R., Hashimoto, D.A., Hoffman, M.M., Huisman, M., Jannin, P., Kahn, C.E., Kainmueller, D., Kainz, B., Karargyris, A., Karthikesalingam, A., Kenngott, H., Kofler, F., Kopp-Schneider, A., Kreshuk, A., Kurc, T., Landman, B.A., Litjens, G., Madani, A., Maier-Hein, K., Martel, A.L., Mattson, P., Meijering, E., Menze, B., Moons, K.G.M., Müller, H., Nichyporuk, B., Nickel, F., Petersen, J., Rajpoot, N., Rieke, N., Saez-Rodriguez, J., Sánchez, C.I., Shetty, S., van Smeden, M., Summers, R.M., Taha, A.A., Tiulpin, A., Tsaftaris, S.A., Calster, B.V., Varoquaux, G., and Jäger, P.F. (2022). Metrics reloaded: Pitfalls and recommendations for image analysis validation. arXiv.org, 2206.01653.
- Malinin, A., Athanasopoulos, A., Barakovic, M., Bach Cuadra, M., Gales, M., Granziera, C., Graziani, M., Kartashev, N., Kyriakopoulos, K., Lu, P.-J., Molchanova, N., Nikitakis, A., Raina, V., La Rosa, F., Sivena, E., Tsarsitalidis, V., Tsompopoulou, E., and Volf, E. (2022). Shifts 2.0: Extending the dataset of real distributional shifts. arXiv.org. doi:10.48550/arXiv.2206.15407.
- McKinley R., Wepfer R., Grunder L., Aschwanden F., Fischer T., Friedli C., Muri R., Rummel C., Verma R., Weisstanner C., Wiestler B., Berger C., Eichinger P., Muehlau M., Reyes M., Salmen A., Chan A., Wiest R., Wagner F. Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence. NeuroImage: Clinical. 2019;25 doi: 10.1016/j.nicl.2019.102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechrez R., Goldberger J., Greenspan H. Patch-based segmentation with spatial consistency: Application to ms lesions in brain MRI. Int. J. Biomed. Imaging. 2016;2016:1–13. doi: 10.1155/2016/7952541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R., Christinck T., Nair T., Bussy A., Premasiri S., Costantino M., Chakravarty M., Arnold D., Gal Y., Arbel T. Propagating Uncertainty Across Cascaded Medical Imaging Tasks For Improved Deep Learning Inference. IEEE Trans. Med. Imaging. 2021;PP:1–1 doi: 10.1109/TMI.2021.3114097. [DOI] [PubMed] [Google Scholar]
- Mei, P., Carneiro, C., Kuroda, M., Fraser, S., Min, L., and Reis, F. (2017). Self-organizing maps as a tool for segmentation of magnetic resonance imaging (MRI) of relapsing-remitting multiple sclerosis. In 2017 12th International Workshop on Self-Organizing Maps and Learning Vector Quantization, Clustering and Data Visualization (WSOM), pages 1–7. doi:10.1109/WSOM.2017.8020005.
- Meier D., Guttmann C., Tummala S., Moscufo N., Cavallari M., Tauhid S., Bakshi R., Weiner H. Dual-sensitivity multiple sclerosis lesion and csf segmentation for multichannel 3t brain MRI: Dual-sensitivity ms lesion and csf segmentation. J. Neuroimaging. 2017;28 doi: 10.1111/jon.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn Z., Pemberton H., Gray J., Goodkin O., Carrasco F., Scheel M., Nawabi J., Barkhof F. Commercial volumetric MRI reporting tools in multiple sclerosis: a systematic review of the evidence. Neuroradiology. 2022;65:1–20. doi: 10.1007/s00234-022-03074-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin M., Soulier T., Hamzaoui M., Yazdan-Panah A., Bodini B., Ayache N., Stankoff B., Colliot O. Online hard example mining vs. fixed oversampling strategy for segmentation of new multiple sclerosis lesions from longitudinal FLAIR MRI. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair T., Precup D., Arnold D., Arbel T. Exploring uncertainty measures in deep networks for Multiple Sclerosis lesion detection and segmentation. Med. Image Anal. 2019;59 doi: 10.1016/j.media.2019.101557. [DOI] [PubMed] [Google Scholar]
- Narayana, P., Coronado, I., Robinson, M., Sujit, S., Datta, S., Sun, X., Lublin, F., Wolinsky, J., and Gabr, R. (2018). Multimodal MRI segmentation of brain tissue and t2-hyperintense white matter lesions in multiple sclerosis using deep convolutional neural networks and a large multi-center image database. In 2018 9th Cairo International Biomedical Engineering Conference (CIBEC), pages 13–16. doi:10.1109/CIBEC.2018.8641800.
