Abstract
In the era of ‘precision medicine’, liquid biopsies based on cell-free DNA (cfDNA) have emerged as a promising tool in the oncology field. cfDNA from cancer patients is a mixture of tumoral (ctDNA) and non-tumoral DNA originated from healthy, cancer and tumor microenvironmental cells. Apoptosis, necrosis, and active secretion from extracellular vesicles represent the main mechanisms of cfDNA release into the physiological body fluids. Focused on HNC, two main types of cfDNA can be identified: the circulating cfDNA (ccfDNA) and the salivary cfDNA (scfDNA). Numerous studies have reported on the potential of cfDNA analysis as potential diagnostic, prognostic, and monitoring biomarker for HNC. Thus, ctDNA has emerged as an attractive strategy to detect cancer specific genetic and epigenetic alterations including DNA somatic mutations and DNA methylation patterns. This review aims to provide an overview of the up-to-date studies evaluating the value of the analysis of total cfDNA, cfDNA fragment length, and ctDNA analysis at DNA mutation and methylation level in HNC patients.
Keywords: Cell-free DNA; Circulating tumor-DNA; DNA methylation; Somatic mutations, Liquid biopsy; Head and neck squamous cell carcinoma
1. Introduction
Head and neck cancer (HNC) is the seventh most common malignancy with an estimated 930,000 new cases and 470,000 deaths occurring in 2020 [1]. HNC involves multiple anatomic subsites like the oral cavity, oropharynx, hypopharynx, and larynx, with squamous cell carcinoma (HNSCC) being the most common histological type [2], [3]. Although cases related to the consumption of tobacco and alcohol are slowly declining in developed countries, over the last decades, human papillomavirus (HPV) status has emerged as a novel risk and a prognostic factor for this malignancy [4], [5]. This variety of locations and etiologies leads to a multifactorial pathogenesis with different deranged molecular pathways; however, up to date, despite the information retrieved by the use of genomics, the precise molecular mechanisms underlying HNC development and progression remain incompletely understood [3], [6].
Application of next-generation sequencing (NGS) in liquid biopsies has raised as an attractive strategy to characterize the molecular profile of solid tumors throughout a minimally invasive procedure, leading to an insightful understanding of the carcinogenesis process [7]. In general, liquid biopsies allow the analysis of different tumor-derived components such as circulating tumor cells (CTCs), cell-free acid nucleics (cfDNA) or extracellular vesicles, which are present in different human body fluids like blood, saliva, urine or cerebrospinal fluid [8], [9], [10]. Nowadays, this minimally-or non-invasive strategy represents an alternative or a complementary approach to solid biopsy for molecular genetic analyses [11] and cancer biomarkers identification.
In the era of ‘precision medicine’, cfDNA analysis supposes the most studied biomarker inside the liquid biopsies because of the valuable information that it offers regarding the patient’s disease situation. CfDNA comprises a highly fragmented nuclear double-stranded and/or mitochondrial DNA released from the cells into physiological body fluids [12]. Mandel and Métais reported the presence of cfDNA in blood of healthy individuals for the first time in 1948 [13] but it was not until 1977 when Leon et al. detected cfDNA in serum from cancer patients [14], suggesting that its concentration could be proportionally related with the stage of the disease and the type of tumor [15], [16].
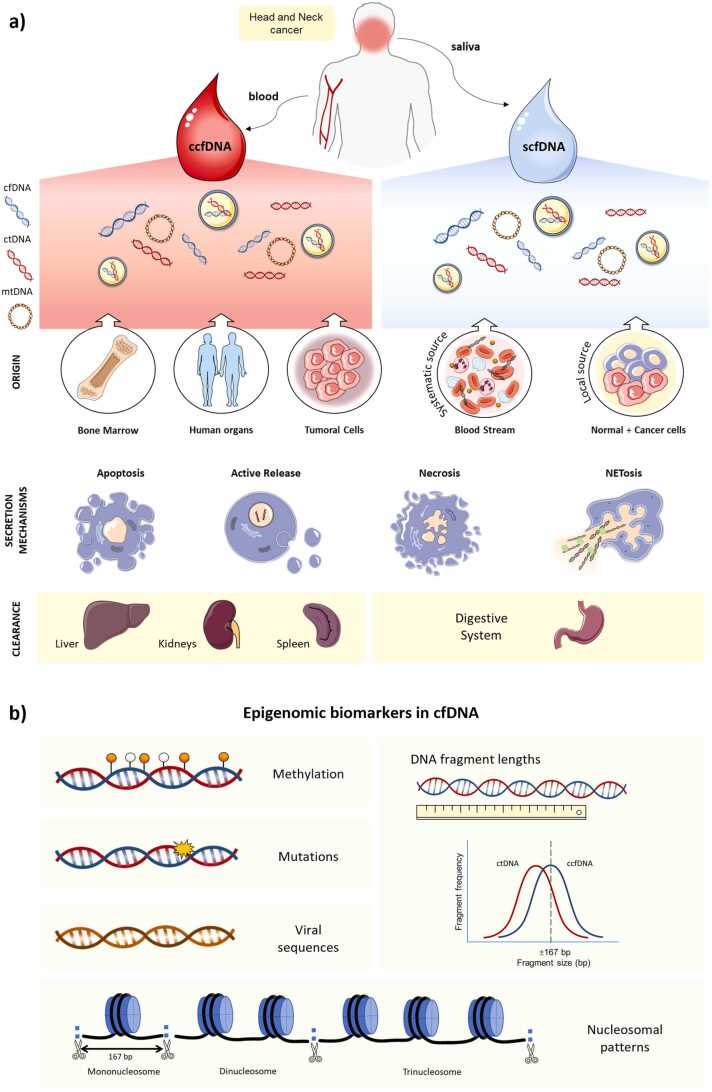
The majority of cfDNA released into plasma from healthy individuals stems from the hematopoietic system [17]. However, in cancer, there is an uncertain small fraction that belongs to tumor-derived cfDNA, known as circulating tumor DNA (ctDNA). This subpopulation of cfDNA varies depending on the type of cancer, the stage, or the biofluid analyzed [18]. Consequently, cfDNA from cancer patients is a mixture of tumoral (ctDNA) and non-tumoral DNA originated from healthy, cancer and tumor microenvironmental cells [12]. Different mechanisms have been suggested for explaining the cfDNA release, including apoptosis, necrosis, and active secretion from extracellular vesicles [19], [20], [21], [22], nonetheless, the precise mechanisms by which cfDNA is released into body fluids remain to be clarified [18]. In plasma, most of the cfDNA is thought to come from an apoptotic cell process, showing a size distribution pattern near 167 bp that corresponds approximately to the length of the DNA that is wrapped around a nucleosome (∼147 bp) plus a linker fragment (∼20 bp). This non-random fragmentation process is due to the cleavage of the internucleosomal chromatin regions by endonucleases [23], [24]. Besides, longer DNA fragments have also been identified in the circulating cell-free DNA (ccfDNA) fraction, which may be released from necrosis [22], NETosis [25] and exosomes [26]. Nevertheless, ctDNA is more fragmented than non-tumor cfDNA [27] and mainly composed of shorter fragments of less than 145 bp [12], [27], [28], [29] (Fig. 1).
Fig. 1.
Overview of cell-free DNA in head and neck cancer: (a) origin, release mechanisms, excretion and (b) epigenomic biomarkers. It includes normal and tumor cell-free DNA (cfDNA), known as ctDNA, which came from cellular processes like apoptosis, necrosis, active secretion, or NETosis. Regarding their origin, circulating cfDNA (ccfDNA) predominantly originates from the bone marrow, organs within the human body, and tumor cells and it is excreted by the liver, the kidneys, and the spleen. CfDNA in blood and saliva can originate from diverse sources. Conversely, in saliva, the main contributors to cfDNA are the epithelial cells scrapped off from the oral cavity, representing the local source and, to a lesser extent, from the systemic circulation throughout the secretion of the salivary glands (systemic source), being the digestive system the one on charge of the excretion of salivary cell-free DNA (scfDNA). (b) Epigenomic biomarkers in cfDNA (mutations, methylation, viral DNA, fragmentomics, and nucleosome patterns) offer valuable insights into various aspects of genomic regulation and pathological processes.
Since ctDNA could reflect the genetic and epigenetic alterations of the tumor, several studies in HNC have explored this biomarker as an opportunity for non-invasive cancer management. Here, we described the different types of cfDNA in HNC and their potential clinical utility in cancer management and tumor characterization, along with its implications in diagnosis, in prognosis, and in monitoring cancer patients' disease progression and therapy response.
2. Types of cell-free DNA in head and neck cancer
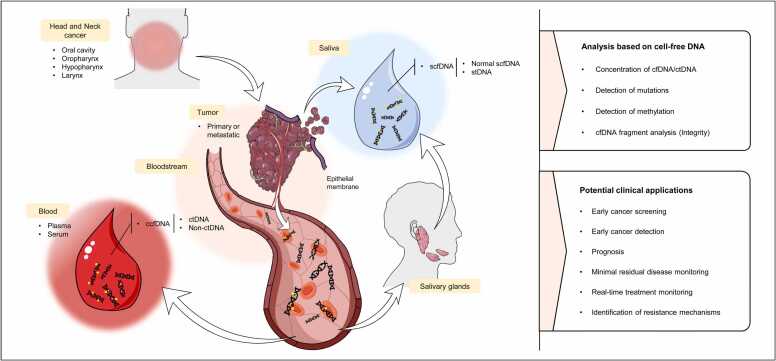
Two main types of cfDNA have been evaluated in HNC: ccfDNA and salivary cfDNA (scfDNA) (Fig. 2). The studies that analyze the ccfDNA can focus either on serum or plasma cfDNA fraction, however, as plasma shows higher levels of ctDNA because it is less diluted than in serum , represents a better option to analyze ctDNA [30], [31]. In recent years, apart from blood, various studies have highlighted the potential utility of salivary DNA for HNC management, including the study of the cfDNA fraction or the overall salivary total DNA. Of note, scfDNA is a mixture of non-tumoral and tumoral cfDNA (coined as salivary tumor DNA–stDNA) that can come directly from oral cells (local source) or from systemic circulation throughout the salivary glands [32].
Fig. 2.
Schematic overview of cell-free DNA release mechanisms, molecular analysis strategies and potential applications in head and neck cancer. ccfDNA, circulating cell-free DNA; ctDNA, circulating tumor DNA; non-ctDNA, non-circulating tumor DNA; scfDNA, salivary cell-free DNA; stDNA, salivary tumor DNA.
