Abstract
Objective:
Molecular detection and co-presence of carbapenem-resistant genes in the isolates of Pseudomonas aeruginosa are less commonly reported from Quetta. In the present study, we determined to highlight the antibiotic sensitivity profile and genetic mechanism of carbapenem resistance.
Methods:
The cross-sectional study was conducted from May to September 2018 at the Hi-tech laboratory, Centre for Advance Studies in Vaccinology and Biotechnology, University of Baluchistan, Quetta. Biochemical and molecular methods were ascertained for the recognition of the isolates and minimum inhibitory concentration was performed using E-test and broth microdilution methods. The molecular basis of carbapenemase activity was determined by identifying carbapenemase genes in the isolates.
Results:
Of the (n=23) P. aeruginosa isolated from pus aspirates obtained from surgical/burn units, we have detected blaIMP (n=7/8) 87.5%, blaNDM-1 (n=5/8) 62.5%, and blaSHV (n=4/8) 50%. The co-existence of multiple antibiotic-resistant genes, blaIMP, blaNDM-1 and blaSHV was found in (n=2/8) 25% isolates. These isolates displayed resistance against a range of antimicrobials from β-lactams, tetracyclines, cephalosporins, quinolones, monobactams, aminoglycosides, sulphonamides, phosphoric acid, macrolides, and polypeptide groups, suggesting extensive-drug resistance.
Conclusion:
The emergence of MBL and ESBL producers is an alarming threat in the region. It is of great importance to determine the resistance mechanism of bacterial bugs. The lack of new antimicrobials particularly against gram-negative bacteria is quite alarming worldwide.
KEYWORDS: Carbapenemase, XDR, blaSHV, blaIMP, blaNDM-1, Quetta
INTRODUCTION
Pseudomonas aeruginosa is an obligate aerobic, saprophytic, non-fermenting, gram-negative bacillus mostly inhabits in humid environment.1 In the recent previous year 2017, the WHO declared 12 bacterial isolates that were the greatest threat to human health, amongst those carbapenem-resistant P. aeruginosa was given critical priority.2 The discovery of antimicrobials was thought a symbol of hope in human fight against infections, but in the developing countries it is still the foremost cause of death. The emergence of resistance is the leading barrier in treating infectious diseases.3
Antimicrobial susceptibilities are classified by the European Centre for Disease Control (ECDC) and the Centers for Disease Control and prevention (CDC), MDR as non-susceptible to at least one antimicrobial agent in three or more groups, XDR as non-susceptible to at least one antimicrobial agent in all but two or fewer classes (i.e., bacterial isolates remain sensitive to only one or two groups), and PDR as non-susceptible to all antibacterial classes.4 Carbapenem antimicrobials are usually considered to be the last-line agents to treat severe infections caused by P. aeruginosa. Though, the recent rise in the occurrence of carbapenem-resistant P. aeruginosa hospital acquired isolates is of great concern.5
There are various techniques reported to determine the MICs for both ESBL positive and ESBL negative pathogenic bacteria that included Vitek-2, micro-scan, sensititre, agar dilution, broth microdilution, broth macrodilution, phoenix automated system, and E-test, that also help in the evaluation of different therapeutic drugs.6 Therefore, the molecular-based confirmatory test also has a dynamic role in the detection of ESBLs. It showed a high level of specificity as well as sensitivity in the identification of specific ESBL derivatives found in clinical species.
With the prevalence of carbapenem resistant genes and their rapid spreading due to mobile genetic elements, these antimicrobials are restricted. Different variants of ESBLs and MBLs such as blaIMP, blaNDM, blaVIM, and blaSHV producing P. aeruginosa have been isolated from different regions of the world.7 Therefore, an extensive survey is necessary to further explore the carbapenemase-producing P. aeruginosa globally. The aim of this research investigation was to identify the distribution of ESBLs and MBLs (carbapenemase) genes of Ambler class amongst the clinical isolates of P. aeruginosa in this region. As P. aeruginosa is the leading nosocomial superbug in the hospital settings and source of greater resistance. Moreover, partial work has been done in the province of Baluchistan on this globally emerging threat. Therefore, the present study was planned which would give guidelines to the physicians to treat the patients in a proper way and anticipated to recommend the patients antibiotics after going through culture and sensitivity testing, thus avoiding the consequences of worse resistance.
