Highlights
-
•
CD2 and CD27 are the core immune genes involved in brain metastases of BC.
-
•
CD2 and CD27 expression are closely associated with brain metastases of BC.
-
•
Reduced CD2 and CD27 are related to the activation of nitrogen metabolic pathways.
-
•
CD2 and CD27 show a negative correlation with the proportion of M2 macrophages.
-
•
Our study identifies potential therapeutic targets for brain metastases of BC.
Keywords: Breast cancer, Brain metastasis, CD2, CD27, Macrophage polarization, Nitrogen metabolic pathway
Abstract
Objective
Our study aimed to reveal the possible molecular mechanisms of CD2 and CD27 in influencing the tumor microenvironment of breast cancer (BC) brain metastasis based on the TCGA (The Cancer Genome Atlas) and SRA (Sequence Read Archive) databases.
Methods
We calculated the proportions of tumor-infiltrating immune cells and the immune and stromal cell scores in 1222 BC samples from the TCGA-BRCA dataset, followed by identification of candidate DEGs. We further screened for BC brain metastasis-related DEGs in the BC brain metastasis dataset SUB12911144 from the SRA database. Finally, we established a mouse breast cancer brain metastasis model for in vivo validation.
Results
We further screened two immune-regulatory DEGs (CD2 and CD27). GSEA analysis showed that the downregulation of CD2 and CD27 expression was closely related to the activation of nitrogen metabolism pathways. CIBERSORT algorithm analysis showed a correlation between the expression of 16 types of tumor-infiltrating immune cells and CD2 and 19 types of tumor-infiltrating immune cells and CD27. In addition, CD2 and CD27 expression were negatively associated with the proportion of M2 macrophages. In vivo experimental results demonstrated that overexpression of CD2/CD27 could suppress the M2 polarization of macrophages and inhibit breast cancer brain metastasis.
Conclusion
In the tumor microenvironment, overexpression of CD2/CD27 inhibited the activation of nitrogen metabolism pathways and suppressed M2 polarization of macrophages, thereby preventing brain metastasis of breast cancer.
Introduction
According to an epidemiological report published by the International Agency for Research on Cancer (IARC), breast cancer (BC) accounts for 7%-10% of all malignant tumors [1]. With approximately 2.3 million new cases and 690,000 deaths worldwide, BC has surpassed lung cancer as the most prevalent malignancy worldwide [2]. Worldwide, BC incidence and mortality are on a significant rise in all regions and age groups, posing a serious threat to global public health [3]. At the same time, the trend towards younger BC incidence also means that it could be detrimental to the female workforce, especially women in socially important positions, thus widening the gender gap in employment and seriously hampering socio-economic development [4,5]. The risks contributing to BC's development include genetic and non-genetic factors. Genetic factors include mutations in susceptibility genes such as BRCA1 and BRCA2 [6,7] non-genetic factors include reproductive and hormonal risks such as early age at menarche, late menopause, short breastfeeding periods, and obesity [[8], [9], [10]].
Although the incidence of BC remains high, the overall 5-year survival rate for women with BC has increased to 90%, largely due to advances in early diagnosis and comprehensive treatment strategies for BC [11]. Breast-conserving surgery (BCS), mastectomy, adjuvant radiotherapy, and chemotherapy are currently the main clinical strategies for BC treatment, with targeted drugs used to refine treatment depending on the type of BC [[12], [13], [14]]. Although technological innovations and improved treatments have led to important clinical advances in the treatment of BC, the high recurrence rate of BC remains a major problem in clinical BC prognosis, mainly caused by BC metastasis. Statistically, about 90% of BC-related deaths are attributed to tumor metastasis. BC is the second most common cause of brain metastases among all solid tumors, with approximately 15–30% of BC patients eventually developing brain metastases [15]. The incidence of brain metastases in patients with advanced Her2+ BC is 25–50%, while the incidence of brain metastases in advanced triple-negative BC is as high as 50%, and the median overall survival of BC patients once brain metastases occur is reduced to 4–6 months [16,17]. In addition, the blood-brain barrier makes it difficult for conventional therapeutic agents to enter the central nervous system, preventing effective treatment of BC brain metastases. Therefore, it is clinically important to investigate the molecular mechanisms of BC brain metastases and identify potential therapeutic targets.
The tumor microenvironment (TME) is the internal environment in which tumor cells are produced and live, and it differs significantly from the normal cell growth environment [18]. Recent studies have shown that TM(E), which includes stromal cells, immune cells, cellular exocytosis, and physical environmental factors, is closely related to tumor malignant tumor proliferation, immune evasion, and drug tolerance, and is the environmental "soil" on which tumors develop, develop and metastasize [19,20]. Tumourigenesis and development initiate micro-environmental remodeling, which further supports tumor progression and abnormal regulation of immune cells, which could contribute to tumor immune evasion and malignant proliferation [[19], [20], [21]]. In TME, tumor-associated macrophages (TAM) are mostly induced to polarize into M2-type macrophages, which have immunosuppressive functions and could suppress the immune response, thus promoting tumor metastasis [[22], [23], [24]]. Therefore, TME and tumor progression are characterized by a synergistic and dynamic development, and overcoming or weakening the TME barrier is of great importance for tumor treatment and prognosis.
In this study, we screened BC brain metastasis-related genes from TCGA and SRA public databases with bioinformatics analysis. We combined the CIBERSORT and ESTIMATE algorithms to calculate the proportion of tumor-infiltrating immune cells and immune cell and stromal cell scores to explore the core factors affecting the involvement of TME in the development of BC brain metastasis. This study will provide insight into the pathogenesis of BC brain metastases and provide potential molecular targets for the early diagnosis and treatment of BC brain metastases.
Methods
TCGA data download and collation
Data download and collation of clinical data and transcriptomic data from the TCGA website BC via the tools gdc_client.exe and Rstudio. The RNA-Seq comparison annotation file (GDC.h38 GENCODE v22 GTF) provided in the TCGA database was also downloaded, and the Ensemble id in the sequencing dataset was converted to gene names via R language, as well as the clinical information of the corresponding patients, including age, gender, TNM stage, pathological stage, and prognosis. One thousand two hundred twenty-two patients were obtained, of which 1109 were BC patient specimens and 113 were paired normal tissue specimens.
Stromal cell score and immune cell score
The ESTIMATE scoring system consists of a stromal and immune cell score. The scores are based on gene expression in the TCGA database of lung adenocarcinoma patients. The scores can be downloaded online at https://bioinformatics.mdanderson.org/estimate/disease. The ESTIMATE algorithm could infer the infiltration of stromal and immune cells in TME by analyzing transcriptomic data from tumor samples.
Survival analysis
BC patients were divided into high and low-expression groups according to the median of each score, and survival analysis was performed using the "survival" and "survminer" packages in R. Kaplan-Meier survival curves were plotted, and log-rank tests were performed. Log-rank test, P<0.05, indicates statistical significance.
Correlation analysis between clinicopathological features and stromal and immune cell scores
Patients were grouped according to their clinicopathological characteristics, and the difference between high and low stromal and immune cell scores in each group was analyzed. χ2 test was used to analyze the correlation between stromal and immune cell scores and clinicopathological characteristics. The median of the scores was used to divide these samples into a high stromal cell score group, a low stromal cell score group, and an immune cell score as before two groups. The Kaplan-Meier method and Log-rank test were used to analyze the difference in overall survival rate between the two groups.
Screening for differentially expressed genes
BC patient samples were divided into two groups, a high stromal (or immune) score group and a low stromal (or immune) score group, based on the median scores of the immune cell score and stromal cell score. Differential analysis was performed using the "limma" package to screen for differentially expressed genes (DEGs). Screening criteria were set: |log2FC|>1, false discovery rate FDR<0.05. Heat maps were created using the R language "pheatmap" package.
Construction of protein-protein interaction (PPI) networks
The STRING tool (https://string-db.org/) was used to construct the interaction network of DEGs and visualize gene interactions using Cytoscape software (version 3.7.2). The molecular complex detection (Mcode) plug-in of Cytoscape was then used to find the most important modules, i.e., the most densely connected regions, based on topological principles. Parameters were set to MCODE score > 5, degree truncation value = 2, node truncation value = 0.2, maximum depth = 100, and k-score = 2.
Functional enrichment analysis of candidate genes
The R language 'ClusterProfiler' package, the 'richplot' package, and the 'ggplot2′ package were used to perform GO and KEGG enrichment analysis of candidate genes, including biological process (BP), molecular function (MF) and cellular component (CC) analysis. KEGG enrichment analysis, including biological process (BP), molecular function (MF), and cellular component (CC) analysis, was performed. Potential and key targets were screened for P < 0.05, and cellular functions and signaling pathways were analyzed according to the P value.
COX regression analysis
The univariate COX regression analysis was performed using the R language 'survival' package and shows the top 16 genes in the univariate COX ranked from smallest to the largest p-value.
GSEA gene enrichment analysis
Gene set enrichment analysis (GSEA) was performed using "c2.cp.kegg.symbols.gmt" as the reference gene set, with P<0.05 indicating a statistically significant difference.
SRA microarray data acquisition and differential gene expression analysis
The BC brain metastasis-related expression profile dataset SUB12911144 was obtained through the SRA database (https://www.ncbi.nlm.nih.gov/sra/), including three BC brain metastasis-free samples and three BC brain metastasis samples.