3. Applications of cell-free DNA in head and neck cancer
3.1. Cell-free DNA concentration
Several studies have determined the ccfDNA concentration in cancer patients and non-cancer controls, generally observing higher cfDNA levels in cancer patients compared to healthy individuals, and intermediate cfDNA levels in patients with benign conditions [33]. Focusing on HNC (Table 1), Mazurek et al. quantified the total cfDNA concentration in plasma of 200 HNC patients and 15 age-matched healthy controls by TERT amplification. An increase in cfDNA levels was observed in HNC patients compared to healthy controls, although this trend was not significant. Interestingly, when the analysis was performed according to the tumor anatomic sites, oropharyngeal cancer patients showed significantly higher cfDNA levels compared to other head and neck locations, which was explained by a greater release of cfDNA to the bloodstream as a consequence of the inflammation process associated to oropharyngeal tumors. Also, significantly high levels of cfDNA were observed in IV stage tumors and in N2–3 nodal disease, indicating its value as a biomarker for tumor progression [34]. Later, Lin et al. found significantly higher plasma cfDNA levels in oral squamous cell carcinoma (OSCC) patients compared to healthy controls using a spectrophotometric analyzer for the quantification of total cfDNA. Also, plasma cfDNA levels were significantly associated with tumor size, TNM stage, and lymphovascular invasion, according to this, those OSCC patients with large tumors, cervical nodal metastasis and advanced TNM stage showed higher levels of cfDNA. Moreover, using a cut-off of 20.2 ng/mL of plasma, the receiver operating characteristic (ROC) analysis yielded an area under the curve (AUC) of 0.69 suggesting the potential of plasma cfDNA for discriminating oral cancer [35]. In this line, Verma et al. detected significantly higher serum cfDNA levels in HNSCC patients compared to healthy individuals. Furthermore, stratified analysis by anatomic tumor subsites and TNM stage revealed an increase of serum cfDNA levels in oral cavity cancers and stage IV tumors, nevertheless, this association was not significant [36]. In contrast, other authors did not find significant differences between the concentrations of cfDNA in cancer patients and non-cancer controls [37]. In Shukla et al. study, plasma cfDNA levels were not significantly increased either in OSCC or in oral potentially malignant disorders compared to healthy individuals, which was explained by the quantification method used [37]. Previously, Coulet et al. evaluated the concentration of cfDNA in 117 HNC patients using a fluorometric method, but no correlation was also observed between plasma DNA levels and tumor stage, tumor location and gender [38]. Different factors could influence on the cfDNA levels such as the clinicopathological characteristics of the study subjects or the methodological characteristics of each investigation including sample processing, storage, nucleic acid isolation methods, and quantification techniques [39]. Furthermore, it is essential to highlight that high levels of cfDNA are not specific to cancer since physiological, [40] as well as other pathological conditions [41], [42], can affect cfDNA concentration. Particularly, various studies have quantified the levels of mitochondrial DNA in the cfDNA showing an increased of cell-free mitochondrial DNA in several malignancies, [43], [44] including HNC [45]. Kumar et al. reported that the cell-free mitochondrial DNA copy number in plasma of HNC patients was significantly higher compared to healthy controls. In addition, the concentration of cell-free mitochondrial DNA was also higher than cell-free nuclear DNA, revealing with the ROC analysis the potential diagnostic performance of cell-free mitochondrial DNA (84% sensitivity and 100% specificity) compared to cell-free nuclear DNA (53% sensitivity and 87% specificity) for discriminating HNC patients from controls [46]. In addition to plasma cfDNA, recently, our research group quantified for the first time the total scfDNA levels of oral cancer patients and healthy individuals using a fluorometric method. Although these findings evidence that oral cancer patients presented higher scfDNA levels compared to healthy individuals, no significant differences were observed, probably because of the limited cohort analyzed [47]. In this line, Sayal et al. quantified the salivary cell-free nuclear DNA levels and salivary cell-free mitochondrial DNA levels by qPCR, observing median scores significantly higher in HNC patients compared control groups. Moreover, ROC curve analysis yielded to AUC values of 0.758 for salivary cell-free nuclear DNA and 0.826 for salivary cell-free mitochondrial DNA, which reflects its value as potential diagnostic biomarkers [45]. Interestingly, high salivary cell-free nuclear DNA levels and salivary cell-free mitochondrial levels were associated with a poor overall survival in HNC patients. Moreover, univariate analysis revealed that the salivary cell-free mitochondrial DNA was an independent predictor of the patient's overall survival which showed the potential application of salivary cell-free mitochondrial DNA analysis for HNC prognosis [48].
Table 1.
Values of cfDNA quantification in saliva and plasma from HNC patients and non-cancer controls.
| Author | Biofluid | Isolation cfDNA method | DNA quantification method | Sample cohort | cfDNA concentration |
|---|---|---|---|---|---|
| Coulet et al. 2000 | Plasma | QIAmp Blood Kit (Qiagen) | Fluorometry (DyNA Quant 200 fluorimeter) | 16 OCSCC 49 OPSCC 32 HPSCC 20 EPSCC |
< 100 ng/mL (68.75%)/100 ng/mL (25%)/> 250 ng/mL (6.25%) < 100 ng/mL (63.26%)/100 ng/mL (26.5%)/> 250 ng/mL (10.20%) < 100 ng/mL (62.5%)/100 ng/mL (28.12%)/> 250 ng/mL (9.37%) < 100 ng/mL (70%)/100 ng/mL (25%)/> 250 ng/mL (5%) |
| Shukla et al. 2013 | Plasma | No isolation | Spectrophotometry (NanoDrop ND-1000 spectrophotometer; Thermo Fisher Scientific) | 150 OSCC 90 OPMD 150 HC 150 OSCC* |
44.36 ± 4.20 ng/μL mean ± SD 41.30 ± 4.32 ng/μL 41.12 ± 3.08 ng/μL 41.99 ± 3.91 ng/μL |
| Mazurek et al. 2016 | Plasma | Genomic Mini AX Body Fluids kit (A&A Biotechnology) |
qPCR (TERT gene amplification) | 200 HNC 15 HC 72 OPSCC 15 NP 20 HP 85 L |
9.22 ± 2.64 ng/mL mean ± SD 5.19 ± 7.96 ng/mL 9.60 ± 6.23 ng/mL 9.02 ± 7.41 ng/mL 8.29 ± 2.74 ng/mL 7.34 ± 4.04 ng/mL |
| Kumar et al. 2017 | Plasma | QIAamp Circulating Nucleic Acid Kit (Qiagen) | qPCR (GAPDH gene amplification for cfnDNA and D-Loop genes amplification for cf-mtDNA) | 54 HNSCC 52 HC |
5451.66/29,103,476.15† GE/mL median 1650.9/9189,312.54† GE/mL |
| Lin et al. 2018 | Plasma | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Spectrophotometry (TapeStation 2200, Agilent Technology) | 121 OSCC 52 HC |
53.1 ± 6.69 ng/mL mean ± SD 24 ± 3.33 ng/mL |
| Verma et al. 2020 | Serum | Charge Switch® gDNA 1 mL Serum Kit (Invitrogen) | SYBR Green qPCR (β-globin gene amplification) | 27 HNSCC 16 HC |
952.67 ± 657.43 ng/mL 60.65 ± 30.42 ng/mL |
| Rapado-González et al. 2022 | Saliva | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Fluorometry (Qubit 4 Fluorometer (Thermo Fisher Scientific) | 19 OSCC 15 HC |
6200 ng/mL (2500 to 11,233) median ± IQR 4333 ng/mL (1080 to 14,467) |
| Sayal et al. 2022 | Saliva | DNeasy Blood and Tissue kit (Qiagen) | qPCR (B2MG gene amplification for cfnDNA and mitochondrial gene amplification for cf-mtDNA) | 102 HNSCC 31 OLK 137 HC |
7.31/5.12† mega copies/mL median 2/1.44† mega copias/mL 2.29/0.92† mega copias/mL |
Abbreviations: cfDNA, cell-free DNA; cfnDNA, cell-free nuclear DNA; cf-mtDNA, cell-free mitochondrial DNA; HNSCC, head and neck squamous cell carcinoma; OCSCC, oral cavity squamous cell carcinoma; OPSCC, oropharynx squamous cell carcinoma; HPSCC, hypopharynx squamous cell carcinoma; EPSCC, endopharynx squamous cell carcinoma; OSCC, oral squamous cell carcinoma; OPMD, oral potentially malignant disorders; HC, healthy controls; NP, nasopharynx; HP, hypopharynx; L, larynx; OLK, oral leukoplakia; SD, standard deviation; IQR, interquartile range; *post-treatment OSCC patients, † cf-mtDNA.
Overall, cfDNA concentration shows potential as a clinical tool for HNC management, however, it is important to keep in mind that most of these studies have a retrospective design [34], [35], [37], [38], [45], [46], [47]. Although a control group was included in various of them to evaluate its potential as diagnostic biomarker [35], [36], [45], [47], the lack of longitudinal follow-up did not allow us to know how the inter- and intra-individual cfDNA levels vary during disease evolution. In addition, since various factors can determine the cfDNA concentration levels, different clinicopathological variables such as tumor stage or tumor location were considered in the correlation analysis [34], [37], [38]. However, the relationship with other variables such as comorbidities that can interfere with cfDNA concentration were not considered in any study.
3.2. Cell-free DNA fragmentomics
As aforementioned, apoptosis has been described as the primary mechanism by which cfDNA is released by cells into circulation, resulting in mono-nucleosome structures or multiples thereof (oligonucleosomes) [49]. However, there are other less predictive sources of cfDNA, such as the process of necrosis, which is more related to tumoral cells and represents a cfDNA font of high molecular-weight DNA fragments [23]. The analysis of the length of cfDNA fragments forms part of the ‘fragmentomics’ field and, based on these concepts, the cfDNA integrity (cfDI) index started to be explored. It consists of a formula that uses repetitive DNA sequences that are found all over the genome [50] for characterizing the fragmentation pattern of the cfDNA based on the ratio between longer, more associated with a necrosis origin, to shorter DNA fragments, which in theory represent the whole amount of cfDNA [51], [52]. Although (cfDI) has been more studied in other types of cancers [51], [53], only a few articles have evaluated its potential as a cancer diagnostic biomarker in HNC. Jiang et al. were one of the first to study the cfDI index, they described that the index was significantly greater in the plasma of a group of 58 HNSCC patients compared to the control subjects. However, no significant difference was found between the pre- and postoperative index values in the plasma [54]. In 2022, Rapado-González et al. demonstrated in a cohort of 19 OSCC patients the median values of salivary cfDI indexes for both ratios used, ALU115/ALU60 and ALU247/ALU60, were significantly higher in OSCC in comparison with healthy controls [47]. These studies highlight the potential of the cfDI index as a screening marker for the detection of cancer, but more studies and standardization in the process is needed.
3.3. Somatic mutations in cell-free DNA
The fraction of ctDNA represents a small percentage of the total ccfDNA (sometimes <0.01%), finding in those metastatic cancer patients higher concentrations of mutant DNA fragments in circulation compared to patients with local cancer disease [16], [55]. Since ctDNA harbors specific tumor genomic alterations and can provide a more thorough profile of tumor heterogeneity [56], several studies have explored this advantage by designing assays that may improve HNC management (Table 2).
Table 2.
