Abstract
De novo heterozygous loss-of-function mutations in phosphatase and tensin homolog (PTEN) are strongly associated with autism spectrum disorders; however, it is unclear how heterozygous mutations in this gene affect different cell types during human brain development and how these effects vary across individuals. Here, we used human cortical organoids from different donors to identify cell-type specific developmental events that are affected by heterozygous mutations in PTEN. We profiled individual organoids by single-cell RNA-seq, proteomics and spatial transcriptomics and revealed abnormalities in developmental timing in human outer radial glia progenitors and deep-layer cortical projection neurons, which varied with the donor genetic background. Calcium imaging in intact organoids showed that both accelerated and delayed neuronal development phenotypes resulted in similar abnormal activity of local circuits, irrespective of genetic background. The work reveals donor-dependent, cell-type specific developmental phenotypes of PTEN heterozygosity that later converge on disrupted neuronal activity.
Introduction
Autism spectrum disorder (ASD) is a childhood-onset neurodevelopmental disorder with cognitive, sensory and motor deficits (1). ASD is characterized by strong clinical heterogeneity and a complex genetic component, with variants in hundreds of loci associated with increased risk of ASD (2–7). Emerging evidence indicates that mutations in specific genes associated with ASD affect the developmental timing of specific cell types (8–10) and can vary in penetrance between genetic backgrounds (8). It remains unclear whether later in development there is convergence across genetic backgrounds on shared functional phenotypes.
To test the relationship between early and late phenotypes and their relationship to genetic backgrounds, we focused on heterozygous loss of function mutations in phosphatase and tensin homolog (PTEN), which are strongly associated with ASD risk (7). Moreover, prior work has associated PTEN with key cell-biological processes in neurodevelopment in mouse models, including roles in neural progenitor proliferation and neuronal structure, function and plasticity (reviewed in (11)). However, how PTEN mutation causes ASD risk remains largely unknown. While complete loss of function of PTEN has been associated with abnormal proliferation (12–14), the impact of heterozygous PTEN loss of function (as found in the context of ASD) in human brain development still needs to be elucidated.
Results
Here, we used reproducible organoid models of the developing human cerebral cortex (15) to investigate the cell-type and dosage-specific phenotypes associated with heterozygous PTEN mutations across different donors (genetic backgrounds). We used CRISPR-Cas9 to generate induced pluripotent stem cell (iPSC) lines carrying a heterozygous protein–truncating frameshift mutation leading to a premature STOP codon in the phosphatase domain of PTEN, a region found mutated in patients (16) (see Materials and Methods, Supplementary Material, Fig. S1A), in two different genetic backgrounds (Mito210 and PGP1), and produced mutant and isogenic control organoids from these lines (Supplementary Fig. 1B). The introduction of a premature STOP codon led to a reduced production of PTEN protein in both genetic backgrounds, as verified by Western blot comparing PTEN protein levels in mutant iPSC cells versus isogenic control lines. Consistent with a reduction in PTEN function, the downstream effectors AKT and ERK (MAPK) showed increased phosphorylation (Supplementary Material, Fig. S1C). No significant differences were observed in PTEN signaling–associated pathways in brain organoid cell types in single-cell RNA-sequencing (scRNA-seq) data, including apoptosis (Supplementary Material, Fig. S2). Immunohistochemistry validated proper initiation of neural development in both PTEN heterozygous and isogenic control organoids (Supplementary Material, Fig. S1D and E).
Because PTEN and its downstream effectors AKT and MAPK1 are ubiquitously expressed across all cortical cell types during control organoid development by scRNA-seq (17) (Supplementary Material, Fig. S1F), we investigated the cell-type specific phenotypes caused by heterozygous mutation in PTEN, using scRNA-seq of individual organoids at 1 month in culture (Fig. 1A and B and Supplementary Material, Fig. S1G and H). Analysis of 64 207 single cells from control and PTEN mutant organoids at 1 month across two independent genetic backgrounds (Mito210 and PGP1, one batch each), showed that PTEN heterozygous organoids had no significant difference in the proportion of any cortical cell type compared with control, in either genetic background (n = 3 single organoids per batch, false discovery rate (FDR) > 0.05, logistic mixed model; Supplementary Material, Fig. S3A and B). Thus, at early stages of development, loss of one copy of PTEN does not cause differences in the proportion of any cortical cell type in human cortical organoids.
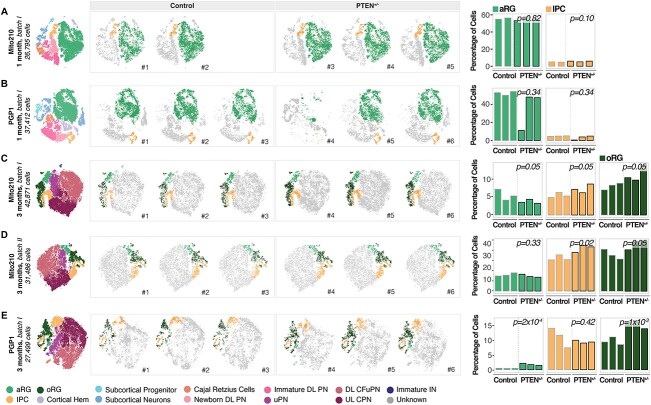
Figure 1.
oRG are consistently affected in PTEN mutant organoids independently of genetic background. (A–E) Left: t-SNE plots of scRNA-seq data, color-coded by cell type, from 1-month Mito210 control and PTEN heterozygous mutant organoids (Mito210 control n = 2, Mito210 heterozygous n = 3) (A), 1-month PGP1 control and PTEN heterozygous mutant organoids (n = 3 single organoids per genotype) (B), 3-month Mito210 control and PTEN heterozygous mutant organoids (n = 3 single organoids per genotype; batch I) (C), 3-month Mito210 control and PTEN heterozygous mutant organoids (n = 3 single organoids per genotype; batch II) (D) and 3-month PGP1 control and PTEN heterozygous mutant organoids (n = 3 single organoids per genotype) (E). Middle: t-SNE plots for individual control and mutant organoids, with cell types of interest highlighted in color: apical radial glia (aRG, light green), intermediate progenitor cells (IPC, yellow) and oRG (dark green). Right: bar charts showing the percentage of cells for the highlighted cell populations in each control and mutant organoid. FDRs for a difference in cell type proportions between control and mutant, based on logistic mixed models (see Materials and Methods) are shown. aRG, apical radial glia; DL, deep layer; UL, upper layer; PNs, projection neurons; oRG, outer radial glia; IPCs, intermediate progenitor cells; CPNs, callosal projection neurons; CFuPNs, corticofugal projection neurons; uPNs, unspecified PN; INs, interneurons.
PTEN loss of function in early mouse development has been reported to affect the spatial organization of the mouse brain (13,18). We therefore investigated the effects of a heterozygous mutation in PTEN on spatial organization of organoid cell types at 2 months of culture, when this brain organoid model shows clear structural organization of progenitors into rosettes around the organoid periphery (17), using spatial transcriptomics (Slide-seq) on heterozygous and isogenic control Mito210 organoids. The spatial organization for most cell types was similar between conditions (Supplementary Material, Fig. S3C), although outer radial glia (oRG), a progenitor cell type associated with human cortical expansion and evolution (19,20), was more disorganized in mutant organoids, with a less well-defined ring-like arrangement of rosettes around the organoid periphery (Supplementary Material, Fig. 3C).
This led us to wonder whether the composition of organoid cell types would be affected at later time points. We therefore profiled 102 216 additional cells with scRNA-seq from three separate differentiation batches at a later stage, 3 months in culture (n = 3 separate organoids per genotype per batch) (Fig. 1C–E, Supplementary Material, Figs S1I and J and 3D), when the highest diversity of progenitor and projection neuron subtypes is found in these organoids (15,17). In both genetic backgrounds, heterozygous PTEN organoids showed a consistent significant increase in oRG (FDR < 0.05, logistic mixed models; Fig. 1C–E and Supplementary Material, Fig. S3F). Certain other progenitor and neuronal populations were mildly affected in different organoid batches: PGP1 PTEN mutant organoids displayed an increase in aRG (FDR = 0.0002; Fig. 1E), one batch of Mito210 PTEN mutant organoids showed an increase in IPCs (FDR = 0.05 for batch I; FDR = 0.02 for batch II; Fig. 1D), and one batch of Mito210 showed an increase in callosal projection neurons (CPNs) (21) (FDR = 0.045; Supplementary Material, Fig. S3E and F). The increase in radial glial cells observed in PTEN mutant organoids was validated by immunohistochemistry using the progenitor marker SOX2 (Supplementary Material, Fig. S3G). However, aside from oRG, none of these cell types showed consistent differences in proportion in all three batches. Together, the data show that although mutations in a single gene can have multiple cell-type specific effects that may individually vary with genetic background, PTEN haploinsufficiency robustly alters oRG progenitors independently of donor of origin.
Figure 4.
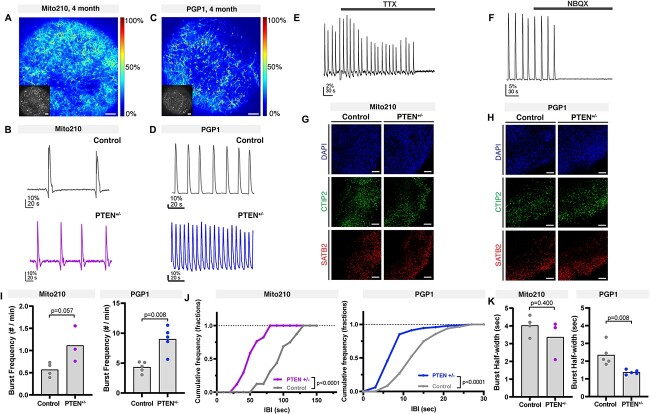
Developmental abnormalities in PTEN mutant organoids converge on altered circuit activity. (A–D) Network bursting in cortical organoids. (A, C) Representative images of intact Mito210 (A) and PGP1 (C) control organoid at 4 months transduced with SomaGCaMP6f2, showing the ΔF/F signal at the peak of a network burst using a pseudocolored scale. Inset: Raw fluorescence image. Scale bar: 100 μm. (B, D) Representative population-averaged calcium transient for control (top) and heterozygous mutant (bottom) organoids generated from Mito210 (B) and PGP1 (D) donor lines. (E, F) Example population-averaged calcium transients in control PGP1 organoids upon bath application of 2 μM TTX (E) and 20 μM NBQX (F). (G, H) Immunohistochemistry for CTIP2 (green) and SATB2 (red) in control and PTEN heterozygous mutant organoids at 3 months, from both Mito210 (G) and PGP1 (H) cell lines. Scale bars: 100 μm. (I) Spontaneous network burst frequency. The dots show the average values per organoid, and the bars show the mean across all organoids. (J) Cumulative frequency distribution of inter-burst interval (IBI) for control and mutant organoids. (K) Spontaneous network burst duration. The dots show the average values per organoid, and the bars show the mean across all organoids. Min, minutes; sec, seconds.