Differential analysis was performed using the R "limma" package (https://www.ncbi.nlm.nih.gov/gds), with |logFC| > 1 and significant P < 0.05 as the filtering criteria, and differentially expressed genes in each dataset were selected. Volcano and heat maps were created using the R package "ggplot2" and "heatmap".
Assessment of immune cell infiltration
The BC dataset from the TCGA database was analyzed using the CIBERSORT algorithm (https://cibersort.stanford.edu/) to filter the appropriate samples based on P < 0.05 and to calculate the percentage of each immune cell in the samples. The "ggpubr" package was used to visualize the infiltration of the 22 immune cells and to analyze the differences in the infiltration of the 22 immune cells. 0.05.
Wayne's analysis
Wayne analysis was performed using the Draw Venn Diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to obtain candidate genes.
Breast cancer brain metastasis model
Twenty-four 4-week-old SPF-grade female BALB/c nude mice (weighing 20–22 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd., China (401) for In vivo experiments. The mice were housed under constant temperature (25–27°C) and humidity (45–50%), with free access to sterilized water and food. The experimental animals were used for medical research, and all procedures were approved by our institution's animal committee (No. GMU-22-20230409). A breast cancer brain metastasis model was established by intracranial injection of GFP/Luc-labeled MDA-MB-231 cells (CBP30106L, purchased from Nanjing KeyGen Biotech Co., Ltd., China, Jiangsu). A 4 µL containing 7.5 × 104 MDA-MB-231 cells was stereotactically injected into the right striatum (2 mm from the outside of bregma, 2.5 mm deep) using a Hamilton syringe. The injection site was sealed with bone wax, and the wound was sutured [25]. On the 6th and 11th days after tumor inoculation, mice were intravenously injected with 100 µL of 1 × 109 TU of lentivirus overexpressing CD2 or CD27 or negative control. In vivo, bioluminescence imaging was performed using the IVIS imaging system to monitor tumor growth. Brain tissues were collected and fixed for slicing when the mice died [26].
RT-qPCR
Total RNA was extracted from tissue samples using the TRIZOL reagent (15596-018, solarbio, USA) according to the manufacturer's instructions. To measure mRNA expression, total RNA was reverse transcribed into cDNA using the PrimeScript™ RT-PCR Kit (TaKaRa, Mountain View, CA, USA). Real-time quantitative reactions were performed on the LightCycler 480 system (Roche Diagnostics, Pleasanton, CA, USA), using SYBR Premix Ex TaqTM (TaKaRa) with β-actin as the internal control. Primers used for amplification were designed and synthesized by Shanghai General Biological Technology Co., Ltd. (Table S1). The relative transcription level of the target gene was calculated using the relative quantification method (2-ΔΔCT method). 2-ΔΔCt represents the fold change in expression of the target gene in the experimental group compared to the control group, where ΔΔCT = ΔCt experimental group - ΔCt control group, and ΔCt = Ct target gene - Ct internal control gene. Ct is the cycle threshold value at which the fluorescence signal reaches a set threshold during the exponential phase of amplification [27]. Each sample was run in triplicate, and each experiment was repeated three times.
Western blot
Total protein was extracted from cell lines or frozen tissue samples using RIPA lysis buffer (R0010, Solarbio, China) mixed with a proteinase inhibitor cocktail (Roche) on ice. The protein concentration of each sample was determined using the BCA assay kit (Pierce, Rockford, IL, USA). Total protein (30 μg) was separated by 10% SDS-PAGE and transferred to a PVDF membrane (Merck Millipore, Billerica, MA, USA). The membrane was blocked with 5% BSA for 1 h, washed with TBST three times for 5 min each time, and then incubated overnight at 4°C with specific primary antibodies, including CD2 (1:1000, rabbit anti-ab131276, Abcam), CD27 (1:1000, rabbit anti-ab131254, Abcam), GAPDH (1:1000, rabbit anti-ab8245, Abcam). The membrane was washed with TBST three times for 5 min each the next day and then incubated with the corresponding secondary antibody goat anti-rabbit IgG (ab6721, 1:500, Abcam). The membrane was washed with TBST three times for 10 min each time and visualized using the SuperSignal West Pico chemiluminescence substrate (Thermo Fisher, Waltham, MA, USA) to detect specific bands. GAPDH antibody was used as an internal control. The relative level of the target protein was expressed as the gray value of the target protein band/internal reference protein band [28]. Each experiment was repeated three times.
H&E staining
The nude mouse tumor tissue was fixed with 4% paraformaldehyde, embedded in paraffin, and sliced (5 µm). The sliced sections were evaluated for pathological changes by H&E staining, first stained with hematoxylin for 9 min and then stained with editing for 4 min [29,30], and observed under a microscope (Nikon, Japan).
Immunohistochemistry staining
For immunohistochemistry (IHC) staining, nude mouse tumor tissues were fixed in 10% neutral formalin (NBF), followed by paraffin embedding. Then the tumor block was cut into 5 μm thick slices and placed on positively charged slides. IHC staining was performed on the Ventana Discovery Ultra (Ventana Medical Systems, Roche Diagnostics, Indianapolis, USA) IHC automatic staining machine. The slides were loaded on the machine and subjected to deparaffinization, rehydration, endogenous peroxidase activity inhibition, and antigen retrieval. Then, the slides were incubated with Ki67 antibody (ab16667, Abcam, UK). IHC tumor sections were visualized using the DISCOVERY-ChromoMap-DAB kit (Ventana), and Suomijin (Ventana) was used for counterstaining [31]. The slides were imaged using the Leica Dmi8 microscope and analyzed using image-pro Premier software.
TUNEL staining
Tumor cell apoptosis in the tumor tissues was detected using the TUNEL apoptosis detection kit (Millipore, Billerica, MA, USA), and the protocol was followed as provided in the instructions manual. Specifically, paraffin-embedded sections of mouse tumor tissues were deparaffinized and rehydrated, and treated with 20 μg/mL proteinase K (ST532, Beyotime, China) without DNase at 20-37°C for 30 min and washed three times with PBS. The sections were then incubated at room temperature in a 3% hydrogen peroxide solution prepared in PBS for 20 min, washed three times with PBS, and incubated at 37°C in a biotinylated solution for 60 min under light avoidance. After that, the sections were washed once with PBS, incubated at room temperature in a stopping solution for 10 min, washed three times with PBS, incubated at room temperature in 50 μL of Streptavidin-HRP working solution for 30 min, and washed three times with PBS. Subsequently, 0.2–0.5 mL of DAB chromogenic solution was added and incubated at room temperature for 5-30 min, then washed three times with PBS. The sections were sealed and observed under a reverse microscope with ten fields randomly selected in each group to count the positive and total cells. The apoptotic cell (%) was calculated using the formula: (number of apoptotic cells/total number of cells) × 100%. Cells with brown-yellow nuclei were considered positive apoptotic cells, whereas cells with blue nuclei were considered normal cells [32,33].
Flow cytometry
Nude mice were anesthetized with excessive isoflurane, and brain tissues were collected and mechanically chopped, followed by digestion with PBS containing 2.5 mg/mL trypsin. The cell suspension was homogenized through a 40 μm nylon membrane (Becton Dickinson Labware, Franklin Lakes; NJ, USA) and centrifuged at 500 g for 10 min. After incubation with AF488-CD206 (321114, 1:50) and APC-CD68 (333810, 1:100) at 4°C for 16 h, the stained cells were washed twice with PBS and analyzed using a flow cytometer (Becton-Dickinson, Mountain View, CA, USA) with Cell-QuestTM software to calculate the cell percentage [25,34].
Detection of glutamine content in tumor tissues
Fresh tumor tissues weighing 50 mg were collected and homogenized in cold 70% ethanol. The content of Glutamine was determined according to the protocol provided by the manufacturer of the Glutamine Assay Kit (ab197011, Abcam, UK). The samples and standard solutions were added to a 96-well plate and incubated for 30 minutes with a hydrolysis mixture. The reaction mixture was then added and incubated for 60 min. Analysis was performed using the Multiskan SkyHigh full-wavelength spectrophotometer (Thermo Fisher, USA).
Statistical analysis
Data were statistically analyzed using SPSS version 24.0. The chi-square test was used to analyze the correlation between the matrix, immune cell score, and clinical pathological characteristics. The t-test and Cytoscape software selected differentially expressed genes (DEGs). The Kaplan-Meier method was used to evaluate the predictive value of key genes for brain metastasis in breast cancer. Quantitative data were expressed as means ± standard deviations. Non-paired t-tests were used to compare cancer tissues and adjacent tissues. One-way analysis of variance was used for multiple group comparisons, followed by Tukey's post-hoc test. For comparisons between different time points, repeated-measures analysis of variance was used, and Bonferroni was used for post-hoc tests. A P-value of less than 0.05 was considered statistically significant.