Somatic gene mutations detected in liquid biopsies from head and neck cancer patients.
| Author | Cohort | Location | Stage | HPV | Type of samples (N) | Technique | % mutated cases/ (total cases) | Genes Analyzed | Main objective |
|---|---|---|---|---|---|---|---|---|---|
| Wang et al. 2015 | 93 HNSCC | 46 OC 34 OP 10 L 3 HP |
20 (I-II) 73 (III-IV) | 30 (+) 63 (–) |
Saliva | ddPCR and Safe-SeqS | 76%/(93) | TP53, PIK3CA, CDKN2A, FBXW7, HRAS, NRAS | To detect somatic mutations and HPV in plasma and saliva from HNSCC patients |
| Plasma | 87%/(47) | ||||||||
| Braig et al. 2016 | 46 HNSCC | 17 OP 12 OC 8 HP 4 L 2 PNS 2 OP/HP 1 HP/L |
3 (II) 42 (III-IV) 1 (UNK) |
5 (+) 41 (-) |
Plasma | Targeted sequencing | 46%/(20) | EGFR (exon 12), KRAS/NRAS (exons 2/3/4), HRAS (exons 2/3) | To identify acquired RAS mutations which could correlated with resistance to cetuximab in plasma samples from HNSCC patients during and after therapy |
| Mazurek et al. 2016 | 200 HNSCC | 72 OP 85 L 20 HP 15 NP 8 UNK |
83 (I-III) 114 (IV) |
28 (+) 172 (-) |
Plasma | PCR | 0/(200) |
KRAS G12C (c .34 G>T) EGFR (p.E746-A750del) |
To analyze the most frequent mutations of KRAS and EGFR in plasma for identifying HNSCC patients for treatment with anti-EGFR monoclonal antibodies and EGFR inhibitors |
| Perdomo et al. 2017 | 36 HNSCC (ARCAGE study) | n.a. | 14 (I-II) 22 (III-IV) |
36 (-) | Plasma | Targeted sequencing | 42%/ (36) | TP53, NOTCH1, CDKN2A, CASP8, PTEN | To evaluate the presence of DNA alterations in plasma ctDNA and oral rinses |
| 37 HNSCC (LA study) | n.a. | 37 (III-IV) | UNK | Plasma | 8.10%/(37) | TP53 | |||
| Oral rinse | 37.84%/(37) | ||||||||
| van Ginkel et al. 2017 | 6 HNSCC | 5 OC 1 OP |
1 (II) 5 (IV) |
6 (-) | Plasma | ddPCR | 100%/(6) | TP53 | To investigate whether low levels of ctDNA in plasma of HNSCC patients can be detected using ddPCR |
| Egyud et al.2018 | 8 HNSCC | n.a. | 1 (I) 7 (IV) |
4 (+) 4 (-) |
Plasma | Targeted sequencing (SiMSen-Seq) | n.a./(8) | TP53, ARID1B, ATM, CDK8, FANCA, RASA1, CSM2D, SIN3A, KRAS, NSD1, SMARCA4, XRCC2, BCL10, RPTOR | To examine the potential role of ctDNA in treatment monitoring and recurrence detection in HNSCC patients based on patient’s tumor specific mutations |
| Schmidt et al. 2018 | 29 HNSCC | 15 OP 10 OC 1 HP 1 L 2 UNK |
29 (III-IV) | 14 (+) 15 (-) |
Plasma | Allele-specific Plex-PCR™ technology | 31.03%/(29) | PIK3CA (p.E545K) | To determine whether Plex-PCR™ technology could be used to detect PIK3CA p.E545K mutation in HNSCC plasma samples |
| Galot et al. 2020 | 39-HNSCC (20 metastatic disease and 19 with recurrent disease) | 22 OP 8 OC 6 HP 6 L 1 UNK |
20 (IV) | OP 5 (+) OP 17 (-) |
Plasma | Targeted sequencing | 51%/(39) | Custom panel of 604 genes | To investigate the feasibility of detecting ctDNA in a prospective cohort of recurrent and/or metastatic HNSCC patients using a tissue-agnostic approach and evaluate the concordance of the mutational landscape between ctDNA and matched tumor |
| Mes et al. 2020 | 40 HNSCC | 5 OC 18 OP 10 HP 5 L 2 UNK |
2 (I) 4(II) 6 (III) 28 (IV) |
10 (+) 9 (-) 1 UNK 20 n.a. |
Plasma | Targeted sequencing | 67%/(27) | Custom panel of 12 genes (AJUBA, CASP8, CDKN2A, FAT1, FBXW7, HRAS, KMT2D, NOTCH1, NSD1, PIK3CA, PTEN, TP53) | To detect somatic mutations in tumor and corresponding plasma and to identify ctDNA without prior knowledge of tumor DNA aberrations |
| Khandelwal et al. 2020 | 22 OPSCC | 22 OP | n.a. | 11 (+) 11 (-) |
Plasma | Targeting sequencing | 50%/(22) | Accel-Amplicon 56 G Oncology Panel v2 (Swift Biosciences) | To explore the potential of ctDNA for detecting tumor somatic mutations and predicting recurrence or persistence disease |
| Burgener et al. 2021 | 30 HNSCC | 1 PNS 23 OC 3 L 3 HP |
4 (I) 2 (II) 5 (III) 19 (IV) |
9 (-) 21 n.a. |
Plasma | Targeted sequencing | 67%/(30) | CAncer Personalized Profiling by deep Sequencing (CAPP-seq) using 42 frequently recurrent genomic alterations in HNSCC from TCGA | To conduct multimodal profiling of mutations and methylation in ctDNA of HNSCC patients |
| Hilke et al. 2020 | 20 HNSCC | 14 OP 4 HP 2 OC |
n.a. | 5 (+) 14 (-) 1 n.a. |
Plasma | Targeted sequencing | 83%/(60) | 127 driver tumor mutations | To explore the capacity of ctDNA to monitor the treatment response during radio-chemotherapy and detect the molecular residual disease post-treatment |
| Porter et al. 2020 | 60 R/M HNSCC* * | 21 OP 12 OC 8 SG 6 L 4 HP 4 T 3 NP 2 UNK |
n.a. | 15 (+) 9 (-) 36 (UNK) | Blood | Targeted sequencing | 76%/(76) | Custom panel of 1021 genes | To characterize the ctDNA mutational profile of advanced HNC and identify actionable mutations |
| Wilson et al. 2020 | 75 HNSCC | 28 OC 22 OP 14 L 7 HP 3 PNS 1 NP |
28 (I-III) 47 (IVA-C) |
20 (+) 33 (-) 22 (UNK) |
Plasma | Targeted sequencing | 76% /(75) | Guardant360™ platform (73 genes) |
Characterization of genomic landscape in ctDNA and tumor DNA of HNSCC patients and assessment the prognostic impact |
| Wu et al. 2021 | 27 HNSCC | 11 L 10 HP 5 OC 1 OP |
19 (I-II) 18 (III-IV) |
27 (-) | Plasma | Targeted sequencing | 70.04%/(27) | Custom panel of 1021 genes | To profile the mutational features of different HNSCC samples including tumour tissues, tumor-adjacent tissue, pre- and post-surgical ctDNA and salivary ctDNA |
| Saliva | 63%/(27) | ||||||||
| Shanmugam et al. 2021 | 121 OSCC | 121 OC | 58 (I-II) 63 (III-IV) |
UNK | Saliva | Targeted sequencing | 95,87%/(121) | CASP8, PIK3CA, FAT1, CDKN2A, NOTCH1, HRAS, TP53 | To detect tumor-specific mutations in saliva of patients with OSCC |
| Cui et at. 2021 | 11 OSCC | 11 OC | 4 (II) 7 (III-IV) |
UNK | Plasma | Targeted sequencing | 27%/(11) | Custom panel of 71 genes | To examine the feasibility of using serial liquid biopsies in detecting minimal residual disease in oral cancer patients |
| Saliva | 91%/(11) | ||||||||
| Flach et al. 2022 (a) | 8 HNSCC | 3 OC 3 OP 1 L 1 HP |
1 (I-II) 7 (III-IV) | 8 (-) | Plasma | Targeted sequencing | 87.5%/(8) | Oncomine™ Comprehensive Assay v3Panel (161 genes) | Characterization of the mutational landscape in tumor, histopathologically negative resection margins and plasma cfDNA |
| Kogo et al. 2022 | 26 HNSCC | 5 OC 3 OP 7 L 3 HP 6 EAC |
6 (I-II) 20 (III-IV) |
22 (-) 4 (+) |
Plasma | Targeted sequencing | n.a./(18) | TP53, PIK3CA, KMT2D, FAT1, FBXW8, NOTCH3, CREBBP | To detect ctDNA candidate genes and performed ctDNA monitoring using ddPCR |
| Flach et al. 2022 (b) | 17 HNSCC | 5 OC 2 OP 7 L 4 HP 1 SPT |
17 (III-IV) | 17 (-) | Plasma | Multiplex PCR and targeted NGS | 100%/(17) | RaDaR™ patient-specific assay/ Personalised RaDaR™ panels (from 34 to 52 variants) | To determine whether post-operative ctDNA detection can act as a biomarker for surgical tumour clearance and to evaluate the potential of personalised ctDNA analysis for early detection of relapse |
| Rapado-González et al. 2022 | 3 HNSCC | 2 OC 1 HP |
3 (IV) | 3 (-) | Plasma | Targeted NGS | 110%/(3) | TruSight Tumor 170 panel (170 genes) | To detect somatic mutations in tumor and cfDNA from locoregional recurrent and/or metastatic HNSCC patients |
| Lin et al. 2022 | 107 OSCC | 107 OC | 21 (I-II) 86 (III-IV) |
n.a. | Plasma | ddPCR | 56.5%/(23) | TP53 | To evaluate the five most frequent coding TP53 mutations by using cancerous tissue and cfDNA in OSCC patients |
Abbreviations: HNSCC, head and neck squamous cell carcinoma; cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; OC, oral cavity; OP, oropharynx; L, larynx; HP, hypopharynx; T, thyroid; SG, salivary gland; PNS, paranasal sinus; UNK, unknown; EAC, external auditory canal; NP, nasopharynx; SPT, secondary primary tumor; PCR, polymerase chain reaction; ddPCR, droplet digital-PCR; HPV, human papilloma virus; n.a., not available.
3.4. Disease diagnosis and genotyping
Nowadays, the molecular tumor characterization using plasma ctDNA assays has been implemented in the clinic routine in some types of cancer [57], [58], representing a great advance to achieve precision oncology. Particularly in HNC, various studies have carried out different assays for testing the application of genomic profiling using liquid biopsies, mainly focusing on plasma cfDNA. In 2015, Wang et al. analyzed the presence of HPV16 DNA sequences and somatic mutations in TP53, PIK3CA, CDKN2A, FBXW7, HRAS, and NRAS in tumor, plasma, and saliva samples from 93 HNSCC patients. Tumor DNA was detected in 76% and 87% of saliva and plasma samples, respectively. Moreover, when both fluids were tested in combination, the detection rate increased to 96%. Interestingly, the detection of tumor DNA was influenced by the anatomic location and the stage of the tumor showing saliva a 100% sensitivity for early-stage oral cavity tumors and a relatively high fraction of mutant DNA (median 0.65%). However, plasma ctDNA was a predictor more sensitive than salivary tumor DNA for oropharynx (47% vs. 91%), hypopharynx (70% vs. 86%), and larynx (67% vs. 100%) tumors, as well as in advanced disease (92% vs. 70%). Furthermore, plasma HPV DNA showed a sensitivity of 86% (21/30 cases) while in saliva, the detection rate was 40% (12/30 cases), indicating the potential of HPV-cfDNA as a biomarker for HNSCC detection [59]. Similarly, Perdomo et al. evaluated the ctDNA performance comparing plasma samples of 36 HNSCC cases with their matched tissue, in which 65 mutations were previously identified by tumor sequencing in 5 genes (TP53, NOTCH1, CDKN2A, CASP8, and PTEN). In contrast with Wang et al. study [59], ctDNA alterations were only detected in 42% (15/36) of cases, which could be explained by cfDNA degradation related to prolonged storage of plasma samples (>10 years). Specifically, a total of 18 mutations (28%) in TP53, CASP8, NOTCH1 and CDKN2A were detected in both, tumor tissue and plasma, from HNC patients. Moreover, by sequencing of the entire coding region of TP53 in 37 III-IV stage patients, they identified 36 mutations in tumor, 3 in plasma and 26 in oral rinse sample. However, only 4 missense mutations in TP53 gene (p.Gly244Cys, p.Arg288Trp, p.Glu286Gly, and p.Val173Leu) were concordant between tumor and oral rinse samples whereas only one missense mutation (TP53 p.Val173Leu) was detected in the three samples, which indicates a low concordance for TP53 mutations among tumor and liquid biopsies. The mutations only detected in oral rinses could reflect the tumor heterogeneity and/or the genetic alterations in the squamous epithelial cells lining the oral cavity as a result of field cancerization, mostly related to tobacco and alcohol consumption. Interestingly, oral cavity and oropharynx tumors showed a higher proportion of TP53 variants in oral rinses compared to larynx tumors, which suggests a lower release rate of tumor DNA into saliva by the HNSCC anatomic locations which are more distant to oral cavity [60]. Recently, Shanmugam et al. developed a targeted NGS panel based on 7 genes (CASP8, PIK3CA, FAT1, CDKN2A, NOTCH1, HRAS, and TP53) to identify somatic mutations at low frequencies in saliva from 121 OSCC patients by ultra-deep sequencing. The design of these gene panel was based on publicly available OSCC datasets by selecting genes in which mutations would represent > 85% of patients with OSCC. This targeted sequencing approach detected 278 variants at ≥ 4% allele frequency in 87.6% of tumor samples (n = 106), of which 48.6% were missense variants and 28.8% non-sense mutations. In addition, mutations were detected in 75.5% of I-II and 97% of III-IV stage tumors, with TP53 remaining as the most mutated gene in tumor and saliva. The sequencing of oral rinses showed 377 variants at ≥ 0.1% allele frequency in 95.86% of patients (n = 116), of which 45.35% were missense variants and 28.9% non-sense mutations. An overall concordance of 93.4% was observed between primary tumor and oral rinse samples, being this rate increased in advanced stages (97%) compared to early stages (87.5%). Moreover, saliva revealed somatic mutations that were not detected in tumor, which evidences the potential of salivary tumor DNA for reflecting the intratumor heterogeneity in real-time of oral cancer [61].