METHODS
Three hundred and fifty (n=350) clinical samples (pus aspirates) were taken aseptically from different units of tertiary care hospitals. Patients of both gender and all age groups were included in the present study. All the samples were obtained aseptically in sterile 5cc syringes (Becton Dickinson, USA) and with sterilized cotton swabs (autoclaved at 121°C at 15 psi for 20 minutes) from surgical wards, outdoor patient departments (OPDs), burns wards, OPD and burns intensive care units (ICUs).
Ethical Approval:
The bioethical approval for the present study was taken from the Institutional Bioethical Committee Bolan Medical Complex Hospital, No. Estt-DA-11 BMCH-AP-1587-8 Quetta, Baluchistan.
Patients with oozing pus from their wounds after several antibiotic therapies were enrolled in the present study. Complete history was taken from the patients about time and duration of infection, pre-existing clinical complications, antibiotic therapy, and area/climate where they were living.
Selection of the patients with antimicrobial-resistant infections was based on complete clinical history as well as several previous antibiotic therapies. All the patients were advised to stop antibiotic therapy for 72 hours before sample collection because, samples contain more bacterial load prior to the antibiotic therapy.8 Samples were labeled and transported to the microbiology laboratory immediately.
Conventional microbiological methods were applied for bacterial identification. All the samples were streaked simultaneously on MacConkey and Cetrimide agar plates (Oxoid, UK) and were incubated aerobically for 24 hours at 37°C.9 The isolates of P. aeruginosa were identified by the analytical profile index (API20NE) system (bioMerieux, France) according to the manufacturer’s instructions.10
Standardized antibiotic sensitivity test, disc diffusion Kirby Bauer method, and 0.5 McFarland turbidity standard were performed against all the isolates of P. aeruginosa. The minimum inhibitory concentrations (MICs) of extensively drug-resistant P. aeruginosa isolates were achieved by commercially available E-test strips (Oxoid, UK and Liofilchem, Italy) and broth microdilution method as previously described.11 The CLSI and FDA breakpoints were followed in the result interpretation. The broth-microdilution method was used to determine the MICs of colistin and polymyxin-B in cation-adjusted Mueller Hinton broth (CAMHB), following the CLSI guidelines12, using polymyxin B and colistin sulfate powders (Sigma-Aldrich, Germany). The broth microdilution panels were incubated at 35°C for 16-20 hours and the results were interpreted as described.13
The gram-negative isolates of P. aeruginosa showing zone of inhibition ≤25mm for ceftriaxone and/or ≤22mm for ceftazidime and/or ≤27mm for cefotaxime were screened for potential ESBL production following CLSI guidelines.12 The isolates of P. aeruginosa were initially screened phenotypically by double disk diffusion synergy method for ESBLs detection.14 This new phenotypic test was performed to detect the carbapenemase activity in P. aeruginosa isolates by inactivating the carbapenem. The mCIM is a very healthy and gainful phenotypic method.12
The genomic DNA of extensive drug-resistant isolates of P. aeruginosa was extracted by a commercially available kit (WizPrep, Korea, lot No. 4A0619-07) following the manufacturer’s directions. The specie-specific primers were used to identify the 16S rRNA gene in the isolates of P. aeruginosa, using the following set of primers, PA-SS- F- GGGGGATCTTCGGACCTCA and PA-SS-R CCTTAGAGTGCCCACCCG.15 The gel was visualized in the gel documentation system (wealtec dolphin-view S # WDV-50710004 USA) for the amplicon size of 956 bp. The plasmid DNA of extensive drug-resistant isolates of P. aeruginosa was extracted by a commercially available plasmid extraction kit (BioLabs, UK) following the manufacturer’s directions.
All the ESBL and MBL (carbapenemase) positive isolates of P. aeruginosa (n=23) based on phenotypic detection were further confirmed by singleplex PCR for the detection of blaNDM-1, blaIMP, and blaSHV using the following sets of primers (Table-I).
Table-I.