- Narayana P., Coronado I., Sujit S., Sun X., Wolinsky J., Gabr R. Are multi-contrast magnetic resonance images necessary for segmenting multiple sclerosis brains? a large cohort study based on deep learning. Magn. Resonance Imaging. 2019;65:8–14. doi: 10.1016/j.MRI.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana P., Govindarajan K., Goel P., Datta S., Lincoln J., Cofield S., Cutter G., Lublin F., Wolinsky J. Regional cortical thickness in relapsing remitting multiple sclerosis: A multi-center study. NeuroImage: Clinical. 2013;2:120–131. doi: 10.1016/j.nicl.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass, M., Bensalah, H., Njeh, I., Slima, M., and BenHamida, A. (2022). Computer aided diagnosis (cad) tool for ms lesions exploration in multimodal brain MRI. In 2022 6th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), pages 1–6. doi:10.1109/ATSIP55956.2022.9805933.
- Nguyen T., Zhang S., Gupta A., Zhao Y., Gauthier S., Wang Y. Fast and robust unsupervised identification of ms lesion change using the statistical detection of changes algorithm. Am. J. Neuroradiol. 2018;39 doi: 10.3174/ajnr.A5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong K.H., Ramachandram D., Mandava R., Shuaib I.L. Automatic white matter lesion segmentation using an adaptive outlier detection method. Magn. Resonance Imaging. 2012;30:807–823. doi: 10.1016/j.MRI.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Opbroek A., Ikram M., Vernooij M., de Bruijne M. Transfer Learning Improves Supervised Image Segmentation Across Imaging Protocols. IEEE Trans. Med. Imaging. 2014;34 doi: 10.1109/TMI.2014.2366792. [DOI] [PubMed] [Google Scholar]
- Page M., Mckenzie J., Bossuyt P., Boutron I., Hoffmann T., Mulrow C., Shamseer L., Tetzlaff J., Akl E., Brennan S., Chou R., Glanville J., Grimshaw J., Hróbjartsson A., Lalu M., Li T., Loder E., Mayo-Wilson E., Mcdonald S., Moher D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2021;372 doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos T., Tripoliti E., Plati D., Zelilidou S., Vlachos K., Konitsiotis S., Fotiadis D. White Matter Lesion Segmentation for Multiple Sclerosis Patients implementing deep learning. Engineering in Medicine & Biology Society (EMBC) 2022;2022:3818–3821. doi: 10.1109/EMBC48229.2022.9871401. [DOI] [PubMed] [Google Scholar]
- Pemberton H., Zaki L., Goodkin O., Das R., Steketee R., Barkhof F., Vernooij M. Technical and clinical validation of commercial automated volumetric MRI tools for dementia diagnosis–a systematic review. Neuroradiology. 2021;63 doi: 10.1007/s00234-021-02746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmadi M., Valdés-Hernández M., Li H., Guerrero R., Meijboom R., Wiseman S., Waldman A., Zhang J., Rueckert D., Wardlaw J., Komura T. Limited One-time Sampling Irregularity Map (LOTS-IM) for Automatic Unsupervised Assessment of White Matter Hyperintensities and Multiple Sclerosis Lesions in Structural Brain Magnetic Resonance Images. Comput. Med. Imaging Graph. 2019;79 doi: 10.1016/j.compmedimag.2019.101685. [DOI] [PubMed] [Google Scholar]