Ongoing advances in NGS technology have allowed the development of gene panels for genomic profiling throughout liquid biopsies. In this line, Porter et al. analyzed the molecular profile of 60 recurrent or metastatic HNSCC using the Guardant360 platform composed by 70 genes for digital sequencing of ctDNA. They found ctDNA alterations in the 83% of patients, mainly in TP53 (68%), PIK3CA (34%), NOTCH1 (20%), and ARID1A (15%) genes. A total of 21 mutations were identified both in tumor and ctDNA (66% of concordance), however, in patients in which tissue NGS was available they detected through the sequencing of their ctDNA that 73% of them had new mutations in plasma, being most of them actionable mutations. Interestingly, ctDNA allowed to identify in 66% of HNSCC patients an off-label option and in 90% of them a trial option, which reflects the clinical value of ctDNA for identifying potentially targetable alterations [62]. Similarly, Wilson et al. characterized the molecular profile of 76 HNSCC patients using the Foundation One platform for tumor DNA sequencing and the Guardant360 platform for plasma ctDNA analysis. They reported tumor and ctDNA alterations in 100% and 76.6% of the patients, respectively, identifying actionable ctDNA mutations in 63.5% of patients. TP53, EGFR, KIT, BRAF, FGFR2, and FGFR3 genes showed a similar number of ctDNA and tumor DNA alterations, whereas ARID1A, ATM, and MET genes presented more alterations in ctDNA. The concordance rate for altered genes between tumor DNA and ctDNA was of 13%. Specifically, TP53 was the most mutated gene (73.3% of patients), showing a total of 127 alterations. Interestingly, ctDNA and, in particularly, TP53 ctDNA alterations were more frequent in recurrent (88% and 64.7, respectively) and metastatic (86% and 63.6%, respectively) patients compared with no evidence of the disease (27% and 9.1%, respectively) at the time of blood collection. Additionally, the presence of ctDNA alterations, TP53 ctDNA alterations, and DNA repair genes (APC, ATM, BRCA1, and/or BRCA2) were significantly associated with a decreased overall survival, which also shows the potential clinical value of ctDNA analysis for predicting HNSCC prognosis [63]. Using a more comprehensive approach, Galot et al. analyzed the feasibility to detect ctDNA in locoregional recurrent and/or metastatic HNSCC patients by targeted sequencing using a custom panel of 604 genes with ≥ 1% allele frequency. They detected ctDNA in 51% of patients (20/39), this probability was increased in patients suffering from metastatic (14/20) compared to locoregional recurrent disease (6/19) (70% vs. 30%). Like Wang et al. study, TP53 (50%) was the gene most frequently mutated in the cfDNA from HPV-negative HNC patients, followed by PIK3CA (15%). Interestingly, a low concordance rate in a cohort of 18 HNSCC patients was observed between ctDNA and tumor variants (19%), while in metastatic patients the rate of solid tumor variants identified in ctDNA was 42%. Moreover, 26% of the plasma variants were not detected in the matched tumor tissue, indicating the potential of cfDNA for providing information about the tumor heterogeneity and increasing the detection of actionable mutations. In addition, multivariate analysis showed that the metastatic status, tumor variant allele frequency, and ctDNA quantity were associated with the likelihood of detecting tumor tissue variants in plasma [64]. Similarly, our research group explored the potential of cfDNA for tumor mutational profiling of HNSCC using the TruSight Tumor 170 panel (TST170). An overall concordance rate of 37.5% was found between somatic mutations identified in ctDNA and matched tumor tissue, whereas a 62.5% of somatic variants were only detected in ctDNA. These findings highlight the possibility to detect somatic mutations in both cfDNA and tumor tissue using this targeted-NGS panel [65]. Recently, Flach et al. profiled the mutational landscape in primary and recurrent tumor tissue throughout the correspondence between resection margins and plasma cfDNA from 8 HNSCC patients using a targeted NGS panel of 161 genes. A total of 24 somatic variants were detected in the primary tumor, of which 5 were also detected in the resection margins (20.8%); 9 in plasma cfDNA (37.5%); and 3 (12.5%) in both resection margin and plasma cfDNA. Once again, TP53 was the most mutated gene in primary tumor and in resection margins, showing the 62.5% of plasma samples the same TP53 variants. Also, same tumor variants were detected in plasma samples for CDKN2A (25%), NF1 (25%), NOTCH1 (12.5%), and CDK12 (12.5%) genes, highlighting the potential clinical application of liquid biopsy for mutational profiling during HNC management [66].
3.5. Disease monitoring
Another potential clinical application of ctDNA analysis is the prediction of therapy response and monitoring of the disease. In this line, Egyud et al. demonstrated the ability of ctDNA preceding local recurrence of oral cavity tumors after surgical resection using tumor-specific profiles. However, tumor -specific mutations were detected in the post-operative ctDNA of two patients that developed distant recurrence which suggested different mutational profile between primary tumor and metastatic lesions [67]. For their part, Wang et al. detected the presence of tumor DNA in plasma and saliva before clinical manifestation of recurrence indicating the clinical value of both liquid biopsies for treatment monitoring in HNSCC patients [59]. In this line, Cui et al. reported the potential of using serial plasma and saliva ctDNA liquid biopsies for detecting minimal residual disease in oral cancer patients throughout a targeted deep sequencing panel. Interestingly, the presence of ctDNA in liquid samples increased the possibility of recurrence in oral cancer patients, being found higher ctDNA levels in saliva than in plasma. Moreover, the concordance rate between salivary cfDNA and tumor tissue DNA was 72.7%, while the rate for plasma cfDNA was 9.1%, highlighting the value of salivary cfDNA as an early tumor recurrence biomarker [68]. Recently, Wu et al. described that the presence of tumor adjacent tissue-specific mutations in post-operative plasma or saliva was indicative of disease relapse. Interestingly, they found out that the detection of tumor-adjacent tissue-specific mutations in addition to tumor-specific mutations in plasma and saliva after surgery allowed to better predict the relapse in comparison with the only detection of tumor-specific mutations, showing sensitivities of 75% for plasma and 87.5% for saliva. Moreover, postoperative ctDNA or salivary tumor DNA-positive patients was associated with a significantly shorter disease-free survival, indicating the potential of both post-operative liquid biopsies for predicting prognosis. Additionally, those patients who post-surgical ctDNA or salivary tumor DNA were detected, relapse was confirmed earlier in comparison with conventional clinical imaging [69]. In the same line, the detection of high concentrations of mutant cell-free mitochondrial DNA in postoperative serum samples was observed in oral cancer patients that suffered recurrence or metastatic disease, which indicates the potential value of cell-free mitochondrial DNA as prognostic biomarker [70], [71]. Moreover, numerous studies have explored the potential of ctDNA for predicting therapy resistance in HNSCC patients. Braig et al. detected RAS mutations during Cetuximab-based treatment in ctDNA in 46% of non-responder HNSCC patients while no RAS mutations were identified in the responder group, indicating that acquisition of RAS mutant clones during treatment significantly correlates with clinical resistance. Importantly, the early detection of RAS mutations during treatment was a predictor of disease progression demonstrating the potential of cfDNA for monitoring therapy response [72]. Later, Khandelwal et al. analyzed, by the sequencing of tumor and cfDNA, the mutational profile of responder and non-responder oropharyngeal cancer patients using a commercially gene panel with a limit of detection of allele frequency of 1%. After sequencing tumor and plasma samples of 22 oropharyngeal cancer patients (11 non-responders vs 11 responders), they identified somatic mutations in 12 tumor and 11 plasma samples. TP53, FBXW7 and PTEN were the genes most frequently mutated in tumor samples, while in plasma were EGFR, TP53 and APC. Importantly, cfDNA sequencing revealed the same tumor somatic variants of TP53, FBXW7 and CDKN2A in 4 patients HPV-negative non-responders and one HPV-positive responder patient, these findings suggest the potential of cfDNA for prediction recurrence in HPV-negative patients. In addition, survival analysis showed that HPV positive status and the detection of ctDNA were associated with a worse overall survival [73]. Hilke et al. carried out, for first time, a longitudinal analysis by ultra-deep sequencing of ctDNA in 20 patients with locally advanced HNC who underwent to radiochemotherapy. They detected ctDNA in 85% of patients, observing that higher ctDNA levels were significantly associated with bigger tumor volumes. Interestingly, they found a significantly negative correlation between the tumor allele fraction in the plasma and the course of treatment, showing a median decreasing from 1% (baseline sample) to 0.01% (6–12 weeks after treatment). In addition, the presence of minimal residual disease was also evaluated, observing recurrence in the 100% of cases with detectable ctDNA after the treatment. Of note, circulating HPV DNA showed a similar dynamic than ctDNA during treatment monitoring, remaining undetectable after treatment, suggesting its potential for diagnosis, monitoring therapy and detection recurrence in HNC [74]. Similarly, Kogo et al. monitored ctDNA levels based on specific tumor mutations in 18 HNSCC patients, observing that cases with positive ctDNA levels after initial curative treatment developed clinical disease recurrence, showing a worse prognosis compared to ctDNA negative patients [75].
Overall, the scientific evidence highlights the potential use of ctDNA mutations for diagnosis and monitoring HNC, however, most of these studies are retrospective and with a small cohort of study from a single institution [47], [62], [63], [64], [66], [67], [68], [74]. Thus, in various studies the number of included patients was under 50 [47], [64], [66], [67], [68], [72], [73], [74], [75]. Further studies with a prospective design and larger sample cohorts will allow to validate the clinical use of ctDNA in HNC. Another limitation in some studies was the lack of sequencing data from peripheral blood leukocytes to explore the role of clonal hematopoiesis mutations [60], [65], [66]. In addition to the heterogenous nature of HNC, the different methodological design of the studies including the sample type (saliva, plasma or serum), the collection and processing time, and the methods for cfDNA isolation and quantification can influence the recovery of cfDNA. In the same line, different detection thresholds for ctDNA sequencing have been reported in the studies. Then, it is still necessary to optimize and standardize different pre-analytical and analytical variables to reach the clinical implementation of this biomarker for HNC management.