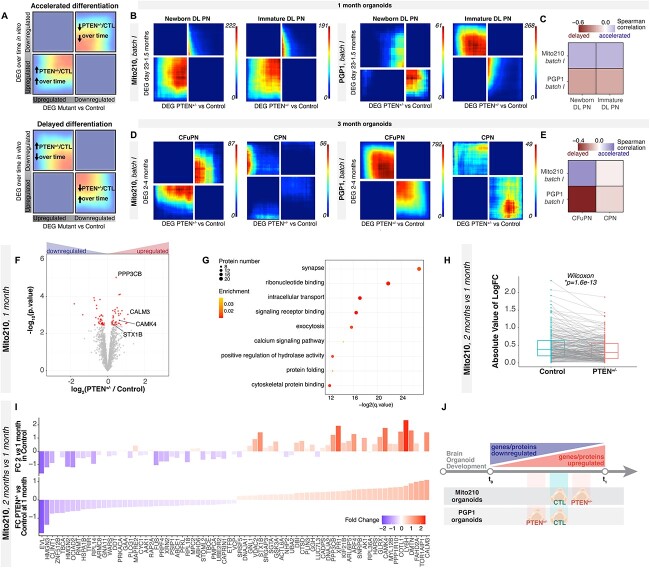
Given the broad expression and function of PTEN, we wondered if later developmental events could also be affected by heterozygosity of this gene. We hypothesized that, despite the lack of consistent quantitative differences in proportions of the various neuronal populations in PTEN mutant organoids at 1 and 3 months, projection neurons may display alterations in their gene expression profiles or developmental speed across genotypes, as we and others have previously reported for other ASD-associated genes (8–10,22). Therefore, we compared the genes affected in PTEN heterozygous organoids to genes that vary in expression over organoid development, to relate transcriptional changes induced by PTEN mutation to developmental progression. We first calculated the differentially expressed genes (DEGs) for each cell type between control and heterozygous mutant PTEN organoids at each time point (i.e. 1 and 3 months in vitro) (see Materials and Methods, Supplementary Material, Table S1). We then used an existing high-resolution developmental atlas of human cortical organoids we previously obtained with the same organoid differentiation protocol (15,17) to define gene changes associated with developmental time in each cell type. We calculated the genes that change over time in individual cell types across cortical organoid development, from all of the cell lines in that dataset, including the two lines used here (Mito210 and PGP1), as well as two others (HUES66 and GM08330), depending on time point. From this analysis, we obtained a list of genes that were upregulated or downregulated over time in specific cell types as cortical organoids develop. We then compared the mutation-induced changes (i.e. DEGs between PTEN heterozygous vs. control organoids in each cell type) at 1 month to the developmentally regulated genes from the atlas around this time point (i.e. DEGs that change over the period from day 23 to 1.5 months in control organoids in each cell type; Supplementary Material, Table S2). Similarly, the DEGs of PTEN mutant vs. control organoids at three months in each cell type were compared with temporally variable genes from the 2- to 4-month time period of the atlas for the corresponding types (Supplementary Material, Table S3). To compare these lists, we used rank–rank hypergeometric overlap (RRHO2) (23,24) (Fig. 2A–E and Supplementary Material, Fig. S4A and C), which allowed us to determine whether the gene sets altered by the PTEN mutation indicated an acceleration or deceleration of development for each cell type: an overlap between genes upregulated during normal organoid development and upregulated cell type–specific DEGs between mutant and control organoids would imply an accelerated development of a given cell population. Conversely, an overlap between genes downregulated during normal organoid development and upregulated cell-type specific DEGs between mutant and control organoids would imply a decelerated development of this cell population.
Figure 2.
Deep-layer neurons show asynchronous development across PTEN mutant organoids at multiple timepoints. (A) Schematic explaining the rank–rank hypergeometric overlap (RRHO2) output plot. Genes from each list are ordered from most upregulated to most downregulated, with the most upregulated gene of each differential gene list in the lower left corner. (B) RRHO2 plots comparing differentially expressed genes (DEG) between control and PTEN heterozygous mutant organoids at 1 month in vitro versus genes changing over time in organoids between 23 days and 1.5 months in vitro, in the newborn DLPN and immature DLPN populations. (C) Spearman correlation of DEG between each cortical cell type within control and PTEN mutant organoids at 1 month, and genes that change within that cell type from 23 days to 1.5 months in control organoids (17). Correlation is calculated on each gene’s signed logFC. (D) RRHO2 plots comparing differentially expressed genes between control and PTEN heterozygous mutant organoids at three month in vitro versus genes changing over time in organoids between two and four months in vitro, in the CFuPN and CPN populations. (E) Spearman correlation of DEG between each cortical cell type within control and PTEN heterozygous mutant organoids at 3 months, and genes that change within that cell type from 2 to 4 months in control organoids (17). Correlation is calculated on each gene’s signed logFC. (F) Volcano plot showing fold change versus FDR of measured proteins in MS experiments comparing PTEN heterozygous versus control organoids cultured for 35 days (n = 4 single organoids per genotype). Significant DEPs are shown in red (FDR < 0.1). Proteins mentioned in the text are highlighted. (G) Enriched GO terms for DEPs between PTEN heterozygous and control organoids cultured for 1 month in vitro. (H) Protein expression changes at 2 vs. 1 month for control and mutant organoids. Gray lines connect values for the same protein in the two genotypes. P-value from a paired signed Wilcoxon rank test. Only significant DEPs are shown; for the same analysis done with all detected proteins, see Supplementary Material, Figure 3. (I) Comparison of protein expression changes in PTEN heterozygous vs. control organoids at 1 month (bottom) versus changes in 2- vs. 1-month control organoids (top). Color and y-axis indicate log2 fold change. Only proteins with significantly differential expression between control and mutant at 1 month are shown (n = 3 single organoids per genotype for 2 months). (J) Schematic summarizing line-specific developmental acceleration/deceleration phenotypes from scRNA-seq and proteomics experiments. DL, deep layer; CPN, callosal projection neurons; CFuPN, corticofugal; CTL, control; FC, fold change.
Strikingly, heterozygous PTEN mutation induced differences in predicted developmental speed across many organoid cell types, more prominently at 3 months in vitro (Supplementary Material, Fig. S4A and C); that is, there was a significantly larger or smaller overlap between the genes up(down)regulated in each cell population in the PTEN heterozygous mutant with the genes up(down)regulated in the same cell population over time than was expected by chance. Notably, the direction of this effect differed between donors: while most of the Mito210 cell types displayed gene expression differences indicating accelerated development in both organoid batches, the PGP1 cell types were predominantly predicted to be delayed, suggesting that genomic context is an important modulator of the PTEN phenotype (Supplementary Material, Fig. S4). Importantly, not all cell types showed this effect, e.g. DEG in heterozygous cortical hem and CPN did not show a correlation with developmentally regulated genes, suggesting cell-type specificity of this phenotype (Supplementary Material, Fig. S4A and C). To confirm this result, we performed bulk RNA-seq profiling on single PTEN heterozygous and isogenic control PGP1 organoids from an independent differentiation batch at 1 month (Supplementary Material, Table S4). Consistent with the scRNA-seq results, DEGs upregulated in heterozygous relative to control organoids were correlated with genes that were downregulated in the organoid atlas between 23 days and 1.5 months, confirming a predicted developmental delay in PTEN heterozygous organoids (Supplementary Material, Fig. S4B).
Among projection neurons, in the 1-month comparison, deep-layer projection neurons (DLPNs) in the Mito210 heterozygous organoids showed gene expression changes suggesting accelerated differentiation in the mutant; in contrast, PGP1 heterozygous organoids indicated delayed differentiation of DLPNs at this stage (Fig. 2B and C). Effects on deep-layer neuron populations were still evident at 3 months: again, Mito210 corticofugal projection neurons (CFuPNs) appeared to be accelerated in their development, while PGP1 CFuPNs were delayed (Fig. 2D and E). This effect was seen in all differentiation batches, at both stages (Supplementary Material, Fig. S4A and C). Gene Ontology (GO) analysis of the genes that deep-layer neurons had in common with genes up(down)regulated over time pointed to changes in metabolic and RNA processing genes, as well as genes involved in forebrain development and synaptic signaling (Supplementary Material, Fig. S4D and E). Other neuronal populations such as CPN did not show significant association with developmental progression, indicating cell-type specificity among projection neurons. Thus, heterozygous PTEN mutation results in changes in predicted developmental timing in both the Mito210 and PGP1 donor cell lines, which is modulated by both cell type and genetic background.
