Figure 1.
SARS-CoV-2 infects a subset of human sensory neurons
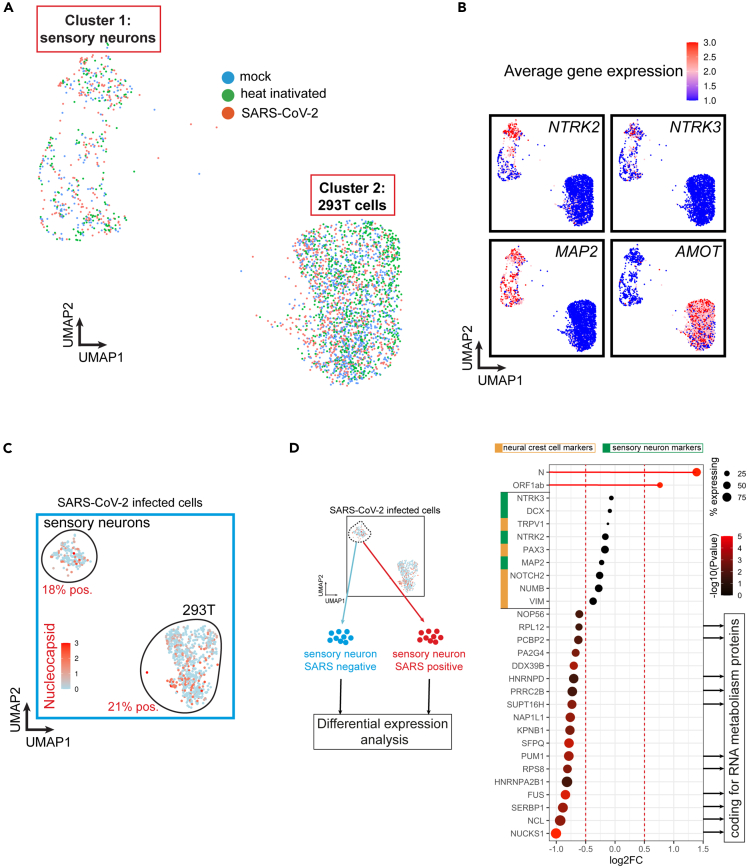
(A) UMAP of single-cell RNA sequencing (scRNA-seq) from hiPSC-sensory neurons co-cultured with HEK293T cells exposed to heat-inactivated virus or SARS-CoV-2 strain WA1/2020 for 48h. Mock is used as a control.
(B) Feature plot of average gene expression for sensory markers NTRK2 (Trk.B), NTRK3 (Trk.C), pan-neuronal marker MAP2 and 293T cell marker AMOT in all samples combined (mock, heat-inactivated and SARS-CoV-2).
(C) Feature plot of average gene expression for SARS-CoV-2’s Nucleocapsid in hiPSC-sensory neurons co-cultured with HEK293T cells exposed to WA1/2020 SARS-CoV-2 for 48h.
(D) Differential gene expression analysis between SARS-CoV-2 positive and negative sensory neurons. The dot size indicates the proportion of sensory neurons expressing the corresponding gene. The color gradient of each dot indicates the p-value for the change in gene expression (more red = more significant). Genes with a negative Log2FC have reduced expression in SARS-CoV-2 positive neurons compared to SARS-CoV-2 negative neurons.