- Rakic M., Vercruyssen S., Van Eyndhoven S., de la Rosa E., Jain S., Huffel S., Maes F., Smeets D., Sima D. icobrain ms 5.1: Combining Unsupervised and Supervised Approaches for Improving the Detection of Multiple Sclerosis Lesions. NeuroImage: Clinical. 2021;31 doi: 10.1016/j.nicl.2021.102707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D.S., Lucchinetti C.F., Calabresi P.A. Multiple sclerosis. N. Engl. J. Med. 2018;378(2):169–180.. doi: 10.1056/NEJMra1401483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinella A., Crispino E., Guarnera F., Giudice O., Ortis A., Russo G., Lorenzo C., Maimone D., Pappalardo F., Battiato S. Boosting multiple sclerosis lesion segmentation through attention mechanism. Comput. Biol. Med. 2023;161 doi: 10.1016/j.compbiomed.2023.107021. [DOI] [PubMed] [Google Scholar]
- Roura E., Oliver A., Cabezas M., Valverde S., Pareto D., Vilanova J.C., Ramió-Torrentà L., Rovira A., Llado X. A toolbox for multiple sclerosis lesion segmentation. Neuroradiology. 2015;57 doi: 10.1007/s00234-015-1552-2. [DOI] [PubMed] [Google Scholar]
- Rovira A., Corral J., Auger C., Valverde S., Vidal-Jordana A., Oliver A., Barros A., Wong Y., Tintorè M., Pareto D., Aymerich X., Montalban X., Llado X., Alonso J. Assessment of automatic decision-support systems for detecting active t2 lesions in multiple sclerosis patients. Multiple Sclerosis J. 2021;28 doi: 10.1177/13524585211061339. 135245852110613. [DOI] [PubMed] [Google Scholar]
- Roy, P., Bhuiyan, A., and Ramamohanarao, K. (2013). Automated segmentation of multiple sclerosis lesion in intensity enhanced flair MRI using texture features and support vector machine. In 2013 IEEE International Conference on Image Processing, ICIP 2013 - Proceedings, pages 4277–4281. doi:10.1109/ICIP.2013.6738881.
- Roy S., Carass A., Prince J., Pham D. Longitudinal patch-based segmentation of multiple sclerosis white matter lesions. In Machine learning in medical imaging. MLMI (Workshop) 2015;volume:9352. doi: 10.1007/978-3-319-24888-2_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghibakhi M., Pourreza H., Mahyar H. Multiple Sclerosis Lesions Segmentation using Attention-Based CNNs in FLAIR Images. IEEE J. Transl. Eng. Health Med. 2022;10:1. doi: 10.1109/JTEHM.2022.3172025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajja B., Datta S., He R., Mehta M., Gupta R., Wolinsky J., Narayana P. Unified Approach for Multiple Sclerosis Lesion Segmentation on Brain MRI. Ann. Biomed. Eng. 2006;34:142–151. doi: 10.1007/s10439-005-9009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M., Cabezas M., Valverde S., Pareto D., Oliver A., Salvi J., Rovira A., Llado X. A supervised framework with intensity subtraction and deformation field features for the detection of new t2-w lesions in multiple sclerosis. NeuroImage: Clinical. 2017;17C(607–615) doi: 10.1016/j.nicl.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M., Ryan M., Oliver A., Hussain K., Llado X. Improving the detection of new lesions in multiple sclerosis with a cascaded 3d fully convolutional neural network approach. Front. Neurosci. 2022;16:1007619. doi: 10.3389/fnins.2022.1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M., Valverde S., Cabezas M., Pareto D., Oliver A., Salvi J., Rovira A., Llado X. Multiple sclerosis lesion synthesis in MRI using an encoder-decoder u-net. IEEE Access. 2019 doi: 10.1109/ACCESS.2019.2900198. PP:1–1. [DOI] [Google Scholar]