To examine the molecular changes induced by PTEN mutation at the protein level, we performed proteomic analysis of control and heterozygous mutant organoids in the Mito210 line (Fig. 2F–J and Supplementary Material, Fig. S4F and G). Whole-proteome mass spectrometry of organoids at 1 month in culture (n = 4 single organoids per genotype) detected more than 4000 proteins, although PTEN could not be identified among them. Differential expression analysis revealed significant differential expression of 75 proteins between genotypes (FDR < 0.1, moderated t-test; Fig. 2F, Supplementary Material, Table S5). Gene set enrichment analysis (GSEA) identified ‘synapse’ as the most enriched biological process, along with exocytosis (e.g. STX1B) and the calcium signaling pathway (e.g. PPP3CB, CALM3 and CAMK4), pointing to a potential effect of the PTEN mutation on neuronal maturation (Fig. 2G). We also performed this whole-proteome analysis at a later time point, 2 months (n = 3 single organoids per genotype). While there were no differences in bulk protein abundance between mutant and control at 2 months (Supplementary Material, Table S5), when we examined the proteins that were differentially expressed between time points (2 months vs. 1 month) in each genotype, the mutant organoids showed significantly smaller expression changes over time than controls did (P = 1.6*10−13, Wilcoxon rank test, Fig. 2H and Supplementary Material, Fig. S4F), indicating that the mutant 1-month organoids resembled their older counterparts more closely than the stage-matched control organoids did. Furthermore, the mutation-driven changes (i.e. the set of proteins that were differentially expressed between genotypes in the 1-month organoids) were correlated with time-driven changes (i.e. the set of proteins that changed in both the control and mutant proteomes through time, 2 months vs. 1 month), indicating that heterozygous loss of function of PTEN affects the expression of developmentally regulated proteins (Pearson’s r = 0.76, P < 10−15, Fig. 2I and J and Supplementary Material, Fig. S4G). This result points to an accelerated development of Mito210 PTEN mutant organoids, consistent with the predicted developmental acceleration observed in the scRNA-seq data from this line.
In humans, different types of mutations and dosages of PTEN result in different diseases: heterozygous loss-of-function of PTEN has been associated with ASD (25–27), while both heterozygous and homozygous PTEN loss-of-function has been associated with tumor syndromes (28–30). We therefore investigated whether different dosages of PTEN may also result in different phenotypes in brain organoids. To test this, we generated homozygous PTEN mutations in the PGP1 parental cell line, using the same gRNA to target both alleles of the gene (see Materials and Methods, Supplementary Material, Fig. S1A). We then generated cortical organoids with the same protocol as before (Supplementary Material, Fig. S5A). Western blot analysis performed in the homozygous line showed a complete absence of PTEN, accompanied by increased phosphorylation of the downstream effectors AKT and ERK (MAPK1), which was much more pronounced than in the heterozygous line (Supplementary Material, Fig. S1C).
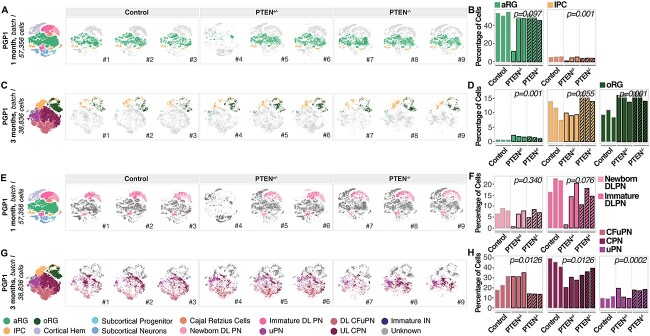
Profiling of 19 944 single cells by scRNA-seq at 1 month (Fig. 3A, Supplementary Material, Fig. S5B) showed that among the cortical cell types, the only significant change in cell type composition was a moderate decrease of IPC in the PTEN homozygous mutant compared with control organoids (n = 3 single organoids per genotype, FDR = 0.001, logistic mixed models; Fig. 3B and Supplementary Material, Fig. S5C and D). Analysis of 11 337 single cells profiled from 3-month-old homozygous mutant PGP1 organoids (Fig. 3C and Supplementary Material, Fig. S5E) revealed an increase in the proportion of oRG (FDR = 0.001), as previously observed in another organoid model grown from PTEN homozygous mutant iPSC (12), and of aRG (FDR = 0.001); these two populations were also increased in the heterozygous mutant (Fig. 3D and Supplementary Material, Fig. S5F and G). This indicates that overproduction of the oRG population is a consistent outcome of reduced PTEN, in both homozygous and heterozygous settings, independent of genetic background. However, we observed differences between the heterozygous and homozygous mutants; assignment of cells to phases of the cell cycle based on expression of related genes revealed a decrease in the proportion of proliferating oRG (i.e. oRG cells assigned to the S/G2/M phases of the cell cycle) (FDR = 3.12*10−15, Supplementary Material, Fig. S5H). In addition, the proportions of neuronal populations showed different effects in the PTEN homozygous mutant at 3 months (Fig. 3C-D and 3G-H). Collectively, the data indicate that gene dosage affects the phenotypic manifestation of PTEN mutations (31), underscoring the importance of studying this gene in the heterozygous state, similar to that found in ASD patients.
Figure 3.
Differences in PTEN dosage result in convergent progenitor phenotypes, but divergent projection neurons abnormalities. (A) Left: t-SNE plot of scRNA-seq data, color-coded by cell type, from 1-month PGP1 control, PTEN heterozygous and PTEN homozygous mutant organoids (n = 3 single organoids per genotype). Right: t-SNE plots for control, PTEN heterozygous, and PTEN homozygous mutant individual organoids, with cell types of interest highlighted in color: aRG (light green), intermediate progenitors cells (IPC, yellow) and oRG (dark green). (B) Bar charts showing the percentage of cells for the highlighted cell populations in a, right, in each control, PTEN heterozygous, and PTEN homozygous mutant organoid at 1 month in vitro. FDRs for a difference in cell type proportions between control and mutant, based on logistic mixed models (see Materials and Methods) are shown. (C) Left: t-SNE plot of scRNA-seq data, color-coded by cell type, from 3-month PGP1 control, PTEN heterozygous and PTEN homozygous mutant organoids (n = 3 single organoids per genotype). Right: t-SNE plots for control, PTEN heterozygous, and PTEN homozygous mutant individual organoids, with cell types of interest highlighted in color: apical radial glia (aRG, light green), IPCs (yellow) and oRG (dark green). (D) Bar charts showing the percentage of cells for the highlighted cell populations in (C), right, in each control, PTEN heterozygous, and PTEN homozygous mutant organoid at 3 months in vitro. FDRs for a difference in cell type proportions between control and mutant, based on logistic mixed models (see Materials and Methods) are shown. (E) Left: t-SNE plot of scRNA-seq data, color-coded by cell type, from 1-month PGP1 control, PTEN heterozygous and PTEN homozygous mutant organoids (n = 3 single organoids per genotype). Right: t-SNE plots for control, PTEN heterozygous, and PTEN homozygous mutant individual organoids, with cell types of interest highlighted in color: newborn DLPNs (light pink), immature DLPNs (dark pink). (F) Bar charts showing the percentage of cells for the highlighted cell populations in (E), right, in each control, PTEN heterozygous, and PTEN homozygous mutant organoid at 1 month in vitro. FDRs for a difference in cell type proportions between control and mutant, based on logistic mixed models (see Materials and Methods) are shown. (G) Left: t-SNE plot of scRNA-seq data, color-coded by cell type, from 3-month PGP1 control, PTEN heterozygous and PTEN homozygous mutant organoids (n = 3 single organoids per genotype). Right: t-SNE plots for control, PTEN heterozygous, and PTEN homozygous mutant individual organoids, with cell types of interest highlighted in color: corticofugal projection neurons (CFuPNs, pink), callosal projection neurons (CPNs, burgundy), unspecified projection neurons (uPNs, purple). (H) Bar charts showing the percentage of cells for the highlighted cell populations in (G), right, in each control, PTEN heterozygous, and PTEN homozygous mutant organoid at 1 month in vitro. FDRs for a difference in cell type proportions between control and mutant, based on logistic mixed models (see Materials and Methods) are shown. aRG, apical radial glia; DL, deep layer; UL, upper layer; PN, projection neurons; oRG, outer radial glia; IPCs, intermediate progenitor cells; CPNs, callosal projection neurons; CFuPNs, corticofugal projection neurons; uPNs, unspecified PNs; INs, interneurons; CTL, control.
Correct function of the cortical local circuit depends on the timely integration and maturation of multiple neuronal subtypes. We hypothesized that the asynchronous development of specific neuronal populations seen in the PTEN heterozygous mutant organoids may affect the function of cortical local circuits. To test this, we performed calcium imaging analysis in intact preparations of 4-month-old heterozygous mutant and control organoids from both genetic backgrounds. We analyzed spontaneous neuronal activity by transducing organoids with adeno-associated viruses bearing the calcium indicator CAG::SomaGCaMP6f2 (32) (see Materials and Methods), and performing acute recordings of intracellular calcium dynamics (Fig. 4). Both control and mutant organoids displayed spontaneous activity (163 +/− 77 active cells from 183 +/− 79 total cells, per imaged section, Fig. 4A–D), with the main form of activity being network bursting (Fig. 4A–D). When organoids were treated with the voltage-gated sodium channel blocker tetrodotoxin (TTX), calcium transients were abolished, suggesting that activity was mediated by action potentials (Fig. 4E, Supplementary Material, Fig. S6A). To verify the participation of excitatory neurons (Fig. 4F–H), we treated organoids with an antagonist of non-NMDA glutamate receptors, 2,3-dioxo-6-nitro-1,2,3,4-tetrahydorbenzo[f]quinoxaline-7-sulfonamide (NBQX), which suppressed spontaneous activity, indicating that synchronized network bursting is driven by functional glutamatergic synapses (Supplementary Material, Fig. S6B).