Primer sequences used for PCR of drug resistance genes in ESBL & MBL (carbapenemase) producers.
| Gene | Oligonucleotide Sequences | Amplicon Size (bp) | References |
|---|---|---|---|
| blaNDM-1 | F-5’-GGGCAGTCGCTTCCAACGGT-3’ | 475 | (16) |
| R-5’-GTAGTGCTCAGTGTCGGCAT-3’ | |||
| blaIMP | F-5’-TGAAAGGCTTATCTGTATTC-3’ | 740 | (17) |
| R-5’- TAGTTGCTTGGTTTTGATG-3’ | |||
| blaSHV | F-5’-AGCCGCTTGAGCAAATTAAAC-3’ | 713 | (18) |
| R-5’- ATCCCGCAGATAAATCACCAC-3’ |
The sequencing of the PCR product of specie-specific 16S rRNA, blaIMP, blaNDM-1, and blaSHV genes in the representative isolates of P. aeruginosa was performed commercially with DNA sequencer using forward primers.
RESULTS
Based on disc diffusion Kirby Bauer method and 0.5 McFarland turbidity standard (data not shown), out of (n=23) P. aeruginosa isolates (n=4) 17.4% were found as non MDR, (n=11) 47.8% MDR, and (n=8) 34.7% XDR (Fig.1). These isolates were resistant against anti-pseudomonal antibiotics belonging to carbapenems, beta-lactam, aminoglycosides, monobactams, fluroquinolones, penicillins, cephalosporins, and lipopeptides groups. The susceptibility patterns such as, non-MDR, MDR, and XDR were categorized as terminologies created by the CDC, and ECDC.4
Fig.1.
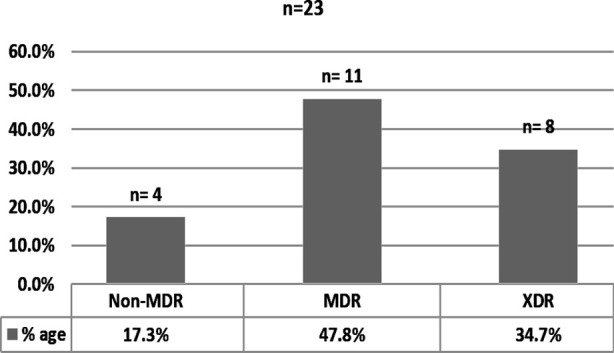
Number & percentage of non-MDR, MDR, & XDR P. aeruginosa.
The MICs data and percentage of XDR isolates of P. aeruginosa against each tested antibiotics has shown in (Table-II & III). Out of isolates (n=23), eight were categorized XDR, as they were only sensitive to one or two families of antimicrobials tested. The results of MICs were compared with disc diffusion method, in disc diffusion susceptibility percentage of tigecycline was S= 12.5%, I=50% and R=37.5% whereas, MICs showed S= 25%, I=12.5% and R=62.5%. The difference was noted in other antimicrobials as well and concluded that disc diffusion is just a screening method, while MICs must be used especially in case of MDR and XDR bacterial isolates.
Table-II.
Susceptibility data for P. aeruginosa isolates MICs (E-test) method.
| Antimicrobial Agents | Breakpoints for resistance (µg/dL) | XDR isolates (n=8) | ||
|---|---|---|---|---|
|
| ||||
| S (%) | I (%) | R (%) | ||
| Imipenem | ≥8 | 00 | 25 | 75 |
| Tigecycline | ≥8 | 25 | 12.5 | 62.5 |
| Amikacin | ≥64 | 12.5 | 00 | 87.5 |
| Ceftazidime | ≥32 | 00 | 00 | 100 |
| Ciprofloxacin | ≥4 | 00 | 00 | 100 |
Table-III.
Susceptibility data for P. aeruginosa isolates MICs broth microdilution method.
| Breakpoints for resistance (µg/mL) | XDR isolates (n=8) | |||
|---|---|---|---|---|
|
| ||||
| S (%) | I (%) | R (%) | ||
| Colistin | ≥8 | 62.5 | 00 | 37.5 |
| Polymyxin-B | ≥8 | 50 | 25 | 25 |
Out of (n=23) P. aeruginosa only MDR and XDR isolates (resistant to 3rd generation cephalosporin), were screened phenotypically for ESBL production. Among which (9/11, 81.8%) were screened as ESBL positive (Table-IV). All the carbapenem-resistant (XDR) isolates of P. aeruginosa were tested for carbapenem inactivation activity. The sensitivity of the test in P. aeruginosa was 75% (Table-IV).
Table-IV.