3.6. DNA methylation in cell-free DNA
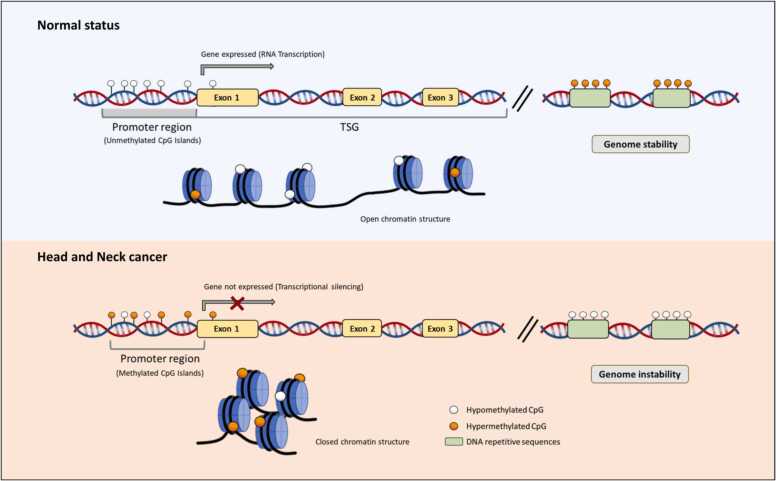
CfDNA fragments harbor not only tumor-specific mutations in their sequences but also tumor-specific epigenetic alterations such as DNA methylation (Fig. 3). Abnormal DNA methylation is recognized as a hallmark of cancer development and progression in which cancer cells are characterized by global loss of methylation (hypomethylation), that promotes genomic instability, and focal gain of methylation (hypermethylation) within the promoter region of specific-tumor suppressor genes, that lead to transcriptional inactivation [76]. In addition, given that DNA methylation alterations occur at early stages during carcinogenesis and some genes seem to acquire tissue-specific DNA methylation [76], [77], its analysis using liquid biopsies based on cfDNA has emerged as an attractive tool in HNC for early diagnosis, prognosis, and real-time monitoring disease (Table 3).
Fig. 3.
DNA methylation alterations in cell-free DNA. In normal cells, tumor suppressor genes (TSG) feature unmethylated CpG islands in their promoter regions, which correlate with an open chromatin structure and gene expression (upper left). However, in cancer cells, there is a focal hypermethylation within the promoter region of these genes that is correlated with a condensed, closed chromatin structure which causes transcriptional gene silencing (bottom left). Furthermore, cancer also exhibits a widespread hypomethylation phenomenon, which contributes to genomic instability (bottom left).
Table 3.
Methylated genes detected in liquid biopsies-based on cell-free DNA on head and neck cancer patients.
| Author | Cohort | Location | Stage | HPV | Type of samples (N) | Technique | Genes Analyzed | % methylated cases/ (total cases) | Main objective |
|---|---|---|---|---|---|---|---|---|---|
| Sanchez-Cespedes et al. 2000 | 95 HNSCC | OC (50) L (15) HP (9) OP (6) PS (3) |
n.a. | n.a. | Serum | MSP | P16 | 31%/(26) | To analyze the promoter hypermethylation pattern of the p16, MGMT, GSTP1, and DAPK genes in tumor and paired serum DNA samples from HNC patients |
| MGMT | 48%/(29) | ||||||||
| DAPK | 18%/(17) | ||||||||
| Wong et al. 2003 | 73 HNSCC | OC (33) HP (21) L (11) OP (8) |
n.a. | n.a. | Plasma | qPCR | P16 | 65%/(20) | To evaluate and quantify p16 and p15 methylation levels in plasma cfDNA samples of 20 HNSCC patients |
| P15 | 60%/(20) | ||||||||
| Nakahara et al. 2006 | 17 OSCC | OC (17) | 11 (I-II) 6 (III-IV) |
n.a. | Serum | MSP | P16 | 54.5%/(17) | To evaluate p16 promoter methylation in serum for detecting recurrent OSCC |
| Carvalho et al. 2008 | 211 HNSCC | n.a. | n.a. | n.a. | Serum | MSP | HIC1 | 31.4%/(70) | To evaluate aberrant promoter hypermethylation of candidate tumor suppressor genes in serum from HNSCC patients |
| PGP9.5 | 7.7%/(52) | ||||||||
| CDH1 | 32.3%/(62) | ||||||||
| CCND2 | 6.4%/(47) | ||||||||
| TIMP3 | 10%/(50) | ||||||||
| TGFBR2 | 8.1%/(37) | ||||||||
| Mydlarz et al. 2016 | 100 HNSCC | OP (46) OC (34) L (14) HP (2) UNK (4) |
20 (I-II) 80 (III-IV) | n.a. | Serum | qMSP | EDNRB | 10%/(100) | To analyze the promoter hypermethylation levels of EDNRB, DCC, and p16 in serum samples from HNSCC patients |
| DCC | 20%/(10) | ||||||||
| P16 | 50%/(2) | ||||||||
| Schröck et al. 2017 | 141 HNSCC | OC (38) OP (41) HP (17) L (33) Others (9) CUP (3) |
n.a. | n.a. | Plasma | qPCR | SEPT9 | n.a. | To explore the value of quantitative SEPT9 and SHOX2 methylation levels in cfDNA for the clinical management of HNSCC patients |
| SHOX2 | n.a. | ||||||||
| De Vos et al. 2017 | 141 HNSCC | OC (38) OP (41) HP (17) L (33) Others (9) CUP (3) |
n.a. | n.a. | Plasma | qPCR | SEPT9 | n.a. | To evaluate SEPT9 and SHOX2 methylation by different quantification algorithms (relative quantification, absolute quantification, quasi-digital PCR) with regard to their clinical performance |
| SHOX2 | n.a. | ||||||||
| De Jesús et al. 2020 | 54 OPSCC | OP (54) | 5 (I-II) 49 (III-IV) |
32 (-) 21 (+) | Plasma | ddPCR | CCNA1 | 63.6%/(11) | To evaluate methylation-based markers in plasma from OP patients as emerging tools for accurate/noninvasive follow-up |
| TIMP3 | 18.18%/(11) | ||||||||
| CDH8 | 9.1%/(11) | ||||||||
| DAPK | 9.1%/(11) | ||||||||
| LY6D | 45.2%/(42) | ||||||||
| Wang et al. 2021 | 202 OSCC | n.a. | n.a. | n.a. | Serum | qMSP | T-cadherin | 30.7%/(202) | To investigate the methylation status of T-cadherin in the sera of OSCC patients and correlated it with various clinicopathological characteristics and patient outcomes |
| Ishikawa et al. 2022 | 5 OSCC | OC (5) | n.a. | n.a. | Plasma | qMSP | OPRL-1 | n.a. | To explore the potential utility of opioid receptor gene methylation in pretreatment and posttreatment ctDNA oral cancer samples |
| OPRM-1 |
Abbreviations: cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; HNSCC, head and neck squamous cell carcinoma; OSCC, oral squamous cell carcinoma; PS, paranasal sinus; OC, oral cavity; OP, oropharynx; HP, hypopharynx; L, larynx; CUP, cancer of unknown primary; MSP, methylation-specific PCR; qMSP, quantitative-MSP; droplet-digital PCR, ddPCR; qPCR, quantitative-PCR; n.a., not available.
3.7. Diagnosis and prognosis
The first study that analyzed aberrant DNA methylation in liquid biopsies in HNC was performed in 2000 by Sanchez-Cespedes et al. This study evaluated, by methylation-specific PCR (MSP), the promoter hypermethylation of 4 genes (p16, MGMT, GSTP1, and DAPK) in 95 HNSCC tumors, detecting aberrant methylation on the genes p16, MGMT and DAPK in the 55% (52/95) of cases. Of the tumor methylated patients, 42% (21/50) displayed the same methylation changes in paired serum samples, with frequencies of p16, MGMT, and DAPK promoter hypermethylation of 36% (8/26), 48% (14/29), and 18% (3/17), respectively. As control group, they analyzed serum samples from 25 HNC patients; aberrant methylation was not found neither in the primary tumor nor in any serum sample. Interestingly, although no association was found between the presence of aberrant methylation in serum DNA and the clinicopathological variables including stage, tumor size, node involvement, or tumor location; serum methylation was more frequently observed in metastatic patients [78]. Along the same line, Wong et al. examined the methylation of p15 and p16 in plasma samples from 20 HNC and 24 healthy controls by real-time PCR. Methylation of p16 and p15 was observed in 65% and 60% of HNC patients respectively, whereas in healthy controls were methylated in 20% and 50%. Although gene methylation was observed in healthy plasma, the mean concentration of p16 and p15 methylated cfDNA was significantly higher in HNC patients compared with normal controls indicating the potential role of both markers for screening high-risk populations for early HNC and monitoring treatment response [79]. In another study, Nakahara et al. also examined the methylation of p16 in tumor and serum of 17 OSCC patients and eight healthy controls by MSP. Hypermethylation of p16 was detected in 64.7% of tumors and 54.5% of paired serum samples, whereas no methylation was observed in healthy controls. Like other authors aforementioned [78], no association was found between serum methylation and the tumor size, presence of lymph-node metastasis, and histological tumor differentiation [80]. Using a more comprehensive approach, Carvalho et al. evaluated a panel of 21 promoter hypermethylation tumor-suppressor genes in serum and saliva samples of 211 HNSCC patients and 527 normal controls by quantitative-MSP. In serum, throughout the combination of six genes (CCND2, TIMP3, HIC1, PGP9.5, TGFBR2, and CDH1) the sensitivities obtained ranged from 38.6% to 81% and specificities from 92.3% to 92.5% whereas in saliva the different combinations of hypermethylated CCNA1, DCC, DAPK, MINT31, MGMT, and p16 genes yielded to sensitivities ranging from 24% to 35.1% and specificities from 90% to 97% [81]. In the same line, Mydlarz et al. evaluated on the one hand, the methylation levels of EDRB, DCC, and p16 genes in serum samples from 100 HNSCC and 50 healthy controls observing amplification of EDNRB in 10% of HNSCC patients but in none of the healthy controls. Moreover, DCC methylation was detected in two patients that also amplified EDNRB, and in one of these patients was also observed p16 methylation. Interestingly, in nine of these 10 patients, promoter EDNRB hypermethylation was detected in saliva rinses, suggesting the ability of both liquid biopsies for reflecting the tumor methylation alterations [82]. Later, Schröck et al. analyzed the methylation status of SEPT9 and SHOX2 in two cohorts of HNSCC and control patients. Based on the results from the validation phase, SEPT9 and SHOX2 ccfDNA methylation levels were significantly higher in cancer patients compared to control group, reporting the combination of both genes a diagnostic accuracy of 0.80. Interestingly, SHOX2 and SEPT9 methylated plasma levels were significantly correlated with tumor and nodal category, and besides, SEPT9 and SHOX2 hypermethylation plasma levels were associated with a higher risk of death, which indicates the clinical value of these markers for diagnosis, molecular staging, and prognosis [83]. In another study, de Jesus et al. quantified the methylation levels of CCNA1, CDH8, DAPK, and TIMP3 genes in tumor and plasma samples from oropharyngeal cancer patients in order to evaluate their diagnostic capacity. The methylation of at least one of these genes was detected in 71% (37/52) of tumor samples, observing tumor-specific methylation frequencies of 48.1% in CCNA1, 40.4% in DAPK, 40.4% in CDH8, and 32.7% in TIMP3. In plasma, the methylation of these genes was detected in 73.3% (11/15) of oropharyngeal cancer patients, whereas no methylated ctDNA was detected in healthy individuals. An overall concordance of 80% was determined between tumor and plasma, and besides, a positive correlation was found between the tumor and plasmatic ctDNA methylation levels. Additionally, ROC analysis yielded to 73.3% sensitivity and 100% specificity (AUC: 0.867), indicating the diagnostic performance of methylated ctDNA for discriminating oropharyngeal cancer patients from healthy controls [84]. Also, aberrant promoter methylation of T-cadherin gene was reported in serum and tissue of OSCC patients. Moreover, methylation of T-cadherin was associated with advanced stage, positive lymph nodes, and tumor recurrence suggesting that loss of this gene by promoter methylation promotes tumor progression and lymph node metastasis through activating the PI3K/AKT/mTOR pathway. Furthermore, T-cadherin methylation was reported as an independent prognostic factor for oral cancer patients [85], which reflects its potential as prognostic biomarker. Recently, Patel et al. characterized the cfDNA methylation profile in pre- and post-surgery plasma samples of HNC patients, identifying by an unsupervised cluster analysis 30 differentially methylated regions that differentiated both groups. Interestingly, the top five validated differentially methylated were located in the promoter region of the genes PENK, NXPH1, ZIK1, TBXT, and CDO1. In addition, they found that the methylation of SFRP4, SOX1, IRF4, and PCDH17 was associated with the overall survival of HNC patients, supporting its utility as prognostic biomarkers [86].