PTEN heterozygous organoids showed an increased level of bursting activity compared with control, in both the Mito210 and PGP1 genetic backgrounds (Mito210 burst frequency P = 0.057, IBI cumulative frequency P < 0.0001, four controls vs. three heterozygous, Mann–Whitney test and Kolmogorov–Smirnov test, respectively; PGP1 burst frequency P = 0.008, IBI cumulative frequency P < 0.0001, n = 5 organoids per condition, Mann–Whitney test and Kolmogorov–Smirnov test, respectively, Fig. 4I and J). In addition, bursts had shorter duration in the heterozygous organoids compared with controls in the PGP1 background (P = 0.008, n = 5 organoids per condition, Mann–Whitney test), and Mito210 organoids followed a similar trend (Fig. 4K). Interestingly, control organoids in the two parental lines displayed a striking difference in the level of spontaneous activity (burst per minute: 0.57 ± 0.16, Mito210 vs. 4.33 ± 0.90, PGP1), suggesting that the degree of spontaneous neural activity may be an idiosyncratic trait that varies between donor lines. To test if there were differences in key developmental pathways at baseline levels, between the two lines, we used the scRNA-seq data to perform DEG analysis of control PGP1 versus Mito210, for each cell type made in the organoids (Supplementary Material, Table S6). When performing GO analysis, the strongest differences observed were related to metabolic pathways. These data also showed differences in terms related to neuronal development, such as axonogenesis, forebrain development and synapse organization (Supplementary Material, Fig. S6C and D). Despite the differences in baseline levels of spontaneous activity, both backgrounds showed the same relative phenotype in the mutant (Fig. 4I), suggesting that these differences in genetic background do not compensate for the mutant phenotype. We previously reported that bursting activity (burst frequency and burst half-width) is altered in organoids with a heterozygous mutation for a different ASD-associated gene, SUV420H1 (8). Interestingly, although both genes affect bursting behavior, they do so in opposite directions, suggesting that mutations in different ASD risk genes may affect cortical circuit activity in different ways. Taken together, the data support the hypothesis that early abnormalities in the development and maturation of specific populations of neurons converge on later defects in circuit physiology, resulting in higher-order phenotypes that are shared across both genetic backgrounds and individual ASD risk genes.
Discussion
Human brain organoids offer an unprecedented opportunity to investigate early phenotypic manifestations associated with risk genes for complex neurodevelopmental disorders, including ASD. Here, we investigated the function of PTEN, a gene previously associated with ASD (7,33,34), in early stages of human cortical development. We reveal effects of PTEN mutation involving different steps of development, including alteration of specific progenitor populations (oRG), asynchronous development of DLPNs, and abnormal circuit activity.
While an increase in oRG had been previously reported in human brain organoids with homozygous loss-of-function of PTEN (12), we also found an increase of oRG in PTEN heterozygous cortical organoids, which is important considering that heterozygous mutations are more commonly found in ASD patients (7). Of note, the oRG phenotype observed was different between cell lines: in the PGP1 genomic context, the phenotype was stronger than in the Mito210. Recent work also reported an increase in oRG in human brain organoids from an ASD patient cell line carrying a PTEN mutation. Interestingly, the same mutation in another genetic background did not lead to this oRG phenotype (35). Combined, these data point to differences in the expressivity of the oRG phenotype dependent on genomic context.
While the oRG phenotype agrees with previously reported results in human brain organoids (12,35), we found a previously undescribed effect of PTEN on human projection neuron development. Interestingly, while the progenitor phenotype is independent of PTEN heterozygosity or homozygosity, changes in neuronal proportion were seen primarily in the homozygous mutant. These results point to the importance of studying risk genes in contexts that recapitulate the gene dosage state found in patients.
Different human genomic contexts have been suggested to modulate the phenotypic expressivity of ASD risk genes in a mutation- and phenotype-dependent manner (8). Here, we further confirm the importance of investigating ASD risk mutations across different genomic contexts; we find that the same PTEN mutation affects the development of the same neuronal population in lines from different donors, but it does so in opposite directions in the two genetic backgrounds. In line with our results, recent work has also shown that, depending on genetic background, the same PTEN mutation can have opposite effects on neurogenesis and neuronal development (35). These results point toward mechanisms that may underlie the variability of clinical manifestations observed in human patients with mutation in the same risk gene. It is intriguing that, despite the divergent molecular and cellular phenotypes observed for different ASD risk genes and in different genetic backgrounds, these effects ultimately may converge on changes in local circuit activity. This supports the hypothesis that the effects of different ASD risk genes may converge on similar features of higher-order processes of circuit activity and function.
Materials and Methods
Pluripotent stem cell culture
All experiments involving human cells were performed according to ISSCR 2016 guidelines and approved by the Harvard University IRB and ESCRO committees. The Mito210 iPSC line was derived by B. Cohen laboratory (McLean Hospital), and the PGP1 iPSC line by G. Church (Harvard University) [Church, 2005 (36)]. Both Mito210 and PGP1 are human male iPSC lines from individuals with no known history of ASD or other psychiatric condition (Mito210 confirmed by structured psychiatric interview, PGP1 with publicly available records). Cell lines were cultured as previously described (15). All human iPSC cultures were maintained below passage 50, were negative for mycoplasma (assayed with MycoAlert PLUS Mycoplasma Detection Kit, Lonza) and karyotypically normal (G-banded karyotype test performed by WiCell Research Institute). The Mito210 line was authenticated by genotyping analysis (Fluidigm FPV5 chip) performed by the Broad Institute Genomics Platform (in 2017). The PGP1 line was authenticated using STR analysis completed by TRIPath (in 2018).
CRISPR guide design
The CRISPR guide for PTEN was designed using the Benchling CRISPR Guide Design Tool (Benchling Biology Software). The guide was designed to maximize on-target efficiency and minimize off-target sites in intragenic regions. It was chosen to induce a stop codon in the catalytic phosphatase domain of PTEN (gRNA: 5’-CCAAATTTAATTGCAGAGGT-3′ (AGG)).
CRISPR-mediated gene editing
The Mito210 PTEN edited line was generated by the Broad Institute Stem Cell Facility. The guide RNA and Cas9 (EnGen Cas9 NLS from New England Biolabs) were transfected by using the NEON transfection system (Thermo Fisher Scientific, 1050 V, 30 ms, two pulses and 2.5 × 105 cells).
The PGP1 PTEN mutant line was generated in collaboration with the Harvard Stem Cell Institute (HSCI) iPS Core Facility. Briefly, parental cells were transfected with the Neon system. For 100 000 cells, 6.25 pmol TrueCut TM Cas9 Protein v2 (Thermo Fisher Cat: A36496) and 12.5 pmol of sgRNA (Synthego) were used. Post transfection, the pools of cells were harvested to test knock-out efficiency. The best pool was then selected for low-density plating. A week later, colonies were picked and clones were screened by PCR and Sanger sequencing. Heterozygous and homozygous clones were expanded, and the genotypes were reconfirmed post expansion.
Sequence confirmation of edits
Insertions/deletions in individual clones were screened via PCR amplification using primers flanking the guide. The Mito210 PTEN mutant clone has a heterozygous 1 nt insertion. The PGP1 PTEN mutant clone has a heterozygous 1 nt insertion. The PGP1 PTEN homozygous clone has a 1 nt insertion in one allele and 11 nt insertion in the second allele. Information about edits in each cell line is listed in Supplementary Material, Table S7.
Organoid differentiation
Dorsally patterned forebrain organoids were generated as previously described (15,37). Briefly, on day 0, human iPSCs or ESCs were dissociated to single cells with Accutase (Gibco), and 9000 cells per well were reaggregated in ultra-low cell-adhesion 96-well plates with V-bottomed conical wells (sBio PrimeSurface plate; Sumitomo Bakelite) in Cortical Differentiation Medium (CDM) I, containing Glasgow-MEM (Gibco), 20% Knockout Serum Replacement (Gibco), 0.1 mM Minimum Essential Medium non-essential amino acids (MEM-NEAA) (Gibco), 1 mM pyruvate (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), 100 U/ml penicillin (ThermoFisher) and 100 μg/ml streptomycin (Corning). For the PGP1 line, cells were plated in the same pluripotent medium in which they were maintained for 1 day to better enable embryoid body formation. From days 0–6, ROCK inhibitor Y-27632 (Millipore) was added (final concentration of 20 μM). From day 0/1–18, Wnt inhibitor IWR1 (Calbiochem) and TGFβ inhibitor SB431542 (Stem Cell Technologies) were added (final concentration of 3 and 5 μM, respectively). From day 18, the aggregates were cultured in ultra-low attachment culture dishes (Corning) under orbital agitation in CDM II, containing DMEM/F12 medium (Gibco), 2 mM Glutamax (Gibco), 1% N2 (Gibco), 1% Chemically Defined Lipid Concentrate (Gibco), 0.25 μg/ml fungizone (Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin. On day 35, cell aggregates were transferred to spinner-flask bioreactors (Corning) and maintained in CDM III (CDM II supplemented with 10% fetal bovine serum (FBS) (GE-Healthcare), 5 μg/ml heparin (Sigma) and 1% Matrigel (Corning)). From day 70, organoids were cultured in CDM IV (CDM III supplemented with a B27 supplement (Gibco) and 2% Matrigel).
Immunohistochemistry
Samples were fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Services) for either 30 min (1-month-old organoids) or 1–3 h (2-month and older organoids). Samples were washed with 1X phosphate-buffered saline (PBS) (Gibco), cryoprotected in a 30% sucrose solution overnight at 4°C, embedded in optimum cutting temperature (OCT) compound (Tissue Tek) and cryosectioned at 14–18 μm thickness. Sections were washed with 0.1% Tween-20 (Sigma) in PBS, blocked for 1 h at room temperature (RT) with 6% donkey serum (Sigma) + 0.3% Triton X-100 (Sigma) in PBS and incubated with primary antibodies overnight diluted with 2.5% donkey serum +0.1% Triton X-100 in PBS. After washing, sections were incubated at RT with secondary antibodies diluted in the same solution as with primary antibodies (1:1000–1:1200; Supplementary Material, Table S8) for 2 h at RT or overnight at 4°C, washed, and incubated with DAPI staining (1:10 000 in PBS + 0.1% Tween-20) for 15 min to visualize cell nuclei. Slices were mounted using Fluoromount-G (Invitrogen).
Microscopy
Immunofluorescence images were acquired with the Lionheart™ FX Automated Microscope (BioTek Instruments), or with an Axio Imager.Z2 (Zeiss), and analyzed with the Gen5 (BioTek Instruments) or Zen Blue (Zeiss) image processing software.