Results
Bioinformatics analysis
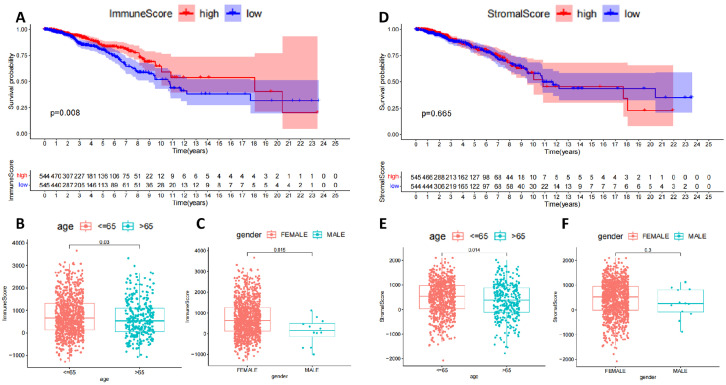
TME is important in mediating tumor metastasis, immune escape, and immunotherapy suppression [35]. The TME was scored based on the ESTIMATE algorithm, and the scores were combined with the survival time and survival status of the corresponding samples to plot survival curves. The results of the ESTIMATE algorithm analysis showed that the survival rate of BC patients gradually decreased over time; by dividing BC patients into high and low subgroups based on the immune cell score, we found that the survival rates of the two groups were significantly We found a statistically significant difference in survival rates between the two groups (Fig. 1A). In addition, there were also statistically significant differences in immune cell scores between BC patients of different ages and genders (Fig. 1B,C). Next, BC patients were also divided into high and low subgroups based on stromal cell scores. Still, there was no significant difference in survival between the two groups (Fig. 1D), no difference in immune cell scores between BC patients by gender (Fig. 1E), and a significant difference in immune cell scores between BC patients by age (Fig. 1F). The above results suggest that the content of immune cells and survival rates in BC samples are more relevant than stromal cells. However, the mechanism of the role of stromal cells in BC still cannot be ignored.
Fig. 1.
Correlation between immune cell score and stromal cell score based on ESTIMATE algorithm and prognosis of BC patients
Note: (A) Plotting survival curves based on the ESTIMATE algorithm for immune cell scores; (B) Analysis of variability between high and low age groups based on immune cell scores; (C) Analysis of variability between genders based on immune cell scores; (D) Plotting survival curves based on the ESTIMATE algorithm for stromal cell scores; (E) Analysis of variability between high and low age groups based on stromal cell scores (E) Differences between high and low age groups based on stromal cell scores; (F) Differences between genders based on stromal cell scores.
Candidate DEGs function primarily in immune response or inflammatory response mechanisms
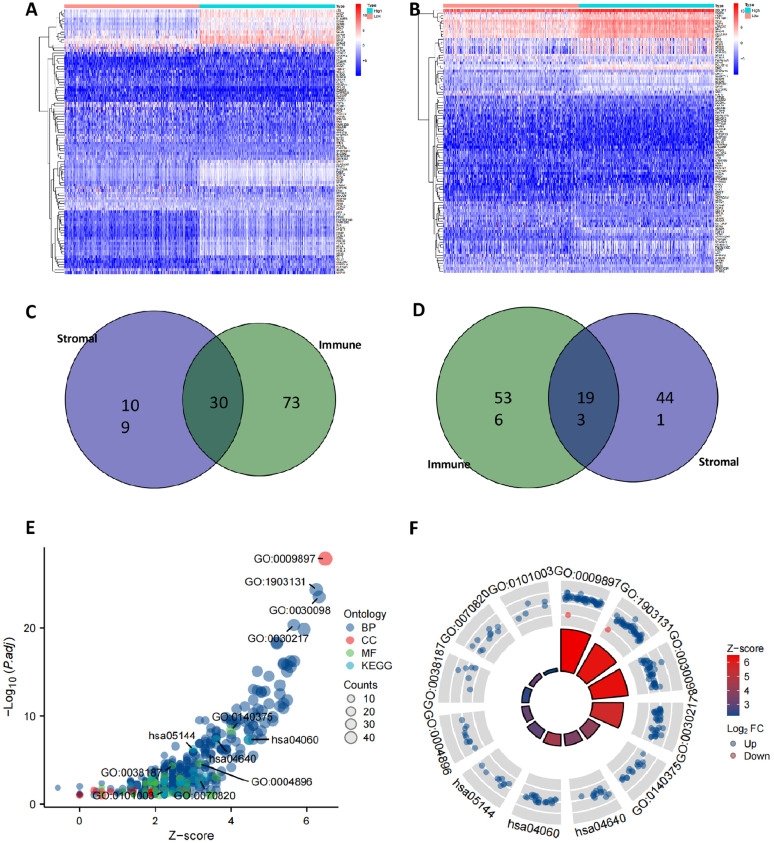
To further investigate the molecular mechanisms involved in BC progression, we screened for DEGs involved in the inflammatory immune response based on immune and stromal cell scores. One hundred thirty-nine up-regulated DEGs and 634 down-regulated DEGs were found in the high stromal cell score group compared to the low stromal cell score group (Fig. 2A); and 103 up-regulated DEGs and 729 down-regulated DEGs were found in the high immune cell score group compared to the low immune cell score group (Fig. 2B). A total of 103 up-regulated DEGs and 729 down-regulated DEGs were found in the high immune cell scoring group compared to the low immune cell scoring group (Fig. 2B). A total of 30 up-regulated DEGs and 193 down-regulated DEGs (Fig. 2C,D) were screened for immune cell and stromal cell-related differential genes by Wayne analysis.
Fig. 2.
Screening of DEGs involved in the inflammatory immune response based on immune cell score and stromal cell score
Note: (A) Heat map of differential genes screened based on stromal cell scores; (B) Heat map of differential genes screened based on immune cell scores; (C) Wayne analysis of up-regulated differential genes associated with immune cells and stromal cells; (D) Wayne analysis of down-regulated differential genes associated with immune cells and stromal cells; (E,F) Candidate DEGs subjected to GO and KEGG functional enrichment analysis.
The 223 candidate DEGs obtained were subjected to GO and KEGG analysis. As shown in Fig. 2E,F, the results of GO functional analysis showed that the candidate DEGs were mainly enriched in mononuclear cell differentiation, leukocyte mediated immunity, leukocyte activation involved in immune response, and cell activation involved in immune response in biological process (BP). Involved in immune response and cell activation involved in immune response; in cellular components (CC), they were mainly enriched in the outer side of the plasma membrane, tertiary granule, and The KEGG pathway was analyzed in terms of the KEGG pathway, and the KEGG pathway was analyzed in terms of the KEGG pathway. The results of the KEGG pathway analysis showed that the candidate DEGs were mainly enriched in molecular pathways related to immune response or inflammatory response. The above results suggest that the 223 candidate DEGs mainly function in immune response or inflammatory response mechanisms.
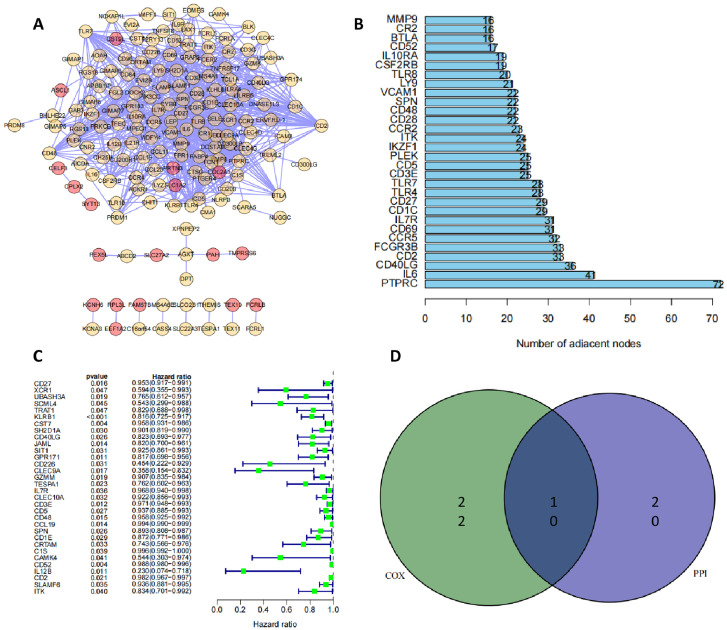
PPI network and single-factor COX regression together screen 10 candidate DEGs
The 223 candidate DEGs were constructed into a protein network interoperation map (Fig. 3A), each node representing a protein. The number of gene nodes connected to the candidate DEGs was displayed in combination with Cytoscape software (Fig. 3B), and the top 30 DEGs in terms of gene nodes were screened as alternative genes for the core DEGs of the network. The 32 genes associated with prognosis in BC were further identified using one-way COX regression and plotted in a forest plot (Fig. 3C). A Wayne analysis of the alternative genes obtained from the PPI network with the prognosis-related genes obtained from one-way COX regression yielded 10 candidate DEGs, namely CD40LG, CD2, IL7R, CD27, CD3E, CD5, ITK, CD48, SPN and CD52 (Fig. 3D).
Fig. 3.
Screening of candidate DEGs by PPI network and one-way COX regression analysis
Note: (A) Protein network interplay map constructed from 223 candidate DEGs; (B) Number of nodes of candidate DEGs linked genes counted; (C) One-way COX regression to identify prognosis-associated genes in BC; (D) Alternative genes obtained from PPI network and prognosis-associated genes obtained from one-way COX regression for Wayne analysis.