- Salem M., Valverde S., Cabezas M., Pareto D., Oliver A., Salvi J., Rovira A., Llado X. A fully convolutional neural network for new t2-w lesion detection in multiple sclerosis. NeuroImage: Clinical. 2020;25 doi: 10.1016/j.nicl.2019.102149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarica B., Seker D. New MS lesion segmentation with deep residual attention gate U-Net utilizing 2D slices of 3D MR images. Frontiers in Neuroscience. 2022;16:912000. doi: 10.3389/fnins.2022.912000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarica B., Seker D., Bayram B. A Dense Residual U-Net for Multiple Sclerosis Lesions Segmentation from Multi-Sequence 3D MR Images. Int. J. Med. Informatics. 2022;170 doi: 10.1016/j.ijmedinf.2022.104965. [DOI] [PubMed] [Google Scholar]
- Schläger S., Li H., Baum T., Zimmer C., Moosbauer J., Byas S., Mühlau M., Wiestler B., Finck T. Longitudinal assessment of multiple sclerosis lesion load with synthetic magnetic resonance imaging-a multicenter validation study. Investig. Radiol. 2022 doi: 10.1097/RLI.0000000000000938. [DOI] [PubMed] [Google Scholar]
- Schmidt P., Biberacher V., Küster P., Meier D., Wuerfel J., Lukas C., Bellenberg B., Zipp F., Groppa S., Sämann P., Weber F., Gaser C., Franke T., Bussas M., Kirschke J., Zimmer C., Hemmer B., Mühlau M. Automated segmentation of changes in FLAIR-hyperintense white matter lesions in multiple sclerosis on serial magnetic resonance imaging. NeuroImage: Clinical. 2019;23 doi: 10.1016/j.nicl.2019.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P., Gaser C., Arsic M., Buck D., Förschler A., Berthele A., Hoshi M., Ilg R., Schmid V., Zimmer C., Hemmer B., Mühlau M. An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis. NeuroImage. 2011;59:3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Senra Filho A.C. A hybrid approach based on logistic classification and iterative contrast enhancement algorithm for hyperintense multiple sclerosis lesion segmentation. Med. Biolog. Eng. Comput. 2017;56:1–14. doi: 10.1007/s11517-017-1747-2. [DOI] [PubMed] [Google Scholar]
- Sepahvand, N., Arnold, D., and Arbel, T. (2020). CNN Detection of New and Enlarging Multiple Sclerosis Lesions from Longitudinal MRI Using Subtraction Images. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pages 127–130. doi:10.1109/ISBI45749.2020.9098554.
- Shahab U., Kamran J., Qaisar S., Rashad J., Haider U. Multiple sclerosis lesion segmentation in brain MRI using inception modules embedded in a convolutional neural network. J. Healthcare Eng. 2021;2021 doi: 10.1155/2021/4138137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitter A., Steenwijk M., Ruet A., Versteeg A., Liu Y., Schijndel R., Pouwels P., Kilsdonk I., Cover K., Dijk B., Ropele S., Rocca M., Yiannakas M., Wattjes M., Damangir S., Frisoni G., Sastre-Garriga J., Rovira A., Enzinger C., Vrenken H. Performance of five research-domain automated wm lesion segmentation methods in a multi-center ms study. NeuroImage. 2017;163 doi: 10.1016/j.neuroimage.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Sousa, I., de Oliveira, M., Lisboa-Filho, P., and Cardoso, J. (2021). Evaluation of the impact of domain adaptation on segmentation of multiple sclerosis lesions in MRI. In 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1786–1790. doi:10.1109/BIBM52615.2021.9669533.