Immunohistochemistry quantifications
Images were acquired with an Axio Imager.Z2 (Zeiss) microscope. SOX2+ nuclei were quantified across 45 regions of interest (ROIs, 195 μm × 195 μm) for PGP1 control and 54 ROIs for PGP1 PTEN heterozygous mutant organoids, from two different sections, seven organoids from two different differentiations batches per condition, and across 18 ROIs for Mito210 control and 24 ROIs for Mito210 PTEN heterozygous mutant organoids, from two different sections, from three WT organoids and four heterozygous organoids, from one differentiation batch. Images were analyzed using the ImageJ software.
Western blotting
Proteins were extracted from iPSCs using an N-PER™ Neuronal Protein Extraction Reagent (Thermo Fisher Scientific) supplemented with protease (cOmplete™ Mini Protease Inhibitor Cocktail, Roche) and phosphatase inhibitor (PhosSTOP, Sigma). Lysates were centrifuged for 10 min at 13 500 rpm at 4°C. Supernatants were transferred to new tubes. Protein concentration was quantified using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Fifteen to twenty micrograms of protein lysates were separated on a NuPAGE™ 4-12%, Bis-Tris Gel (Invitrogen) or Mini-PROTEAN 4–15% Gels (Bio-Rad) and transferred onto a PVDF membrane. Blots were blocked in 5% non-fat dry milk (Bio-Rad) and incubated with primary antibodies overnight (Supplementary Material, Table S8). Afterward, the blots were washed and incubated at RT with secondary horseradish peroxidase–conjugated antibodies (Abcam) for 1 h. Blots were developed using SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) or ECL™ Prime Western Blotting System (Millipore), and ChemiDoc System (Bio-Rad).
Dissociation of brain organoids and scRNA-seq
Individual brain organoids were dissociated into a single-cell suspension using the Worthington Papain Dissociation System kit (Worthington Biochemical). Detailed descriptions of the dissociation protocol have been published, with adaptations depending on age and size (37,38). We resuspended dissociated cells in ice-cold PBS containing 0.04% BSA (Sigma, PN-B8667), counted them with Cellometer K2 (Nexcelom Bioscience). Cells were loaded onto a Chromium™ Single Cell B or G Chip (10x Genomics, PN-1000153, PN-1000120), and processed through the Chromium Controller to generate single cell GEMs (Gel Beads in Emulsion). scRNA-seq libraries were generated with the Chromium™ Single Cell 3’ Library & Gel Bead Kit v3 or v3.1 (10x Genomics, PN-1000075, PN-1000121, Supplementary Material, Table S9). We pooled libraries from different samples based on molar concentrations and sequenced them on a NextSeq 500 or NovaSeq instrument (Illumina) with 28 bases for read 1, 55 bases for read 2 and 8 bases for Index 1. If necessary, after the first round of sequencing, we repooled libraries based on the actual number of cells in each and resequenced with the goal of producing ~20 000 reads per cell for each sample.
scRNA-seq data analysis
Reads from scRNA-seq were aligned to the GRCh38 human reference genome, and cell-by-gene count matrices were produced with the Cell Ranger pipeline (v3.0.2, 10x Genomics) (39). Default parameters were used, except for the ‘–cells’ argument. In Mito210 1-month organoids, a control organoid was excluded because not enough cells could be recovered from that 10x channel. Data were analyzed using the Seurat R package v3.2.2 (40) using R v3.6. Cell profiles expressing at least 500 genes were kept, and UMI counts were normalized for each cell by the total expression, multiplied by 106, and log-transformed. Variable genes were found using the ‘mean.var.plot’ method, and the ScaleData function was used to regress out variation owing to differences in total UMIs per cell. Principal component analysis (PCA) was performed on the scaled data for the variable genes, and top principal components were chosen based on Seurat’s ElbowPlot (at least 15 PCs were used in all cases). Cells were clustered in PCA space using Seurat’s FindNeighbors on top principal components with default parameters, followed by FindClusters with resolution = 1.0. Variation in the cells was visualized by t-SNE (t-distributed stochastic neighbor embedding) on the top principal components.
For each dataset, upregulated genes in each cluster were identified using the VeniceMarker tool from the Signac package v0.0.7 from BioTuring (https://github.com/bioturing/signac). Cell types were manually assigned to each cluster by looking at the top most significant upregulated genes and comparing them to canonical markers used in literature to identify different cell types and to reference gene lists established in (17). In a few cases, clusters were further subclustered to assign identities at higher resolution. Specifically,
Mito210 1 month, cluster 18 was split into subcortical neurons and Cajal Retzius.
Mito210 3 months batch 1, cluster 5 was split into CPN and unspecified PN, clusters 9 and 12 were each split into CFuPN and CPN, clusters 13 and 14 were each split into oRG and IP and cluster 12 was split into CFuPN and CPN.
Mito210 3 months batch 2, clusters 4, 9 and 10 were each split into CPN and CFuPN, and clusters 15 and 16 were each split into the oRG and IP.
PGP1 3 months, clusters 13 and 15 were each split into oRG and IP, clusters 11 and 17 into CPN and CFuPN and cluster 10 into PN and aRG.
See metadata on Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP1964), or Seurat Objects in Synapse, to view cluster numbers.
To compare cell type proportions between control and mutant organoids, for each cell type present in a dataset, the glmer function from the R package lme4 v1.1-23 (41) was used to estimate a mixed-effect logistic regression model. The output was a binary indicator of whether cells belong to this cell type, the control or mutant state of the cell was a fixed predictor, and the organoid that the cell belonged to was a random intercept. Another model was fit without the control-versus-mutant predictor, and the anova function from the stats core R package v3.6.3 was used to compare the two model fits. The FDR was calculated with the Benjamini–Hochberg approach (42).
To compare the proportion of proliferating oRG between conditions, Seurat’s CellCycleScoring function was used with default parameters, and any cells with an S score or G2M score above 0.5 was marked as proliferating. The significance of any difference in proportion was evaluated using a mixed-effect logistic regression model as before.
To assess PTEN signaling–associated pathway expression genes belonging to the PI3K-AKT signaling, p53 signaling, mTOR signaling and phosphatidylinositol signaling pathways were downloaded from Kyoto Encyclopedia of Genes and Genomes (KEGG) (43), and genes belonging to the apoptosis pathway were downloaded from MSigDB (44). For each of these gene sets, module scores were calculated using Seurat’s AddModuleScore function. Violin plots were created using the module scores from cells in each cell type, and the average module score per individual organoid is plotted as a point. Adjusted P-values of differential module scores were calculated using a mixed model in order to take organoid-to-organoid variability into account (17), but no pathways were found to have significant differences based on PTEN status in any cell type.
Differential expression analysis
Differential genes between control and mutant organoids and between cell lines were assessed by subsetting each dataset into cells belonging to each cell type. Reads were then summed across cells of each type in each organoid (to create a pseudobulk per cell type). Genes with <10 total reads were excluded, and DESeq2 v1.26 (45) was used to calculate DEGs, with each organoid as a sample (45). GO analysis was performed with the clusterProfiler R package (46), using the enrichGO function and the compareCluster function to highlight processes the gene sets might have in common. For enrichGO, the Biological Process Ontology was used, and the lists were then simplified as in clusterProfiler with a cutoff of 0.5.
Comparison of DEGs between control and mutant organoids to genes changing over time in organoid development
Organoid DEG signatures (mentioned before) were compared with genes changing in control organoids over time. To calculate the latter list, data from (17) were utilized (many of the control organoids from this paper are also included in that dataset, see Supplementary Material, Table S9). As in that paper, data from each time point (23 days, 1, 1.5, 2, 3 and 4 months) across all cell lines and organoids used in that paper were corrected for ambient RNA expression using the decontX function from the Celda R package, version 1.6.1 (47), with each organoid treated as its own sequencing batch. For each cell type, DESeq2 was used to identify genes that are differentially expressed across organoid age, in days. Two lists were made: one using data from 23 days and 1 and 1.5 months, to compare to the PTEN organoids at 1 month, and a second DEG list from the 2-, 3- and 4-month timepoints, to compare to the PTEN organoids at 3 months.
DEG lists from control vs. PTEN mutant organoids were compared with DEG lists across time using two methods. In both cases, DEG lists were ranked by degree of differentiation [−log10(P-value) * sign(effect)] and then compared. First, they were compared using the improved rank–rank hypergeometric overlap test (RRHO2 R package v1.0) (23,24). Second, they were compared using a Spearman correlation via the cor.test function from the stats core R package v3.6.3, and the rho statistic was reported.
Slide-seqV2
For Slide-seqV2, Mito210 organoids at 2 months were embedded in OCT (Tissue Tek) and cryosectioned at 10 μm thickness. Sections were transferred to a custom-made array of densely packed barcoded beads (termed ‘pucks’) for Slide-seqV2 experiments. Library construction was performed as described (48). Briefly, first-strand synthesis was performed by incubating the puck with tissue sections in the reverse-transcription solution followed by tissue digestion, second-strand synthesis, library amplification and purification. The Slide-seqV2 libraries were sequenced on a NovaSeq with an estimated ~100 million reads per puck. See Supplementary Material, Table S9 for sequencing information. The data pertaining to control organoids have been reported previously (17).
Slide-seq V2 data analysis
Sequencing reads were aligned to the GRCh38 human reference genome and analyzed by the Slide-seq pipeline (https://github.com/MacoskoLab/slideseq-tools) as previously described (48). Since organoid sizes were much smaller than the Slide-seq pucks, images were manually cropped to the edges of the organoids and beads outside of the images were excluded.
Cell type decomposition was performed by robust cell type decomposition (RCTD) (49). Briefly, cell type profiles learned from control organoid scRNA-seq data corresponding to the same time point and cell line (17) were used to decompose mixtures from the Slide-seqV2 data. First, for each timepoint, a reference was made by using the age-matched scRNA-seq dataset. Next, the RTCD package fit a statistical model to estimate the mixture and cell type identities at each bead of the Slide-seqV2 data. We restricted our analysis to beads with more than 200 UMIs. Finally, gene set expression of the top 50 genes from each cell type signature as defined in (17) was visualized by plotting 100× the sum of the reads for all genes in the gene set that appeared in the Slide-seqV2 data, normalized by the number of UMI for each bead.