Single gene differential analysis, single gene pairwise differential analysis, and survival analysis screened for 5 core DEGs
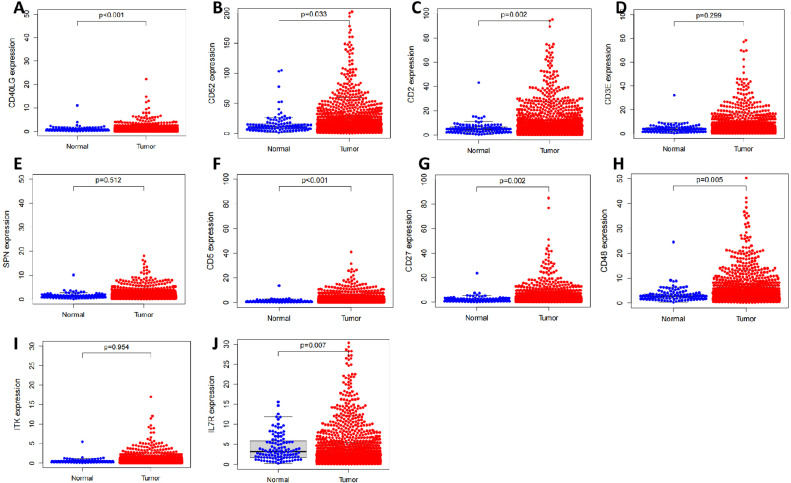
Next, we further examined the expression levels of these 10 candidate DEGs in the TCGA-BRCA dataset. The single gene differential analysis results showed that CD40LG, CD2, IL7R, CD27, CD5, CD48, and CD52 were differentially highly expressed in BC tissues compared to normal breast tissues adjacent to cancer. At the same time, the expression levels of CD3E, SPN, and ITK were not significantly different (Fig. 4). Single pairwise gene difference analysis showed that CD2, CD27, CD5, CD48, and CD52 were significantly more highly expressed in their corresponding BC tissues compared to normal breast tissues adjacent to cancer. In contrast, CD3E, IL7R, CD40LG, SPN, and ITK expression levels were not significantly different (Fig. S1). Finally, the results of the survival analysis curves showed that the expression levels of all 10 candidate DEGs were strongly correlated with the prognosis of BC patients (Fig. S2).
Fig. 4.
Single-gene differential analysis of candidate DEGs
Note: (A) Differential expression of CD40LG in BC tissue and normal breast tissue next to cancer; (B) Differential expression of CD52 in BC tissue and normal breast tissue next to cancer; (C) Differential expression of CD2 in BC tissue and normal breast tissue next to cancer; (D) Differential expression of CD3E in BC tissue and normal breast tissue next to cancer; (E) Differential expression of SPN in BC tissue and normal breast tissue next to cancer; (F) Differential expression of CD5 in BC tissue and normal breast tissue next to cancer; (G) Differential expression of CD27 in BC tissue and normal breast tissue next to cancer; (G) Differential expression of CD27 in BC tissue and normal breast tissue next to cancer. (E) Differential expression of SPN in BC and normal breast tissues; (F) Differential expression of CD5 in BC and normal breast tissues next to cancer; (G) Differential expression of CD27 in BC and normal breast tissues next to cancer; (H) Differential expression of CD48 in BC and normal breast tissues next to cancer; (I) Differential expression of ITK in BC and normal breast tissues next to cancer (I) Differential expression of ITK in BC and normal breast tissues; (J) Differential expression of IL-7R in BC and normal breast tissues.
Combining these results, we screened five core DEGs that were differentially highly expressed in BC tissues, involved in immunomodulatory mechanisms, and closely associated with the prognosis of BC patients.
CD2 and CD27 may be key genes involved in the development of BC brain metastases
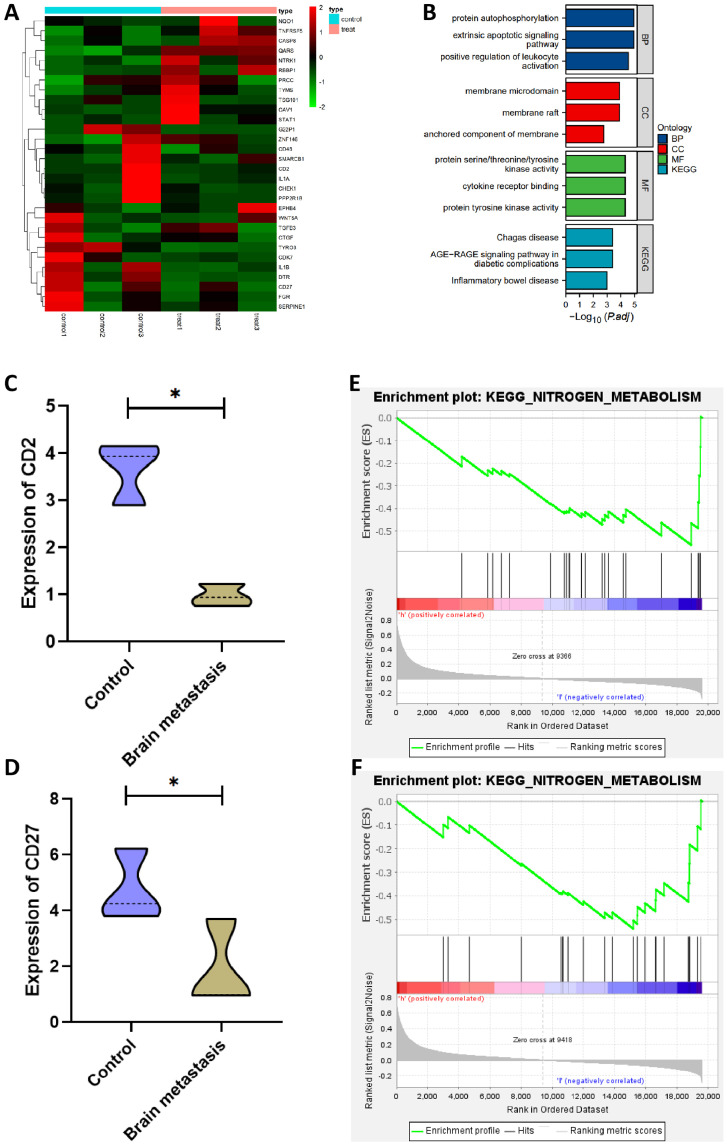
First, 31 DEGs were obtained from the BC brain metastasis dataset, of which 10 were up-regulated, and 21 were down-regulated (Fig. 5A). GO and KEGG pathway analysis revealed that the differential genes were extensively involved in the development of immune responses (Fig. 5B). Further examination of the expression levels of core DEGs in the BC brain metastasis dataset showed that the expression levels of CD2 and CD27 were significantly lower in the brain metastasis group compared to the control group (Fig. 5C,D). The above results suggest that the levels of CD2 and CD27 gradually decreased with the progression of the tumor, which correlated with the size of the primary foci and the extent of adjacent tissue involvement in BC, and that the expression of CD2 and CD27 was closely associated with distant brain metastases in BC.
Fig. 5.
Screening of BC brain metastasis-associated DEGs
Note: (A) Heat map of differential expression in the BC brain metastasis dataset; (B) GO and KEGG pathway analysis; (C,D) CD2 and CD27 expression level levels in the brain metastasis group; (E,F) GSEA analysis of the signaling pathways involved in CD2 and CD27.
It has been shown that nitrogen metabolism could support tumor cell and immune cell growth through rapid conversion and that deletion of specific amino acids in TME could act as a metabolic checkpoint for anti-tumor immunity. That acquisition of nitrogen sources from the extracellular environment could be involved in tumor metastasis [36]. The results of the GSEA analysis showed that the down-regulation of CD2 and CD27 expression was closely associated with the activation of nitrogen metabolic pathways (Fig. 5E,F).
CD2 and CD27 influence the immune activity of the immune microenvironment
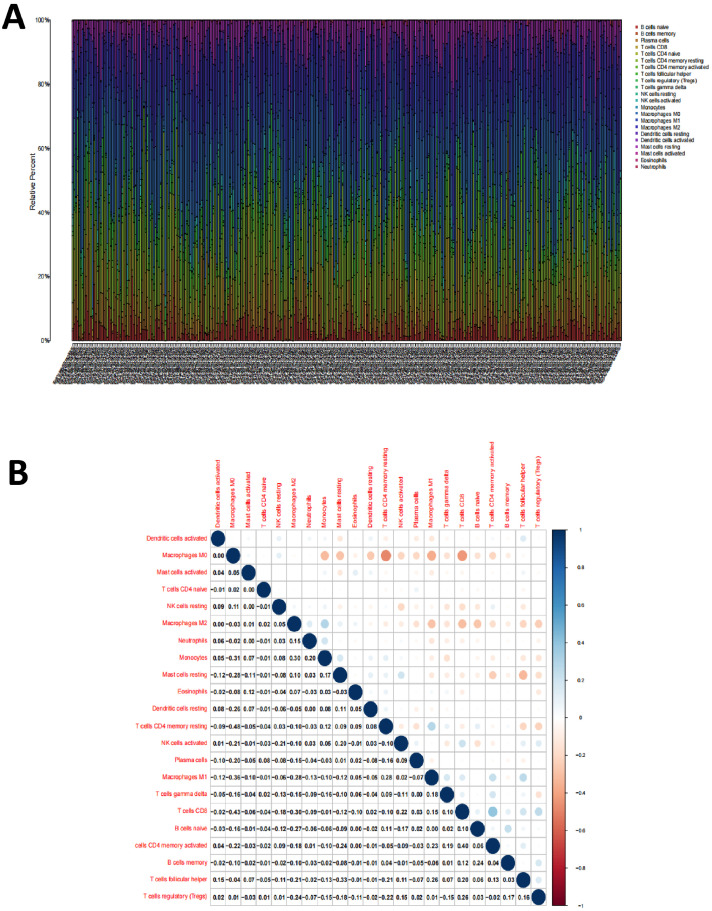
To further validate the correlation between CD2 and CD27 expression levels and the immune micro-environment, we analyzed the proportion of tumor-infiltrating immune subpopulations using the CIBERSORT algorithm. We constructed a profile of 22 immune cell types in BC samples. The proportion of 22 tumor-infiltrating immune cells in the BC samples was shown in Fig. 6A. Pearson correlation analysis was performed to examine the correlation between these 22 tumor-infiltrating immune cells (Fig. 6B).