Taking into account the scientific evidence, different gene-specific methylation biomarkers has been found in liquid biopsies from HNC patients. However, the performance of these biomarkers could be affected by the different methodological design of each study as well as the different preanalytical and postanalytical factors. Thus, the size of study cohort represents one of the most frequent limitations in the investigations [80], [84], [86], [87], so a further validation in an independent cohort is required to validate the performance of methylation biomarkers. Regarding the patient clinicopathological characteristics, the TNM stage was specified in only three studies [80], [82], [84] whereas the HPV status was described in one research [84]. Moreover, different tumor anatomic locations were included in the studies [78], [79], [82], [83] which could complicate the comparison and evaluation of the methylation biomarkers across the investigations. Since promoter methylation can be associated with the age, race, tobacco and alcohol consumption, the control group must be closely matched to the patient group as we can only observe in some investigations [81], [82]. In addition, any included studies in the present review evaluated the potential clinical performance of these methylation biomarkers in a benign control group, which could contribute to understand the role of these methylation markers in the early stages of head and neck carcinogenesis. Overall, a group of specific gene methylation markers were tested in liquid biopsies from HNC patient’s trough different techniques which could influence the sensitivity levels of the evaluated markers [78], [79], [82], [84]. Further larger-scale methylation technologies applied to liquid biopsies will allow to discover novel methylation markers and develop novel methylation signatures with clinical potential for HNC management.
3.8. Disease monitoring
Today, there are few studies in HNC that have reported the potential of cfDNA methylation for predicting recurrence and monitoring disease evolution. Nakahara et al. demonstrated the potential of serum cfDNA methylation analysis after surgery for detecting disease recurrence at early stage. They evaluated the methylation of p16 in serum samples collected 2 months post-surgery treatment and at the time of recurrence, detecting serum methylation of p16 in 3 of 4 patients at the recurrence of the disease [80]. In another prospective study, plasma methylation levels of SEPT9 and SHOX9 during patient´s follow-up were indicative of disease progression in 92% of patients who had a first positive result during the monitoring. In addition, plasma methylation levels above the cutoff were found in 47% of patients with locoregional recurrence or distant metastasis, within which in 78%, positive methylation could be detected in plasma up to 377 days earlier with respect to clinicopathological confirmation of the tumor progression. This showed the clinical value of both methylated plasma cfDNA markers for early detection of recurrence or metastatic disease in HNC patients [83]. Also, the detection of high plasma cfDNA methylation levels of CCNA1 gene after treatment were related with disease recurrence, suggesting the clinical usefulness of methylated ccfDNA for disease monitoring [84]. Recently, Misawa et al. identified a significant association between the hypermethylation of ATP2A1, CALML5, DNAJC5G, GNMT, GPT, LY6D, LYNX1, MAL, MGC16275, and MRGPRF, linked as well with the increase of recurrence in oropharyngeal cancer patients. Further methylation ctDNA analysis revealed that CALML5, DNAJC5G, and LY6D were methylated in 73.8%, 45.2%, and 45.2% of tumor samples, respectively, showing a high ability for discriminating HPV-associated oropharyngeal patients from healthy individuals. Importantly, the serial ctDNA analysis of 8 patients demonstrated that methylated CALML5, DNAJC5G, and LY6D can be detected at pre-treatment samples in 100%, 87.5%, and 87.5% of cases, respectively. Then, the ctDNA methylation levels of these markers were monitored at different time points during follow-up which demonstrated the potential value of these methylated markers to assess the clinical evolution of the disease in oropharyngeal cancer patients who underwent different treatments [88]. Similarly, Ishikawa et al. observed significantly higher OPRL1 gene levels in pretreatment samples with respect to post-treatment ctDNA samples, reflecting the potential utility of ctDNA methylation-based detection in the clinical management of oral cancer [87].
3.9. Multimodal profiling in cell-free DNA
An advantage of cfDNA is the possibility of analyzing simultaneously different genetic alterations such as somatic mutations or epigenetic aberrations, like DNA methylation, which provides a more comprehensive overview of the molecular landscape of head and neck tumors. Recently, Burgener et al. carried out a multimodal profiling of plasma cfDNA without prior molecular characterization of the tumor in 30 HNSCC patients and 20 risk-matched healthy individuals. On the one hand, mutational profiling of cfDNA conducted by CAncer Personalized Profiling sequencing (CAP-seq) and matched with genomic DNA from peripheral blood leukocytes revealed ctDNA in 20 patients and allowed to identify 43 tumor mutations with a minor allele frequency ranging from 0.14% to 4.83%. Importantly, plasma somatic mutations of driver genes such as TP53, PIK3CA, FAT1, and NOTCH1 showed similar mutational frequencies compared to head and neck tumors from The Cancer Genome Atlas (TCGA). Moreover, mutations in non-driver cancer genes such as GRIN1 and MYC were also detected in plasma, which could contribute to increasing the sensitivity of ctDNA detection. On the other hand, cfDNA methylome profiling by cfMeDIP-seq revealed 941-ctDNA derived hypermethylated regions enriched for CpG islands. Moreover, most of these regions overlapped with others that were hypermethylated in head and neck primary tumors in comparison with the adjacent normal tissue, describing HNSCC specific methylation patterns that indicate that many plasma hypermethylated regions are derived from tumor hypermethylated regions. Interestingly, a decrease in ctDNA’s fragment length in HNSCC patients was associated with ctDNA abundance, with a significant correlation between mutation- and methylation-based ctDNA abundance. The detection of ctDNA in baseline plasma samples detected using both mutation- and methylation- based methods was found to be a predictor of poor overall survival; on the contrary, the tumor stage was not associated with survival, demonstrating the prognostic value of multimodal cfDNA profiling in cancer. In addition, the measurement of ctDNA abundance by cfMeDIP-seq was applied for assessing the response after definitive treatment, this approach allowed to identify patients at high risk of disease recurrence, evidencing its clinical utility for monitoring tumors with few recurrent or clonal mutations in serial samples [89]. In another study, Mes et al. (2020) detected different genetic alterations in plasma cfDNA, including copy number aberrations (CNAs) and HPV DNA, through low-coverage whole genome sequencing in addition to somatic mutations by deep targeted sequencing of 12 cancer driver genes (AJUBA, CASP8, CDKN2A, FAT1, FBXW7, HRAS, KMT2D, NOTCH1, NSD1, PIK3CA, PTEN, and TP53) in a cohort of 40 HNC patients. They found CNAs and somatic mutations from patients with known tumor mutation data in 52% (14/27) and 67% (18/27) of the plasma samples, respectively. Like in tumor analysis, HPV was detected in plasma cfDNA on 100% HPV-positive HNC patients. Importantly, the combined analysis of CNA, HPV DNA, and somatic mutations in plasma cfDNA increased the ctDNA detection rate to 78%. Moreover, a positive correlation was found between TNM stage, and the detection of CNAs or mutations found in plasma, whereas the location of the primary tumor and HPV-status were not associated with the detection of ctDNA [90].
4. Cell-free DNA and imaging in head and neck cancer
Liquid biopsy has showed a great potential in the era of precision medicine due to its ability to capture the tumor genomic landscape of the disease which can influence in the decision making for personalized treatments in cancer. By liquid biopsy analysis is possible to track the genomic alterations detected in ctDNA during disease evolution. CtDNA levels correlate with the tumor volume measured by computed tomography (CT) and positron emission tomography (PET) imaging representing an attractive and minimally invasive strategy for the longitudinal treatment monitoring and detection of minimal residual disease after curative intent therapy [91]. Thus, ctDNA dynamic changes are associated with treatment outcomes and could be predictive of subsequent radiographic results in the disease follow-up [92]. In HNC, several studies have demonstrated the potential of monitoring specific somatic mutations by ctDNA analysis for predicting disease recurrence in patients with positive ctDNA levels [59], [74], [75]. Also, longitudinal monitoring of plasma circulating tumor HPV during post-treatment was indicative of disease recurrence earlier than the routine clinical follow-up [93]. In addition, the molecular cfHPV16 testing could complement the imaging-based assessment (MRI, CT, or 18 F-FDG PET-CT) for early identification of treatment failure in HPV-positive oropharyngeal cancer patients, allowing for more effective salvage therapy [94]. These results suggest that ctDNA could be complementary to radiological assessments providing a more rapid evaluation of tumor response than traditional imaging alone. In this line, future studies will provide more evidence about the synergize of imaging and liquid biopsy as an integrated approach for HNC management.
5. Application of cell-free DNA assays for cancer management
The ongoing advances in the detection and characterization of ctDNA have allowed the design of single- and multigene assays to detect genetic alterations in plasma cfDNA for using it as companion diagnostics and selecting molecular targeted therapies. In this line, various gene specific ctDNA tests are already being used into clinical practice for cancer management like the Cobas® EGFR mutation test V2 (Roche), that allows to screen for EGFR mutations in plasma cfDNA from patients with advanced- stage non-small cell lung cancer, and the Therascreen PIK3CA RGQ PCR kit (Qiagen), that is designed to detect PIK3CA mutations in tumor tissue or plasma from patients with advanced-stage hormone receptor (HR)+ /HER2 − breast cancer [95]. Additionally, high-throughput NGS-based multigene liquid biopsy tests have been approved by the FDA for comprehensive ctDNA testing such as the Guardant360 CDx (Guardant) and the FoundationOne Liquid CDx (F1LCDx), that allows a broad cancer genotyping and the identification of clinically actionable alterations that can guide the use of molecularly targeted therapies in different solid tumor entities [96], [97]. In HNC, numerous assays have showed the potential clinical utility of ctDNA testing for HNC profiling and disease monitoring as we have described in the present review; however, any specific commercial ctDNA assay has been designed for HNC management. Despite these advances, the application of liquid biopsy based on ctDNA assays in HNC is in its infancy and more research efforts are needed to develop and validate tests based on ctDNA than can reliably detect and monitor this disease and confidently incorporate them into routine clinical care.
6. Conclusion
Modern high-throughput genomic approaches have allowed liquid biopsies-based biomarkers to gain more attention in oncology care. In special, the study of cfDNA has proven that it is a dynamic and minimally non-invasive biomarker that provides real-time molecular information about tumor disease. Several studies have highlighted the potential utility of cfDNA analysis in HNC, mainly focusing on genetic and epigenetic biomarkers, for tumor detection, prognosis, and therapy monitoring of these patients. However, although liquid biopsies based on cfDNA have demonstrated to be a powerful tool in HNC management as we have exposed in this review, large-scale prospective studies need to be performed to further demonstrate their clinical utility as a biomarker for HNC which would enable a more effective and personalized treatment of these patients.