Bulk RNA-seq
For bulk RNA-seq, 1-month PGP1 PTEN organoids (n = 3 separate organoids for heterozygous mutant and isogenic control) were used. We extracted total RNA using Quick-RNA Mini Prep (Zymo Research) and followed the vendor protocol with a DNase treatment step. We constructed RNA-seq libraries using the Smart-seq2 protocol (50), with minor modifications as follows: (1) we used 2 ng total RNA as input, and (2) we used 0.1 ng cDNA as input and made the NexteraXT (Illumina) libraries using half of the standard volume. We pooled libraries from different samples based on molar concentration and sequenced them on a Nextseq500 instrument (Illumina) with 50 bases for read 1, 25 bases for read 2 and 8 bases each for Index 1 and 2. We performed the same experiment with Mito210 mutant and control organoids; however, the number of DEGs between mutant and control was not sufficient to allow the RRHO2 analysis. Therefore, we did not proceed with the analysis of the Mito210 experiment.
For comparison of DEGs between control and mutant organoids to genes changing over time in organoid development, binary base call (BCL) files were converted to FASTQ files using the bcl2fastq (RRID:SCR_015058) package version 2.20.0. The reads were then aligned to human GRCh38 reference, and the genes were quantified through the RSEM (51) version 1.3.0 pipeline using the STAR (52) version 2.7.10a aligner (using the following parameters: —star, —star-gzipped-read-file, —num-threads 4, −paired-end). DEGs between control and the PTEN heterozygous mutant were identified using R (v4.0.3) package DESeq2 (v1.30.1) (53). Gene quantification results were loaded with the tximport function (tximport package v1.18.0) (54) and converted to DESeq data (DESeqDataSetFromTximport function from DESeq2). A negative binomial Wald test was applied to find DEG between the two conditions (DESeq function with the default settings). To compare whether the obtained DEGs show concordance with accelerated or delayed differentiation, we identified DEGs that vary in expression across time, using all cells in a pseudobulk, and used RRHO2 to compare DEG lists, as in the section Comparison of Differentially Expressed Genes Between Control and Mutant Organoids to Genes Changing Over Time in Organoid Development.
Cell lysis and filter-aided sample preparation digestion for mass spectrometry
For 1-month (35 d.i.v.) and 2-month (70 d.i.v.) Mito210 PTEN organoids, four heterozygous mutant and four control and three heterozygous mutant and three control organoids were used, respectively. Cells were placed into microTUBE-15 (Covaris) microtubes with TPP buffer (Covaris) and lysed using a Covaris S220 Focused-ultrasonicator instrument with 125 W power over 180 s, at 10% max peak power. Upon precipitation with chloroform/methanol, extracted proteins were weighed and digested according to the filter-aided sample preparation (FASP) protocol (100 μg). Briefly, the 10 K filter was washed with 100 μl of triethylammonium bicarbonate (TEAB). Each sample was added and centrifuged, and the supernatant was discarded. Then, 100 μl of Tris(2-carboxyethyl) phosphine (TCEP) 37 was added for 1 h and centrifuged, and the supernatant was discarded. While shielding from light, 100 μl IAcNH2 was added for 1 h followed by centrifugation, and the supernatant was discarded. Next, 150 μl 50 mM TEAB + Sequencing Grade Trypsin (Promega) was added and left overnight at 38°C, upon which the samples were centrifuged and supernatant collected. Lastly, 50 μl 50 mM TEAB was added to the samples, followed by spinning and supernatant collection. The samples were then transferred to HPLC.
Tandem mass tag mass tagging protocol peptide labeling
The tandem mass tag (TMT) label reagents were equilibrated to RT and resuspended in anhydrous acetonitrile or ethanol (for the 0.8 mg vials, 41 μl were added; for the 5 mg vials, 256 μl were added). The reagent was dissolved for 5 min with occasional vortexing. A TMT Label Reagent (41 μl, equivalent to 0.8 mg) was added to each 100–150 μg sample. The reaction was incubated for 1 h at RT. Reaction was quenched by adding 8 μl of 5% hydroxylamine to the sample and incubating for 15 min. Samples were combined, dried in a SpeedVac (Eppendorf) and stored at −80°C.
Hi-pH separation and mass spectrometry analysis
Before submission to LC–MS/MS (liquid chromatography with tandem mass spectrometry), each experiment sample was separated on a Hi-pH column (Thermo Fisher Scientific) according to the vendor’s instructions. After separation into 40 fractions, each fraction was submitted for a single LC–MS/MS experiment, performed on a Lumos Tribrid (Thermo Fisher Scientific) equipped with a 3000 Ultima Dual nanoHPLC pump (Thermo Fisher Scientific). Peptides were separated onto a 150 μm inner-diameter microcapillary trapping column, packed first with ~3 cm of C18 Reprosil resin (5 μm, 100 Å, Dr Maisch GmbH) followed by a PharmaFluidics micropack analytical column 50 cm. Separation was achieved through applying a gradient from 5 to 27% ACN in 0.1% formic acid over 90 min at 200 nl per minute. Electrospray ionization was enabled through applying a voltage of 1.8 kV using a homemade electrode junction at the end of the microcapillary column and sprayed from stainless-steel tips (PepSep). The Lumos Orbitrap was operated in a data-dependent mode for the MS methods. The MS survey scan was performed in the Orbitrap in the range of 400–1800 m/z at a resolution of 6 × 104, followed by the selection of the 20 most intense ions (TOP20) for CID-MS2 fragmentation in the Ion trap using a precursor isolation width window of 2 m/z, an AGC setting of 10 000, and a maximum ion accumulation of 50 ms. Singly charged ion species were not subjected to CID fragmentation. Normalized collision energy was set to 35 V and an activation time of 10 ms. Ions in a 10 ppm m/z window around ions selected for MS2 were excluded from further selection for fragmentation for 90 s. The same TOP20 ions were subjected to an HCD MS2 event in the Orbitrap part of the instrument. The fragment ion isolation width was set to 0.8 m/z, AGC was set to 50 000, the maximum ion time was 150 ms, normalized collision energy was set to 34 V with an activation time of 1 ms for each HCD MS2 scan.
Mass spectrometry data generation
Raw data were submitted for analysis in Proteome Discoverer 2.4 (Thermo Fisher Scientific) software. Assignment of MS/MS spectra was performed using the Sequest HT algorithm by searching the data against a protein sequence database including all entries from the Human Uniprot database (55,56) and other known contaminants such as human keratins and common laboratory contaminants. Sequest HT searches were performed using a 10 ppm precursor ion tolerance and requiring each peptide’s N-/C-termini to adhere with trypsin protease specificity, while allowing up to two missed cleavages. 16-plex TMTpro tags on peptide N-termini and lysine residues (+304.207 Da) was set as static modifications, while methionine oxidation (+15.99492 Da) was set as variable modification. A MS2 spectra assignment FDR of 1% on the protein level was achieved by applying the target-decoy database search. Filtering was performed using Percolator (64 bit version) (57). For quantification, a 0.02 m/z window centered on the theoretical m/z value of each of the six reporter ions and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were exported in the result file of Proteome Discoverer 2.4 search engine as Excel tables. The total signal intensity across all peptides quantified was summed for each TMT channel, and all intensity values were normalized to account for potentially uneven TMT labeling and/or sample handling variance for each labeled channel.
Mass spectrometry data analysis
Potential contaminants were filtered out and proteins supported by at least two unique peptides were used for further analysis. We kept proteins that were missing in at most one sample per condition. Data were transformed and normalized using variance-stabilizing normalization using the DEP package of Bioconductor (58). To perform statistical analysis, data were imputed for missing values using random draws from a Gaussian distribution with width 0.3 and a mean that is down-shifted from the sample mean by 1.8. To detect statistically significant differential protein abundance between conditions, we performed a moderated t-test using the LIMMA package of Bioconductor (59), employing an FDR threshold of 0.1. GSEA was performed using the GSEA software (44). GO and KEGG pathway annotation was utilized to perform functional annotation of the significantly regulated proteins. GO terms and KEGG pathways with FDR q-values < 0.05 were considered statistically significant. Correlation between the mutant effect (e.g. PTEN+/− vs. control at 1 month) and the time effect (e.g. 2 vs. 1 month in control) was calculated using Pearson correlation. For PTEN 2 vs. 1 month, changes in protein levels in heterozygous and control organoids were compared with one another with a signed paired Wilcoxon rank test, using stat_compare_means from the ggpubr R package (https://rpkgs.datanovia.com/ggpubr/).
Calcium imaging
Organoids were transduced with pAAV-CAG-SomaGCaMP6f2 (Addgene, #158757) by pipetting 0.2 μl of stock virus to 500 μl of Cortical Differentiation Medium IV (CDM IV, without Matrigel) in 1 well of a 24-well dish containing a single organoid. On the next day, each organoid was transferred to a 6-well plate filled with 2 ml of fresh medium. On the third day after transduction, organoids were transferred to low-attachment 10 cm plates, and on the seventh day, medium was switched to BrainPhys (5790 STEMCELL Technologies) supplemented with 1% N2 (17502-048 Thermo Fisher), 1% B27 (17 504 044 Thermo Fisher), GDNF (20 ng/ml, Cat. No. 78139 STEMCELL Technologies), BDNF (20 ng/ml, 450-02 Peprotech), cAMP (1 mM, 100-0244 Stemcell Technologies), ascorbic acid (200 nM, Cat. No. 72132 STEMCELL Technologies) and laminin (1 μg/ml, 23 017 015 Life Technologies). Organoids were cultured in BrainPhys for at least 2 weeks before imaging.