No and percentage of ESBL producing & carbapenemase inactivation by mCIM in MDR & XDR isolates of P. aeruginosa.
| Organism | ESBL Positive (DDST) | ESBL Negative | mCIM Positive | mCIM Negative |
|---|---|---|---|---|
| P. aeruginosa | 9/11 (81.8%) | 2/11 (18.1%) | (6/8) 75% | (2/8 )25% |
We have detected ESBL and carbapenemase genes in the extensively resistant clinical isolates of P. aeruginosa (n=8), among which blaIMP genes were (n=7/8) 87.5%, blaNDM-1 (n=5/8) 62.5%), and blaSHV were (n=4/8) 50%. The co-existence of multiple antibiotic-resistant genes blaSHV, blaIMP, and blaNDM was found in (n=2/8) 25% isolates.
DISCUSSION
Antibiotic resistance causes a major public health threat across the globe and is increasing gradually, mostly in developing countries. It is the irrational use of antibiotics that has given rise to resistance against a range of antimicrobials. Infections caused by bacterial superbugs are challenging and difficult to treat. This challenge cuts across the developed and developing countries of the world because there is a narrow therapeutic option left, as very few antibacterial agents such as carbapenems, tigecycline, and colistin are available.19
All the clinical isolates of P. aeruginosa included in the present study were from hospital settings. This was in accordance with the results reported by,20 in which it was documented that P. aeruginosa is medically important because it causes nosocomial infections throughout the world. The movement of genes and other sequences of the DNA plays an active role in the spread of drug resistance.
In the present study (n=350) clinical isolates were included for the detection of multi and extensive, drug-resistant P. aeruginosa. Among the gram-negative bacilli (n=157) P. aeruginosa were (n=23) 14.6 %. The persistent spread of ESBLs mediated resistance has intensely increased in both hospitals and the community. It was observed in our isolates that ESBL producers were (n=9/11) 81.8%, which were higher than the results reported by21 (69.5%) and almost doubled than the earlier study reported by,22 in which (46.8%) of the isolates were ESBL producers phenotypically. This increasing percentage is very alarming in short period. Similarly, MBL (carbapenemases) (6/8) 75% were detected in the present study. It was observed that among the isolates of P. aeruginosa (n=8) tested by the mCIM, one isolate was detected as false negative. The false-negative result missed by mCIM was linked to blaIMP (1/8) carbapenemases. The missed isolate was confirmed as PCR positive for the relevant gene. In contrary, lower incidence of MBL (carbapenemases) producers was documented by,23 in Egypt (64.8%), and24, 37.6%. This rapid increase is again alarming in the region.
Our results showed variable prevalence for MBL (carbapenemase) genes, among which the most prevalent genotype was, blaIMP (n=7/8) 87.5% followed by blaNDM-1 (n=5/8) 62.5%, and for ESBL genes like blaSHV (n=4/8) 50%, this was higher than the results reported from Kampala, Uganda, at Mulago hospital in which the frequency of carbapenemase genes like blaIMP were (9/25) 36%, blaVIM (8/25) 32%, blaNDM-1 (1/25) 4% in carbapenem-resistant P. aeruginosa.25
The co-existence of multiple antibiotic-resistant genes blaIMP, blaNDM-1 and blaSHV, in the present study was found in (n=2/8) 25% isolates, in contrary, our results showed a high percentage (25%) of combination genes than reported by23, (8.5%) in the combination of five genes, and (2.1%) in a combination of three genes like, blaNDM, blaIMP, and blaTEM, but blaTEM was reported in combination instead of blaSHV. Similarly, combination of two genes blaCTX-M/blaSHV is reported in the isolates of Klebsiella pneumonia from tertiary care hospitals in Lahore, Pakistan.26
This higher prevalence is the indication of extensive drug resistance against multiple antimicrobials. The spread dynamics of antibiotic resistance is more complex and need more studies in-depth together with whole genome sequencing or multilocus sequence typing to investigate epidemiological evidence of transmission of ESBL and MBL (carbapenemases) in the region. Moreover, the present study is clinically more significant, as it will create awareness among the physicians and common people and help in careful selection of antimicrobials. Furthermore, infection control measures and over the counter usage of antimicrobials must be prevented by health authorities following implementation of strict rules and regulations.
Limitations:
Although it was a multi-setting study from different tertiary care hospitals of Quetta city that treat the patients even from peripheries of the province, the results cannot be generalized to the patients of the other provinces. Further studies are required to limit the increasing and alarming antimicrobial resistance.