- Spies L., Tewes A., Suppa P., Opfer R., Buchert R., Winkler G., Raji A. Fully automatic detection of deep white matter t1 hypointense lesions in multiple sclerosis. Phys. Med. Biol. 2013;58:8323–8337. doi: 10.1088/0031-9155/58/23/8323. [DOI] [PubMed] [Google Scholar]
- Steenwijk M., Pouwels P., Daams M., van Dalen J.W., Caan M., Richard E., Barkhof F., Vrenken H. Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs) NeuroImage: Clinical. 2013;3:462–469. doi: 10.1016/j.nicl.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M., Lee J., Chin B., Chin M., Commowick O., Tran H., Markovic-Plese S., Jewells V., Warfield S. 3D Segmentation in the Clinic: A Grand Challenge II: MS lesion segmentation. MIDAS J. 2008 doi: 10.54294/lmkqvm. [DOI] [Google Scholar]
- Subbanna N., Precup D., Arnold D., Arbel T. IMaGe: Iterative multilevel probabilistic graphical model for detection and segmentation of multiple sclerosis lesions in brain MRI. International Conference on Information Processing in Medical Imaging. 2015;24:514–526. doi: 10.1007/978-3-319-19992-4_40. [DOI] [PubMed] [Google Scholar]
- Sweeney E., Shinohara R., Shiee N., Mateen F., Chudgar A., Cuzzocreo J., Calabresi P., Pham D., Reich D., Crainiceanu C. OASIS is Automated Statistical Inference for Segmentation, with applications to multiple sclerosis lesion segmentation in MRI. NeuroImage: Clinical. 2013;2:402–413. doi: 10.1016/j.nicl.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney E., Vogelstein J., Cuzzocreo J., Calabresi P., Reich D., Crainiceanu C., Shinohara R. A comparison of supervised machine learning algorithms and feature vectors for ms lesion segmentation using multimodal structural MRI. PloS One. 2014;9 doi: 10.1371/journal.pone.0095753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur S., Schindler M., Bilello M., Bakas S. Clinically deployed computational assessment of multiple sclerosis lesions. Front. Med. 2022;9 doi: 10.3389/fmed.2022.797586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G., Correale J., Fazekas F., Filippi M., Freedman M.S., Fujihara K., Galetta S.L., Hartung H.P., Kappos L., Lublin F.D., Marrie R.A., Miller A.E., Miller D.H., Montalban X., Mowry E.M., Sorensen P.S., Tintoré M., Traboulsee A.L., Trojano M., Uitdehaag B.M.J., Vukusic S., Waubant E., Weinshenker B.G., Reingold S.C., Cohen J.A. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17(2):162–173. doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Todea, A.R., Melie-Garcia, L., Barakovic, M., Cagol, A., Rahmanzadeh, R., Galbusera, R., Lu, P.-J., Weigel, M., Ruberte, E., Radue, E.-W., Schaedelin, S., Benkert, P., Oezguer, Y., Sinnecker, T., Müller, S., Achtnichts, L., Vehoff, J., Disanto, G., Findling, O., Chan, A., Salmen, A., Pot, C., Lalive, P., Bridel, C., Zecca, C., Derfuss, T., Remonda, L., Wagner, F., Vargas, M., Du Pasquier, R., Pravata, E., Weber, J., Gobbi, C., Leppert, D., Wuerfel, J., Kober, T., Marechal, B., Corredor-Jerez, R., Psychogios, M., Lieb, J., Kappos, L., Cuadra, M.B., Kuhle, J., Granziera, C., and for the Swiss MS Cohort Study (2023). A Multicenter Longitudinal MRI Study Assessing LeMan-PV Software Accuracy in the Detection of White Matter Lesions in Multiple Sclerosis Patients. Journal of Magnetic Resonance Imaging, n/a(n/a). doi:10.1002/jMRI.28618. [DOI] [PubMed]
- Tomas-Fernandez X., Warfield S. A Model of Population and Subject (MOPS) Intensities With Application to Multiple Sclerosis Lesion Segmentation. IEEE Trans. Med. Imaging. 2015;34 doi: 10.1109/TMI.2015.2393853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P., Thoprakarn U., Gourieux E., dos Santos C.L., Cavedo E., Guizard N., Cotton F., Krolak-Salmon P., Delmaire C., Heidelberg D., Pyatigorskaya N., Ströer S., Dormont D., Martini J.-B., Chupin M. Automatic segmentation of white matter hyperintensities: validation and comparison with state-of-the-art methods on both Multiple Sclerosis and elderly subjects. NeuroImage: Clinical. 2022;33 doi: 10.1016/j.nicl.2022.102940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripoliti, E., Zelilidou, S., Vlahos, K., Konitsiotis, S., and Fotiadis, D. (2019). ProMiSi Architecture - A Tool for the Estimation of the Progression of Multiple Sclerosis Disease using MRI. In 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), pages 284–287. doi:10.1109/BIBE.2019.00058.