Brain organoids were randomly selected and transferred to a recording chamber containing BrainPhys. Imaging was performed using a confocal scanner (CSU-W1, Andor confocal unit attached on an inverted microscope [Ti-Eclipse, Nikon]), while the organoids were kept at 37°C using a heating platform and a controller (TC-324C, Warner Instruments). We used a 10× objective (Plan Apo λ, 10×/0.45), resulting in a field of view of 1.3 × 1.3 mm2 and a pixel size of 0.6 μm. Imaging took place in fast-time-lapse mode, with an exposure time of 100 ms, resulting in an acquisition rate of ~8 frames/s. Spontaneous activity was recorded in three different z-planes, for at least 22 min of baseline activity in total (with no pharmacology treatment).
Stock solutions of 2,3-dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f] quinoxaline-7-sulfonamide disodium salt (NBQX disodium salt, Abcam; 100 mM) and tetrodotoxin citrate (TTX, Abcam; 10 mM) were prepared in ddH2O. Bath application of NBQX (antagonist of AMPA/kainate glutamate receptors) and TTX (voltage-gated sodium-channel antagonist) was applied to achieve a final bath concentration of 20 and 2 μM, respectively.
Data were converted from nd2 format to tiff, and automated motion correction and cell segmentation were performed using Suite2p (60), followed by manual curation of segmented cells (we examined the spatial footprint and temporal characteristics of each candidate cell, as well as manually adding neurons with clear cell-body morphology). Then, mean raw fluorescence for each cell was measured as a function of time.
Analysis of calcium imaging data
Analysis was done using in-house MATLAB scripts. Raw calcium signals for each cell, F(t), were converted to represent changes from baseline level, ΔF/F(t), defined as (F(t) − Fo(t))/Fo(t). The time-varying baseline fluorescence, Fo(t), for each cell was a smoothed fluorescence trace obtained after applying a 10-second-order median filter centered at t. Calcium events elicited by action potentials were detected based on a threshold value given by their peak amplitude (5 times the standard deviation of the noise value) and their first time derivative (2.5 times the standard deviation of the noise value).
The analysis of network bursting was performed based on the population-averaged calcium signal along all segmented cells. A peak in the population signal was considered a network burst if it met the following criteria: (1) the peak amplitude was >10 times the standard deviation of the noise value, (2) a set of bursting cells composed of at least 20% of total cells were active during that population calcium transient and (3) a cell was considered part of the set of bursting cells only if it participated in at least 50% of the network bursts.
The peaks of the network bursts were used to measure the inter-burst interval (IBI), and the frequency was obtained from the average IBI. The burst half-width was also measured from the population-averaged calcium signal by calculating the width of the transient at 50% of the burst peak amplitude.
Supplementary Material
Acknowledgements
We thank J. R. Brown for editing the manuscript, and the entire Arlotta Laboratory for support and insightful discussions; V. Vuong, C. Abbate and S.N. Smith for technical support in organoid culture and characterization; N. Haywood for scRNA-seq experiments; A. Shetty for help with scRNA-seq cell type classifications; S. Simmons for revising the manuscript; the Broad Genomics Platform for sequencing; the Cohen laboratory for the Mito 210 line; L.M. Daheron at the Harvard Stem Cell Facility for expanding edited lines and generating the PGP1 PTEN mutant lines; A. Podury for helping to establish the calcium imaging protocols; and B. Budnik at the Harvard Center for Mass Spectrometry for conducting proteomics experiments.
Contributor Information
Martina Pigoni, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Ana Uzquiano, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Bruna Paulsen, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Amanda J Kedaigle, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Sung Min Yang, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Panagiotis Symvoulidis, McGovern Institute for Brain Research, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA.
Xian Adiconis, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Silvia Velasco, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Rafaela Sartore, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Kwanho Kim, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Ashley Tucewicz, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Sarah Yoshimi Tropp, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA.
Kalliopi Tsafou, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Xin Jin, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Society of Fellows, Harvard University, Cambridge, MA 02138, USA.
Lindy Barrett, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Fei Chen, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Edward S Boyden, McGovern Institute for Brain Research, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA; MIT Center for Neurobiological Engineering, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA; Harvard-MIT Health Sciences & Technology Program (HST), Harvard Medical School, Boston, MA 02115, USA; Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA; Howard Hughes Medical Institute, MIT, Cambridge, MA 02138, USA; Department of Brain of Cognitive Sciences, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA; Department of Media Arts and Sciences, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA; Department of Biological Engineering, Massachusetts Institute of Technology (MIT), Cambridge, MA 02139, USA.
Aviv Regev, Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Joshua Z Levin, Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Paola Arlotta, Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA 02138, USA; Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Funding
This work was supported by grants from the Chan Zuckerberg Initiative to P.A., the Stanley Center for Psychiatric Research to P.A. and J.Z.L., the Broad Institute of MIT and Harvard to P.A. and J.Z.L., the National Institutes of Health (R01MH112940 to P.A. and J.Z.L., and P50MH094271, U01MH115727 and RF1MH123977 to P.A.), the Klarman Cell Observatory to J.Z.L. and A.R. and the Howard Hughes Medical Institute to A.R. A.R. was a Howard Hughes Medical Institute and a Koch Institute extramural member while conducting this study.
Conflict of Interest statement: P.A. is a SAB member at Herophilus, Rumi Therapeutics and Foresite Labs and is a co-founder of Vesalius. A.R. is a founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics and, until August 31, 2020, was a SAB member of Syros Pharmaceuticals, Neogene Therapeutics, Asimov and Thermo Fisher Scientific. From August 1, 2020, A.R. is an employee of Genentech and has equity in Roche. From September 1, 2021, M.P. is an employee of Roche.
Authors’ Contributions
P.A., M.P., A.U. and B.P. conceived the experiments. M.P., R.S. and A.T. generated, cultured and characterized all organoids, and P.A. supervised their work. A.U., A.T. and S.Y.T. performed immunohistochemistry experiments. X.A. performed bulk and scRNA-seq experiments with help from M.P., A.U., R.S. and S.V.; X.J. performed Slide-seq experiments, with the supervision of F.C. A.J.K. and J.Z.L. performed scRNA-seq analysis, and J.Z.L. and A.R. supervised the work. K.K. performed the bulk RNA-seq analysis. M.P., A.U., B.P., S.V. and A.J.K. worked on cell type assignments and data analysis. K.T., M.P. and A.J.K. performed proteomics analysis, supervised by K.L.; S.M.Y. and P.S. performed the calcium imaging experiments and analysis, supervised by E.S.B. and with the help of R.S. L.B. generated the Mito210 PTEN mutant line; P.A., M.P., A.U., B.P. and A.J.K. wrote the manuscript with contributions from all authors. All authors read and approved the final manuscript.
Data Availability
Count and meta data from scRNA-seq and Slide-seq will be available on the Single Cell Portal, at https://singlecell.broadinstitute.org/single_cell/study/SCP1964. Raw data, proteomics and bulk RNA-seq data will be deposited into a restricted access database before publication. Code used during data analysis will be available at https://github.com/AmandaKedaigle/PTENBrainOrganoids.
References
- 1. Lord, C., Brugha, T.S., Charman, T., Cusack, J., Dumas, G., Frazier, T., Jones, E.J.H., Jones, R.M., Pickles, A., State, M.W. et al. (2020) Autism spectrum disorder. Nat. Rev. Dis. Primers, 6, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geschwind, D.H. and Levitt, P. (2007) Autism spectrum disorders: developmental disconnection syndromes. Autism spectrum disorders: developmental disconnection syndromes. Curr. Opin. Neurobiol., 17, 103–111. [DOI] [PubMed] [Google Scholar]
- 3. Grove, J., Ripke, S., Als, T.D., Mattheisen, M., Walters, R.K., Won, H., Pallesen, J., Agerbo, E., Andreassen, O.A., Anney, R. et al. (2019) Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet., 51, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rosenberg, R.E., Law, J.K., Yenokyan, G., McGready, J., Kaufmann, W.E. and Law, P.A. (2009) Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. Pediatr. Adolesc. Med., 163, 907–914. [DOI] [PubMed] [Google Scholar]
- 5. Ruzzo, E.K., Pérez-Cano, L., Jung, J.Y., Wang, L.K., Kashef-Haghighi, D., Hartl, C., Singh, C., Xu, J., Hoekstra, J.N., Leventhal, O. et al. (2019) Inherited and de novo genetic risk for autism impacts shared networks. Cell, 178, 850–866.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sanders, S.J., Murtha, M.T., Gupta, A.R., Murdoch, J.D., Raubeson, M.J., Willsey, A.J., Ercan-Sencicek, A.G., Di Lullo, N.M., Parikshak, N.N., Stein, J.L. et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature, 485, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Satterstrom, F.K., Kosmicki, J.A., Wang, J., Breen, M.S., De Rubeis, S., An, J.Y., Peng, M., Collins, R., Grove, J., Klei, L. et al. (2020) Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell, 180, 567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paulsen, B., Velasco, S., Kedaigle, A.J., Pigoni, M., Quadrato, G., Deo, A.J., Adiconis, X., Uzquiano, A., Sartore, R., Yang, S.M. et al. (2022) Autism genes converge on asynchronous development of shared neuron classes. Nature, 602, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Villa, C.E., Cheroni, C., Dotter, C.P., López-Tóbon, A., Oliveira, B., Sacco, R., Yahya, A.Ç., Morandell, J., Gabriele, M., Tavakoli, M.R. et al. (2022) CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep., 39, 110615. [DOI] [PubMed] [Google Scholar]