Fig. 6.
Profile and correlation analysis of tumor-infiltrating immune cells in BC samples
Note: (A) 22 immune cell profiles were constructed in BC samples; (B) Pearson correlation analysis between 22 tumor-infiltrating immune cells.
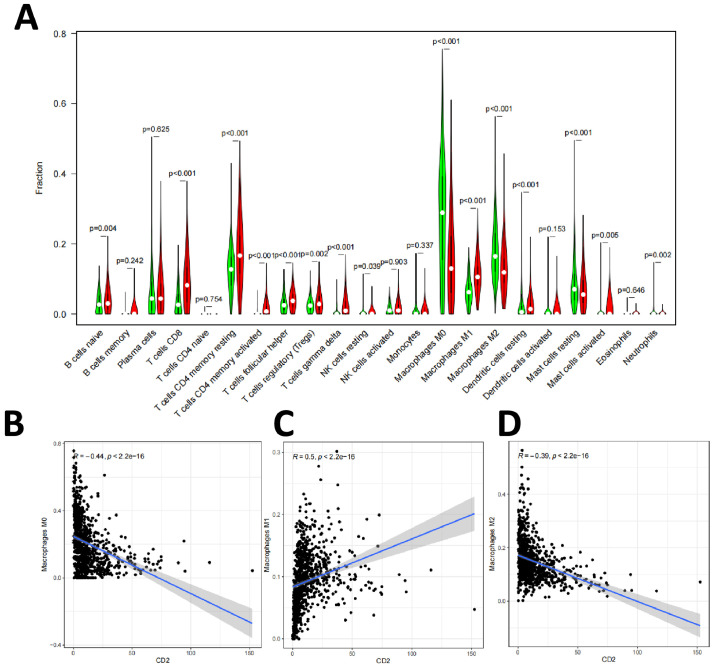
Next, we divided the BC samples into CD2 and CD27 high/low expression groups and detected the expression differences of 22 immune cells relative to the median expression of CD2 and CD27, respectively. The differential analysis results showed that a total of 15 types of tumor-infiltrating immune cells were correlated with the expression level of CD2 (Fig. 7A), including B cells naïve, T cells CD8, T cells CD4 memory resting, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), T cells gamma delta, NK cells resting, Macrophages M0, Macrophages M1, Macrophages M2, Dendritic cells resting, Mast cells resting, Mast cells activated, and Neutrophils. Pearson correlation analysis was used to test the correlation between the proportion of tumor-infiltrating immune cells and CD2 expression, which showed that 16 types of tumor-infiltrating immune cells were correlated with CD2 expression, including M0, M1, and M2 macrophages (Fig. 7B–D).
Fig. 7.
Correlation between the proportion of tumor-infiltrating immune cells and CD2 expression
Note: (A) Correlation analysis of the expression levels of tumor-infiltrating immune cells and CD2; (B) Correlation of M0 macrophage ratio with CD2 expression; (C) Correlation of M1 macrophage ratio with CD2 expression; (D) Correlation of M2 macrophage ratio with CD2 expression.
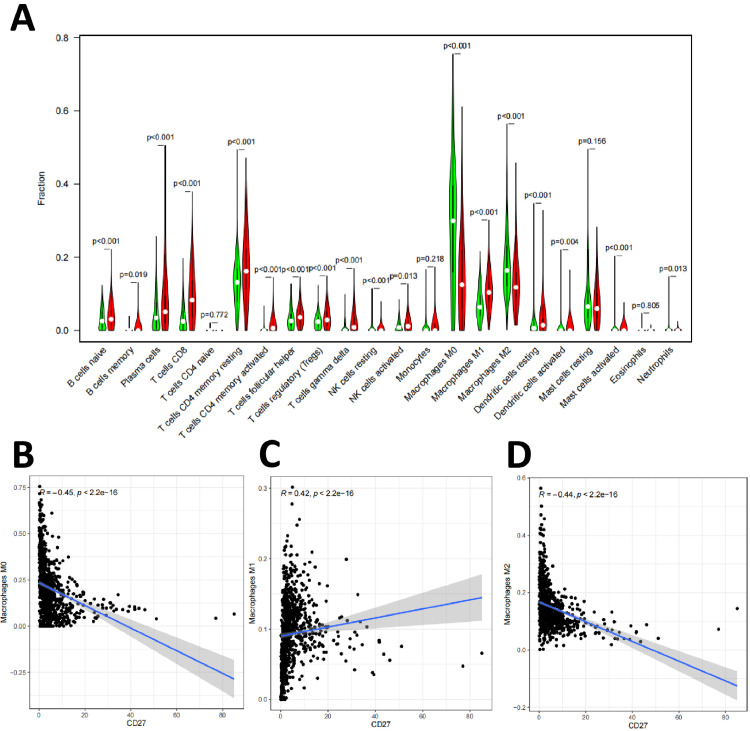
Similarly, the differential analysis results showed that a total of 18 types of tumor-infiltrating immune cells were correlated with the expression level of CD27 (Fig. 8A), including B cells naïve, B cells memory, Plasma cells, T cells CD8, T cells CD4 memory resting, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), T cells gamma delta, NK cells resting, NK cells activated, Macrophages M0, Macrophages M1, Macrophages M2, Dendritic cells resting, Dendritic cells activated, Mast cells activated, and Neutrophils. Pearson correlation analysis was used to test the correlation between the proportion of tumor-infiltrating immune cells and CD27 expression, which showed that 19 types of tumor-infiltrating immune cells were correlated with CD27 expression, including M0, M1, and M2 macrophages (Fig. 8B–D).
Fig. 8.
Correlation between the proportion of tumor-infiltrating immune cells and CD27 expression
Note: (A) Correlation analysis of the expression levels of tumor-infiltrating immune cells and CD27; (B) Correlation of M0 macrophage ratio with CD27 expression; (C) Correlation of M1 macrophage ratio with CD27 expression; (D) Correlation of M2 macrophage ratio with CD27 expression.
Amino acids such as spermine in nitrogen metabolism are closely associated with the polarization of M2 macrophages [37]. Our study also observed that CD2 and CD27 expression showed a negative correlation with the proportion of M2 macrophages, which are closely associated with tumor metastasis. Therefore, CD2 and CD27 may regulate nitrogen metabolic pathways and influence M2 macrophage polarization, and thus in BC brain metastasis.
Overexpression of CD2/CD27 could inhibit M2 polarization of macrophages and thus inhibit brain metastasis of breast cancer
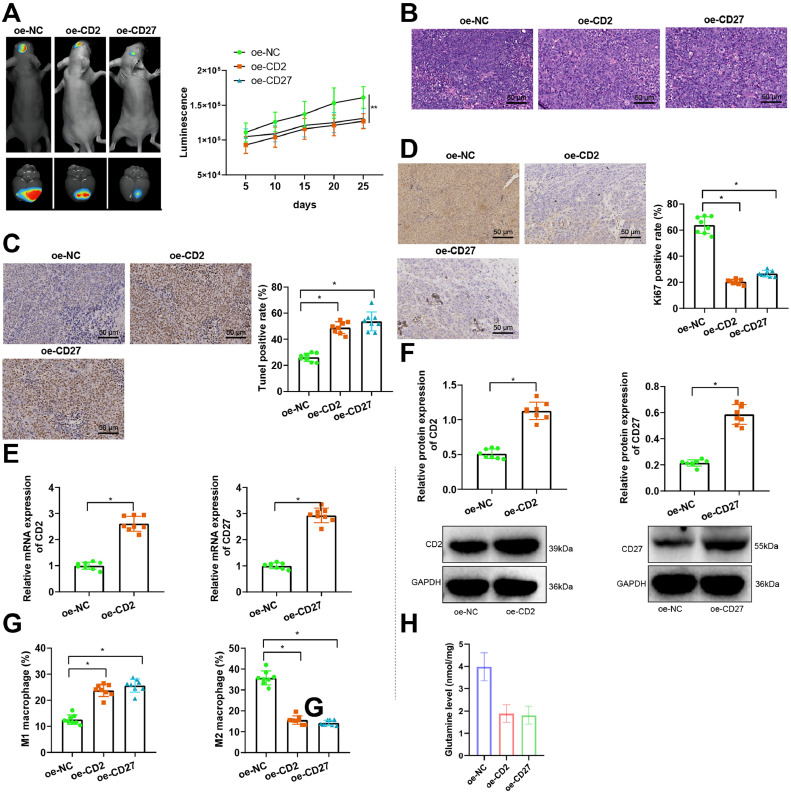
A brain metastasis model of nude mice with MDA-MB-231 cells carrying GFP/Luc markers was established by intracranial injection, and the overexpression of CD2 or CD27 lentivirus was injected for treatment. In vivo, bioluminescence imaging was performed using the IVIS spectral system to monitor tumor growth. Results showed that tumor formation was significantly inhibited after treatment with CD2 or CD27 lentivirus (Fig. 9A).