- Valcarcel A., Linn K., Khalid F., Vandekar S., Tauhid S., Satterthwaite T., Muschelli J., Martin M., Bakshi R., Shinohara R. A dual modeling approach to automatic segmentation of cerebral t2 hyperintensities and t1 black holes in multiple sclerosis. NeuroImage: Clinical. 2018;20 doi: 10.1016/j.nicl.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel A., Linn K., Vandekar S., Satterthwaite T., Muschelli J., Calabresi P., Pham D., Martin M., Shinohara R. MIMoSA: An Automated Method for Intermodal Segmentation Analysis of Multiple Sclerosis Brain Lesions. J. Neuroimaging. 2018;28 doi: 10.1111/jon.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel A., Muschelli J., Pham D., Martin M., Yushkevich P., Brandstadter R., Patterson K., Schindler M., Calabresi P., Bakshi R., Shinohara R. TAPAS: A Thresholding Approach for Probability Map Automatic Segmentation in Multiple Sclerosis. NeuroImage: Clinical. 2020;27 doi: 10.1016/j.nicl.2020.102256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia L., Clèrigues A., Valverde S., Salem M., Oliver A., Rovira A., Llado X. Evaluating the use of synthetic t1-w images in new t2 lesion detection in multiple sclerosis. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.954662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde S., Cabezas M., Roura E., Gonzalez-Villa S., Pareto D., Vilanova J.C., Ramio-Torrenta L., Rovira A., Oliver A., Llado X. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage. 2017;155 doi: 10.1016/j.neuroimage.2017.04.034. [DOI] [PubMed] [Google Scholar]
- Van Hecke W., Costers L., Descamps A., Ribbens A., Nagels G., Smeets D., Sima D. A Novel Digital Care Management Platform to Monitor Clinical and Subclinical Disease Activity in Multiple Sclerosis. Brain Sci. 2021;11:1171. doi: 10.3390/brainsci11091171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang, Y.S., Cao, Y., Chang, P.D., Chow, D.S., Brandt, A.U., Paul, F., Scheel, M., and Xie, X. (2020). SynergyNet: A Fusion Framework for Multiple Sclerosis Brain MRI Segmentation with Local Refinement. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pages 131–135. doi:10.1109/ISBI45749.2020.9098610.
- Varoquaux G., Cheplygina V. Machine learning for medical imaging: methodological failures and recommendations for the future. npj Digital Medicine. 2022;5:48. doi: 10.1038/s41746-022-00592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokinger K., Feuerriegel S., Kesselheim A. Mitigating bias in machine learning for medicine. Communications Medicine. 2021;1 doi: 10.1038/s43856-021-00028-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton C., King R., Rechtman L., Kaye W., Leray E., Marrie R.A., Robertson N., Rocca N.L., Uitdehaag B., van der Mei I., Wallin M., Helme A., Napier C.A., Rijke N., Baneke P. Rising prevalence of multiple sclerosis worldwide: Insights from the Atlas of MS. Multiple Sclerosis J. 2020;26(14):1816–1821. doi: 10.1177/1352458520970841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss N., Rueckert D., Rao A. Multiple sclerosis lesion segmentation using dictionary learning and sparse coding. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2013;16:735–742. doi: 10.1007/978-3-642-40811-3_92. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Lacheret C., Fukutomi H., Kamraoui R., Denat L., Zhang B., Prevost V., Zhang L., Ruet A., Triaire B., DOUSSET V., Coupé P., Tourdias T. Validation of a Denoising Method Using Deep Learning-Based Reconstruction to Quantify Multiple Sclerosis Lesion Load on Fast FLAIR Imaging. American Journal of Neuroradiology. 2022;43 doi: 10.3174/ajnr.A7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., George A., Wu A., Niu X., Lin J., Adusumilli G., Naismith R., Cross A., Sun P., Song S.-K. Deep learning with diffusion basis spectrum imaging for classification of multiple sclerosis lesions. Ann. Clin. Transl. Neurol. 2020;7 doi: 10.1002/acn3.51037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim, M. and Dandil, E. (2021a). DeepMSWeb: A Web-Based Decision Support System via Deep Learning for Automatic Detection of MS Lesions. In 2021 2nd International Informatics and Software Engineering Conference (IISEC), pages 1–6. doi:10.1109/IISection 54230.2021.9672360.