CONCLUSION
The MBL and ESBL producers are an emerging threat in the region. Resistance of the commonly used antibiotics against carbapenem-resistant bacterial species has been developed. It is of great importance to determine the resistance mechanism and eliminate its root cause. Culture and sensitivity must be considered as an essential elements before going towards antibiotic therapy. The spread of ESBL and MBL, antimicrobial resistance, and the lack of new antimicrobials particularly against gram-negative bacteria is quite alarming globally.
Authors’ Contribution:
MD and MA: Conceived the presented idea, planning of the work and designed the basis of the manuscript. Contributed to PCR and lab work, prepared the manuscript and provided data.
SRL: Analyzed the data, and developed the theoretical part.
MA: Analyzed and interpreted the data.
MD: Is responsible for the accuracy and integrity of the work.
All authors contributed equally to this work and they provided critical feedback, helped shape the research and approved the final version of the manuscript for publication.
Footnotes
Conflict of Interest: None.
Source of Funding: None.
REFERENCES
- 1.Dubois V, Arpin C, Melon M, Melon B, Andre C, Frigo C, et al. Nosocomial outbreak due to a multiresistant strain of Pseudomonas aeruginosa P12:efficacy of cefepime-amikacin therapy and analysis of β-lactam resistance. J Clin Microbiol. 2001;39(6):2072–2078. doi: 10.1128/JCM.39.6.2072-2078.2001. doi: 10.1128/JCM.39.6.2072-2078.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) WHO publishes list of bacteria for which new antibiotics are urgently needed. [(Accessed 2022)]. Available at: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/ en/
- 3.Kapoor G, Saigal S, Elongavan A. Action and resistance mechanisms of antibiotics:A guide for clinicians. J anaesthesiol Clin Pharmacol. 2017;33(3):300. doi: 10.4103/joacp.JOACP_349_15. doi: 10.4103/joacp. JOACP_349_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basak S, Singh P, Rajurkar M. Multidrug resistant and extensively drug resistant bacteria:a study. J Pathog. 2016;2016 doi: 10.1155/2016/4065603. doi: 10.1155/2016/4065603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Chen XL, Huang AW, Liu SL, Liu WJ, Zhang N, et al. Mortality attributable to carbapenem-resistant Pseudomonas aeruginosa bacteremia:a meta-analysis of cohort studies. Emerg Microbes Infect. 2016;5(1):1–6. doi: 10.1038/emi.2016.22. doi: 10.1038/emi.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Jayol A, Nordmann P. Polymyxins:antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–596. doi: 10.1128/CMR.00064-16. doi. 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCracken MG, Adam HJ, Blondeau JM, Walkty AJ, Karlowsky JA, Hoban DJ, et al. Characterization of carbapenem-resistant and XDR Pseudomonas aeruginosa in Canada:results of the CANWARD 2007–16 study. J Antimicrob Chemother. 2019;74(Supplement_4):iv32–8. doi: 10.1093/jac/dkz285. doi: 10.1093/jac/dkz285. [DOI] [PubMed] [Google Scholar]
- 8.Harris AM, Bramley AM, Jain S, Arnold SR, Ampofo K, Self WH, et al. In Open forum Infect Dis. 4. Oxford University Press: 2017. Influence of antibiotics on the detection of bacteria by culture-based and culture-independent diagnostic tests in patients hospitalized with community-acquired pneumonia; p. 1. doi: 10.1093/ofid/ofx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggal S, Khatri PK, Parihar RS, Arora R. Antibiogram of various bacterial isolates from pus samples in a tertiary care centre in Rajasthan. Int J Sci Res. 2015;4(5):1580–1584. [Google Scholar]
- 10.Pirkani GS, Awan MA, Abbas F, Din M. Culture and PCR based detection of bacteria causing urinary tract infection in urine specimen. Pak J Med Sci. 2020;36(3):391. doi: 10.12669/pjms.36.3.1577. doi: 10.12669/pjms.36.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Din M, Babar KM, Ahmed S, Aleem A, Shah D, Ghilzai D, et al. Prevalence of extensive drug resistance in bacterial isolates harboring blaNDM-1 in Quetta Pakistan. Pak J Med Sci. 2019;35(4):1155. doi: 10.12669/pjms.35.4.372. doi: 10.12669/pjms.35.4.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. Perfor CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute. 2018. Retrieved from www.clsi.orgmance Standards for Antimicrobial Susceptibility Testing .