Fig. 9.
CD2/CD27 could influence M2 polarization of macrophages and participate in breast cancer brain metastasis.
Note: (A) IVIS spectral system is used for in vivo bioluminescence imaging to monitor tumor growth; (B) H&E staining is used to observe the morphology of nude mouse tumor tissue (× 200); (C) TUNEL staining is used to detect cell apoptosis in nude mouse tumor tissue (× 200); (D) Ki67 immunohistochemical staining is used to detect Ki67 protein expression in nude mouse tumor tissue (× 200); (E) RT-qPCR is used to detect the expression of CD2 and CD27 mRNA in nude mouse tumor tissue; (F) WB is used to detect the expression of CD2 and CD27 protein in nude mouse tumor tissue; (G) Flow cytometry is used to detect the proportion of macrophages in each group of nude mouse tumor tissue. (H) The Glutamine content in tumor tissues of each group was measured using the Glutamine Assay Kit. * indicates P<0.05 compared with the oe-NC group, with 8 nude mice in each group.
H&E staining, TUNEL, and Ki67 immunohistochemical staining results showed that the apoptosis of tumor cells was significantly increased and the proliferation of cells was effectively reduced after treatment with CD2 or CD27 lentivirus (Fig. 9B–D).
The expression of CD2 and CD27 in nude mouse tumor tissue was detected by Western blot and PCR. The results showed that the expression of CD2 in the tumor tissue of the oe-CD2 group was significantly elevated. The expression of CD2 in the tumor tissue of the oe-CD27 group was also significantly elevated compared to the oe-NC group (Fig. 9E,F).
Flow cytometry was used to detect the proportion of macrophages in the tumor tissue of each group of nude mice. The results showed that the proportion of M1 macrophages in the tumor tissue of the oe-CD2 and oe-CD27 groups was significantly increased. In contrast, the proportion of M2 macrophages was significantly inhibited compared to the oe-NC group (Fig. 9G). In addition, compared to the oe-NC group, the oe-CD2 and oe-CD27 groups exhibited a significant reduction in Glutamine content in the tumor tissues (Fig. 9H).
The results mentioned above demonstrate that the overexpression of CD2/CD27 can inhibit nitrogen metabolism and M2 polarization of macrophages, thereby suppressing brain metastasis in breast cancer.
Discussion
In our study, we screened the TCGA database for tumor-infiltrating immune cell-related genes associated with prognostic survival and clinicopathological features of BC patients based on the CIBERSORT and ESTIMATE algorithms, and combined PPI network, one-way COX regression analysis, single-gene difference analysis, single-gene pairwise difference analysis, and survival analysis to screen for five core DEGs. The BC brain metastasis dataset was further used to screen for CD2 and CD27, key genes involved in developing BC brain metastasis. CD2 and CD27 are mainly involved in immune activities, play a crucial role in the BC immune micro-environment, and may be potential indicators of the immune status of tumor infiltration in BC patients [[38], [39], [40], [41]]. Furthermore, we found that the proportion of immune and stromal components in tumor-infiltrating immune cells was strongly correlated with BC progression. These results surface the importance of exploring the interactions between tumor cells and immune cells to provide new ideas for developing more effective treatment options.
CD2 is a transmembrane glycoprotein of the immunoglobulin superfamily expressed on the surface of T cells, NK cells, thymocytes, and dendritic cells [42]. The binding partners of CD2 are lymphocyte-associated antigens expressed on the surface of B cells, T cells, monocytes, granulocytes, and thymic epithelial cells [43]. CD2 has multiple roles; intracellularly, CD2 influences the rearrangement of the agonist cytoskeleton and activates cell signaling; CD2 is an important component of immune synaptic assembly following T cell-APC binding and contributes to T cell signaling; and plays an important role in thymocyte development and NK cell activation [44]. In our study, GSEA analysis showed that the high CD2 expression group was mainly enriched in the B-cell receptor signaling pathway, T-cell receptor signaling pathway, apoptosis, chemokine signaling pathway, JAK-STAT signaling pathway, and natural killer cell-mediated cytotoxicity; in addition, the low CD2 expression group was mainly enriched in nitrogen metabolism, potassium propionate metabolism, potassium butyrate metabolism, and other metabolic pathways.
CD27 and its ligand CD70 are two important components of the TNF-TNFR superfamily, and their interaction regulates the proliferation and differentiation of T, B, and NK cells [45]. During T cell activation, CD27 expression increases transiently but is down-regulated when T cells undergo several rounds of differentiation. CD27 is also expressed in B cells through the activation of antigen receptors and is a typical marker of memory B cells [[46], [47], [48]]. In addition, CD27 is also expressed in NK cells, and its expression could be induced by IL-2 activation [49,50]. Therefore, we further analyzed the relationship between CD27 and tumor immune infiltrating cells.GSEA analysis showed that the high CD27 expression group was mainly enriched in chemokine signaling pathways, cytokine-cytokine receptor interactions, natural killer cell-mediated cytotoxicity, T-cell receptor signaling pathways, and B-cell receptor signaling pathways. However, the CD27 low-expression group was mainly enriched in metabolic pathways such as nitrogen metabolism, potassium propionate metabolism, and potassium butyrate metabolism.
Our studies have found that CD2 and CD27 are associated with nitrogen metabolism, which plays a regulatory role in tumorigenesis. Nitrogen is required for the ab initio synthesis of various biomolecules, including nucleotides, amino acids, polyamines, hexosamine, glutathione, porphyrins, ammonia, creatine, nitric oxide, and many other important biological compounds [51,52]. Amino acids and nucleotides are the main nitrogen sources in cells and support tumor and immune cell growth through rapid turnover [53,54]. The reduced state of nitrogen is obtained mainly through amino acids, which account for most of the biomass required by proliferating cells, are the main reservoir of cellular nitrogen, and play a key role in the growth of BC tumor cells.
TAMs are among the most dominant, common, and numerous tumor-infiltrating immune cell populations in TME. They are the main natural immune component of the tumor mesenchyme, consisting mainly of macrophages residing in surrounding tissues and circulating monocytes newly recruited to inflammation and tissue injury [55]. There is growing evidence that TAMs play an important role in tumourigenesis, progression, and intra-tumor immunotherapy and that high-density TAMs are associated with poor prognosis, drug resistance, enhanced angiogenesis, and metastasis in cancer [[56], [57], [58], [59]]. The Mills team proposed that TAMs are mainly divided into anti-tumour M1-type macrophages (classical activation state) and pro-tumour M2-type macrophages (selective activation state), which respond to the Th1-Th2 polarisation of T cells, respectively [60]. TAMs are generally biased towards pro-tumor M2-type polarisation, can participate in tumor invasion and metastasis, and promote epithelial-mesenchymal transition (Epithelial transition). They also play an important role in promoting epithelial-mesenchymal transformation (EMT), angiogenesis, and immunosuppression [61,62]. In addition, M2-TAMs contribute to poor prognosis by suppressing CD8+ T cell function as a factor affecting the efficacy of chemotherapy and radiotherapy and promoting tumor progression.
In our study, we found that CD2 and CD27 expression showed a negative correlation with the proportion of M2 macrophages. The possible reason for this is that tumor immunity is abnormal as the tumor stage progresses, leading to a gradual decrease in CD2 and CD27 expression, and their expression correlates with the extent of BC primary foci and adjacent tissue involvement. In conclusion, the outcome of CD2 and CD27 in BC tissues is complex, exerting a tumor-promoting or tumor-suppressing role as the tumor progresses.
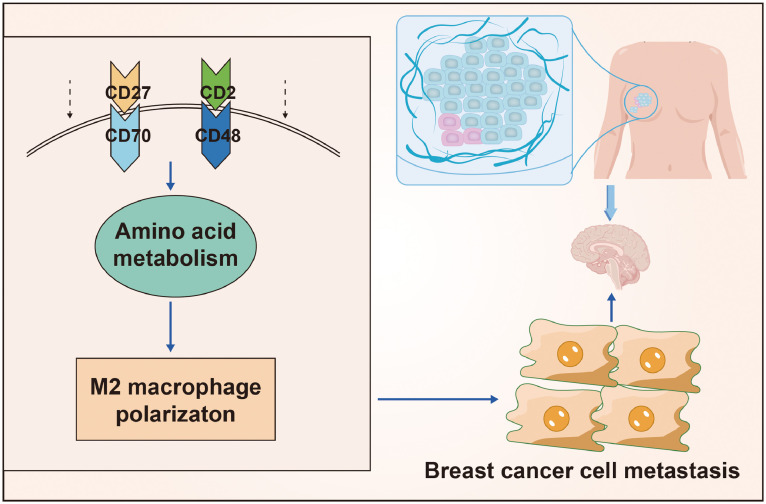
Conclusion
Summing up the results, we could tentatively conclude that in TME, overexpression of CD2/CD27 could inhibit the activation of nitrogen metabolism pathways and suppress M2 polarization of macrophages, thereby preventing brain metastasis of breast cancer (Fig. 10). Our study provides a theoretical basis for a deeper understanding of the mechanisms underlying the development of BC brain metastasis and the search for potential diagnostic and therapeutic targets. Further in vivo and in vitro studies are needed to investigate the specific molecular mechanisms by which CD2 and CD27 regulate the nitrogen metabolic pathway to promote macrophage M2 polarization and thus accelerate BC brain metastasis as the tumor progresses.