- Yildirim, M.S. and Dandil, E. (2021b). Automated Multiple Sclerosis Lesion Segmentation on MR Images via MaskR-CNN. In 2021 5th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), pages 570–577. doi:10.1109/ISMSIT52890.2021.9604593.
- Zangeneh, D. and Yazdi, M. (2016). Automatic segmentation of multiple sclerosis lesions in brain MRI using constrained GMM and genetic algorithm. In 2016 24th Iranian Conference on Electrical Engineering (ICEE), pages 832–837. doi:10.1109/IranianCEE.2016.7585635.
- Zeng C., Gu L., Liu Z., Zhao S. Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI. Front. Neuroinform. 2020;14 doi: 10.3389/fninf.2020.610967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Song, Y., Liu, S., Lill, S., Wang, C., Tang, Z., You, Y., Gao, Y., Klistorner, A., Barnett, M., and Cai, W. (2018). MS-GAN: GAN-Based Semantic Segmentation of Multiple Sclerosis Lesions in Brain Magnetic Resonance Imaging. In 2018 Digital Image Computing: Techniques and Applications (DICTA), pages 1–8. doi:10.1109/DICTA.2018.8615771.
- Zhang, H., Valcarcel, A., Bakshi, R., Chu, R., Bagnato, F., Shinohara, R., Hett, K., and Oguz, I. (2019). Multiple Sclerosis Lesion Segmentation with Tiramisu and 2.5D Stacked Slices, volume 11766, pages 338–346. Medical Image Computing and Computer Assisted Intervention - MICCAI 2019. doi:10.1007/978-3-030-32248-9_38. [DOI] [PMC free article] [PubMed]
- Zhang H., Zhang J., Li C., Sweeney E., Spincemaille P., Nguyen T., Gauthier S., Wang Y. ALL-Net: Anatomical Information Lesion-wise Loss Function Integrated into Neural Network for Multiple Sclerosis Lesion Segmentation. NeuroImage: Clinical. 2021;32 doi: 10.1016/j.nicl.2021.102854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Duan Y., Wang X., Zhuo Z., Haller S., Barkhof F., Liu Y. A deep learning algorithm for white matter hyperintensity lesion detection and segmentation. Neuroradiology. 2022;64 doi: 10.1007/s00234-021-02820-w. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Duan Y., Wang X., Zhuo Z., Haller S., Barkhof F., Liu Y. A deep learning algorithm for white matter hyperintensity lesion detection and segmentation. Neuroradiology. 2022;64 doi: 10.1007/s00234-021-02820-w. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Utriainen D., Wang Y., Kang Y., Haacke M. Automated White Matter Hyperintensity Detection in Multiple Sclerosis Using 3D T2 FLAIR. Int. J. Biomed. Imaging. 2014;2014 doi: 10.1155/2014/239123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Zhang, J., Wang, R., Zhang, Q., Gauthier, S., Spincemaille, P., Nguyen, T., and Wang, Y. (2021b). Geometric Loss For Deep Multiple Sclerosis Lesion Segmentation. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), pages 24–28. doi:10.1109/ISBI48211.2021.9434085.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.