- 13.Simar S, Sibley D, Ashcraft D, Pankey G. Colistin and polymyxin B minimal inhibitory concentrations determined by Etest found unreliable for Gram-negative bacilli. Ochsner J. 2017;17(3):239–242. [PMC free article] [PubMed] [Google Scholar]
- 14.Abrar S, Vajeeha A, Ul-Ain N, Riaz S. Distribution of CTX-M group I and group III β-lactamases produced by Escherichia coli and klebsiella pneumoniae in Lahore, Pakistan. Microb Pathog. 2017;103:8–12. doi: 10.1016/j.micpath.2016.12.004. doi: 10.1016/j.micpath.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Wang J, Hu J, Zhang H, Cai C, Shen J, et al. Improved PCR primers to amplify 16S rRNA genes from NC10 bacteria. Appl Microbiol Biotechnol. 2016;100(11):5099–5108. doi: 10.1007/s00253-016-7477-9. [DOI] [PubMed] [Google Scholar]
- 16.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, et al. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother. 2011;66(9):2002–2005. doi: 10.1093/jac/dkr226. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 17.Franco MR, Caiaffa-Filho HH, Burattini MN, Rossi F. Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian university hospital. Clinics. 2010;65:825–829. doi: 10.1590/S1807-59322010000900002. doi: 10.1590/S1807-59322010000900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 19.Pragasam AK, Vijayakumar S, Bakthavatchalam YD, Kapil A, Das BK, Ray P, et al. Molecular characterisation of antimicrobial resistance in Pseudomonas aeruginosa and Acinetobacter baumannii during 2014 and 2015 collected across India. Indian J Med Microbiol. 2016;34(4):433–441. doi: 10.4103/0255-0857.195376. doi: 10.4103/0255-0857.195376. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Ma J, Jia W, Li W. Antimicrobial resistance and molecular characterization of gene cassettes from class 1 integrons in Pseudomonas aeruginosa strains. Microb Drug Resist. 2020;26(6):670–676. doi: 10.1089/mdr.2019.0406. doi: 10.1089/mdr.2019.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbas G, Khan I, Mohsin M, Younas T, Ali S. High rates of CTX-M group-1 extended-spectrum β-lactamases producing Escherichia coli from pets and their owners in Faisalabad, Pakistan. Infect Drug Resist. 2019;12:571. doi: 10.2147/IDR.S189884. doi: 10.2147/IDR. S189884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abd El-Baky RM, Ibrahim RA, Mohamed DS, Ahmed EF, Hashem ZS. Prevalence of virulence genes and their association with antimicrobial resistance among pathogenic E coli isolated from Egyptian patients with different clinical infections. Infect Drug Resist. 2020;13:1221. doi: 10.2147/IDR.S241073. doi: 10.2147/IDR.S241073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masoud SM, Abd El-Baky RM, Aly SA, Ibrahem RA. Co-existence of certain ESBLs, MBLs and plasmid mediated quinolone resistance genes among M DR E coli isolated from different clinical specimens in Egypt. Antibiotics. 2021;10(7):835. doi: 10.3390/antibiotics10070835. doi: 10.3390/antibiotics10070835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim Y, Sani Y, Saleh Q, Saleh A, Hakeem G. Phenotypic detection of extended spectrum beta lactamase and carbapenemase co-producing clinical isolates from two tertiary hospitals in Kano, North West Nigeria. Ethiop J Health Sci. 2017;27(1):3–10. doi: 10.4314/ejhs.v27i1.2. doi: 10.4314/ejhs.v27i1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kateete DP, Nakanjako R, Namugenyi J, Erume J, Joloba ML, Najjuka CF, et al. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago hospital in Kampala, Uganda (2007–2009) Springerplus. 2016;5(1):1–1. doi: 10.1186/s40064-016-2986-7. doi: 10.1186/s40064-016-2986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gondal AJ, Saleem S, Choudhry N, Ahmad FJ, Bukhari H, Yasmin N, Jahan S. Emergence of multidrug-resistant ST11 blaKPC-2 producing Klebsiella pneumoniae coharboring blaCTX-M and blaSHV in Pakistan. J Infect Dev Ctries. 2023;17(02):210–217. doi: 10.3855/jidc.17041. doi: 10.3855/jidc.17041. [DOI] [PubMed] [Google Scholar]