Fig. 10.
Molecular mechanisms by which CD2 and CD27 influence the tumor microenvironment in the development of brain metastases from breast cancer.
Availability of data and materials
The article's data will be shared on reasonable request to the corresponding author.
Ethics approval and consent to participate
The experimental animals were used for medical research, and all procedures were approved by our institution's animal committee (No. 2201142).
Consent for publication
Not applicable.
Fig. S1. Single gene pairwise differential analysis of candidate DEGs
Note: (A) Analysis of CD2 single gene pairing difference between BC tissue and normal breast tissue next to cancer; (B) Analysis of CD3E single gene pairing difference between BC tissue and normal breast tissue next to cancer; (C) Analysis of CD5 single gene pairing difference between BC tissue and normal breast tissue next to cancer; (D) Analysis of IL7R single gene pairing difference between BC tissue and normal breast tissue next to cancer (E) Analysis of single gene pairing differences between BC tissue and normal breast tissue next to cancer in SPN; (F) Analysis of single gene pairing differences between BC tissue and normal breast tissue next to cancer in CD52; (G) Analysis of single gene pairing differences between BC tissue and normal breast tissue next to cancer in CD27; (H) Analysis of single gene pairing differences between BC tissue and normal breast tissue next to cancer in CD48; (I) Analysis of ITK single gene pairing difference between BC tissue and adjacent normal breast tissue; (J) Analysis of the single gene pairing difference between BC tissue and normal breast tissue next to cancer for CD40LG.
CRediT authorship contribution statement
Guanyou Huang: Conceptualization, Methodology, Investigation, Funding acquisition, Project administration, Resources, Writing – original draft, Writing – review & editing. Yujuan Wu: Software, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Hongchuan Gan: Data curation, Formal analysis, Visualization, Writing – review & editing. Liangzhao Chu: Validation, Supervision, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
Funding
This work is supported by the Science and Technology Foundation of Guizhou Provincial Health Commission (gzwkj2022-348).
Acknowledgment
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101768.
Appendix. Supplementary materials
Fig. S2. Survival analysis of candidate DEGs
Note: (A) Survival analysis of the prognostic association between CD2 expression and BC patients; (B) Survival analysis of the prognostic association between CD3E expression and BC patients; (C) Survival analysis of the prognostic association between SPN expression and BC patients; (D) Survival analysis of the prognostic association between CD48 expression and BC patients; (E) Survival analysis of the prognostic association between CD27 expression and BC patients; (F) Survival analysis of the prognostic association between CD5 expression and BC patients; (G) survival analysis of the prognostic association between CD40LG expression and BC patients; (H) survival analysis of the prognostic association between ITK expression and BC patients; (I) survival analysis of the prognostic association between CD42 expression and BC patients; (J) survival analysis of the prognostic association between IL7R expression and BC patients.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Parkin D.M., Pineros M., Znaor A., et al. Cancer statistics for the year 2020: an overview. Int. J. Cancer. 2021 doi: 10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 2.Zubair M., Wang S., Ali N. Advanced approaches to breast cancer classification and diagnosis. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.632079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson L., Gathani T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022;95(1130) doi: 10.1259/bjr.20211033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sopik V. International variation in breast cancer incidence and mortality in young women. Breast Cancer Res. Treat. 2021;186(2):497–507. doi: 10.1007/s10549-020-06003-8. [DOI] [PubMed] [Google Scholar]
- 5.Konat-Baska K., Matkowski R., Blaszczyk J., Blaszczyk D., Staszek-Szewczyk U., Pilat-Norkowska N., et al. Does breast cancer increasingly affect younger women? Int. J. Environ. Res. Publ. Health. 2020;17(13) doi: 10.3390/ijerph17134884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romagnolo A.P., Romagnolo D.F., Selmin O.I. BRCA1 as target for breast cancer prevention and therapy. Anticancer. Agents Med. Chem. 2015;15(1):4–14. doi: 10.2174/1871520614666141020153543. [DOI] [PubMed] [Google Scholar]
- 7.Narod S.A., Salmena L. BRCA1 and BRCA2 mutations and breast cancer. Discov. Med. 2011;12(66):445–453. [PubMed] [Google Scholar]
- 8.Rozenberg S., Di Pietrantonio V., Vandromme J., Gilles C. Menopausal hormone therapy and breast cancer risk. Best Pract. Res. Clin. Endocrinol. Metab. 2021;35(6) doi: 10.1016/j.beem.2021.101577. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Aparicio A., Schmidt-RioValle J., Garcia P.A., Gonzalez-Jimenez E. Short breastfeeding duration is associated with premature onset of female breast cancer. Clin. Nurs. Res. 2022;31(5):901–908. doi: 10.1177/10547738211069725. [DOI] [PubMed] [Google Scholar]
- 10.Lee K., Kruper L., Dieli-Conwright C.M., Mortimer J.E. The Impact of obesity on breast cancer diagnosis and treatment. Curr. Oncol. Rep. 2019;21(5):41. doi: 10.1007/s11912-019-0787-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 12.Montagna G., Morrow M. Breast-conserving surgery without radiation therapy for invasive cancer. Clin. Breast Cancer. 2021;21(2):112–119. doi: 10.1016/j.clbc.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Moo T.A., Sanford R., Dang C., Morrow M. Overview of breast cancer therapy. PET Clin. 2018;13(3):339–354. doi: 10.1016/j.cpet.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisusi F.A., Akala E.O. Drug combinations in breast cancer therapy. Pharm. Nanotechnol. 2019;7(1):3–23. doi: 10.2174/2211738507666190122111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosonaga M., Saya H., Arima Y. Molecular and cellular mechanisms underlying brain metastasis of breast cancer. Cancer Metastasis. Rev. 2020;39(3):711–720. doi: 10.1007/s10555-020-09881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takada M., Toi M. Neoadjuvant treatment for HER2-positive breast cancer. Chin. Clin. Oncol. 2020;9(3):32. doi: 10.21037/cco-20-123. [DOI] [PubMed] [Google Scholar]
- 17.Medina M.A., Oza G., Sharma A., Arriaga L.G., Hernandez Hernandez J.M., Rotello V.M., et al. Triple-negative breast cancer: a review of conventional and advanced therapeutic strategies. Int. J. Environ. Res. Public Health. 2020;17(6) doi: 10.3390/ijerph17062078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arneth B. Tumor microenvironment. Medicina. 2019;56(1) doi: 10.3390/medicina56010015. (Kaunas) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Y., Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021;221 doi: 10.1016/j.pharmthera.2020.107753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vitale I., Manic G., Coussens L.M., Kroemer G., Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30(1):36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Wu T., Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 22.Choi J., Gyamfi J., Jang H., Koo J.S. The role of tumor-associated macrophage in breast cancer biology. Histol. Histopathol. 2018;33(2):133–145. doi: 10.14670/HH-11-916. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Yang P., Wang J., Zhang J., Ma Q., Jiang Y., et al. HLF regulates ferroptosis, development and chemoresistance of triple-negative breast cancer by activating tumor cell-macrophage crosstalk. J. Hematol. Oncol. 2022;15(1):2. doi: 10.1186/s13045-021-01223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassetta L., Fragkogianni S., Sims A.H., Swierczak A., Forrester L.M., Zhang H., et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35(4):588–602. doi: 10.1016/j.ccell.2019.02.009. e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S.Y., Sharma S., Wu K., Tyagi A., Zhao D., Deshpande R.P., et al. Tamoxifen suppresses brain metastasis of estrogen receptor-deficient breast cancer by skewing microglia polarization and enhancing their immune functions. Breast Cancer Res. 2021;23(1):35. doi: 10.1186/s13058-021-01412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Jiang T., Zhang H., Gou X., Han C., Wang J., et al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat. Cell Biol. 2020;22(10):1276–1285. doi: 10.1038/s41556-020-00586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X., Xie Y., Dong M. Cancer-associated fibroblasts derived extracellular vesicles promote angiogenesis of colorectal adenocarcinoma cells through miR-135b-5p/FOXO1 axis. Cancer Biol. Ther. 2022;23(1):76–88. doi: 10.1080/15384047.2021.2017222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu P.N., Yan M.D., Lai H.C., Huang R.L., Chou Y.C., Lin W.C., et al. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int. J. Cancer. 2014;134(3):542–551. doi: 10.1002/ijc.28399. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Xue B., Zhang Y., Guo G., Duan X., Dou D. Up-regulated circBACH2 contributes to cell proliferation, invasion, and migration of triple-negative breast cancer. Cell Death Dis. 2021;12(5):412. doi: 10.1038/s41419-021-03684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Qiao K., Zhu K., Li X., Zhao C., Li J., et al. Long noncoding RNA HCG18 promotes malignant phenotypes of breast cancer cells via the HCG18/miR-103a-3p/UBE2O/mTORC1/HIF-1alpha-positive feedback loop. Front Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.675082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S., Wu H., Wu X., You Z., Jiang Y., Liang X., et al. Silencing of PGK1 promotes sensitivity to paclitaxel treatment by upregulating XAF1-mediated apoptosis in triple-negative breast cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.535230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv H., Li J., Che Y. miR-31 from adipose stem cell-derived extracellular vesicles promotes recovery of neurological function after ischemic stroke by inhibiting TRAF6 and IRF5. Exp. Neurol. 2021;342 doi: 10.1016/j.expneurol.2021.113611. [DOI] [PubMed] [Google Scholar]