Conflict of interest
R.L.-L. reports other from Nasasbiotech, during the develop of the study; grants and personal fees from Merck, grants and personal fees from AstraZeneca, personal fees from Bayer, personal fees from Roche, personal fees and non-financial support from BMS, personal fees from Leo, personal fees from Pharmamar, outside the submitted work. The rest of the authors have nothing to disclose.
Funding
This work was supported by the Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union (PI20/01449). O.R.-G. is funded by a postdoctoral fellowship from Axencia Galega de Innovacion (GAIN), Programa de ayudas a la etapa posdoctoral de la Xunta de Galicia (IN606B-2022/007). A. R.-C. is funded by a predoctoral fellowship from Axencia Galega de Innovación (GAIN), Programa de ayudas a la etapa predoctoral de la Xunta de Galicia (IN606A-2021/009).
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chow L.Q.M. Head and neck cancer. New Engl J Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 3.Marur S., Forastiere A.A. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Leemans C.R., Braakhuis B.J.M., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 5.Sabatini M.E., Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer. 2020;122:306–314. doi: 10.1038/s41416-019-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 7.Fettke H., Kwan E.M., Azad A.A. Cell-free DNA in cancer: current insights. Cell Oncol. 2019;42:13–28. doi: 10.1007/s13402-018-0413-5. [DOI] [PubMed] [Google Scholar]
- 8.Li S., Yi M., Dong B., Tan X., Luo S., Wu K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer. 2021;148:2640–2651. doi: 10.1002/ijc.33386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loreth D., Schuette M., Zinke J., Mohme M., Piffko A., Schneegans S., et al. CD74 and CD44 expression on CTCs in cancer patients with brain metastasis. Int J Mol Sci. 2021;22:6993. doi: 10.3390/ijms22136993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapado-González Ó., López-López R., López-Cedrún J.L., Triana-Martínez G., Muinelo-Romay L., Suárez-Cunqueiro M.M. Cell-free microRNAs as potential oral cancer biomarkers: from diagnosis to therapy. Cells. 2019;8:1653. doi: 10.3390/cells8121653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikanjam M., Kato S., Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131. doi: 10.1186/s13045-022-01351-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thierry A.R., el Messaoudi S., Gahan P.B., Anker P., Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastas Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel P., Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 14.Leon S.A., Shapiro B., Sklaroff D.M., Yaros M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 15.Zill O.A., Banks K.C., Fairclough S.R., Mortimer S.A., Vowles J.V., Mokhtari R., et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res. 2018;24:3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 16.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattox A.K., Bettegowda C., Zhou S., Papadopoulos N., Kinzler K.W., Vogelstein B. Applications of liquid biopsies for cancer. Sci Transl Med. 2019:11. doi: 10.1126/scitranslmed.aay1984. [DOI] [PubMed] [Google Scholar]
- 19.Mulcahy H.E., Croke D.T., Farthing M.J. Cancer and mutant DNA in blood plasma. Lancet. 1996;348:628. doi: 10.1016/S0140-6736(05)65067-2. [DOI] [PubMed] [Google Scholar]
- 20.Stroun M., Lyautey J., Lederrey C., Olson-Sand A., Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 21.Schwarzenbach H., Hoon D.S.B., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 22.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D., et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 23.Heitzer E., Auinger L., Speicher M.R. Cell-free DNA and apoptosis: how dead cells inform about the living. Trends Mol Med. 2020;26:519–528. doi: 10.1016/j.molmed.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Alix-Panabières C., Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 2021;11:858–873. doi: 10.1158/2159-8290.CD-20-1311. [DOI] [PubMed] [Google Scholar]
- 25.Ronchetti L., Terrenato I., Ferretti M., Corrado G., Goeman F., Donzelli S., et al. Circulating cell free DNA and citrullinated histone H3 as useful biomarkers of NETosis in endometrial cancer. J Exp Clin Cancer Res. 2022;41:151. doi: 10.1186/s13046-022-02359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahlert C., Melo S.A., Protopopov A., Tang J., Seth S., Koch M., et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Underhill H.R., Kitzman J.O., Hellwig S., Welker N.C., Daza R., Baker D.N., et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabon J.J., Hamilton E.G., Kurtz D.M., Esfahani M.S., Moding E.J., Stehr H., et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–251. doi: 10.1038/s41586-020-2140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marass F., Stephens D., Ptashkin R., Zehir A., Berger M.F., Solit D.B., et al. Fragment size analysis may distinguish clonal hematopoiesis from tumor-derived mutations in cell-free DNA. Clin Chem. 2020;66:616–618. doi: 10.1093/clinchem/hvaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pittella-Silva F., Chin Y.M., Chan H.T., Nagayama S., Miyauchi E., Low S.K., et al. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin Chem. 2020;66:946–957. doi: 10.1093/clinchem/hvaa103. [DOI] [PubMed] [Google Scholar]
- 31.Jung M., Klotzek S., Lewandowski M., Fleischhacker M., Jung K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem. 2003;49:1028–1029. doi: 10.1373/49.6.1028. [DOI] [PubMed] [Google Scholar]
- 32.Rapado-González Ó., Muinelo-Romay L., Suárez-Cunqueiro M.M. Vol. 112. 2021. Letter to the editor: “Liquid biopsy based on saliva cell-free DNA as a potential biomarker for head and neck cancer. (Oral Oncol). [DOI] [PubMed] [Google Scholar]
- 33.Szpechcinski A., Chorostowska-Wynimko J., Struniawski R., Kupis W., Rudzinski P., Langfort R., et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer. 2015;113:476–483. doi: 10.1038/bjc.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazurek A.M., Rutkowski T., Fiszer-Kierzkowska A., Małusecka E., Składowski K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016;54:36–41. doi: 10.1016/j.oraloncology.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Lin L.H., Chang K.W., Kao S.Y., Cheng H.W., Liu C.J. Increased plasma circulating cell-free DNA could be a potential marker for oral cancer. Int J Mol Sci. 2018;19:3303. doi: 10.3390/ijms19113303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma T., Kumari S., Mishra S., Rastogi M., Tiwari V., Agarwal G.R., et al. Circulating free DNA as a marker of response to chemoradiation in locally advanced head and neck squamous cell carcinoma. Indian J Pathol Microbiol. 2020;63:521–526. doi: 10.4103/IJPM.IJPM_28_20. [DOI] [PubMed] [Google Scholar]
- 37.Shukla D., Kale A.D., Hallikerimath S., Yerramalla V., Subbiah V. Can quantifying free-circulating DNA be a diagnostic and prognostic marker in oral epithelial dysplasia and oral squamous cell carcinoma? J Oral Maxillofac Surg. 2013;71:414–418. doi: 10.1016/j.joms.2012.04.039. [DOI] [PubMed] [Google Scholar]
- 38.Coulet F., Blons H., Cabelguenne A., Lecomte T., Lacourreye O., Brasnu D., et al. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2000;60:707–711. [PubMed] [Google Scholar]
- 39.Bryzgunova O.E., Konoshenko M.Y., Laktionov P.P. Concentration of cell-free DNA in different tumor types. Expert Rev Mol Diagn. 2021;21:63–75. doi: 10.1080/14737159.2020.1860021. [DOI] [PubMed] [Google Scholar]
- 40.Hummel E.M., Hessas E., Müller S., Beiter T., Fisch M., Eibl A., et al. Cell-free DNA release under psychosocial and physical stress conditions. Transl Psychiatry. 2018;8:236. doi: 10.1038/s41398-018-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzano A., Israr M.Z., Garcia D.F., Middleton L., D’Assante R., Marra A.M., et al. Circulating cell-free DNA levels are associated with adverse outcomes in heart failure: testing liquid biopsy in heart failure. Eur J Prev Cardiol. 2021;28:e28–e31. doi: 10.1177/2047487320912375. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y., Song Y., Chang J., Zhou X., Qi Q., Tian X., et al. High levels of circulating cell-free DNA are a biomarker of active SLE. Eur J Clin Invest. 2018;48 doi: 10.1111/eci.13015. [DOI] [PubMed] [Google Scholar]
- 43.Linke C., Hunger R., Reinwald M., Deckert M., Mantke R. Quantification of mitochondrial cfDNA reveals new perspectives for early diagnosis of colorectal cancer. BMC Cancer. 2023;23:291. doi: 10.1186/s12885-023-10748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng X., Schwarzenbach H., Yang Y., Müller V., Li N., Tian D., et al. Circulating mitochondrial DNA is linked to progression and prognosis of epithelial ovarian cancer. Transl Oncol. 2019;12:1213–1220. doi: 10.1016/j.tranon.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayal L., Hamadah O., Almasri A., Idrees M., Thomson P., Kujan O. Saliva-based cell-free DNA and cell-free mitochondrial DNA in head and neck cancers have promising screening and early detection role. J Oral Pathol Med. 2023;52:29–36. doi: 10.1111/jop.13392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar M., Srivastava S., Singh S.A., Das A.K., Das G.C., Dhar B., et al. Cell-free mitochondrial DNA copy number variation in head and neck squamous cell carcinoma: a study of non-invasive biomarker from Northeast India. Tumor Biol. 2017;39 doi: 10.1177/1010428317736643. 101042831773664. [DOI] [PubMed] [Google Scholar]
- 47.Rapado‐González Ó., López‐Cedrún J.L., Lago‐Lestón R.M., Abalo A., Rubin‐Roger G., Salgado‐Barreira Á., et al. Integrity and quantity of salivary cell‐free DNA as a potential molecular biomarker in oral cancer: a preliminary study. J Oral Pathol Med. 2022;51:429–435. doi: 10.1111/jop.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayal L., Hamadah O., AlMasri A., Idrees M., Kassem I., Habbal W., et al. Salivary-based cell-free mitochondrial DNA level is an independent prognostic biomarker for patients with head and neck squamous cell carcinoma. J Pers Med. 2023;13:301. doi: 10.3390/jpm13020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwarzenbach H. Circulating nucleic acids as biomarkers in breast cancer. Breast Cancer Res. 2013;15:211. doi: 10.1186/bcr3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gemmell N.J. Repetitive DNA: genomic dark matter matters. Nat Rev Genet. 2021;22(342–342):8. doi: 10.1038/s41576-021-00354-8. [DOI] [PubMed] [Google Scholar]
- 51.Umetani N., Giuliano A.E., Hiramatsu S.H., Amersi F., Nakagawa T., Martino S., et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–4276. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 52.Kustanovich A., Schwartz R., Peretz T., Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20:1057–1067. doi: 10.1080/15384047.2019.1598759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinha S., Brown H., Tabak J., Fang Z., Tertre M.C., du, McNamara S., et al. Multiplexed real-time polymerase chain reaction cell-free DNA assay as a potential method to monitor stage IV colorectal cancer. Surgery. 2019;166:534–539. doi: 10.1016/j.surg.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Jiang W.W., Zahurak M., Goldenberg D., Milman Y., Park H.L., Westra W.H., et al. Increased plasma DNA integrity index in head and neck cancer patients. Int J Cancer. 2006;119:2673–2676. doi: 10.1002/ijc.22250. [DOI] [PubMed] [Google Scholar]