- 10. Lalli, M.A., Avey, D., Dougherty, J.D., Milbrandt, J. and Mitra, R.D. (2020) High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation. Genome Res., 30, 1317–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skelton, P.D., Stan, R.V. and Luikart, B.W. (2019) The role of PTEN in neurodevelopment. Complex Psychiatry, 5, 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li, Y., Muffat, J., Omer, A., Bosch, I., Lancaster, M.A., Sur, M., Gehrke, L., Knoblich, J.A. and Jaenisch, R. (2017) Induction of expansion and folding in human cerebral organoids. Cell Stem Cell, 20, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groszer, M., Erickson, R., Scripture-Adams, D.D., Lesche, R., Trumpp, A., Zack, J.A., Kornblum, H.I., Liu, X. and Wu, H. (2001) Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science, 152, 2186–2189. [DOI] [PubMed] [Google Scholar]
- 14. Gregorian, C., Nakashima, J., Le Belle, J., Ohab, J., Kim, R., Liu, A., Smith, K.B., Groszer, M., Garcia, A.D., Sofroniew, M.V. et al. (2009) Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J. Neurosci., 29, 1874–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Velasco, S., Kedaigle, A.J., Simmons, S.K., Nash, A., Rocha, M., Quadrato, G., Paulsen, B., Nguyen, L., Adiconis, X., Regev, A. et al. (2019) Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature, 570, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orrico, A., Galli, L., Buoni, S., Orsi, A., Vonella, G. and Sorrentino, V. (2009) Novel PTEN mutations in neurodevelopmental disorders and macrocephaly. Clin. Genet., 75, 195–198. [DOI] [PubMed] [Google Scholar]
- 17. Uzquiano, A., Kedaigle, A.J., Pigoni, M., Paulsen, B., Adiconis, X., Kim, K., Faits, T., Nagaraja, S., Antón-Bolaños, N., Gerhardinger, C. et al. (2022) Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell, 185, 3770–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li, L., Liu, F. and Ross, A.H. (2003) PTEN regulation of neural development and CNS stem cells. PTEN regulation of neural development and CNS stem cells. J. Cell. Biochem., 88, 24–28. [DOI] [PubMed] [Google Scholar]
- 19. Pinson, A. and Huttner, W.B. (2021) Neocortex expansion in development and evolution—from genes to progenitor cell biology. Curr. Opin. Cell Biol., 73, 73–79. [DOI] [PubMed] [Google Scholar]
- 20. Del-Valle-anton, L. and Borrell, V. (2022) Folding brains: from development to disease modeling. Physiol.l Rev., 102, 511–550. [DOI] [PubMed] [Google Scholar]
- 21. Paulsen, B., Velasco, S., Kedaigle, A.J., Pigoni, M., Quadrato, G., Deo, A., Adiconis, X., Uzquiano, A., Kim, K., Simmons, S.K. et al. Human brain organoids reveal accelerated development of cortical neuron classes as a shared feature of autism risk genes. BioRxiv. [Google Scholar]
- 22. Birtele, M., Del Dosso, A., Xu, T., Nguyen, T., Wilkinson, B., Urenda, J.-P., Knight, G., Moore, R., Sharma, R., Pirrotte, P. et al. The autism-associated gene SYNGAP1 regulates human cortical neurogenesis. BioRxiv. [Google Scholar]
- 23. Cahill, K.M., Huo, Z., Tseng, G.C., Logan, R.W. and Seney, M.L. (2018) Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci. Rep., 8, 9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Plaisier, S.B., Taschereau, R., Wong, J.A. and Graeber, T.G. (2010) Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res., 38, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Butler, M.G., Dazouki, M.J., Zhou, X.P., Talebizadeh, Z., Brown, M., Takahashi, T.N., Miles, J.H., Wang, C.H., Stratton, R., Pilarski, R. et al. (2005) Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet., 42, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frazier, T.W., Embacher, R., Tilot, A.K., Koenig, K., Mester, J. and Eng, C. (2015) Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol. Psychiatry, 20, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Busch, R.M., Srivastava, S., Hogue, O., Frazier, T.W., Klaas, P., Hardan, A., Martinez-Agosto, J.A., Sahin, M., Eng, C., Warfield, S.K. et al. (2019) Neurobehavioral phenotype of autism spectrum disorder associated with germline heterozygous mutations in PTEN. Transl. Psychiatry, 9, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang, S.I., Parsons, R. and Ittmann, M. (1998) Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin. Cancer Res., 4, 811–815. [PubMed] [Google Scholar]
- 29. Krohn, A., Diedler, T., Burkhardt, L., Mayer, P.S., De Silva, C., Meyer-Kornblum, M., Kötschau, D., Tennstedt, P., Huang, J., Gerhäuser, C. et al. (2012) Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am. J. Pathol., 181, 401–412. [DOI] [PubMed] [Google Scholar]
- 30. Serebriiskii, I.G., Pavlov, V., Tricarico, R., Andrianov, G., Nicolas, E., Parker, M.I., Newberg, J., Frampton, G., Meyer, J.E. and Golemis, E.A. (2022) Comprehensive characterization of PTEN mutational profile in a series of 34,129 colorectal cancers. Nat. Commun., 13, 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trotman, L.C., Niki, M., Dotan, Z.A., Koutcher, J.A., Di Cristofano, A., Xiao, A., Khoo, A.S., Roy-Burman, P., Greenberg, N.M., Van Dyke, T. et al. (2003) Pten dose dictates cancer progression in the prostate. PLoS Biol., 1, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shemesh, O.A., Linghu, C., Piatkevich, K.D., Goodwin, D., Celiker, O.T., Gritton, H.J., Romano, M.F., Gao, R., Yu, C.C. et al. (2020) Precision calcium imaging of dense neural populations via a cell-body-targeted calcium indicator. Neuron, 107, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanders, S.J., He, X., Willsey, A.J., Ercan-Sencicek, A.G., Samocha, K.E., Cicek, A.E., Murtha, M.T., Bal, V.H., Bishop, S.L., Dong, S. et al. (2015) Insights into autism Spectrum disorder genomic architecture and biology from 71 risk loci. Neuron, 87, 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stessman, H.A.F., Xiong, B., Coe, B.P., Wang, T., Hoekzema, K., Fenckova, M., Kvarnung, M., Gerdts, J., Trinh, S., Cosemans, N. et al. (2017) Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet., 49, 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu, S., Bury, L.A.D., Eum, J. and Wynshaw-Boris, A. (2023) Autism-specific PTEN p.Ile135Leu variant and an autism genetic background combine to dysregulate cortical neurogenesis. Am. J. Hum. Genet., 110, 826–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Church GM. The personal genome project. Mol Syst Biol. 2005;1:2005.0030. 10.1038/msb4100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Velasco, S., Paulsen, B. and Arlotta, P. (2019) Highly reproducible human brain organoids recapitulate cerebral cortex cellular diversity. Protoc, Exch.
- 38. Quadrato, G., Sherwood, J.L. and Arlotta, P. (2017) Long-term culture and electrophysiological characterization of human brain organoids. Protoc. Exch. [Google Scholar]
- 39. Zheng, G.X.Y., Terry, J.M., Belgrader, P., Ryvkin, P., Bent, Z.W., Wilson, R., Ziraldo, S.B., Wheeler, T.D., McDermott, G.P., Zhu, J. et al. (2017) Massively parallel digital transcriptional profiling of single cells. Nat. Commun., 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W.M., Hao, Y., Stoeckius, M., Smibert, P. and Satija, R. (2019) Comprehensive integration of single-cell data. Cell, 177, 1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bates, D., Mächler, M., Bolker, B.M. and Walker, S.C. (2015) Fitting linear mixed-effects models using lme4. J. Stat. Softw., 67, 1–48. [Google Scholar]
- 42. Benjamini, Y. and Hochberg, Y. (1995) Benjamini Y, Hochberg Y. controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B, 57, 289–300. [Google Scholar]
- 43. Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto Encyclopedia of genes and genomes. KEGG: Kyoto Encyclopedia of Genes and Genomes., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee, S., Ebert, B.L., Gillette, M.A., Paulovich, A., Pomeroy, S.L., Golub, T.R., Lander, E.S. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A., 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lun, A.T.L. and Marioni, J.C. (2017) Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics, 18, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu, G., Wang, L.G., Han, Y. and He, Q.Y. (2012) ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang, S., Corbett, S.E., Koga, Y., Wang, Z., Johnson, W.E., Yajima, M. and Campbell, J.D. (2020) Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol., 21, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stickels, R.R., Murray, E., Kumar, P., Li, J., Marshall, J.L., Di Bella, D.J., Arlotta, P., Macosko, E.Z. and Chen, F. (2021) Highly sensitive spatial transcriptomics at near-cellular resolution with slide-seqV2. Nat. Biotechnol., 39, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cable, D.M., Murray, E., Zou, L.S., Goeva, A., Macosko, E.Z., Chen, F. and Irizarry, R.A. (2022) Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol., 40, 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Picelli, S., Faridani, O.R., Björklund, Å.K., Winberg, G., Sagasser, S. and Sandberg, R. (2014) Full-length RNA-seq from single cells using smart-seq2. Nat. Protoc., 9, 171–181. [DOI] [PubMed] [Google Scholar]
- 51. Li, B. and Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dobin, A., Davis, C.A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T.R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Love, M.I., Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soneson, C., Love, M.I. and Robinson, M.D. (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res, 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bairoch, A. and Apweiler, R. (1999) The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucleic Acids Res., 27, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bateman, A., Martin, M.J., O’Donovan, C., Magrane, M., Alpi, E., Antunes, R., Bely, B., Bingley, M., Bonilla, C., Britto, R. et al. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res., 45, 158–169. [Google Scholar]
- 57. Käll, L., Storey, J.D., MacCoss, M.J. and Noble, W.S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res., 7, 29–34. [DOI] [PubMed] [Google Scholar]
- 58. Zhang, X., Smits, A.H., Van Tilburg, G.B.A., Ovaa, H., Huber, W. and Vermeulen, M. (2018) Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc., 13, 530–550. [DOI] [PubMed] [Google Scholar]
- 59. Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W. and Smyth, G.K. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res., 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pachitariu, M., Stringer, C., Dipoppa, M., Schröder, S., Rossi, L.F., Dalgleish, H., Carandini, M. and Harris, K.D. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. BioRxiv. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Count and meta data from scRNA-seq and Slide-seq will be available on the Single Cell Portal, at https://singlecell.broadinstitute.org/single_cell/study/SCP1964. Raw data, proteomics and bulk RNA-seq data will be deposited into a restricted access database before publication. Code used during data analysis will be available at https://github.com/AmandaKedaigle/PTENBrainOrganoids.