- 33.Ye C., Qi W., Dai S., Zou G., Liu W., Yu B., et al. microRNA-223 promotes autophagy to aggravate lung ischemia-reperfusion injury by inhibiting the expression of transcription factor HIF2alpha. Am. J. Physiol. Lung. Cell Mol. Physiol. 2020;319(1):L1–L10. doi: 10.1152/ajplung.00009.2020. [DOI] [PubMed] [Google Scholar]
- 34.Xue J., Sharma V., Hsieh M.H., Chawla A., Murali R., Pandol S.J., et al. Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat. Commun. 2015;6:7158. doi: 10.1038/ncomms8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumari N., Choi S.H. Tumor-associated macrophages in cancer: recent advancements in cancer nanoimmunotherapies. J. Exp. Clin. Cancer Res. 2022;41(1):68. doi: 10.1186/s13046-022-02272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masid M., Ataman M., Hatzimanikatis V. Analysis of human metabolism by reducing the complexity of the genome-scale models using redHUMAN. Nat. Commun. 2020;11(1):2821. doi: 10.1038/s41467-020-16549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latour Y.L., Gobert A.P., Wilson K.T. The role of polyamines in the regulation of macrophage polarization and function. Amino Acids. 2020;52(2):151–160. doi: 10.1007/s00726-019-02719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quastel M., Dustin M. The CD58-CD2 axis in cancer immune evasion. Nat. Rev. Immunol. 2022;22(7):409. doi: 10.1038/s41577-022-00738-9. [DOI] [PubMed] [Google Scholar]
- 39.Demetriou P., Abu-Shah E., Valvo S., McCuaig S., Mayya V., Kvalvaag A., et al. A dynamic CD2-rich compartment at the outer edge of the immunological synapse boosts and integrates signals. Nat. Immunol. 2020;21(10):1232–1243. doi: 10.1038/s41590-020-0770-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starzer A.M., Berghoff A.S. New emerging targets in cancer immunotherapy: CD27 (TNFRSF7) ESMO Open. 2020;4(3) doi: 10.1136/esmoopen-2019-000629. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burugu S., Dancsok A.R., Nielsen T.O. Emerging targets in cancer immunotherapy. Semin. Cancer Biol. 2018;52(Pt 2):39–52. doi: 10.1016/j.semcancer.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Binder C., Cvetkovski F., Sellberg F., Berg S., Paternina Visbal H., Sachs D.H., et al. CD2 immunobiology. Front Immunol. 2020;11:1090. doi: 10.3389/fimmu.2020.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Meng Z., Zhang L., Liu F. CD2 is a novel immune-related prognostic biomarker of invasive breast carcinoma that modulates the tumor microenvironment. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.664845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh A., Marques-Piubelli M.L., Wang X., Sheu T.G., Cheng J., Khan K., et al. CD2-negative lymphoma-associated T-cells: a potential mechanism of immune-evasion in diffuse large B-cell lymphoma. Virchows Arch. 2022;481(4):659–663. doi: 10.1007/s00428-022-03348-x. [DOI] [PubMed] [Google Scholar]
- 45.Flieswasser T., Van den Eynde A., Van Audenaerde J., De Waele J., Lardon F., Riether C., et al. The CD70-CD27 axis in oncology: the new kids on the block. J. Exp. Clin. Cancer Res. 2022;41(1):12. doi: 10.1186/s13046-021-02215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han B.K., Olsen N.J., Bottaro A. The CD27-CD70 pathway and pathogenesis of autoimmune disease. Semin. Arthritis Rheum. 2016;45(4):496–501. doi: 10.1016/j.semarthrit.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Benhamouda N., Sam I., Epaillard N., Gey A., Phan L., Pham H.P., et al. Plasma CD27, a surrogate of the intratumoral CD27-CD70 interaction, correlates with immunotherapy resistance in renal cell carcinoma. Clin. Cancer Res. 2022;28(22):4983–4994. doi: 10.1158/1078-0432.CCR-22-0905. [DOI] [PubMed] [Google Scholar]
- 48.Seyfrid M., Maich W.T., Shaikh V.M., Tatari N., Upreti D., Piyasena D., et al. CD70 as an actionable immunotherapeutic target in recurrent glioblastoma and its microenvironment. J. Immunother. Cancer. 2022;10(1) doi: 10.1136/jitc-2021-003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugita K., Robertson M.J., Torimoto Y., Ritz J., Schlossman S.F., Morimoto C. Participation of the CD27 antigen in the regulation of IL-2-activated human natural killer cells. J. Immunol. 1992;149(4):1199–1203. [PubMed] [Google Scholar]
- 50.Desbois M., Beal C., Charrier M., Besse B., Meurice G., Cagnard N., et al. IL-15 superagonist RLI has potent immunostimulatory properties on NK cells: implications for antimetastatic treatment. J. Immunother. Cancer. 2020;8(1) doi: 10.1136/jitc-2020-000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt M., Pearson A.N., Incha M.R., Thompson M.G., Baidoo E.E.K., Kakumanu R., et al. Nitrogen metabolism in pseudomonas putida: functional analysis using random barcode transposon sequencing. Appl. Environ. Microbiol. 2022;88(7) doi: 10.1128/aem.02430-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scribani Rossi C., Barrientos-Moreno L., Paone A., Cutruzzola F., Paiardini A., Espinosa-Urgel M., et al. Nutrient sensing and biofilm modulation: the example of L-arginine in pseudomonas. Int. J. Mol. Sci. 2022;23(8) doi: 10.3390/ijms23084386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aigner S., Glaser K., Arc E., Holzinger A., Schletter M., Karsten U., et al. Adaptation to aquatic and terrestrial environments in chlorella vulgaris (Chlorophyta) Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.585836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosios A.M., Hecht V.C., Danai L.V., Johnson M.O., Rathmell J.C., Steinhauser M.L., et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell. 2016;36(5):540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gordon S., Pluddemann A. The mononuclear phagocytic system. Generation of diversity. Front Immunol. 2019;10:1893. doi: 10.3389/fimmu.2019.01893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dallavalasa S., Beeraka N.M., Basavaraju C.G., Tulimilli S.V., Sadhu S.P., Rajesh K., et al. The role of tumor-associated macrophages (TAMs) in cancer progression, chemoresistance, angiogenesis and metastasis - current status. Curr. Med. Chem. 2021;28(39):8203–8236. doi: 10.2174/0929867328666210720143721. [DOI] [PubMed] [Google Scholar]
- 57.Lopez-Yrigoyen M., Cassetta L., Pollard J.W. Macrophage targeting in cancer. Ann. NY Acad. Sci. 2021;1499(1):18–41. doi: 10.1111/nyas.14377. [DOI] [PubMed] [Google Scholar]
- 58.Pittet M.J., Michielin O., Migliorini D. Clinical relevance of tumor-associated macrophages. Nat. Rev. Clin. Oncol. 2022;19(6):402–421. doi: 10.1038/s41571-022-00620-6. [DOI] [PubMed] [Google Scholar]
- 59.Mehraj U., Dar A.H., Wani N.A., Mir M.A. Tumor microenvironment promotes breast cancer chemoresistance. Cancer Chemother. Pharmacol. 2021;87(2):147–158. doi: 10.1007/s00280-020-04222-w. [DOI] [PubMed] [Google Scholar]
- 60.Biswas S.K., Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Q., Huang F., Yao Y., Wang J., Wei J., Wu Q., et al. interaction of transforming growth factor-beta-Smads/microRNA-362-3p/CD82 mediated by M2 macrophages promotes the process of epithelial-mesenchymal transition in hepatocellular carcinoma cells. Cancer Sci. 2019;110(8):2507–2519. doi: 10.1111/cas.14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeNardo D.G., Ruffell B. Macrophages as regulators of tumor immunity and immunotherapy. Nat. Rev. Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S2. Survival analysis of candidate DEGs
Note: (A) Survival analysis of the prognostic association between CD2 expression and BC patients; (B) Survival analysis of the prognostic association between CD3E expression and BC patients; (C) Survival analysis of the prognostic association between SPN expression and BC patients; (D) Survival analysis of the prognostic association between CD48 expression and BC patients; (E) Survival analysis of the prognostic association between CD27 expression and BC patients; (F) Survival analysis of the prognostic association between CD5 expression and BC patients; (G) survival analysis of the prognostic association between CD40LG expression and BC patients; (H) survival analysis of the prognostic association between ITK expression and BC patients; (I) survival analysis of the prognostic association between CD42 expression and BC patients; (J) survival analysis of the prognostic association between IL7R expression and BC patients.
Data Availability Statement
The article's data will be shared on reasonable request to the corresponding author.