- 55.Powles T., Assaf Z.J., Davarpanah N., Banchereau R., Szabados B.E., Yuen K.C., et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–437. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 56.Vandekerkhove G., Lavoie J.-M., Annala M., Murtha A.J., Sundahl N., Walz S., et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat Commun. 2021;12:184. doi: 10.1038/s41467-020-20493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Sáez O., Chic N., Pascual T., Adamo B., Vidal M., González-Farré B., et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020;22:45. doi: 10.1186/s13058-020-01284-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujii H., Nagakura H., Kobayashi N., Kubo S., Tanaka K., Watanabe K., et al. Liquid biopsy for detecting epidermal growth factor receptor mutation among patients with non-small cell lung cancer treated with afatinib: a multicenter prospective study. BMC Cancer. 2022;22:1035. doi: 10.1186/s12885-022-10135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Springer S., Mulvey C.L., Silliman N., Schaefer J., Sausen M., et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015:7. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perdomo S., Avogbe P.H., Foll M., Abedi-Ardekani B., Facciolla V.L., Anantharaman D., et al. Circulating tumor DNA detection in head and neck cancer: evaluation of two different detection approaches. Oncotarget. 2017;8:72621–72632. doi: 10.18632/oncotarget.20004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shanmugam A., Hariharan A.K., Hasina R., Nair J.R., Katragadda S., Irusappan S., et al. Ultrasensitive detection of tumor‐specific mutations in saliva of patients with oral cavity squamous cell carcinoma. Cancer. 2021;127:1576–1589. doi: 10.1002/cncr.33393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porter A., Natsuhara M., Daniels G.A., Patel S.P., Sacco A.G., Bykowski J., et al. Next generation sequencing of cell free circulating tumor DNA in blood samples of recurrent and metastatic head and neck cancer patients. Transl Cancer Res. 2020;9:203–209. doi: 10.21037/tcr.2019.12.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson H.L., D’Agostino R.B., Meegalla N., Petro R., Commander S., Topaloglu U., et al. The prognostic and therapeutic value of the mutational profile of blood and tumor tissue in head and neck squamous cell carcinoma. Oncologist. 2021;26:e279–e289. doi: 10.1002/onco.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galot R., van Marcke C., Helaers R., Mendola A., Goebbels R.M., Caignet X., et al. Liquid biopsy for mutational profiling of locoregional recurrent and/or metastatic head and neck squamous cell carcinoma. Oral Oncol. 2020;104 doi: 10.1016/j.oraloncology.2020.104631. [DOI] [PubMed] [Google Scholar]
- 65.Rapado-González Ó., Brea-Iglesias J., Rodríguez-Casanova A., Bao-Caamano A., López-Cedrún J.L., Triana-Martínez G., et al. Somatic mutations in tumor and plasma of locoregional recurrent and/or metastatic head and neck cancer using a next-generation sequencing panel: a preliminary study. Cancer Med. 2022:6615–6622. doi: 10.1002/cam4.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flach S., Kumbrink J., Walz C., Hess J., Drexler G., Belka C., et al. Analysis of genetic variants of frequently mutated genes in human papillomavirus‐negative primary head and neck squamous cell carcinoma, resection margins, local recurrences and corresponding circulating cell‐free DNA. J Oral Pathol Med. 2022;51:738–746. doi: 10.1111/jop.13338. [DOI] [PubMed] [Google Scholar]
- 67.Egyud M., Sridhar P., Devaiah A., Yamada E., Saunders S., Ståhlberg A., et al. Plasma circulating tumor DNA as a potential tool for disease monitoring in head and neck cancer. Head Neck. 2019;41:1351–1358. doi: 10.1002/hed.25563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui Y., Kim H.S., Cho E.S., Han D., Park J.A., Park J.Y., et al. Longitudinal detection of somatic mutations in saliva and plasma for the surveillance of oral squamous cell carcinomas. PLoS One. 2021;16 doi: 10.1371/journal.pone.0256979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu P., Xie C., Yang L., Liu Y., Zeng J., Li X., et al. The genomic architectures of tumour-adjacent tissues, plasma and saliva reveal evolutionary underpinnings of relapse in head and neck squamous cell carcinoma. Br J Cancer. 2021;125:854–864. doi: 10.1038/s41416-021-01464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uzawa K., Kasamatsu A., Baba T., Kimura Y., Nakashima D., Higo M., et al. Quantitative detection of circulating tumor-derived mitochondrial NADH subunit variants as a potential prognostic biomarker for oral cancer. Int J Oncol. 2015;47:1077–1083. doi: 10.3892/ijo.2015.3083. [DOI] [PubMed] [Google Scholar]
- 71.Uzawa K., Baba T., Uchida F., Yamatoji M., Kasamatsu A., Sakamoto Y., et al. Circulating tumor-derived mutant mitochondrial DNA: a predictive biomarker of clinical prognosis in human squamous cell carcinoma. Oncotarget. 2012;3:670–677. doi: 10.18632/oncotarget.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Braig F., Voigtlaender M., Schieferdecker A., Busch C.-J., Laban S., Grob T., et al. Liquid biopsy monitoring uncovers acquired RAS-mediated resistance to cetuximab in a substantial proportion of patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7:42988–42995. doi: 10.18632/oncotarget.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khandelwal A.R., Greer A.H., Hamiter M., Fermin J.M., McMullen T., Moore‐Medlin T., et al. Comparing cell‐free circulating tumor DNA mutational profiles of disease‐free and nonresponders patients with oropharyngeal squamous cell carcinoma. Laryngoscope Invest Otolaryngol. 2020;5:868–878. doi: 10.1002/lio2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hilke F.J., Muyas F., Admard J., Kootz B., Nann D., Welz S., et al. Dynamics of cell-free tumour DNA correlate with treatment response of head and neck cancer patients receiving radiochemotherapy. Radio Oncol. 2020;151:182–189. doi: 10.1016/j.radonc.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 75.Kogo R., Manako T., Iwaya T., Nishizuka S., Hiraki H., Sasaki Y., et al. Individualized circulating tumor DNA monitoring in head and neck squamous cell carcinoma. Cancer Med. 2022;11:3960–3968. doi: 10.1002/cam4.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kulis M., Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 77.Constâncio V., Nunes S.P., Henrique R., Jerónimo C. DNA methylation-based testing in liquid biopsies as detection and prognostic biomarkers for the four major cancer types. Cells. 2020;9:624. doi: 10.3390/cells9030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez-Cespedes M., Esteller M., Wu L., Nawroz-Danish H., Yoo G.H., Koch W.M., et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892–895. [PubMed] [Google Scholar]
- 79.Wong T.S., Man M.W.L., Lam A.K.Y., Wei W.I., Kwong Y.L., Yuen A.P.W. The study of p16 and p15 gene methylation in head and neck squamous cell carcinoma and their quantitative evaluation in plasma by real-time PCR. Eur J Cancer. 2003;39:1881–1887. doi: 10.1016/s0959-8049(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 80.Nakahara Y., Shintani S., Mihara M., Hino S., Hamakawa H. Detection of p16 promoter methylation in the serum of oral cancer patients. Int J Oral Maxillofac Surg. 2006;35:362–365. doi: 10.1016/j.ijom.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 81.Carvalho A.L., Jeronimo C., Kim M.M., Henrique R., Zhang Z., Hoque M.O., et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 82.Mydlarz W.K., Hennessey P.T., Wang H., Carvalho A.L., Califano J.A. Serum biomarkers for detection of head and neck squamous cell carcinoma. Head Neck. 2016;38:9–14. doi: 10.1002/hed.23842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schröck A., Leisse A., de Vos L., Gevensleben H., Dröge F., Franzen A., et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin Chem. 2017;63:1288–1296. doi: 10.1373/clinchem.2016.270207. [DOI] [PubMed] [Google Scholar]
- 84.Jesus L.M., Reis M.B., Carvalho R.S., Scapulatempo Neto C., Almeida G.C., Laus A.C., et al. Feasibility of methylated ctDNA detection in plasma samples of oropharyngeal squamous cell carcinoma patients. Head Neck. 2020;42:3307–3315. doi: 10.1002/hed.26385. [DOI] [PubMed] [Google Scholar]
- 85.Wang Q., Chen Y., Chen Y., Jiang J., Song X., Zhang L., et al. Aberrant promoter methylation of T-cadherin in sera is associated with a poor prognosis in oral squamous cell carcinoma. Neoplasma. 2021;68:528–534. doi: 10.4149/neo_2021_201110N1203. [DOI] [PubMed] [Google Scholar]
- 86.Patel K.B., Padhya T.A., Huang J., Hernandez-Prera J.C., Li T., Chung C.H., et al. Plasma cell-free DNA methylome profiling in pre- and post-surgery oral cavity squamous cell carcinoma. Mol Carcinog. 2023;62:493–502. doi: 10.1002/mc.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishikawa R., Imai A., Mima M., Yamada S., Takeuchi K., Mochizuki D., et al. Novel prognostic value and potential utility of opioid receptor gene methylation in liquid biopsy for oral cavity cancer. Curr Probl Cancer. 2022;46 doi: 10.1016/j.currproblcancer.2021.100834. [DOI] [PubMed] [Google Scholar]
- 88.Misawa K., Imai A., Matsui H., Kanai A., Misawa Y., Mochizuki D., et al. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene. 2020;39:4741–4755. doi: 10.1038/s41388-020-1327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burgener J.M., Zou J., Zhao Z., Zheng Y., Shen S.Y., Huang S.H., et al. Tumor-naïve multimodal profiling of circulating tumor DNA in head and neck squamous cell carcinoma. Clin Cancer Res. 2021;27:4230–4244. doi: 10.1158/1078-0432.CCR-21-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mes S.W., Brink A., Sistermans E.A., Straver R., Oudejans C.B.M., Poell J.B., et al. Comprehensive multiparameter genetic analysis improves circulating tumor DNA detection in head and neck cancer patients. Oral Oncol. 2020;109 doi: 10.1016/j.oraloncology.2020.104852. [DOI] [PubMed] [Google Scholar]
- 91.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C.W., Modlin L.A., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Siravegna G., Mussolin B., Venesio T., Marsoni S., Seoane J., Dive C., et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30:1580–1590. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 93.Chera B.S., Kumar S., Beaty B.T., Marron D., Jefferys S., Green R., et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin Cancer Res. 2019;25:4682–4690. doi: 10.1158/1078-0432.CCR-19-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rutkowski T.W., Mazurek A.M., Śnietura M., Hejduk B., Jędrzejewska M., Bobek-Billewicz B., et al. Circulating HPV16 DNA may complement imaging assessment of early treatment efficacy in patients with HPV-positive oropharyngeal cancer. J Transl Med. 2020;18:167. doi: 10.1186/s12967-020-02330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamadeh L.N., Farhat L., Hilal L., Assi H., Nasr F., Chahine G., et al. Frequency and mutational spectrum of PIK3CA gene mutations in breast cancer patients: largest and first report from Lebanon. Gene. 2023;871 doi: 10.1016/j.gene.2023.147433. [DOI] [PubMed] [Google Scholar]
- 96.Caputo V., De Falco V., Ventriglia A., Famiglietti V., Martinelli E., Morgillo F., et al. Comprehensive genome profiling by next generation sequencing of circulating tumor DNA in solid tumors: a single academic institution experience. Ther Adv Med Oncol. 2022;14 doi: 10.1177/17588359221096878. 17588359221096878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Benavides M., Alcaide-Garcia J., Torres E., Gil-Calle S., Sevilla I., Wolman R., et al. Clinical utility of comprehensive circulating tumor DNA genotyping compared with standard of care tissue testing in patients with newly diagnosed metastatic colorectal cancer. ESMO Open. 2022;7 doi: 10.1016/j.esmoop.2022.100481. [DOI] [PMC free article] [PubMed] [Google Scholar]